Abstract
Although daunorubicin (DNR) is the most widely used anthracycline to treat acute myeloid leukemia (AML), resistance to this drug remains a critical problem. The aim of this study was to investigate the relationship between AML resistance to daunorubicin and susceptibility to natural killer (NK) cell-mediated cell lysis, and the putative expression of miRs. For this purpose, we used the parental AML cell lines U-937 and KG-1 and their equivalent resistant U937(R) and KG-1(R) cell lines. We demonstrate for the first time that the acquisition of resistance to DNR by the parental cell lines resulted in the acquisition of cross-resistance to NK cell-mediated cytotoxicity. miR microarray analysis revealed that this cross-resistance was associated with miR-181a downregulation and the subsequent regulation of MAP3K10 and MAP2K1 tyrosine kinases and the BCL-2 (BCL-2 and MCL-1) family. Overexpression of miR-181a in AML blasts resulted in the attenuation of their resistance to DNR and to NK-cell-mediated killing. These data point to a determinant role of miR-181a in the sensitization of leukemic resistant cells to DNR and NK cells and suggest that miR-181a may provide a promising option for the treatment of immuno- and chemo-resistant blasts.
Keywords: AML, miR-181a, NK lysis
Abbreviations
- ALL
acute lymphocytic leukemia
- AML
acute myeloid leukemia
- BM
bone marrow
- DNR
daunorubicin
- E: T
effector/target
- GrB
granzyme B
- HSCT
hematopoietic stem cell transplantation
- IL-2
interleukin-2
- miRNA
microRNAs
- NK
natural killer
- NKL
NK-cell line
- PB
peripheral blood
Introduction
AML is a genetically and biologically heterogeneous hematologic malignancy defined by accumulation of malignant precursors of the myeloid lineage in the bone marrow (BM) and peripheral blood (PB) circulation due to the failure of these cells to differentiate into normal hematopoietic cells.1-3 Typically, the anthracycline DNR is the first chemotherapeutic drug used in the treatment of both acute lymphocytic leukemia (ALL) and AML.4-10 Cytotoxicity mediated by DNR is generally reported to result from interference with cellular nucleic acid metabolism.11 Despite the cytotoxicity of DNR and its efficiency in modulating different signaling pathways in leukemia cells, acquisition of drug resistance by leukemic blasts reduces the likelihood of overall cure in 30%–40% of patients.12-14
Currently, allogeneic hematopoietic stem cell transplantation (HSCT) is the only curative treatment for many intermediate and high-risk leukemias.15,16
The acquisition of drug resistance by leukemia cells has been shown to affect their interaction with effector cells of the immune system. While few studies indicate that drug resistance acquisition decreases sensitivity of leukemic cells to cytotoxic immune cells, other reports make evident the enhanced susceptibility of resistant leukemia cells as compared to sensitive ones.17-26
NK cells of the innate immune system operate as a first-line defense against infections and malignancies by direct cytolysis of infected or transformed cells through the secretion of potent immune mediators. They also play key roles in transplant rejection and the development of tolerance. Due to their spontaneous and highly cytolytic activity against tumor cells, NK cells have been proposed as promising effectors for adoptive immunotherapy of cancer. Indeed, recent studies demonstrating the efficiency of immunotherapeutic approaches against hematologic malignancies have led to clinical trials investigating the adoptive transfer of NK cells.27-30 The abnormal expression of microRNAs (miRNA) in leukemia disorders has also been reported to play a critical role in leukemia development and clinical outcome.31-41 Although the various and powerful roles of miRNAs in tumorigenesis, metastasis and drug resistance acquisition are well documented, their roles in immune responses in leukemia remain unknown. Here, we reveal a new and key regulatory mechanism of miR-181a in the leukemia cross-resistance to DNR and NK-mediated lysis, and suggest that in DNR resistant patients, miR-therapy could be a new option to achieve complete remission.
Results
The acquisition of resistance to DNR correlates with decreased susceptibility to NK-mediated lysis through an intrinsic mechanism
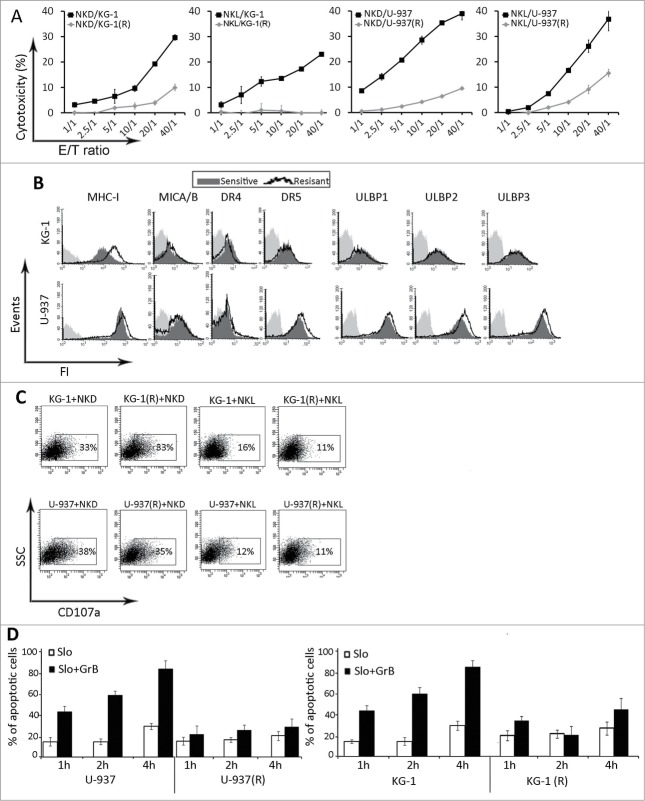
The DNR-resistant AML cell sublines KG-1 (R) and U-937 (R) were established by exposing parental cells to increasing concentrations of the drug (data Fig. S1). To examine the susceptibility of DNR-resistant cells to NK-mediated cytotoxicity, we used the NK-cell line (NKL) and NK cells isolated from healthy donors (NKD). As shown in Fig. 1A, both DNR-resistant cell lines were more resistant to NKL and NKD cell killing as compared to parental DNR-sensitive cells at all effector/target (E:T) ratios tested. To gain insight into the mechanisms associated with decreased susceptibility of DNR resistant cells to NK-mediated lysis, we analyzed the expression pattern of ligands for activating and inhibitory receptors of NK cells in parental and DNR-resistant cell lines. Our results depicted in Fig. 1B showed that only the expression of MHC-I in KG-1 (R) was increased in resistant KG-1 cells as compared to parental sensitive cell line. We next performed the degranulation assay by measuring CD107a expression on NK cells after 4-h of coculture with leukemia cells at a 2:1 effector/target ratio. As shown in Fig. 1C, no difference in the expression of CD107a at the surface of NKL and NKD cells was observed following coculture with both resistant and sensitive leukemia cells. Treatment of leukemia cells with recombinant granzyme B (GrB) at different time points revealed that the resistant cells were also resistant to GrB-mediated cell death (Fig. 1D). Taken together, these results indicate that the resistance of leukemia cells to NK-mediated killing is not related to a modification in the expression of NK ligands but rather to the activation of an intrinsic resistance mechanism.
Figure 1.
