Abstract
Tumor suppressor p53 directly regulated the abundance of the miR-34b/c. The interaction might contribute to certain cancer. We hypothesized that rs4938723 in the promoter region of pri-miR-34b/c and TP-53 Arg72Pro may be related to the risk of papillary thyroid carcinoma (PTC).
A total of 784 patients with PTC and 1006 healthy controls were recruited to participate in this study. The variants were discriminated using a polymerase chain reaction–restriction fragment length polymorphism method (PCR-RFLP). Additionally, the relative expression levels of miR-34b/c and TP-53 in 44 paired samples were revealed by quantitative reverse transcription polymerase chain reaction (qRT-PCR).
A significantly increased risk of PTC was observed in the miR-34b/c rs4938723 CT, CC, and CT/CC genotypes compared with the TT genotype (CT vs TT: adjusted odds ratio [OR] = 1.51, 95%confidence interval [CI] = 1.23–1.85; CC vs TT: adjusted OR = 1.89, 95%CI = 1.39–2.63; CT/CC vs TT: adjusted OR = 1.59, 95%CI = 1.30–1.92, respectively). Significantly increased PTC susceptibility was also associated with the TP-53 Arg72Pro CC and CG/CC genotypes compared with the GG genotype (CC vs GG: adjusted OR = 2.04, 95%CI = 1.54–2.70; CG/CC vs GG: adjusted OR = 1.35, 95%CI = 1.11–1.67, respectively). Stratification analysis revealed that patients carrying the TP-53 Arg72Pro C allele and CC genotype had a significantly increased risk for developing N1 (C vs. G: OR = 1.27, 95%CI = 1.03–1.56; CC vs. GG: OR = 1.62, 95%CI = 1.07–2.46, respectively). Combined analysis showed that the genotypes of rs4938723 CT/CC + TP-53CG/CC increased the risk of PTC compared with rs4938723TT + TP-53GG (OR = 2.25, 95%CI = 1.67–3.03). Additionally, level of miR-34b was significantly upregulated in PTC patients.
These findings indicate that the miR-34b/c rs4938723 and TP-53 Arg72Pro polymorphisms may contribute to the susceptibility of PTC.
INTRODUCTION
Thyroid cancer is the most prevalent type of endocrine cancer with the incidence rates of 4 and 12 per 100,000 in human.1 Papillary thyroid carcinoma (PTC) represents virtually 80% of all thyroid cancers, which is the fifth leading malignancy in female patients.2 Although the exact etiology of PTC is not fully clear, numerous pedigree and genomic association studies have indicated that genetic susceptibilities may contribute to PTC.3–5
MicroRNA (miRNA) are non-encoding small RNAs existing extensively in animals, plants, and viruses, at an approximate length of 21 to 23 nt and highly conserved.6 Mature miRNAs can result in the degradation or translation suppression of mRNA by binding to the 3′untranslated region of target gene.6,7 miRNAs are involved in the regulation of multiple critical biological activities, including cell proliferation,8 apoptosis,9 and tumor genesis.10 Altered expression of miR-34 has been found in thyroid tumors, suggesting that miR-34 may have an important role in thyroid carcinogenesis.11,12
Recently, an rs4938723C/T polymorphism in the promoter region of pri-miR-34b/c gene has been discovered, which is located in the CpG island. The variation of rs4938723 C to T may affect a predicted GATA-X transcription factor binding and then affect the expression and carcinogenesis.13–15
It is well known that p53 can regulate the expression of miRNAs, especially the miR-34 family members (i.e., miR-34a, miR-34b, and miR-34c). The 3 mature miRNAs are encoded by 2 different primary miRNAs. miR-34a is encoded by its own transcript, whereas miR-34b and miR-34c shared a common primary transcript (pri-miR-34b/c).6 The promoter region of miR-34b/c transcripts contains p53-binding sites.16 Moreover, the TP-53 gene has an important function in cell cycle control, apoptosis, and maintenance of DNA integrity.17 The importance of p53 in cell cycle regulation and DNA integrity is such critical that it has been called the “guardian of the genome.”18 Codon 72 (Arg72Pro) in exon 4 of the TP-53 gene is a frequently functional single nucleotide polymorphism (SNP) that leads to a methionine proline conversion.19,20 The TP-53 Arg72Pro SNP results in a change in its protein structure, and this SNP exists only in humans.20
Given the potential role of the TP-53 Arg72Pro in tumorigenesis, several studies have reported the association between the TP-53 Arg72Pro polymorphism and PTC risk in different populations.21–24 However, inconsistent results were obtained. Additionally, there is no report of the polymorphism in Chinese PTC patients. To date, no study has been performed to examine the relationship between the miR-34b/c rs4938723 polymorphism and PTC risk. In this study, we selected the rs4938723 and TP-53 Arg72Pro polymorphisms to investigate the correlation between the 2 polymorphisms and PTC risk in a Chinese population by conducting a case–control study.
