Abstract
Formation of a fully functional four-chambered heart involves an intricate and complex series of events that includes precise spatial–temporal regulation of cell specification, proliferation, and migration. The formation of the ventricular septum during mid-gestation ensures the unidirectional flow of blood, and is necessary for postnatal viability. Notably, a majority of all congenital malformations of the cardiovascular system in humans involve septal abnormalities which afflict 1 out of 100 newborn children in the United States. Thus, a clear understanding of the precise mechanisms involved in this morphogenetic event will undoubtedly reveal important therapeutic targets. The final step in valvuloseptal morphogenesis occurs, in part, by directed movement of flanking myocytes into the cushion mesenchyme. In order to identify the molecular mechanisms that regulate this critical myocyte function, we have developed two in vitro methodologies; a transwell assay to assess population changes in motility and a single-cell tracking assay to identify signals that drive the coordinated movement of these cells. These methods have proven effective to identify focal adhesion kinase (FAK) as an intracellular component that is critical for myocyte chemotaxis.
Keywords: Congenital heart disease, Ventricular septation, Cardiomyocytes, Chemotaxis, Motility, Directional persistence, FAK
1. Introduction
Chemotaxis requires a series of dramatic and coordinated changes in the actin cytoskeleton that enables the cell to polarize toward a chemoattractant, protrude a stable leading edge lamellipodium, and generate the tension required to translocate the remaining cell body (1). One can imagine that this process is even more complex in cardiomyocytes, in which the actin cytoskeleton is aligned in a rigid fashion best suited for isotonic contraction. However, it is becoming clear that cardiomyocyte motility does occur in vivo, particularly during muscularization of the proximal outflow tract (OFT) septum between E11.4 and E15.5 in mice (2, 3). During this developmental window, a subset of cardiomyocytes that flank the cushion mesenchyme dissociates from their existing cell–cell interactions and protrudes into the cushion mesenchyme. Though myocyte chemotaxis is thought to be critical for proper cardiac septation and for alignment of the heart with the great vessels, surprisingly little is known regarding the molecular mechanisms that control this process.
Recent genetic evidence indicates that the integrin class of fibronectin-binding adhesion receptors (α5β1 and others) can regulate both the form and function of the heart (4–10). One of the central proteins involved in the integrin intracellular signaling cascade is the 125 kDa non-receptor protein tyrosine kinase, FAK, which is strongly and rapidly activated by various growth factors and by ligation of two-thirds of the known integrin receptors (11). FAK is expressed at relatively high levels in the mouse myocardium from E8 onward, and we recently generated a line of mice with conditional deletion of myocardial FAK by crossing fakflox/flox mice with an nkx2.5Cre line that directs recombination in cardiac primordia (termed FAKnk mice). We reported that FAKnk mice died shortly after birth and that FAK was essential for appropriate ventricular septation and OFT alignment (12). Interestingly, the defects we observed in the FAKnk hearts resemble some of the most common congenital malformations in humans that involve inappropriate muscularization of the conus (such as DiGeorge Syndrome and Tetralogy of Fallot). Interestingly, we showed that a large number of MF20-positive cardiomyocytes were present in the septal and parietal conal ridges of control, but not FAKnk hearts by E13.5. To determine whether the septal abnormality was a result of impaired myocyte chemotaxis, we developed an in vitro system to monitor the motility of isolated embryonic cardiomyocytes. Herein, we describe in detail two methods to evaluate myocyte chemotaxis in vitro: a transwell migration assay and a single-cell tracking analysis from cardiac explants.
2. Materials
2.1. Cardiomyocyte Cell Isolation
Phosphate buffered saline (PBS): Add 8.0 g of NaCl, 0.2 g of in 1 L distilled KCl, 1.15 g Na2HPO4·7 H2O, 0.2 g of KH2PO4 water, adjust to pH 7.4 with NaOH and sterilize by autoclaving.
Penicillin/streptomycin (PS) stock solution (10,000 U/mL Penicillin, 10,000 μg/mL Streptomycin).
Ice cold PBS + PS (0.5%).
Trypsin stock solution: Dissolve 0.25 g porcine trypsin in PBS.
Trypsin digestion buffer: Add 2 μL of trypsin stock to 998 μL PBS.
Dulbecco’s minimum essential media (DMEM) containing 4.5 g/L D-Glucose, L-Glutamine, and 110 mg/mL sodium pyruvate.
Media 199 (M199) with Earle salts and L-Glutamine.
Prepare DMEM:M199 [4:1] containing PS (0.5%) with or without 15% fetal bovine serum (FBS).
Fibronectin (1 mg/mL solution).