Effects of daunorubicin on leukemia cells. (A) NK-cell mediated cytotoxicity of KG-1 and U937 sensitive and resistant (R) leukemic cells. Target cells were co-incubated with NKD and NKL cells at the indicated E:T ratios for 4 hours at 37°C. (B) The expression of ligands for inhibitory and activating receptors of NK cells in sensitive and resistant KG-1 and U-937 cells (representative plot of n = 3). (C) Flow cytometric based assay for the expression of CD107a of NK cells cocultured for 4 h with DNR sensitive or resistant (R) KG-1 or U937 cells at 2:1 E:T ratio (representative plot of n = 3). (D) Analysis of GrB-induced apoptosis in sensitive or resistant (R) KG-1 or U-937 cells. Cells were incubated with SLO (10 U/μL) alone or in combination with recombinant human GrB (5 ng/μL) for the indicated times. Apoptosis was assessed by flow cytometry using annexin 5 and 7AAD (Representative graph of n = 2).
The acquisition of leukemia cell resistance to DNR is associated with decreased expression of miR-181a and miR-146a
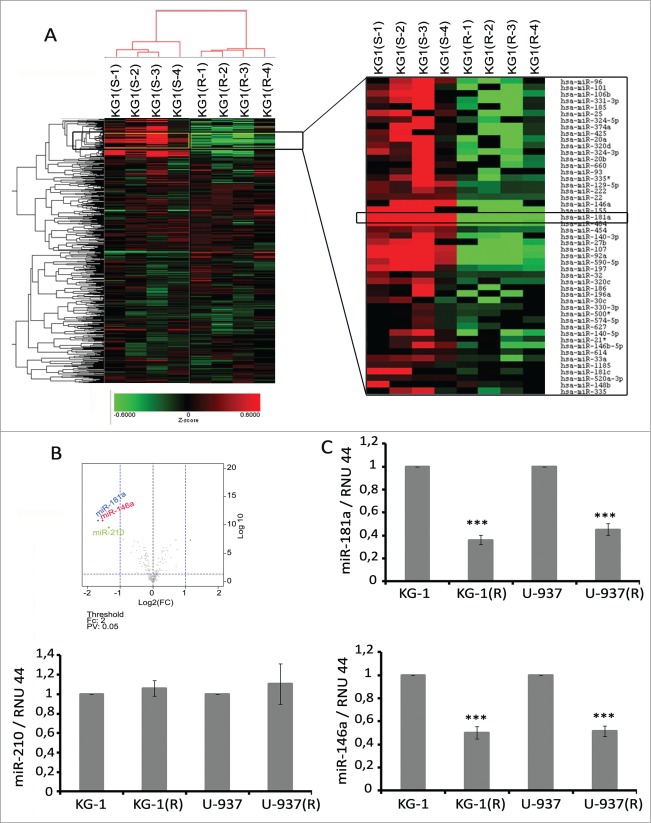
To identify miRNAs that are modified in DNR-resistant leukemia cells, we performed genome-wide miRNA expression profiling of KG-1 and KG-1(R) cells (Fig. 2A). The Volcano plot analysis revealed that miR-181a, miR-146a, and miR-210 were the most highly (more than two-fold change) and significantly (p value < 0.05) downregulated miRs in KG-1(R) cells as compared to parental cells (Fig. 2B). We next measured the expression levels of miR-181a, miR-146a, and miR-210 in KG-1(R) and U-937(R) cells by TaqMan qRT-PCR. Our results confirm the downregulated expression of miR-181a and miR-146a, but not miR-210, in KG-1(R) and U-937(R) cells compared to parental cells (Fig. 2C).
Figure 2.
Microarray analysis of the effects of daunorubicin resistance acquisition by leukemia cells on miRNA expression. (A) Heat map of regulated miRNAs in KG-1 sensitive (S-1–S-4) or resistant (R-1–R-4) cells compared to parental cells. Red signifies upregulation, green downregulation. ArrayExpress accession: E-MTAB-2837, Username: Reviewer_E-MTAB-2837, Password: yikpqsrb. (B) Volcano plot of miRNA expression (log2 fold change) and adjusted P values for KG-1(R). (C) miR-181a, miR-146a and miR-210 expression in DNR-sensitive or -resistant (R) KG1 and U937 cells. Expression levels of RNU44 were used as endogenous control. One representative of at least three separate experiments is shown.
miR-181a is involved in the regulation of AML susceptibility to NK cell-mediated killing
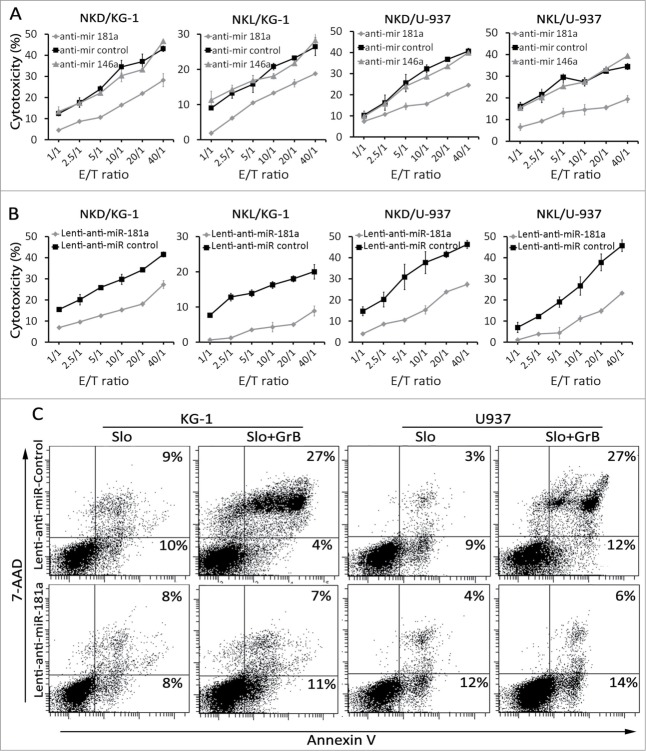
To delineate the potential role of miR-181a and miR-146a in the regulation of leukemia cell susceptibility to NK-mediated lysis, KG-1 and U-937 parental cells were transfected with either anti-miR-181a or -146a The cytotoxicy assay revealed that, unlike miR-146a, the inhibition of miR-181a in KG-1 and U-937 cells was associated with a significant decrease in NKL and NKD cell lysis (Fig. 3A). We next investigate the functional consequence of miR181a inhibition in leukemia cell susceptibility to NK-mediated lysis by using a lentiviral-based miRNA inhibitor, which enabled us to establish KG-1 and U-937 cells lines displaying stably inhibited miR-181a. Cytotoxicity assays revealed that inhibition of miR-181a by lentiviral vector dramatically decreased the susceptibility of AML cells to NK cell-mediated lysis by NKL and NKD as compared to control cells transduced with empty vector (Fig. 3B). To assess the mechanism underlying the decreased susceptibility to NK cell-mediated lysis by anti-miR-181a transduced cells, the expression pattern of NK cell receptor ligands was analyzed. The results revealed that cell surface expression of ligands including ULBP1/2/3, MHC-I, DR4/5, and MICA/B in miR-181a expressing cells was similar to those expressing empty vector (data not shown). Finally, to determine the effect of inhibition of miR-181a on intrinsic mechanisms of resistance to apoptosis, the anti-miR-181a and empty vector-transduced cells were treated with recombinant GrB protein. Data depicted in Fig. 3C indicate that inhibition of miR-181a in KG-1 and U-937 AML cells resulted in enhanced resistance to GrB-induced apoptosis. Collectively, these results demonstrate that inhibition of miR-181a in AML cells induces resistance to apoptosis through intrinsic mechanisms.