MATERIALS AND METHODS
Study Population
The study population was composed of 784 cases of PTC and 1006 controls. Patients were consecutively recruited from the West China Hospital of Sichuan University between May 2012 and June 2015. The diagnosis of PTC was confirmed by histopathological analysis. Clinical information was obtained from surgical and pathological records, including age, gender, invasion, tumor node metastasis status, clinical stages, and multifocality. Baseline profiles of the study population are summarized in Table 1. The controls were selected from healthy volunteers who visited the hospital for medical examination during the same period. We excluded individuals who had a history of thyroid disease or antecedents of malignancy from the control group. The study was approved by the institutional review board of the hospital, and informed consent was obtained from each participant.
TABLE 1.
Demographic and Clinical Characteristics of Study Subjects

Genotyping
Genomic DNA was isolated from EDTA-anticoagulated peripheral blood using a commercial extraction kit (Biobeke, Beijing, China) according to the manufacturer's instructions. The 2 SNPs were genotyped by polymerase chain reaction (PCR)-restriction fragment length polymorphism assay. For the rs4938723, the primers were as follows: 5’-CTCTGGGAACCTTCTTTGACCCAT-3’ (forward); 5’-TGAGATCAAGGCCATACCATTCAAGA-3’ (reverse). The PCR products were digested with Bcc I (New England BioLabs, Beverly, MA, USA), yielding one band of 147 bp for T allele and two bands of 118 and 29 bp for C allele. For the TP-53 Arg72Pro, the primer sequences, PCR conditions and restriction enzymes used were previously established.15,25–27 To classify the genotypes, digested PCR products were separated on polyacrylamide gels and stained with 1.0 g/L argent nitrate. For quality control, 2 researchers read the gel pictures independently. The samples were reanalyzed and verified by DNA sequencing if conflict results occurred. In addition, about 10% of all samples were randomly selected to be confirmed by DNA sequencing, and the results were 100% consistent.
Tissue Samples
Paired tumor and adjacent non-tumor tissue specimens were obtained with informed consent from 44 PTC patients who underwent therapy at the West China Hospital of Sichuan University. All samples, taken during surgery, were immediately frozen in liquid nitrogen for further analysis. All the tumor and non-tumor tissue specimens were diagnosed histopathologically.
RNA Extraction and Quantitative Reverse transcription-PCR Assay
Total RNA was extracted and purified from tissue samples using TRIzol Reagent according to the manufacturer's protocol (Roche, Indianapolis, IN). Reverse transcription-PCR (RT-PCR) was performed by using commercial kits according to the manufacturer's instructions (Ribobio, Guangzhou, China; Thermo Scientific, Vilnius, Lithuania). Quantitative real-time PCR was carried out using SYBR green Master Mix (Qiagen, Hilden, Germany) and samples were amplified in a thermocycler as follows: 95 °C for 10 minutes (1 cycle), 95 °C for 10 seconds, 60 °C for 20 seconds, 70 °C for 10 seconds (40 cycles), and melting curve. Data were normalized using comparative threshold cycle method (U6 for miR-34b/c and GAPDH for TP-53). The primers used were as follows: TP-53: CCAGCCAAAGAAGAAACCAC (forward), TGAGTTCCAAGGC CTCATTC (reverse).28GAPDH: GAAGGTGAAGGTCGGAGTC (forward), GAAGATGGTGATGGGATTTC (reverse). Duplicate Ct values were averaged and miR-34b/c and TP-53 mRNA relative expression levels were determined as 2−ΔΔCt.29
Methylation-Sepecific PCR
A total of 100 genomic DNA samples (including 50 cases and 50 controls) were treated with bisulfite according to the Herman's protocol.30 The PCR primers used for the detection of CpG-methylation of the mir-34b/c promoter have been described previously.31
Statistical Analysis
The age of cases and controls was compared using the student's t-test. Hardy–Weinberg equilibrium was tested using Chi-square test. The χ2 test, with odds ratio (OR) and respective 95% confidence intervals (CIs), were used to estimate the differences of genotype and allele distribution. MiR-34b/c and TP-53 mRNA expression levels were evaluated using Wilcoxon rank sum test. The statistical analyses were performed using the SPSS for windows 13.0 (SPSS Inc, Chicago, IL). Moreover, genotypic association tests in a case–control pattern, assuming codominant, dominant, recessive, and overdominant genetic models were calculated using SNPstats.32 A P level of <0.05 was regarded as statistically significant.
RESULTS
Totally, 784 patients with PTC and 1006 anonymous controls were enrolled in this study. Demographic information and clinical parameters for all subjects are summarized in Table 1. We have determined the prevalence of the miR-34b/c rs4938723 and TP-53 Arg72Pro polymorphisms in PTC patients and controls in order to evaluate its association with the risk of PTC (Fig. 1). Both polymorphisms followed the Hardy–Weinberg equilibrium in the control group (P = 0.43 for rs4938723 and P = 0.11 for TP-53 Arg72Pro). The Quanto software (version 1.2.3) was used to calculate the statistical power. Our study had more than 80% power to detect the association between miR-34b/c rs4938723 and TP-53 Arg72Pro polymorphisms and the risk of PTC if OR = 1.4 under a dominant model (87% for rs4938723 and 83% for TP-53 Arg72Pro).
