Abstract
The instability of the CAG repeat size of the HD gene when transmitted intergenerationally has critical implications for genetic counseling practices. In particular, CAG repeats between 27 and 35 have been the subject of debate based on small samples. To address this issue, we analyzed allelic instability in the Venezuelan HD kindreds, the largest and most informative families ascertained for HD. We identified 647 transmissions. Our results indicate that repeats in the 27–35 CAG range are highly stable. Out of 69 transmitted alleles in this range, none expand into any penetrant ranges. Contrastingly, 14% of alleles transmitted from the incompletely penetrant range (36–39 CAGs) expand into the completely penetrant range, characterized by alleles with 40 or more CAG repeats. At least 12 of the 534 transmissions from the completely penetrant range contract into the incompletely penetrant range of 36–39 CAG repeats. In these kindreds, none of the individuals with 27–39 CAGs were symptomatic, even though they ranged in age from 11 to 82 years. We expect these findings to be helpful in updating genetic counseling practices.
Keywords: trinucleotide repeat, Huntington’s disease, genetic counseling
The length of the CAG repeat sequence in the Huntington’s disease (HD) gene underlies the pathogenesis of the condition. The instability of the CAG repeat length during transmission from parent to child can alter the penetrance of the allele and is of paramount importance for genetic counseling, including prenatal counseling. An especially challenging issue for families and counselors is ascertaining the frequency with which CAG repeat lengths less than the CAG threshold for complete penetrance (40 or greater) expand during transmission [Goldberg et al., 1993a,b; McGlennen et al., 1995; Nance et al., 1998; Maat-Kievit et al., 2001].
HD CAG repeat allele lengths between 36 and 39 repeats have been associated with clinically documented cases of HD. It has been indicated that the stability of alleles with greater than 35 repeats is correlated with the length of the originating repeat length [Wheeler et al., 2007]. While alleles with repeats in the range 36–39 CAGs are therefore expected to have a greater stability than alleles with longer repeats, repeat lengths in this range have been noted to frequently expand during transmission into the penetrant range of 40 repeats. HD alleles in the 27–35 CAG repeat range present a challenge. There are no cases of HD convincingly associated with this repeat range [Kenney et al., 1997]. The frequency of such alleles is estimated to be 1.9% in the general population [Goldberg et al., 1995].
In developing counseling guidelines, it was noted that there is a low risk that a 27–35 repeat allele increases in length upon transmission. However, some findings have led to a tempering of this statement. Specifically Goldberg et al. [1995] reported that intergenerational changes in allele length were observed in new mutation families with allele lengths in the 27–35 range. They claimed that these allele length changes were more frequent in families with new mutations compared to similar sized allele changes within the general population.
A further study using single sperm analysis of four individuals showed that two males in new mutation families had an apparent higher mutation frequency in sperm than the two males from the general population with the same CAG sizes [Chong et al., 1997]. This study estimated that the risk of inheriting an allele that mutates into the affected range, even in a new mutation family, is still extremely low (the risk of expansion of an allele to greater than 35 CAGs was reported to be 2.25% for a sibling of a person with a new mutation). Nevertheless, current counseling guidelines suggest that individuals who have 27–35 CAG repeats in their HD allele be advised that they have a significant risk of transmitting a penetrant allele to their children [Nance et al., 1998].
A more complete analysis of the frequency and characteristics of expansions from the 27–35 repeat range of HD alleles into the affected range would clearly benefit genetic counseling for HD.
The 18,149 member 10-generational Venezuelan kindreds [Wexler et al., 2004] offer a unique potential to address the question of repeat instability in this range. These kindreds were critical to the studies in which the HD gene was originally localized [Gusella et al., 1983] and subsequently isolated [HDCRG, 1993]. The Venezuelans’ average age of onset is 34.35 ± 10.07 years in contrast to the mean age of onset for Americans (37.47 ± 13.28) and Canadians (40.36 ± 12.97). Aside from a somewhat younger age of onset and, being a primarily Hispanic population, the Venezuelan kindreds are otherwise remarkably phenotypically similar to other HD kindreds worldwide. Findings that pertain to this population should generalize worldwide [Wexler et al., 2004].