Sterile 1.5-mL polypropylene tubes.
Sterile 100-mL petri dishes.
Sterile dissection tools (razor blades, Dumont #5 and #7 fine forceps, large scissors).
Wide bore sterile plastic transfer pipettes.
Cell strainers (70 μm Nylon).
2.2. Transwell Assay (with Isolated Cardiomyocytes)
Control transwell inserts (8 μm pore; 24-well plate). Coat the evening before myocyte isolation with fibronectin by applying 300 μL of 10 μg/mL of fibronectin diluted in PBS per well (see Note 1) and placing chamber on a rocker overnight at 4°C.
PBS (see step 1, Subheading 2.1 above).
DMEM:M199 [4:1] containing PS (0.5%) with or without 15% FBS.
Paraformaldehyde (EM grade): Prepare a 4% (w/v) of solution in PBS (see Note 2).
Cotton-tipped wooden applicators.
Inverted microscope with fluorescent capacity.
2.3. Single-Cell Tracking Analysis from Cardiac Explants
12-well tissue culture plate. Coat the evening before myocyte isolation with fibronectin by applying 1 mL of 10 μg/mL of fibronectin diluted in PBS per well. Rock gently at 4°C overnight.
Inverted fluorescent microscope with temperature, humidity, and CO2 control that is equipped with a motorized programmable x–y stage.
Digital CCD camera and image collection software. Details provided herein for use of OpenLab software (PerkinElmer).
2.4. Quantification of Cell Speed and Directional Persistence
Cell tracking image analysis software. Details provided herein for use of IMARIS imaging software (ANDOR Technology).
3. Methods
Our initial experiments using either isolated cells or cardiac explants from FAKnk and genetic control hearts proved problematic in distinguishing myocytes from non-myocytes in three-dimensional migration assays. To overcome this hurdle, we crossed the fakflox/flox and nkx2-5Cre mice to a novel line of mice in which nucleus-targeted GFP is expressed under the control of a truncated β-MHC promoter (hereafter referred to as β-GFP mice). FAK-containing and FAK-null cardiomyocytes isolated from these mice were easily identified by nuclear GFP expression, and these cells exhibited comparable well-defined sarcomeric actin organizations (12) (see Figs. 1 and 2). Importantly, nuclear GFP was not observed in non-myocyte cells that frequently contaminate these cultures.
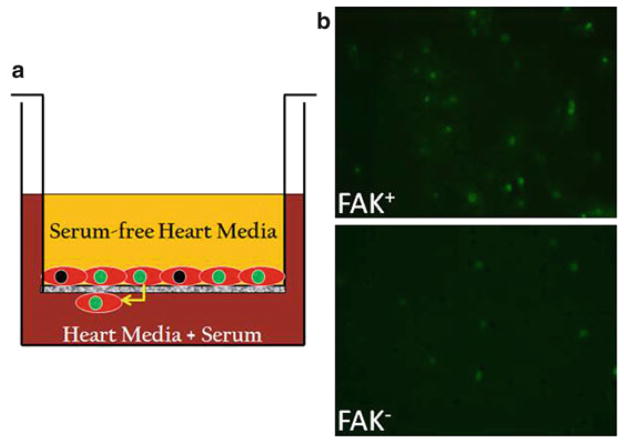
Fig. 1.
FAK depletion impairs cardiomyocyte motility. (a) Schematic depicting the method for determining chemotactic responses using a Transwell assay. (b ) Cells isolated from E13.5 β-GFP genetic control and βGFP/FAKnk hearts were plated on transwell chambers pre-coated with fibronectin (10 μg/mL) as described in Subheading 3.2. Representative images of FAK-containing (FAK+ ) and FAK-null (FAK− ) GFP-positive cells that migrated to the undersurface of the insert are shown (10× magnification). Note significant reduction in numbers of migrated FAK− cells in comparison to controls.
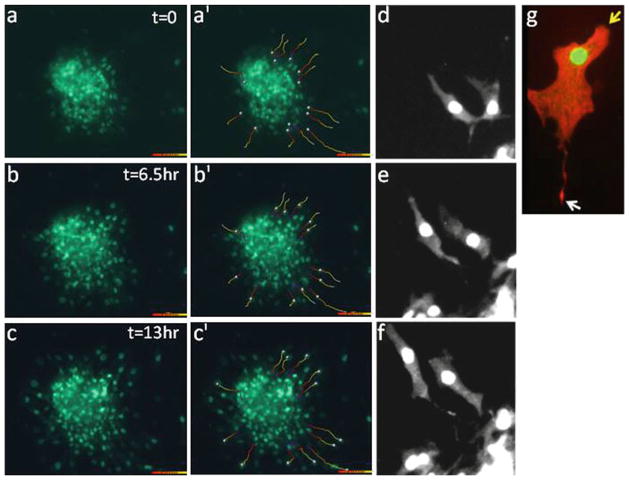
Fig. 2.