FIGURE 1.
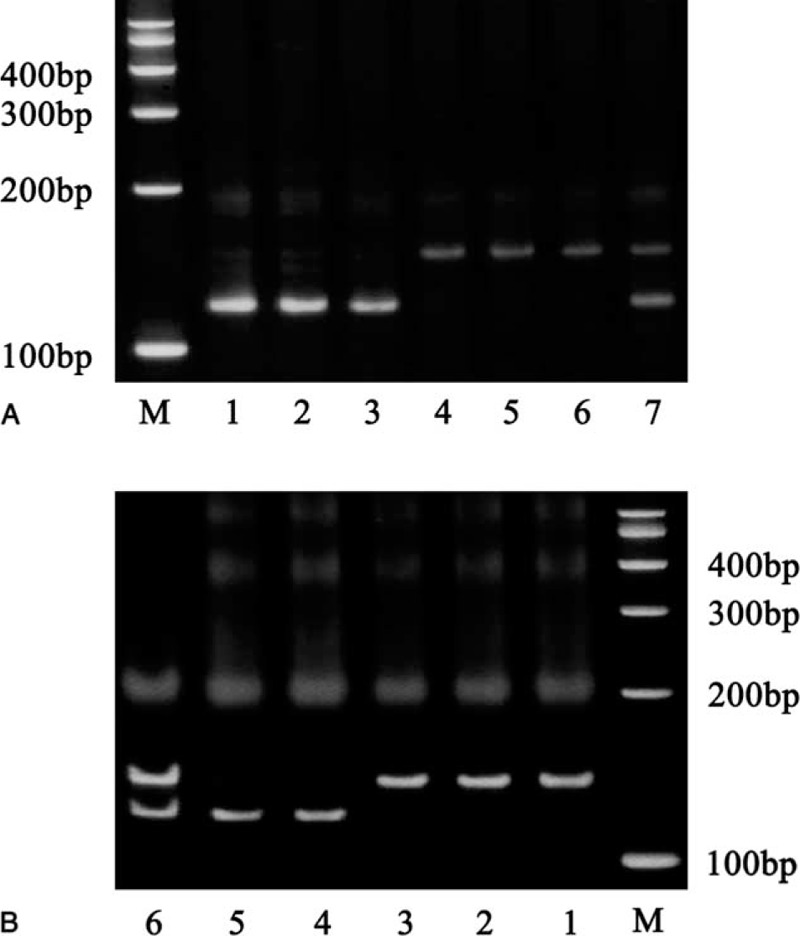
Genotypes of miR-34b/c rs4938723 and TP-53 Arg72Pro. (A) Genotypes of miR-34b/c rs4938723 polymorphisms. M: marker; 1 to 3: CC genotype; 4 to 6: TT genotype; 7: CT genotype. (B) Genotypes of TP-53 Arg72Pro polymorphisms. M: marker; 1 to 3: GG genotype; 4 to 5: CC genotype; 6: CG genotype.
As shown in Table 2, distributions of the miR-34b/c rs4938723 were significantly different between patients and control subjects. Compared with TT genotype, a significantly increased PTC risk was found to be associated with CT (adjusted OR = 1.51, 95%CI = 1.23–1.85, P < 0.001) and CC (adjusted OR = 1.89, 95%CI = 1.39–2.63, P < 0.001) genotype in the codominant model, while CT/CC carriers had a 1.59-fold increased PTC risk (95%CI = 1.30–1.92, P < 0.001) in the dominant model. Compared with TT/CT genotype, CC genotype carriers had a 1.51-fold increased PTC risk (95%CI = 1.14–2.04, P = 0.005) in a recessive model. When compared with TT/CC genotype, CT genotype carriers had a 1.30-fold increased PTC risk (95%CI = 1.07–1.56, P = 0.006).
TABLE 2.