For this study we divided Venezuelan kindred members into four groups with varying lengths of CAG repeats in their HD alleles defined as follows. Individuals with 26 CAG repeats and fewer (range I) are classified normal. Neither they nor any of their offspring have been observed to exhibit HD symptoms. Individuals having alleles between 27 and 35 CAG repeats (range II) also have no known risk of developing symptoms. Individuals with 36–39 CAG repeats (range III) display incomplete penetrance. Only a small number of individuals in this range manifest HD symptoms, usually with later onset. For individuals carrying 40 CAG repeats or more (range IV), the HD allele demonstrates complete penetrance, which invariably gives rise to disease over an average lifespan.
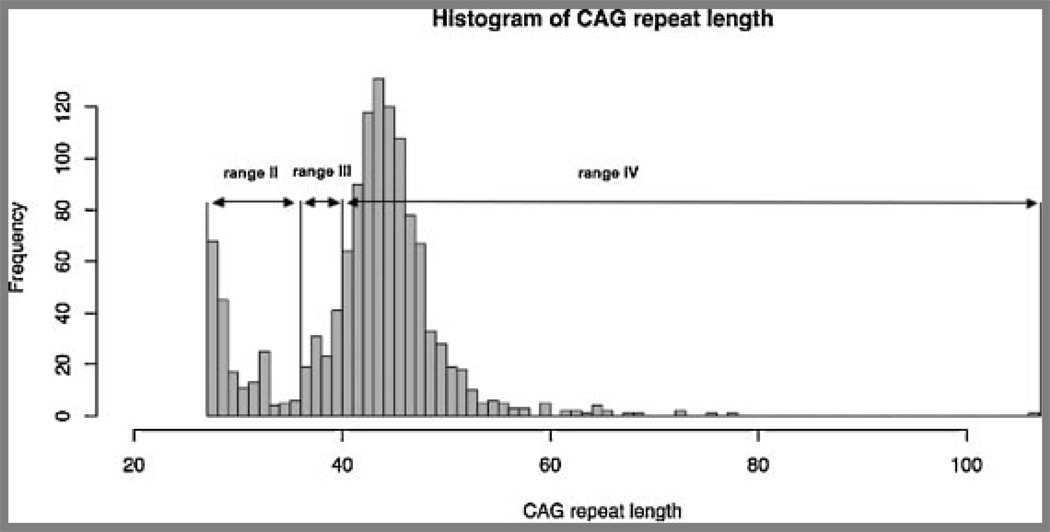
Figure 1 displays the distribution of CAG repeat lengths on both chromosomes in the ranges II, III, and IV for individuals in the Venezuelan kindreds. Range I is excluded for clarity. The distribution of CAG repeat lengths in these kindreds has previously been described as bimodal, representing the combination of separate curves, where one peak corresponds to the normal range of repeats and the other to the fully penetrant range [Wexler et al., 2004]. These two distributions slightly overlap for repeat lengths in the low 30s. Because the Venezuelan kindreds have a high prevalence of HD, the frequency of alleles in range IV is higher than would be found in other populations.
FIG. 1.
Frequency distribution of trinucleotide repeat lengths in normal and HD chromosomes. Repeat lengths are defined as follows: Range I has fewer than 26 CAGs. As no changes were noted, Range I is not included for clarity. Range II has between 27and 35 CAGs and is the particular emphasis of our study. Range III has between 36 and 39 CAGs and is incompletely penetrant. Range IV has 40 or more CAGs and is completely penetrant.
Intergenerational instability was measured as any change in CAG repeat length when transmitted from parent to offspring. This was calculated by subtracting the size of the parent’s CAG allele from the size of the CAG repeat length in the offspring to which it was transmitted. Alleles that could not be traced unambiguously due to homozygosity for the expanded allele in either the parent or child, or both parents carrying the expanded allele were omitted from the analysis. We traced a total of 647 allelic transmissions across the generations. The majority, 534, originated from range IV, 44 from range III, and 69 from range II.
We found no expansions of alleles with fewer than 27 repeats into other ranges. Individuals with alleles in this range (Range I) are therefore excluded in the following analyses.
We especially focused on individuals with 27–39 CAG repeats (ranges II and III) to clarify the frequency with which their alleles expand into the penetrant range and cause symptoms. We analyzed repeat length instability when these alleles are transmitted from generation to generation.