Figure 3.
Effects of miR-181a inhibition on leukemia cells susceptibility to NK cells. (A) NK-cell cytotoxicity of KG-1 and U-937 cells after miR inhibition with anti- miR-181a, anti-miR-146a, and anti-miR control. Cell viability 24 h post transfection was around 80%. NKL and NKD cells used as effectors (Representative curve of n = 5) (B) NK- cell mediated lysis of KG-1 and U-937 cells after transduction with anti-miR 181-a or empty vector transduced cells as negative control. NKL and NKD cells used as effectors (Representative curve of n = 5). (C) KG-1 and U-937 cells were transduced stably with Lenti-anti-mir-181a Lenti-anti-mir-control and cells were incubated with SLO (10 U/μL) alone or in combination with recombinant human GrB (5 ng/μL) for the indicated times. Apoptosis was assessed by flow cytometry using annexin 5 and 7AAD (Representative blot of n = 2).
miR-181a interferes with leukemia cell susceptibility to NK-cell mediated lysis by targeting different members of Bcl-2 and tyrosine kinase families
In order to identify the potential target genes of miR-181a, microarray-based global transcriptional profiling was performed. The results shown in Fig. 4A indicate that KG-1 and U-937 cells transduced with anti-miR-181a had significantly different gene expression signatures. The Volcano plot analysis showed that in KG-1 cells stably trasduced with anti-miR-181a, 499 genes were upregulated and 338 genes were downregulated as compared to KG-1 cells transduced with empty vector. In U-937 cells transduced with anti-miR-181a, 640 genes were upregulated and 986 genes were downregulated compared to U-937 cells transduced with empty vector (Fig. 4B). Quantitative analysis using the miRNA target data base (http://www.targetscan.org) showed that all candidate targets of miR-181a that could have an impact on NK cell-mediated lysis were selected. RT-qPCR results demonstrated that in KG-1 cells transduced with lenti-anti-miR-181a, the expression of MCL-1 and MAP2K1 were increased compared to control cells. In U-937 cells transduced with lenti-anti-miR-181a, the expression of Bcl-2 and MAP3K10 were upregulated compared to control cells. We next attempted to identify the impact of these genes on tumor susceptibility of NK cell-mediated killing. Results of Fig. 5A showed that silencing of miR-181a target genes (Bcl-2 and MAP3K10 for U-937 and MCL-1 and MAP2K1 for KG-1) by specific siRNAs significantly decreased leukemia cells susceptibility to NK mediated-lysis. MCL-1 and Bc--2 are the well-known targets of miR-181a and their modification by miR-181a has been described in multiple studies. To validate MAP3K10 and MAP2K1 as new miR-181a targets we inserted miR-181a binding site sequences from the 3′ UTR of MAP3K10 and MAP2K1 mRNA into the 3′ UTR of psi-check2 vector (Fig. 5B) and conducted luciferase reporter assay (Fig. 5C). We demonstrate that pre-miR-181a decreased the luciferase activation of MAP3K10 and MAP2K1, as compared with pre-miR control, confirming that MAP3K10 and MAP2K1 target sites directly mediate repression of luciferase activity through seed-specific binding.
Figure 4.
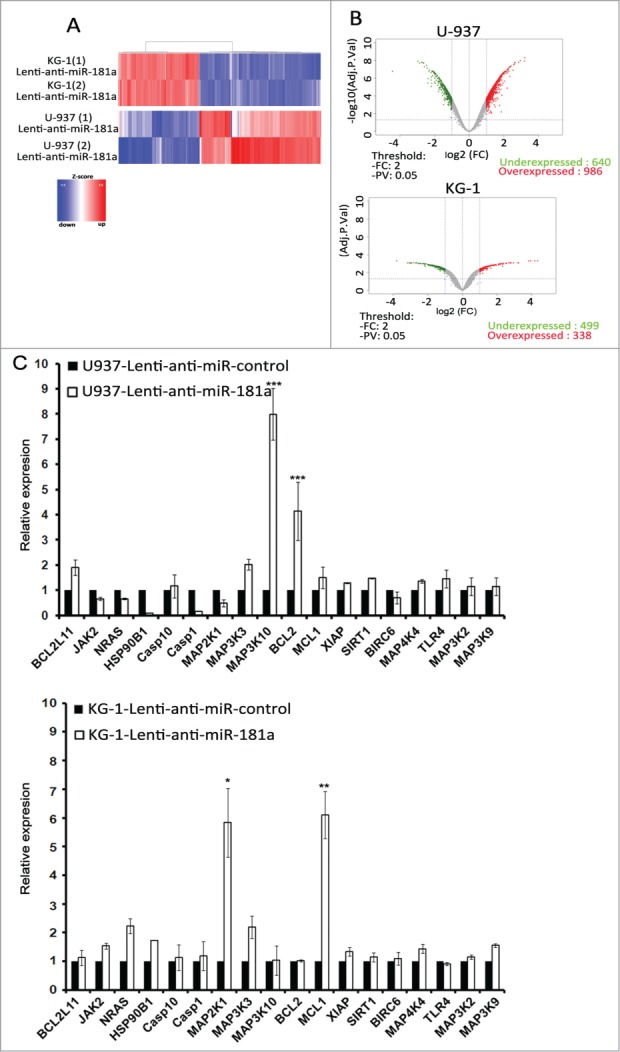
miR-181a target genes. (A) Genome-wide analysis of target genes regulated by miR-181a. KG-1 and U-937 leukemia cells were transduced by Lenti-anti-miR-181a. Shown are gene expression data from 2 independent experiments using Agilent Human Whole Genome Microarray. (B) Volcano plot of gene expression (log2 fold change) and adjusted p values for KG-1 and U-937 transduced cells. (C) SYBR-GREEN qRT-PCR measurement of indicated potential targets of miR-181a in KG-1 and U-937 cells transduced by Lenti-anti-miR-181a or Lenti-anti-miR control (representative graph of n = 3). Expression level of GAPDH was used as endogenous control.
Figure 5.