Association Between the miR-34b/c rs4938723 and TP-53 Arg72Pro Polymorphisms and Risk of PTC
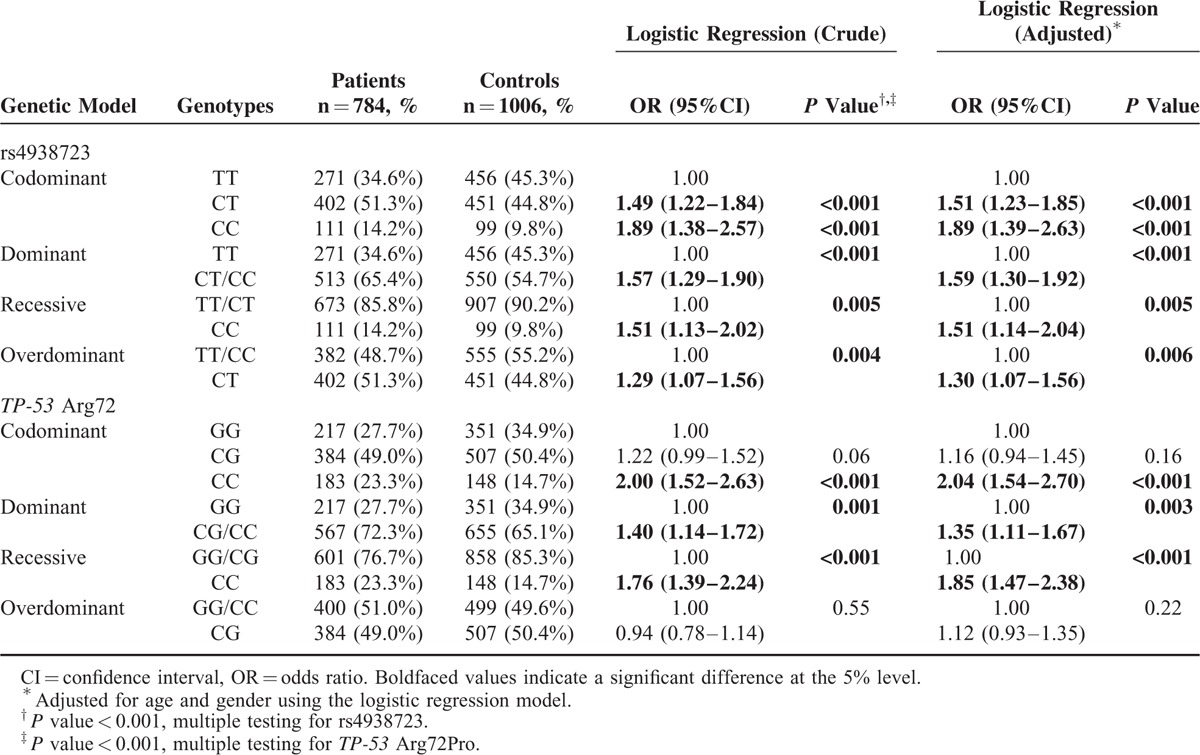
Significant differences were also observed for genotypes of the TP-53 Arg72Pro polymorphisms. Compared with GG genotype, CC and CG/CC genotype carriers had a 2.04- and 1.35-fold increased PTC risk (CC vs GG: 95%CI = 1.54–2.70, P < 0.001; CG/CC vs GG: 95%CI = 1.11–1.67, P = 0.003, respectively) Furthermore, compared with GG/CG genotypes, CC genotype carriers had a 1.85-fold increased PTC risk (95%CI = 1.47–2.38, P < 0.001) in the recessive model.
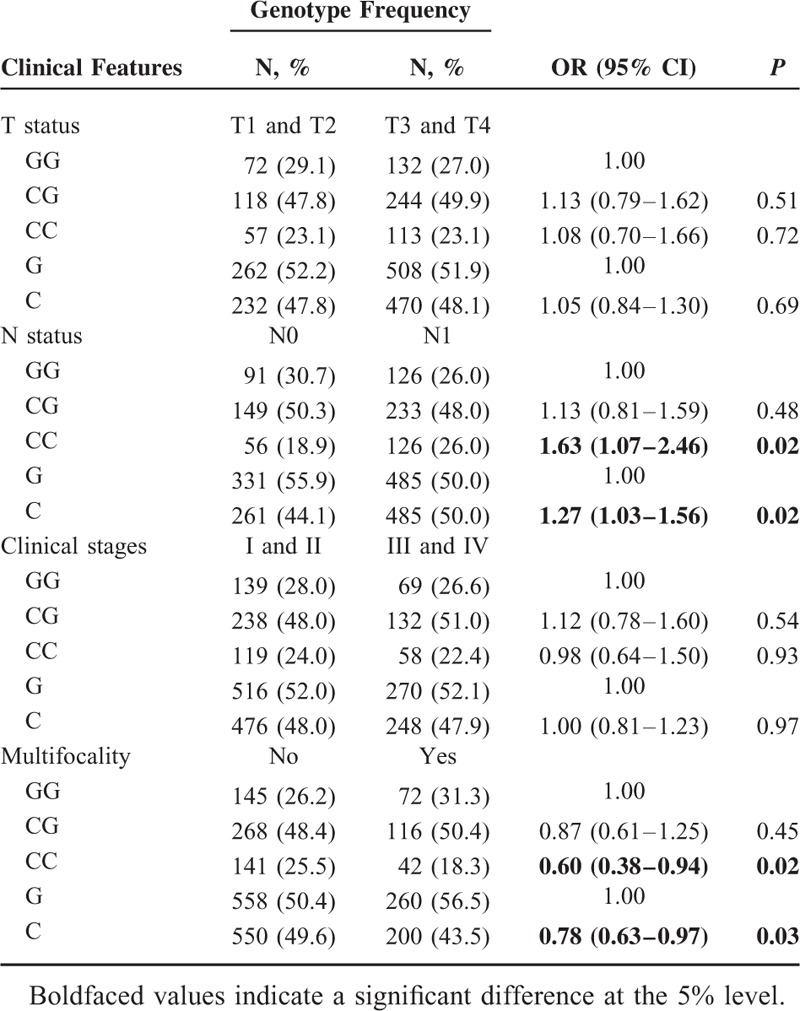
Furthermore, we divided the cases by T status, N status, clinical stages, and multifocality (Tables 3 and 4). The patients carrying the TP-53 Arg72Pro C allele and CC genotype had a significantly increased risk for developing N1 when compared with patients carrying the G allele and GG genotype (C vs G: OR = 1.27, 95%CI = 1.03–1.56, P = 0.02; CC vs GG: OR = 1.63, 95%CI = 1.07–2.46, P = 0.02, respectively). Additionally, the frequencies of the TP-53 Arg72Pro C allele and CC genotype in patients with multifocality were lower than other patients (C vs G: OR = 0.78, 95%CI = 0.63–0.97, P = 0.03; CC vs GG: OR = 0.60, 95%CI = 0.38–0.94, P = 0.02, respectively) (Table 4).