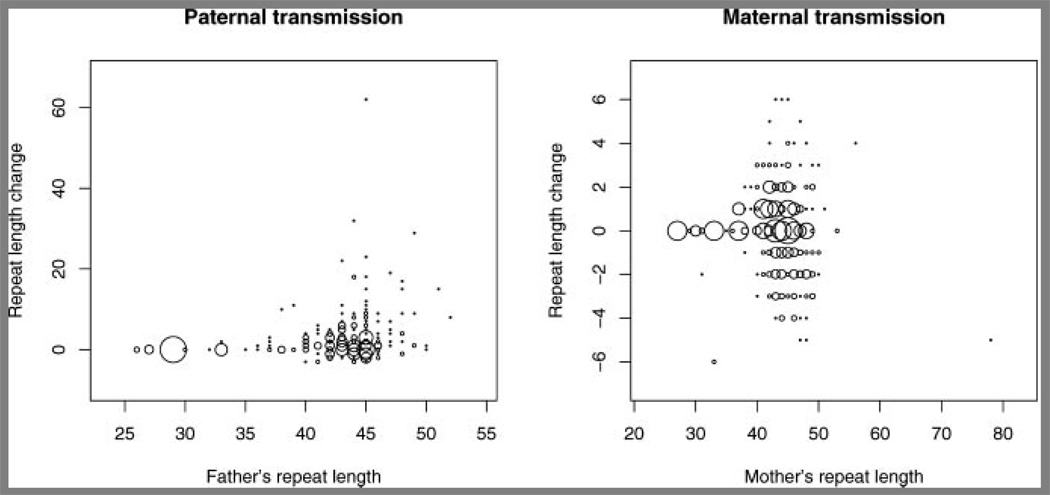
Figure 2 demonstrates that over all ranges, there is a strong parental sex effect on the magnitude of repeat instability during allelic transmission. A total of 268 paternal transmissions and 379 maternal transmissions were traced. Frequencies of intergenerational changes in the number of allelic CAG repeats described in the three ranges are summarized in Table I. Chi-square-tests of independence were performed to compare frequencies of changes for each pair of ranges. Transmission changes in all pairs of ranges were statistically different, with the most significant difference found between ranges II and IV (chi-square = 125.0, P < 0.001). Significant differences in stability were also found between ranges III and IV (chi-square = 26.2, P < 0.001), and between ranges II and III (chi-square = 20.0, P < 0.001).
FIG. 2.
Plot of repeat length change on transmission versus the originating parental repeat length, for paternal and maternal transmissions of alleles with 27 or more CAG repeats. The radius of the circle is proportional to the number of transmissions observed at each point. The smallest circle represents one observation and the largest circle represents 15 observations. A total of 268 paternal transmissions and 379 maternal transmissions were traced. Note that the scales of the plots for paternal and maternal transmissions differ.
TABLE I.
Transmission Counts for Alleles in Each of Three CAG Repeat Rang
| Range II 27–35 CAG repeats |
Range III 36–39 CAG repeats |
Range IV ≥40 CAG repeats |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Male | Female | Total | Male | Female | Total | Male | Female | |
| Transmissions | 69 | 32 | 37 | 44 | 15 | 29 | 534 | 221 | 313 |
| No change | 65 | 31 | 34 | 26 | 9 | 17 | 122 | 33 | 89 |
| Expansions | 1 | 1 | 0 | 17 | 6 | 11 | 264 | 145 | 118 |
| Contractions | 3 | 0 | 3 | 1 | 0 | 1 | 148 | 42 | 106 |
| Expansions into other ranges | 0 | 0 | 0 | 6 | 3 | 3 | Not applicable | Not applicable | Not applicable |
| Contractions into other ranges | 0 | 0 | 0 | 0 | 0 | 0 | 12 | 2 | 10 |
| Mean change | −0.17 | 0.06 | −0.38 | 0.91 | 1.87 | 0.41 | 1.33 | 3.22 | −0.01 |
Range I (26 CAGs or fewer) was omitted for clarity as there were no changes into any penetrant range. Table represents the total number of transmissions; transmissions of alleles with and without changes in repeat length; the mean change of the repeat length; and the number of expansions and contractions into other ranges. Statistics are broken down by sex. Bold text highlights totals in each range.