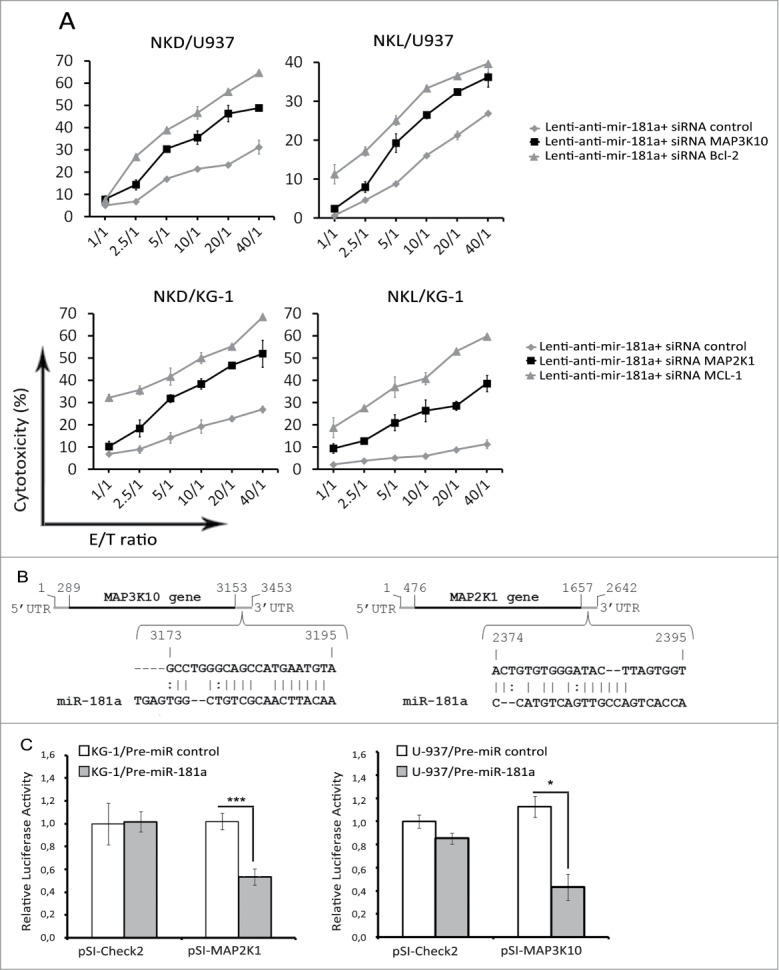
miR-181a modulates susceptibility of leukemia cells to NK lysis through regulation of genes from the Bcl-2 family and the Tyrosine kinase family. (A) U-937 expressing anti-miR-181a was transfected with siRNAs specific for Bc--2 and MAP3K10 or luciferase as control. KG-1 transduced cells expressing anti-miR-181a were transfected with siRNAs specific for MCL-1 and MAP2K1 or luciferase as control. NKL and NKD were used as effector cells. Data represent three independent experiments. (B) Schematic view of predicted miR‑181a binding sites in MAP3K10 and MAp2K1 3′ UTRs, which has been cloned into the psiCHECK2 vector. (C) U-937 and KG-1 cells were co-transfected with 10 nmol/L pre-miR control or pre-miR-181a and either psi-Check empty vector (pSI-Check2) or psi-Check vector encoding 3′UTR of MAP2K1 (pSI-MAP2K1) or MAP3K10 (pSI-MAP3K10). After 48 h, cells were harvested and luciferase activities analyzed. All Renilla luciferase activities were normalized to the firefly luciferase activity. pSI-MAP3K10 and pSI-MAP2K1 correspond to distinct fragments of MAP3K10 and MAP2K1 3′ UTR containing miR-181a putative binding sites. Cell viability 24 h post transfection was around 80%.
Taken together, the results provide evidence that miR-181a modulates tumor cell susceptibility to NK-mediated lysis by targeting different genes in different leukemia cell lines.
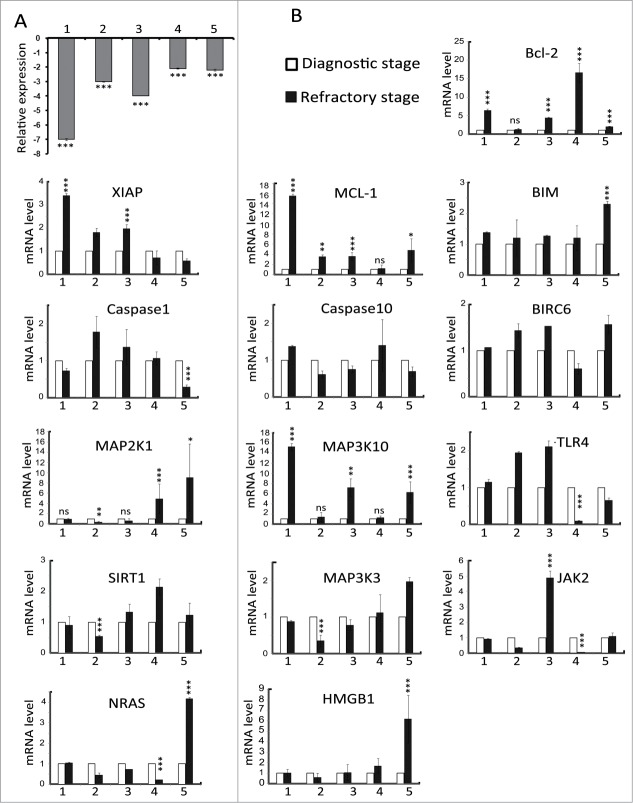
miR-181a is downregulated in refractory stage of AML primary blasts
Expression of miR-181a in cells from 5 AML patients at both the diagnostic and refractory stage was measured and compared. The results indicated that expression of miR-181a was downregulated in all refractory stage samples when compared to those from the diagnostic stage (Fig. 6A). Next, we investigate whether the expression of universal miR-181a target genes was modified in AML patients. qRT-PCR results showed that Mcl-1 and MAP3K10 were the most frequently modified genes (in four patients) and none of the potential target genes were modified in all patients (Fig. 6B). Collectively these results provide evidence that miR-181a is downregulated in drug-resistant blasts but that the target genes differ among AML patients, and that there is not a universal target for miR-181a.
Figure 6.
miR-181a and candidate target gene expression in LAM primary cells. (A) miR-181a expression was measured at diagnostic and refractory stages in five AML patients. Expression levels of RNU44 were used as endogenous control (Representative graph of n = 3). (B) mRNA level of indicated potential targets of miR-181a were measured by SYBR-GREEN qRT-PCR at diagnostic and refractory stage in five AML patients. Expression level of GAPDH was used as endogenous control.
miR-181a induces cross-resistance in refractory AML blasts to NK cells
To determine the impact of miR-181a in primary cell susceptibility to NK lysis, blasts from two AML patients at refractory stage were transfected with pre-miR-181a and subsequently used as target cells in cytotoxic assay. As shown in Fig. 7A, overexpression of miR-181a in primary blasts resulted in an increase in blast sensitivity to NK cell lysis compared to control cells. We next investigate whether miR-181a is capable to modulate drug resistance and whether its induction could reverse AML blasts chemoresistance to DNR. To address this issue, AML blasts were transfected with pre-miR181 or pre-miR control and incubated with DNR for two days. Our results shown in Fig. 7B indicate that cells became partially sensitive to DNR treatment in pre-miR181a transfected cells as compared with control cells, indicating that miR-181 plays a crucial role in the cross resistance of leukemia cells.
Figure 7.