TABLE 3.
Association Between the miR-34b/c rs4938723 Polymorphism and Clinical Features of PTC Patients
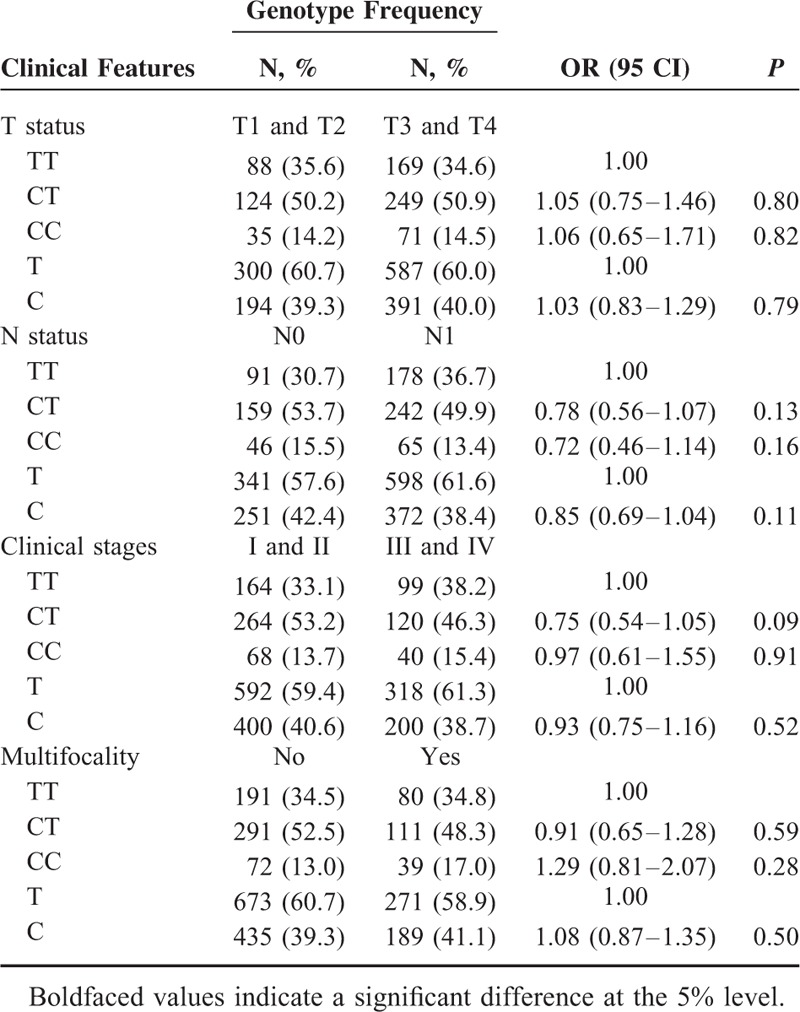
TABLE 4.
Association Between the TP-53 Arg72Pro Polymorphism and Clinical Features of PTC Patients
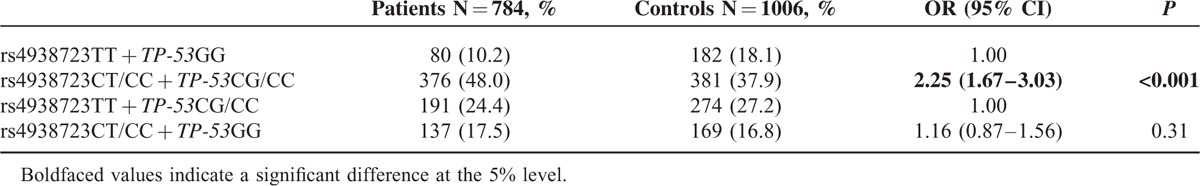
We also examined the combined effects of the miR-34b/c rs4938723 and TP-53 Arg72Pro variants on PTC risk. As shown in Table 5, carriers with the combined genotypes of rs4938723CT/CC and TP-53 Arg72Pro CG/CC had a 2.25-fold increased PTC risk (95%CI = 1.67–3.03, P < 0.001) compared with those carrying the rs4938723TT and TP-53 Arg72ProGG genotypes.
TABLE 5.