Alleles in range IV displayed a high level of instability with 77% (95% confidence interval [73%, 81%]) changing in the number of repeats on transmission. Two hundred sixty four were expansions (49%, 95% confidence interval [45%, 54%]) and 148 were contractions (28%, 95% confidence interval [24%, 32%]). Twelve alleles contracted from range IV to range III. These include two from 41 to 38 CAGs, one from 41 to 39 CAGs, two from 42 to 39 CAGs, one from 43 to 39 CAGs, and one from 40 to 37 CAGs. There were five other instances of contractions for which we could not determine the transmission due to homozygosity in the parent or the child, or both parents having a penetrant allele.
In range III, we observed 44 transmissions of alleles. Twenty six alleles were stable and did not change in size upon transmission. Six alleles crossed over from range III into range IV. Three of these expansions occurred as a result of maternal transmissions, changing from 38 to 40 CAGs, from 39 to 40 CAGs, and from 39 to 41 CAG repeats. Three others were paternal expansions, one in which an allele changed from 37 to 40 CAG repeats. The other two paternal expansions involved more marked changes from 39 to 50 CAG repeats and from 38 to 48 CAG repeats.
We also observed 12 intergenerational changes of alleles which did not lead to any crossover between ranges. Ten alleles expanded by a single CAG, and one expanded by two CAGs. One allele contracted during a paternal transmission from 38 to 37 CAG repeats. Our results demonstrate the relative instability of repeat lengths in range III (36 to 39 CAG repeats).We advocate counseling individuals with alleles in this CAG repeat range that if the allele is transmitted to their offspring there is a significant probability (6/44 = 0.14, 95% confidence interval [0.05, 0.27]) of the allele expanding into the fully penetrant range.
Of the 69 transmissions assessed for instability in range II (27 to 35 CAGs), only four alleles changed in size. A single expansion (1/69 = 0.01,95% confidence interval [0.00, 0.08]) was traced in which a 33 CAG repeat allele increased in length to a 35 CAG repeat allele. This occurred during a transmission of a father to his son. Two of the other changes occurred during maternal transmissions, both of which were relatively large contractions of six CAG repeats, going from 33 to 27 CAG repeats. Both contractions were transmissions from the same mother. The fourth change was a maternal contraction from 31 to 29 CAG repeats. No expansions were detected for repeat lengths with fewer that 33 CAG repeats. In fact, no instance of instability was detected for allele lengths between 27 and 30 CAG repeats. Finally and importantly, no alleles with fewer than 27 CAG repeats were observed to expand into any other ranges.
We observed no instances (0/69) of expansions from range II (27 to 35 CAG repeats) into either the fully penetrant or incompletely penetrant ranges. We do not observe the rare occurrence of such an expansion reported elsewhere [Kelly et al., 1999]. Our findings indicate that individuals with repeat lengths in range II have an low probability of transmitting an allele that expands into any penetrant range.
We also find that none of the 232 individuals in our kindreds with repeat lengths in the 27–39 CAG range have been diagnosed with symptomatic HD. They range in age from 11 to 82 years of age, and 100 of them are older than 34, the average age of onset in the kindreds.
Current guidelines are based on limited data, and could be misleading. They advise that there is a significant risk of expansion of alleles transmitted from the range of 27–35 CAGs, but also state that more clinical research is required to document the true risk. While we are not providing specific risk figures, based on the data presented here, we believe that it is appropriate to reassure families that the risk of an HD allele containing 27–35 CAG repeats expanding into either an incompletely penetrant or fully penetrant range is extremely low. Regardless of the sex of the transmitting parent, this low risk of expansion holds true. Collecting a larger data set in the future might help establish, with greater precision, the parameters of this risk of expansion.
We strongly recommend that information based on the data presented here be included in revising and updating counseling and practice guidelines [Nance et al., 1998]. Health professionals and family members alike have difficulty contending with the possibility that an HD allele in the 27–35 CAG range might expand upon transmission to the next generation. They are understandably worried that an expansion from this range could even potentially lead to full penetrance and disease in later generations. In the largest study to date, we analyzed 647 transmissions in all ranges, of which 69 are in this critical 27–35 CAG range. We can document no instances of any expansions into any penetrant ranges whatsoever. We are confident that individuals and members of families with HD will find our data reassuring.