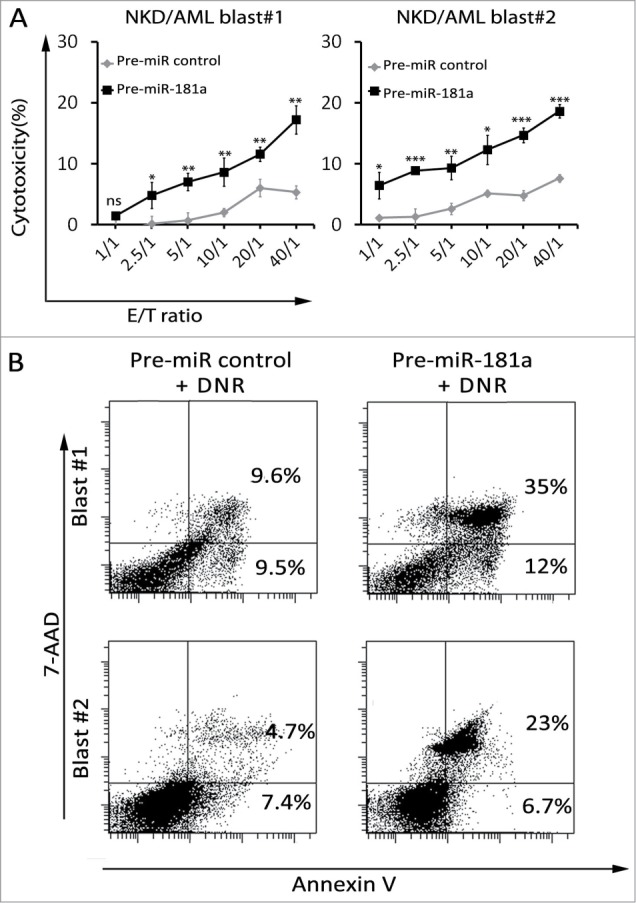
Effects of forced expression of miR-181a on AML primary cells susceptibility to NK cells and DNR treatment. (A) AML blasts (#1 and #2) of patients at refractory stage were transfected with pre-miR181a or pre-miR control and cytotoxic assay was performed at E: T indicated ratios. NKD cells were used as effector cells. (B) The percentage of apoptosis of leukemia blasts after transfection with pre-miR181a or pre-miR control. Leukemia blasts were incubated with DNR for 48 h and the percentage of apoptosis was measured by flow cytometry using an Annexin V/7AAD detection kit.
Discussion
Beneficial effects of donor-derived NK cells in allogenic hematopoietic stem cell transplantation have been demonstrated in different trials. Not only they can eliminate leukemic cells (graft-vs.-leukemia effect) and contribute to prevention of relapse, but they can also destroy T lymphocytes and dendritic cells in the recipient, thereby blocking graft-vs.-host-disease. Thus, the anti-leukemic potential of alloreactive NK cells may offer an ideal approach to cell-based immunotherapy; therefore, adoptive transfer of ex vivo activated NK cells is under investigation in phase I and II clinical trials.43 However, one of the major obstacles that could minimize the positive effect of NK cell administration is the development of cross-resistance. In fact, AML blasts could become resistant to an anti-leukemia drug because of extended chemotherapy, and chemoresistance could also influence their susceptibility to immune effector cells.
In this study, we investigated the molecular basis of the cross-resistance mechanism. Our results indicate that in vitro the acquisition of DNR resistance resulted in the induction of cross-resistance to NK-mediated lysis. More importantly, we demonstrate that NK cell resistance is not due to the modification of NK receptor ligands on the leukemia cell surface, but is associated with an intrinsic mechanism, conferring resistance to GrB treatment as well.
While recent studies have revealed the importance of miRNAs in leukemia disorders and more specifically in drug resistance, their potential involvements in immune response are not well understood. In this study, we showed that miR-181a and miR-146a are both downregulated in DNR-resistant cells. Our findings further indicate that inhibition of miR-181a in leukemia cells impacted their susceptibility to NK-mediated lysis. Although emerging studies indicate the potential involvement of miR-146a in cell proliferation and drug resistance,44,45 blocking miR-146a in our experimental model had no effect on NK-cell mediated lysis. Therefore, we hypothesized that miR-146a is involved in the molecular mechanisms that contribute to drug resistance but not to resistance to NK-mediated lysis. We next attempted to identify the potential genes targets for miR-181a. DNA micro-array analysis performed on cells stably transduced with anti-miR-181a provided surprising evidence that target genes of miR-181a could vary from one cell line to another, despite the capacity of miRNA to induce resistance to NK-cell mediated lysis and GrB treatment. Nevertheless, our results indicate that miR-181a can modify multiple target genes from the Bcl-2 family and tyrosine kinase family. It should be noted that most of the studies about the role of miR-181a in DNR-resistance in leukemia or other models have been performed in only one cell line46-49 and the effects of inhibition have not been compared in different models, except in one recent study where the inhibition of miR-181a (by transfection but not transduction) resulted in the modification of tree members of Bcl-2 family (Bcl-2, Mcl-1, and XIAP) in the all tested leukemia blasts.50 The current studies clearly indicate the importance of miR-181a in the acquisition of drug resistance and in its potential to modulate a vast variety of target genes from different pathways. We have also attempted to validate our observations in primary blasts. It should be emphasized that we were limited in the selection of patients given the difficulty of obtaining blasts from patients at both drug-resistant and sensitive stages. The five patients studied were selected from among 200 candidates for follow-up experiments. The miR-181a expression and all potential target genes of miR-181a were analyzed. We observed that expression of miR-181a was downregulated in all tested patients, although we did not find a gene consistently modified in all samples. Recently, in a similar study, it has been demonstrated miR-181b (another family of this microRNA) increases drug sensitivity in AML by targeting HMGB1 and MCL-1.51 In this report, the authors found modification of these genes in all three tested patients. AML comprises a heterogeneous group of malignancies with diverse origins. Based on our results, we strongly believe that this heterogeneity could describe the diversity of target genes in different patients and leukemia cell lines. DNR is a powerful anti-leukemia agent with the capacity to modulate multiple cellular signaling pathways.52 However, it remains unclear how DNR modifies the expression of miR itself. Preliminary results indicate that NF-κB can interfere with the expression of miR-181a (data not shown).Finally, and most importantly, we showed the ectopic expression of miR-181a sensitize cells to NK cell mediate killing and DNR treatment. Recently Marcucci and colleagues identified a new mechanism to deliver miRNAs into the primary cells.53 This procedure will be tested in our model in order to optimize the outcomes.
The results presented in this paper strongly support the potential of miR-targeted therapy as a novel and promising anti-leukemia approach against “onco-immuno” resistant cells where traditional chemotherapy or immunotherapy protocols with highly cytotoxic effector cells alone may be insufficiently effective.
Materials and Methods
Culture of cell lines
Human AML cell lines were grown in RPMI 1640 medium supplemented with 10% FCS (Seromed) and 1% penicillin-streptomycin. Human NK cells were grown in RPMI 1640 medium supplemented with 15% FCS (Seromed), 1% penicillin-streptomycin, and 300 IU/mL interleukin-2 (IL-2).
Cytotoxicity assay
The cytotoxic activity of the NK cells was measured by a conventional 4-h 51Cr release assay by using triplicate cultures in round-bottom 96-well plates. Different E: T ratios were used as described earlier.23
Abs, reagents
Monoclonal antibodies (mAb) recognizing DR4 (TRAIL-R1), DR5 (TRAIL-R2), and Fas (CD95) were purchased from Biolegend. mAbs directed against ULBP-1,ULBP-2, and ULBP-3 were purchased from R&D system. Anti-CD107a coupled to CyChrome mAb was provided by Becton Dickinson. MHC class I (MHC-I) (W6/32) antibody, obtained from biolegend. Abs against Bcl-2 and MCL-1, were from Cell Signaling Technology.
Flow cytometry analysis
Flow cytometry analysis was performed using a FACSCalibur flow and LSRII cytometer. Data were processed using CellQuest software (BD Biosciences).
NK degranulation assay
NK cells were cocultured with LAM target cells for 4 h at a 2:1 E:T ratio. Degranulation of NK cells was analyzed by flow cytometric analysis of CD107a expression.
miRNA microarray analysis
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RNA samples were prepared from two types of cells, KG-1 and KG-1(R) cells. RNA quality was confirmed using an Agilent 2100 Bioanalyzer (Agilent Technologies). Each microarray includes approximately 15 k features containing probes sourced from the miRBASE public database, release 13.0 (Griffiths-Jones, NAR 2008). The probes are 40-60-mer oligonucleotides directly synthesized on the array, with a special design to cope with the small size of miRNA. In this study we used Human microRNA microarray v3.0, which contains 851 human and 88 human viral miRNAs, each replicated 16 times. The array also contains a set of positive and negative controls that are replicated a variety of times.