The Combined Genotypes Frequencies of the miR-34b/c rs4938723 and TP-53 Arg72Pro Between Patients With PTC and Controls
We analyzed the differential expression of miR-34b/c and TP-53 mRNA in 44 paired PTC tissues and adjacent non-tumor tissues. Level of miR-34b was significantly upregulated in PTC patients (P = 0.02). However, there was no significant difference of the miR-34c and TP-53 mRNA levels between PTC tissues and adjacent non-tumor tissues (P > 0.05) (Fig. 2). Furthermore, no significant association between miR-34b/c rs4938723 polymorphism and miR-34b/c methylation level has been found (data not shown).
FIGURE 2.
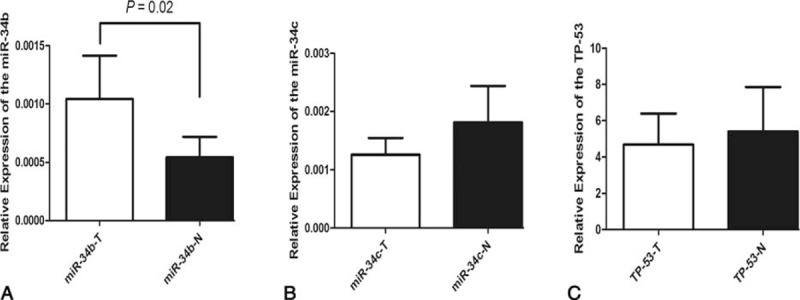
Relative expression of miR-34b/c and TP-53 in 44 paired PTC tumor tissues samples and adjacent normal specimens determined by qRT-PCR. The expression level was normalized using comparative threshold cycle method (U6 for miR-34b/c and GAPDH for TP-53). (A) miR-34b was found significantly differentially upregulated in tumor specimens (P = 0.02). No significant difference of miR-34c (B) and TP-53 mRNA (C) was found.
DISCUSSION
Growing evidence has shown that miR-34b/c is downregulated in a series of cancers, such as colorectal cancer, prostate cancer, gastric cancer, and aggressive PTC, which makes it as a candidate target of anticancer therapy.33–35 In this study, however, we found that the relative expression of miR-34b was significantly upregulated in PTC patients.
Previously, the T to C variation of pri-miR-34b/c CpG island has been predicted to create a GATA-binding site, using the web-based SNP analysis tool.15 To extend the finding, we used methylation-specific PCR to determine whether the miR-34b/c rs4938723 influences the methylation status of miR-34b/c. However, no significant difference between the miR-34b/c rs4938723 polymorphism and methylation status was found. Some association studies have reported that the miR-34b/c rs4938723 CT/CC genotypes were associated with a significantly increased risk of hepatocellular carcinoma.15,36 The similar results were also observed in nasopharyngeal carcinoma, osteosarcoma, and renal cell cancer.26,37,38 In contrast, Gao et al27 reported that the miR-34b/c rs4938723CC genotype had a 0.56-fold decreased risk of colorectal cancer in a Chinese population. The decreased risks were also observed in esophageal squamous cell carcinoma and gastric cancer.25,39,40 Data obtained in the present work indicated that the rs4938723CT and CC genotypes significantly increased PTC risk. The conflicting results further supported the findings that the impacts of genetic polymorphisms may be quite different according to the types of cancer.41
TP-53 Arg72Pro is the most intensively investigated polymorphism in the TP-53 gene. Previously, several groups have investigated the TP-53 Arg72Pro polymorphism in PTC. Boltze et al21 firstly found a small number of CG genotype and no CC genotype in German PTC patients. However, only 21 PTC cases were enrolled in the above study, and the result is not convincing. Granja et al,22 on the contrary, demonstrated a higher frequency of CC genotype among patients with differentiated thyroid cancer in a Brazilian population. Similarly, in Russian and Ukrainian populations, GG genotype was associated with the significantly lower risk of PTC.23 Further meta-analysis supported the result that CC genotype is a risk factor for thyroid cancer.24 In consistent with the positive result, we found that the frequency of CC genotype was significantly higher in patients with PTC than general population.
Stratification analysis was also performed, and we found that the frequencies of the TP-53 Arg72Pro C allele and CC genotype in patients with N1 were higher when compared with that in patients in patients with N0, while lower frequencies of C allele and CC genotype of TP-53 Arg72Pro polymorphism in patients with multifocality were found. The previous study revealed significant correlation between P53 protein overexpression and presence of lymph node metastasis in PTC patients.42 The consistent results proposed that the TP-53 Arg72Pro may be a useful genetic marker for lymph node recurrences.
Previous definitive evidence has demonstrated that the abundance of the miR-34b/c is directly regulated by p53 in cell lines and tissues. Cheng et al43 indicated that miR-34 cooperates with p53 in suppression of prostate cancer. These findings suggest that their interaction might contribute to certain cancers. In this study, we performed a combined analysis which showed that the combined genotypes of rs4938723CT/CC + TP-53CG/CC had a tendency to increase the susceptibility for PTC, indicating that the rs4938723 and TP-53 Arg72Pro may jointly contribute to the risk of PTC.