ACKNOWLEDGMENTS
We are indebted to the Venezuelan families who have participated in this project throughout all these years. Their generous collaboration has been critical to the success of this project and many others. We also acknowledge support from the Hereditary Disease Foundation, NINDS, NIH, grant numbers: NS22031-15, EY-12562, W.M. Keck Foundation and the Wellcome Trust.
Grant sponsor: Hereditary Disease Foundation; Grant sponsor: NINDS; Grant sponsor: NIH; Grant numbers: NS22031-15, EY-12562; Grant sponsor: Keck Foundation; Grant sponsor: Wellcome Trust.
References
- Chong SS, Almqvist E, Telenius H, La Tray L, Nichol K, Bourdelat-Parks B, Goldberg YP, et al. Contribution of DNA sequence and CAG size to mutation frequencies of intermediate alleles for Huntington disease: Evidence from single sperm analyses. Hum Mol Genet. 1997;6:301–330. doi: 10.1093/hmg/6.2.301. [DOI] [PubMed] [Google Scholar]
- Goldberg YP, Kremer B, Andrew SE, et al. Molecular analysis of new mutations for Huntington’s disease: Intermediate alleles and sex of origin effects. Nat Genet. 1993;5:174–179. doi: 10.1038/ng1093-174. [DOI] [PubMed] [Google Scholar]
- Goldberg YP, Andrew SE, Theilmann J, et al. Familial predisposition to recurrent mutations causing Huntington’s disease: Genetic risk to sibs of sporadic cases. J Med Gen. 1993;30:987–990. doi: 10.1136/jmg.30.12.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg YP, McMurray CT, Zeisler J, Almqvist E, Sillence D, Richards F, Gacy AM, Buchanan J, Telenius H, Hayden MR. Increased instability of intermediate alleles in families with sporadic Huntington disease compared to similar sized intermediate alleles in the general population. Hum Mol Genet. 1995;4:1911–1918. doi: 10.1093/hmg/4.10.1911. [DOI] [PubMed] [Google Scholar]
- Gusella J, Wexler N, Conneally P, et al. A polymorphic DNA marker genetically linked to Huntington’s disease. Nature. 1983;306:234–238. doi: 10.1038/306234a0. [DOI] [PubMed] [Google Scholar]
- Kelly TE, Allinson P, McGlennen RC, Baker J, Bao Y. Expansion of a 27 CAG repeat allele into a symptomatic Huntington disease-producing allele. Am J Med Gen. 1999;87(1):91–92. [PubMed] [Google Scholar]
- Kenney C, Hunter C, Davidson A, Jankovic A. Autopsy-proven Huntington’s disease with 29 trinucleotide repeats. Mov Disord. 1997;22(1):127–130. doi: 10.1002/mds.21195. [DOI] [PubMed] [Google Scholar]
- Maat-Kievit A, Losekoot M, Van Den Boer-Van Den Berg H, et al. New problems in testing for Huntington’s disease: The issue of intermediate and reduced penetrance alleles. J Med Genet. 2001;38:E12. doi: 10.1136/jmg.38.4.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlennen RC, Allinson ME, Matthias-Hagen VL, Parker TL, Lovell MA, Kelley T. Evidence of an unstable paternal 27 CAG repeat allele in the huntingtin gene giving rise to clinically overt Huntington disease in a patient with the genotype (17/38) Am J Hum Gen(A246) 1995;57:246. [Google Scholar]
- Nance M, Seltzer W, Ashizawa T, Bennett R, McIntosh N, Myers RH, Potter NT, Shea DK. ACMG/ASHG statement. Laboratory guidelines for Huntington disease genetic testing. Am J Hum Genet. 1998;62:1243–1247. [PMC free article] [PubMed] [Google Scholar]
- The Huntington’s Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Wexler NS, Lorimer J, Porter J, et al. Venezuelan kindreds reveal that genetic and environmental factors modulate Huntington’s disease age of onset. Proc Natl Acad Sci. 2004;101:3498–3503. doi: 10.1073/pnas.0308679101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler VC, Persichetti F, McNeil SM, Mysore JS, Mysore SS, MacDonald ME, Myers RH, Gusella JF, Wexler NS The US Venezuela Collaborative Research Group. Factors associated with HDCAG repeat instability in Huntington’s disease. J Med Genet. 2007;44(11):695–701. doi: 10.1136/jmg.2007.050930. [DOI] [PMC free article] [PubMed] [Google Scholar]