RNA silencing
Specific BCL-2 siRNA (sc-61899), MCL-1 siRNA (sc-35877) and negative control siRNA (sc-37007) were purchased from Santa Cruz Biotechnology, Inc.. Specific MAP2K1 and MAP3K10 siRNA were obtained from Sigma. siRNA was transfected into U937 or KG-1 cells by electroporation (Amaxa, Gaithersburg, MD) following the manufacturer's instructions.
RNA isolation and quantitative analysis for miR-181a
For extraction of miRNAs, TRIzol (Invitrogen) was used. DNase I-treated total RNA (8 ng) was subjected to qRT-PCR analysis using TaqMan miR Reverse Transcription Kit (Applied Biosystems). The miR-181a was detected and quantified by using specific miRNA primers from Ambion. Expression levels of mature miRNAs were evaluated using comparative Ct method (2- ΔCt). Transcript levels of RNU44 were used as endogenous control.
Lentiviral constructs and stably transduced cells
Lentiviral miRNA Inhibitor for miR-181a and empty vector as negative control were purchased from abm (Richmond, Canada). KG-1 and U-937 cells were infected with 1–105 recombinant lentivirus-transducing units in the presence of 6 mg/mL polybrene (Sigma). GFP positive cells isolated by FACSAria III Cell Sorter and stable cells were established.
miR-181a blockade and overexpression
Transfections were realized with anti-miR-181a (miR inhibitor, Ambion) and pre-miR-181a (miR precursor, Ambion) in leukemia cell lines as described earlier. Leukemia primary cells of two patients at refractory relapsed stage transfected in two different ways: NeoFx Transfection Agent (NeoFx; Ambion), and electroporation (Amaxa Reagent T, program U-15 and S-04) used according to manufacturer's instructions.
Gene expression microarray
DNA microarray (Agilent Human microRNA microarray v3.0 (design 021827) 4 × 44 k Microarray (AMADID 39494 - 60,000 spots)) was performed to assess and compare overall gene expression profiles of KG-1 lenti-anti-miR-CT and U937 lenti-anti-miR-CT cells compared respectively to KG-1 lenti-anti-miR-181a and U937 lenti-anti-miR-181a. Analysis was performed to identify genes differentially expressed after miR-181a knockdown in AML target cells with a fold change >2.
Loading of GrB
Analysis of tumor cell sensitivity to GrB was performed as previously described.42
AML sampels
AML blasts of five patients at diagnostic stage (prior to any chemotherapy) and their blasts at refractory relapsed stage (resistance to chemotherapy) were chosen for further experiments.
Statistical analyses
Data were analyzed with GraphPad Prism. The Student t test was used for single comparisons. Data were considered statistically significant when p < 0.05. (*, p < 0.05; **, p < 0.005; and ***, p < 0.0005).
Authorship
Contributions: A.N. and G.V. established the cell lines. A.N performed the experiments. A.N., S.C., and J-H.B designed the study and wrote the manuscript and all authors critically reviewed the manuscript and gave their final approval.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Institut National du Cancer, Association Laurette Fugain, and the Qatar Foundation.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Estey E, Dohner H. Acute myeloid leukaemia. Lancet 2006; 368:1894-907; PMID:17126723; http://dx.doi.org/ 10.1016/S0140-6736(06)69780-8 [DOI] [PubMed] [Google Scholar]
- 2. Lowenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Eng J Med 1999; 341:1051-62; PMID:10502596; http://dx.doi.org/ 10.1056/NEJM199909303411407 [DOI] [PubMed] [Google Scholar]
- 3. Shipley JL, Butera JN. Acute myelogenous leukemia. Exp Hematol 2009; 37:649-58; PMID:19463767; http://dx.doi.org/ 10.1016/j.exphem.2009.04.002 [DOI] [PubMed] [Google Scholar]
- 4. Berman E, Arlin ZA, Gaynor J, Miller W, Gee T, Kempin SJ, Mertelsmann R, Andreeff M, Reich L, Nahmias N, et al. Comparative trial of cytarabine and thioguanine in combination with amsacrine or daunorubicin in patients with untreated acute nonlymphocytic leukemia: results of the L-16M protocol. Leukemia 1989; 3:115-21; PMID:2911205 [PubMed] [Google Scholar]
- 5. Forman SJ, Nademanee AP, O'Donnell MR, Fahey JL, Snyder DS, Farbstein MJ, Lazar GS, DuBois NJ, Blume KG. High-dose cytosine arabinoside and daunomycin as primary therapy for adults with acute nonlymphoblastic leukemia: a pilot study. Semin Oncol 1985; 12:114-6; PMID:4012336 [PubMed] [Google Scholar]
- 6. Hansen OP, Pedersen-Bjergaard J, Ellegaard J, Brincker H, Boesen AM, Christensen BE, Drivsholm A, Hippe E, Jans H, Jensen KB, et al. Aclarubicin plus cytosine arabinoside versus daunorubicin plus cytosine arabinoside in previously untreated patients with acute myeloid leukemia: a Danish national phase III trial. The Danish Society of Hematology Study Group on AML, Denmark. Leukemia 1991; 5:510-6; PMID:2056774 [PubMed] [Google Scholar]
- 7. Rai KR, Holland JF, Glidewell OJ, Weinberg V, Brunner K, Obrecht JP, Preisler HD, Nawabi IW, Prager D, Carey RW, et al. Treatment of acute myelocytic leukemia: a study by cancer and leukemia group B. Blood 1981; 58:1203-12; PMID:6946847 [PubMed] [Google Scholar]
- 8. Weick JK, Kopecky KJ, Appelbaum FR, Head DR, Kingsbury LL, Balcerzak SP, Bickers JN, Hynes HE, Welborn JL, Simon SR, et al. A randomized investigation of high-dose versus standard-dose cytosine arabinoside with daunorubicin in patients with previously untreated acute myeloid leukemia: a Southwest Oncology Group study. Blood 1996; 88:2841-51; PMID:8874180 [PubMed] [Google Scholar]
- 9. Weil M, Glidewell OJ, Jacquillat C, Levy R, Serpick AA, Wiernik PH, Cuttner J, Hoogstraten B, Wasserman L, Ellison RR, et al. Daunorubicin in the therapy of acute granulocytic leukemia. Cancer Res 1973; 33:921-8; PMID:4512953 [PubMed] [Google Scholar]
- 10. Wolff SN, Herzig RH, Fay JW, Phillips GL, Lazarus HM, Flexner JM, Stein RS, Greer JP, Cooper B, Herzig GP. High-dose cytarabine and daunorubicin as consolidation therapy for acute myeloid leukemia in first remission: long-term follow-up and results. J Clin Oncol 1989; 7:1260-7; PMID:2769327 [DOI] [PubMed] [Google Scholar]
- 11. Richardson DS, Johnson SA. Anthracyclines in haematology: preclinical studies, toxicity and delivery systems. Blood Rev 1997; 11:201-23; PMID:9481450; http://dx.doi.org/ 10.1016/S0268-960X(97)90020-5 [DOI] [PubMed] [Google Scholar]