There were still limitations in our study. One is that some potential confounding factors were not considered in this study, such as iodine added salt, expose to radiations, smoking and drinking status. We cannot rule out that these factors may influence the results. Another is that the molecular mechanism is not involved in the study, which should be characterized in the future. Finally, the PCR-restriction fragment length polymorphism method may be subject to false positive results. To minimize the false positive rate, we have taken quality control.
In summary, we evaluated the impact of the genetic variability of the miR-34b/c rs4938723 and TP-53 Arg72Pro on PTC risk, and for the first time, we found that the 2 polymorphisms were associated with a significantly increased risk of PTC in the Chinese population. Further studies in different populations are warranted to confirm these findings.
Acknowledgments
The authors thank grants from the Special Research Foundation of Doctoral Priority to the Development of Field Project (No. 20110181130013), National Natural Science Foundation of China (No. 81302149, 81202387), the Science & Technology Pillar Program of Sichuan Province (2014SZ0001, 2013JY0013), Distinguished Young Scientist of Sichuan University (No. 2013SCU04A38), and the Ph.D. Programs Foundation of Ministry of Education of China (No. 20130181120011).
Footnotes
Abbreviations: CI = confidence interval, miRNA = microRNA, OR = odds ratio, PCR = polymerase chain reaction, PTC = papillary thyroid carcinoma, SNP = single nucleotide polymorphism.
PC and RS contributed equally to this work.
This work was supported by grants from the Special Research Foundation of Doctoral Priority to the Development of Field Project (No. 20110181130013), National Natural Science Foundation of China (No. 81302149, 81202387), the Science & Technology Pillar Program of Sichuan Province (2014SZ0001, 2013JY0013), Distinguished Young Scientist of Sichuan University (No. 2013SCU04A38), and the Ph.D. Programs Foundation of Ministry of Education of China (No. 20130181120011).
The authors have no conflict of interest to disclose.
REFERENCES
- 1.Trovisco V, Soares P, Preto A, et al. Molecular genetics of papillary thyroid carcinoma: great expectations. Arq Bras Endocrinol Metabol 2007; 51:643–653. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013; 63:11–30. [DOI] [PubMed] [Google Scholar]
- 3.Hemminki K, Li X. Familial risk of cancer by site and histopathology. Int J Cancer 2003; 103:105–109. [DOI] [PubMed] [Google Scholar]
- 4.Fallah M, Pukkala E, Tryggvadottir L, et al. Risk of thyroid cancer in first-degree relatives of patients with non-medullary thyroid cancer by histology type and age at diagnosis: a joint study from five Nordic countries. J Med Genet 2013; 50:373–382. [DOI] [PubMed] [Google Scholar]
- 5.Adjadj E, Schlumberger M, de Vathaire F. Germ-line DNA polymorphisms and susceptibility to differentiated thyroid cancer. Lancet Oncol 2009; 10:181–190. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 7.Moreno-Moya JM, Vilella F, Simon C. MicroRNA: key gene expression regulators. Fertil Steril 2014; 101:1516–1523. [DOI] [PubMed] [Google Scholar]
- 8.Cheng AM, Byrom MW, Shelton J, et al. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res 2005; 33:1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tili E, Michaille JJ, Calin GA. Expression and function of micro-RNAs in immune cells during normal or disease state. Int J Med Sci 2008; 5:73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 2004; 101:2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Abdel-Mageed AB, Mondal D, et al. MicroRNA expression profiles in differentiated thyroid cancer, a review. Int J Clin Exp Med 2013; 6:74–80. [PMC free article] [PubMed] [Google Scholar]
- 12.Yip L, Kelly L, Shuai Y, et al. MicroRNA signature distinguishes the degree of aggressiveness of papillary thyroid carcinoma. Ann Surg Oncol 2011; 18:2035–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bossard P, Zaret KS. GATA transcription factors as potentiators of gut endoderm differentiation. Development (Cambridge, England) 1998; 125:4909–4917. [DOI] [PubMed] [Google Scholar]
- 14.Chou J, Provot S, Werb Z. GATA3 in development and cancer differentiation: cells GATA have it!. J Cell Physiol 2010; 222:42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y, Liu L, Liu J, et al. A potentially functional polymorphism in the promoter region of miR-34b/c is associated with an increased risk for primary hepatocellular carcinoma. Int J Cancer 2011; 128:412–417. [DOI] [PubMed] [Google Scholar]
- 16.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature 2007; 447:1130–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bommer GT, Gerin I, Feng Y, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol 2007; 17:1298–1307. [DOI] [PubMed] [Google Scholar]