- 12. Funato T, Harigae H, Abe S, Sasaki T. Assessment of drug resistance in acute myeloid leukemia. Exp Rev Mol Diagnost 2004; 4:705-13; PMID:15347263; http://dx.doi.org/ 10.1586/14737159.4.5.705 [DOI] [PubMed] [Google Scholar]
- 13. Schiller GJ. Treatment of resistant disease. Leukemia 1998; 12 Suppl 1:S20-4; PMID:9452028 [PubMed] [Google Scholar]
- 14. Shaffer BC, Gillet JP, Patel C, Baer MR, Bates SE, Gottesman MM. Drug resistance: still a daunting challenge to the successful treatment of AML. Drug Resist Updat 2012; 15:62-9; PMID:22409994; http://dx.doi.org/ 10.1016/j.drup.2012.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lussana F, Rambaldi A, Finazzi MC, van Biezen A, Scholten M, Oldani E, Carobbio A, Iacobelli S, Finke J, Nagler A, et al. Allogeneic hematopoietic stem cell transplantation in patients with polycythemia vera or essential thrombocythemia transformed to myelofibrosis or acute myeloid leukemia: a report from the MPN Subcommittee of the Chronic Malignancies Working Party of the European Group for Blood and Marrow Transplantation. Haematologica 2014; 99:916-21; PMID:24389309; http://dx.doi.org/ 10.3324/haematol.2013.094284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Platzbecker U. Who benefits from allogeneic transplantation for myelodysplastic syndromes?: new insights. Hematology Am Soc Hematol Educ Program 2013; 2013:522-8; PMID:24319227 [DOI] [PubMed] [Google Scholar]
- 17. Classen CF, Falk CS, Friesen C, Fulda S, Herr I, Debatin KM. Natural killer resistance of a drug-resistant leukemia cell line, mediated by up-regulation of HLA class I expression. Haematologica 2003; 88:509-21; PMID:12745270 [PubMed] [Google Scholar]
- 18. Classen CF, Fulda S, Friesen C, Debatin KM. Decreased sensitivity of drug-resistant cells towards T cell cytotoxicity. Leukemia 1999; 13:410-8; PMID:10086732; http://dx.doi.org/ 10.1038/sj.leu.2401335 [DOI] [PubMed] [Google Scholar]
- 19. Debatin KM. Cytotoxic drugs, programmed cell death, and the immune system: defining new roles in an old play. J Natl Cancer Ins 1997; 89:750-1; PMID:9182966; http://dx.doi.org/ 10.1093/jnci/89.11.750 [DOI] [PubMed] [Google Scholar]
- 20. Friesen C, Fulda S, Debatin KM. Deficient activation of the CD95 (APO-1/Fas) system in drug-resistant cells. Leukemia 1997; 11:1833-41; PMID:9369415; http://dx.doi.org/ 10.1038/sj.leu.2400827 [DOI] [PubMed] [Google Scholar]
- 21. Komada Y, Zhou YW, Zhang XL, Chen TX, Tanaka S, Azuma E, Sakurai M. Fas/APO-1 (CD95)-mediated cytotoxicity is responsible for the apoptotic cell death of leukaemic cells induced by interleukin-2-activated T cells. British J Haematol 1997; 96:147-57; PMID:9012700; http://dx.doi.org/ 10.1046/j.1365-2141.1997.8742505.x [DOI] [PubMed] [Google Scholar]
- 22. Micheau O, Solary E, Hammann A, Martin F, Dimanche-Boitrel MT. Sensitization of cancer cells treated with cytotoxic drugs to fas-mediated cytotoxicity. J Nl Cancer Ins 1997; 89:783-9; PMID:9182976; http://dx.doi.org/ 10.1093/jnci/89.11.783 [DOI] [PubMed] [Google Scholar]
- 23. Nanbakhsh A, Pochon C, Mallavialle A, Amsellem S, Bourhis JH, Chouaib S. c-Myc regulates expression of NKG2D ligands ULBP1/2/3 in AML and modulates their susceptibility to NK-mediated lysis. Blood 2014; 123:3585-95; PMID:24677544; http://dx.doi.org/ 10.1182/blood-2013-11-536219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Posovszky C, Friesen C, Herr I, Debatin KM. Chemotherapeutic drugs sensitize pre-B ALL cells for CD95- and cytotoxic T-lymphocyte-mediated apoptosis. Leukemia 1999; 13:400-9; PMID:10086731; http://dx.doi.org/ 10.1038/sj.leu.2401327 [DOI] [PubMed] [Google Scholar]
- 25. Renvoize C, Roger R, Moulian N, Bertoglio J, Breard J. Bcl-2 expression in target cells leads to functional inhibition of caspase-3 protease family in human NK and lymphokine-activated killer cell granule-mediated apoptosis. J Immunol 1997; 159:126-34; PMID:9200447 [PubMed] [Google Scholar]
- 26. Treichel RS, Bunuan M, Hahn N, Wee K. Altered conjugate formation and altered apoptosis of multidrug-resistant human leukemia cell line affects susceptibility to killing by activated natural killer (NK) cells. Int J Cancer 2004; 108:78-85; PMID:14618619; http://dx.doi.org/ 10.1002/ijc.11555 [DOI] [PubMed] [Google Scholar]
- 27. Curti A, Ruggeri L, D'Addio A, Bontadini A, Dan E, Motta MR, Trabanelli S, Giudice V, Urbani E, Martinelli G, et al. Successful transfer of alloreactive haploidentical KIR ligand-mismatched natural killer cells after infusion in elderly high risk acute myeloid leukemia patients. Blood 2011; 118:3273-9; PMID:21791425; http://dx.doi.org/ 10.1182/blood-2011-01-329508 [DOI] [PubMed] [Google Scholar]
- 28. Miller JS, Soignier Y, Panoskaltsis-Mortari A, McNearney SA, Yun GH, Fautsch SK, McKenna D, Le C, Defor TE, Burns LJ, et al. Successful adoptive transfer and in vivo expansion of human haploidentical NK cells in patients with cancer. Blood 2005; 105:3051-7; PMID:15632206; http://dx.doi.org/ 10.1182/blood-2004-07-2974 [DOI] [PubMed] [Google Scholar]
- 29. Rubnitz JE, Inaba H, Ribeiro RC, Pounds S, Rooney B, Bell T, Pui CH, Leung W. NKAML: a pilot study to determine the safety and feasibility of haploidentical natural killer cell transplantation in childhood acute myeloid leukemia. J Clin Oncol 2010; 28:955-9; PMID:20085940; http://dx.doi.org/ 10.1200/JCO.2009.24.4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stern M, Passweg JR, Meyer-Monard S, Esser R, Tonn T, Soerensen J, Paulussen M, Gratwohl A, Klingebiel T, Bader P, et al. Pre-emptive immunotherapy with purified natural killer cells after haploidentical SCT: a prospective phase II study in two centers. Bone Marrow Transplant 2013; 48:433-8; PMID:22941380; http://dx.doi.org/ 10.1038/bmt.2012.162 [DOI] [PubMed] [Google Scholar]
- 31. Chen Y, Jacamo R, Konopleva M, Garzon R, Croce C, Andreeff M. CXCR4 downregulation of let-7a drives chemoresistance in acute myeloid leukemia. J Clin Invest 2013; 123:2395-407; PMID:23676502; http://dx.doi.org/ 10.1172/JCI66553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell 2010; 140:652-65; PMID:20211135; http://dx.doi.org/ 10.1016/j.cell.2010.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Garzon R, Pichiorri F, Palumbo T, Visentini M, Aqeilan R, Cimmino A, Wang H, Sun H, Volinia S, Alder H, et al. MicroRNA gene expression during retinoic acid-induced differentiation of human acute promyelocytic leukemia. Oncogene 2007; 26:4148-57; PMID:17260024; http://dx.doi.org/ 10.1038/sj.onc.1210186 [DOI] [PubMed] [Google Scholar]
- 34. Garzon R, Volinia S, Liu CG, Fernandez-Cymering C, Palumbo T, Pichiorri F, Fabbri M, Coombes K, Alder H, Nakamura T, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood 2008; 111:3183-9; PMID:18187662; http://dx.doi.org/ 10.1182/blood-2007-07-098749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Havelange V, Ranganathan P, Geyer S, Nicolet D, Huang X, Yu X, Volinia S, Kornblau SM, Andreeff M, Croce CM, et al. Implications of the miR-10 family in chemotherapy response of NPM1-mutated AML. Blood 2014; 123:2412-5; PMID:24596420; http://dx.doi.org/ 10.1182/blood-2013-10-532374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hickey CJ, Schwind S, Radomska HS, Dorrance AM, Santhanam R, Mishra A, Wu YZ, Alachkar H, Maharry K, Nicolet D, et al. Lenalidomide-mediated enhanced translation of C/EBPalpha-p30 protein up-regulates expression of the antileukemic microRNA-181a in acute myeloid leukemia. Blood 2013; 121:159-69; PMID:23100311; http://dx.doi.org/ 10.1182/blood-2012-05-428573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marcucci G, Maharry KS, Metzeler KH, Volinia S, Wu YZ, Mrozek K, Nicolet D, Kohlschmidt J, Whitman SP, Mendler JH, et al. Clinical role of microRNAs in cytogenetically normal acute myeloid leukemia: miR-155 upregulation independently identifies high-risk patients. J Clin Oncol 2013; 31:2086-93; PMID:23650424; http://dx.doi.org/ 10.1200/JCO.2012.45.6228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marcucci G, Mrozek K, Radmacher MD, Garzon R, Bloomfield CD. The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood 2011; 117:1121-9; PMID:21045193; http://dx.doi.org/ 10.1182/blood-2010-09-191312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marcucci G, Radmacher MD, Maharry K, Mrozek K, Ruppert AS, Paschka P, Vukosavljevic T, Whitman SP, Baldus CD, Langer C, et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Eng J Med 2008; 358:1919-28; PMID:18450603; http://dx.doi.org/ 10.1056/NEJMoa074256 [DOI] [PubMed] [Google Scholar]
- 40. Mims A, Walker AR, Huang X, Sun J, Wang H, Santhanam R, Dorrance AM, Walker C, Hoellerbauer P, Tarighat SS, et al. Increased anti-leukemic activity of decitabine via AR-42-induced upregulation of miR-29b: a novel epigenetic-targeting approach in acute myeloid leukemia. Leukemia 2013; 27:871-8; PMID:23178755; http://dx.doi.org/ 10.1038/leu.2012.342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schwind S, Maharry K, Radmacher MD, Mrozek K, Holland KB, Margeson D, Whitman SP, Hickey C, Becker H, Metzeler KH, et al. Prognostic significance of expression of a single microRNA, miR-181a, in cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol 2010; 28:5257-64; PMID:21079133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hamai A, Meslin F, Benlalam H, Jalil A, Mehrpour M, Faure F, Lecluse Y, Vielh P, Avril MF, Robert C, et al. ICAM-1 has a critical role in the regulation of metastatic melanoma tumor susceptibility to CTL lysis by interfering with PI3K/AKT pathway. Cancer Res 2008; 68:9854-64; PMID:19047166; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-0719 [DOI] [PubMed] [Google Scholar]
- 43. Locatelli F, Moretta F, Brescia L, Merli P. Natural killer cells in the treatment of high-risk acute leukaemia. Seminars in immunology 2014; 26:173-9; PMID:24613727; http://dx.doi.org/ 10.1016/j.smim.2014.02.004 [DOI] [PubMed] [Google Scholar]
- 44. Chen G, Umelo IA, Lv S, Teugels E, Fostier K, Kronenberger P, Dewaele A, Sadones J, Geers C, De Greve J. miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PloS one 2013; 8:e60317; PMID:23555954; http://dx.doi.org/ 10.1371/journal.pone.0060317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pogribny IP, Filkowski JN, Tryndyak VP, Golubov A, Shpyleva SI, Kovalchuk O. Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int J Cancer 2010; 127:1785-94; PMID:20099276; http://dx.doi.org/ 10.1002/ijc.25191 [DOI] [PubMed] [Google Scholar]
- 46. Cao YX, Dai CW, Zhang GS. Screening for drug resistance related microRNAs in K562 and K562/A02 cell lines. Zhonghua xue ye xue za zhi = Zhonghua xueyexue zazhi 2010; 31:361-5. [PubMed] [Google Scholar]
- 47. Dahlhaus M, Schult C, Lange S, Freund M, Junghanss C. MicroRNA 181a influences the expression of HMGB1 and CD4 in acute Leukemias. Anticancer Res 2013; 33:445-52; PMID:23393335 [PubMed] [Google Scholar]
- 48. Li H, Hui L, Xu W. miR-181a sensitizes a multidrug-resistant leukemia cell line K562/A02 to daunorubicin by targeting BCL-2. Acta Biochim Biophys Sin 2012; 44:269-77; PMID:22285729; http://dx.doi.org/ 10.1093/abbs/gmr128 [DOI] [PubMed] [Google Scholar]
- 49. Ouyang YB, Lu Y, Yue S, Giffard RG. miR-181 targets multiple Bcl-2 family members and influences apoptosis and mitochondrial function in astrocytes. Mitochondrion 2012; 12:213-9; PMID:21958558; http://dx.doi.org/ 10.1016/j.mito.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhu DX, Zhu W, Fang C, Fan L, Zou ZJ, Wang YH, Liu P, Hong M, Miao KR, Liu P, et al. miR-181a/b significantly enhances drug sensitivity in chronic lymphocytic leukemia cells via targeting multiple anti-apoptosis genes. Carcinogenesis 2012; 33:1294-301; PMID:22610076; http://dx.doi.org/ 10.1093/carcin/bgs179 [DOI] [PubMed] [Google Scholar]
- 51. Lu F, Zhang J, Ji M, Li P, Du Y, Wang H, Zang S, Ma D, Sun X, Ji C. miR-181b increases drug sensitivity in acute myeloid leukemia via targeting HMGB1 and Mcl-1. Int Oncol 2014; 45:383-92; PMID:24756163 [DOI] [PubMed] [Google Scholar]
- 52. Laurent G, Jaffrezou JP. Signaling pathways activated by daunorubicin. Blood 2001; 98:913-24; PMID:11493433; http://dx.doi.org/ 10.1182/blood.V98.4.913 [DOI] [PubMed] [Google Scholar]
- 53. Huang X, Schwind S, Yu B, Santhanam R, Wang H, Hoellerbauer P, Mims A, Klisovic R, Walker AR, Chan KK, et al. Targeted delivery of microRNA-29b by transferrin-conjugated anionic lipopolyplex nanoparticles: a novel therapeutic strategy in acute myeloid leukemia. Clin Cancer Res 2013; 19:2355-67; PMID:23493348 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.