- 18.Botchkarev VA. Flores ER. p53/p63/p73 in the epidermis in health and disease. Cold Spring Harbor Perspect Med 2014; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Storey A, Thomas M, Kalita A, et al. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature 1998; 393:229–234. [DOI] [PubMed] [Google Scholar]
- 20.Dumont P, Leu JI, Della Pietra AC, 3rd, et al. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet 2003; 33:357–365. [DOI] [PubMed] [Google Scholar]
- 21.Boltze C, Roessner A, Landt O, et al. Homozygous proline at codon 72 of p53 as a potential risk factor favoring the development of undifferentiated thyroid carcinoma. Int J Oncol 2002; 21:1151–1154. [PubMed] [Google Scholar]
- 22.Granja F, Morari J, Morari EC, et al. Proline homozygosity in codon 72 of p53 is a factor of susceptibility for thyroid cancer. Cancer Lett 2004; 210:151–157. [DOI] [PubMed] [Google Scholar]
- 23.Rogounovitch TI, Saenko VA, Ashizawa K, et al. TP53 codon 72 polymorphism in radiation-associated human papillary thyroid cancer. Oncol Rep 2006; 15:949–956. [PubMed] [Google Scholar]
- 24.Wu B, Guo D, Guo Y. Association between p53 Arg72Pro polymorphism and thyroid cancer risk: a meta-analysis. Tumour Biol 2014; 35:561–565. [DOI] [PubMed] [Google Scholar]
- 25.Pan XM, Sun RF, Li ZH, et al. Pri-miR-34b/c rs4938723 polymorphism is associated with a decreased risk of gastric cancer. Genet Test Mol Biomark 2015; 19:198–202. [DOI] [PubMed] [Google Scholar]
- 26.Li L, Wu J, Sima X, et al. Interactions of miR-34b/c and TP-53 polymorphisms on the risk of nasopharyngeal carcinoma. Tumour Biol 2013; 34:1919–1923. [DOI] [PubMed] [Google Scholar]
- 27.Gao LB, Li LJ, Pan XM, et al. A genetic variant in the promoter region of miR-34b/c is associated with a reduced risk of colorectal cancer. Biol Chem 2013; 394:415–420. [DOI] [PubMed] [Google Scholar]
- 28.Siprashvili Z, Webster DE, Kretz M, et al. Identification of proteins binding coding and non-coding human RNAs using protein microarrays. BMC Genom 2012; 13:633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 30.Herman JG, Graff JR, Myohanen S, et al. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A 1996; 93:9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki H, Yamamoto E, Nojima M, et al. Methylation-associated silencing of microRNA-34b/c in gastric cancer and its involvement in an epigenetic field defect. Carcinogenesis 2010; 31:2066–2073. [DOI] [PubMed] [Google Scholar]
- 32.Sole X, Guino E, Valls J, et al. SNPStats: a web tool for the analysis of association studies. Bioinformatics (Oxford, England) 2006; 22:1928–1929. [DOI] [PubMed] [Google Scholar]
- 33.Hagman Z, Larne O, Edsjo A, et al. miR-34c is downregulated in prostate cancer and exerts tumor suppressive functions. International journal of cancer. J Int Cancer 2010; 127:2768–2776. [DOI] [PubMed] [Google Scholar]
- 34.Tsai KW, Wu CW, Hu LY, et al. Epigenetic regulation of miR-34b and miR-129 expression in gastric cancer. International journal of cancer. J Int Cancer 2011; 129:2600–2610. [DOI] [PubMed] [Google Scholar]
- 35.Toyota M, Suzuki H, Sasaki Y, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res 2008; 68:4123–4132. [DOI] [PubMed] [Google Scholar]
- 36.Son MS, Jang MJ, Jeon YJ, et al. Promoter polymorphisms of pri-miR-34b/c are associated with hepatocellular carcinoma. Gene 2013; 524:156–160. [DOI] [PubMed] [Google Scholar]
- 37.Zhang S, Qian J, Cao Q, et al. A potentially functional polymorphism in the promoter region of miR-34b/c is associated with renal cell cancer risk in a Chinese population. Mutagenesis 2014; 29:149–154. [DOI] [PubMed] [Google Scholar]
- 38.Tian Q, Jia J, Ling S, et al. A causal role for circulating miR-34b in osteosarcoma. Eur J Surg Oncol 2014; 40:67–72. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, Huang X, Xiao J, et al. Pri-miR-124 rs531564 and pri-miR-34b/c rs4938723 polymorphisms are associated with decreased risk of esophageal squamous cell carcinoma in Chinese populations. PloS One 2014; 9:e100055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin J, Wang X, Zheng L, et al. Hsa-miR-34b/c rs4938723 T>C and hsa-miR-423 rs6505162 C>A polymorphisms are associated with the risk of esophageal cancer in a Chinese population. PLoS One 2013; 8:e80570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yi DH, Wang BG, Zhong XP, et al. Pri-miR-34b/c rs4938723 TC heterozygote is associated with increased cancer risks: evidence from published data. Tumour Biol 2014; 35:11967–11975. [DOI] [PubMed] [Google Scholar]
- 42.Morita N, Ikeda Y, Takami H. Clinical significance of p53 protein expression in papillary thyroid carcinoma. World J Surg 2008; 32:2617–2622. [DOI] [PubMed] [Google Scholar]
- 43.Cheng CY, Hwang CI, Corney DC, et al. miR-34 cooperates with p53 in suppression of prostate cancer by joint regulation of stem cell compartment. Cell Rep 2014; 6:1000–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]