Assessment of cardiomyocyte motility in cardiac explant cultures. (a–c) Time-lapse imaging was performed on cardiac explants from E14.5 β GFP mice at 10× magnification. Individual cell tracks were determined using the Imaris program (as described in Subheadings 3.3 and 3.4) and are shown for each time-point in frames (a′–c′ ). (d–g ) High-power images of motile myocytes at 20 (d–f ) and 40× (g) magnification reveal directional and polarized cardiomyocyte motility. Cell in (g) was stained with phalloidin (red) to detect polymerized actin. Arrows demarcate the leading (yellow ) and trailing edges (white ).
We next utilized these GFP-targeted cells to examine cardio-myocyte migration in vitro using a Boyden transwell system (see Fig. 1a). Using methods described in detail below, we found that 15% serum-containing media stimulated a robust chemotactic response in cardiomyocytes isolated from genetic controlgfp hearts, while chemotaxis was dramatically reduced in FAKnk/gfp cardiomyocytes (12) (see Fig. 1b).
3.1. Cardiomyoctye Isolation
Prepare the trypsin digestion buffer, heart medium, and chilled PBS containing PS, before starting.
Place pregnant dame at embryonic day 13.5 of gestation in isoflurane chamber for 3–4 min or until mouse stops breathing. Euthanize by cervical dislocation.
Wash lower abdomen with 70% ethanol. Using forceps and a large scissor make an incision through the skin of the carcass at the base of the abdomen and then peel back the skin with forceps to the level of the sternum (to expose the underlying membrane).
Wash exposed membrane with small amount of 70% ethanol (to remove any hair). Cut through membrane and move aside intestines and fat to expose uterus. Grab leftmost end of uterus with forceps and remove entire string while trimming away attached muscle, fat, and blood vessels. Place uterus in 50-mL beaker with 20 mL of chilled PBS + PS and swirl to remove excess blood.
Prepare sufficient number of collection tubes containing chilled PBS + PS for harvested hearts.
Transfer uterus to a petri dish filled with chilled PBS + PS and place under dissecting scope (with 1.25× magnification).
Using Dumont #3 and #7 forceps, remove uterine muscle from embryonic string and separate embryos from yolk sac.
Transfer one embryo at a time to a separate petri dish (containing chilled PBS + PS). Isolate heart using 2× magnification. First, remove head of embryo, then while immobilizing embryo with #3 forceps positioned on either side of heart, gently peel away chest membrane with #7 forceps. Move forceps to the base of the heart (under the atria) and pinch off the ventricles.
Transfer ventricles to sterile 6-well plate(s) on ice (containing chilled PBS + PS) and repeat step 8 for remaining embryos.
All remaining steps should be performed under a Laminar Flow Hood. Mince ventricles with a sterile razor blade or fine scissors. Transfer minced tissue (using wide bore transfer pipette) to 1.5-mL microfuge tube containing 500 μL chilled PBS.
Wash minced tissue by brief centrifugation (2 min at 500 × g). Carefully aspirate PBS and repeat. Next, add 200 μL of trypsin digestion buffer (increase to 500 μL if using late embryonic or neonatal hearts) and incubate (while rocking) at 37°C. The incubation time should be 10 min to obtain cardiac explants for single-cell tracking (see Subheading 3.3) or 20 min to obtain single-cell suspensions suitable for Transwell assays (see Subheading 3.2).
Gently titrate cell suspension using a 200-μL pipette approximately ten times to disaggregate tissue. Add 500 μL of serum-containing heart media to each tube and mix by inverting tube five to six times. If obtaining cardiac explants, proceed to step 14.
If performing Transwell assay, samples can be pooled if embryos are syngeneic. Triturate cell suspension using a 1-mL pipette an additional ten times to obtain a single-cell suspension. Pass cell suspension through a 70-μm nylon cell strainer into a sterile 50 mL conical to remove tissue residue. Proceed with Transwell assay (see Subheading 3.2).
For cardiac explants, the dispersed extract is plated directly onto a 12-well tissue culture plastic dish pre-coated with fibronectin (see Subheading 2.3). The plate is then placed in a CO2 incubator and left undisturbed for at least 18 h. Proceed with Subheading 3.3.
3.2. Transwell Assay (with Isolated Cardiomyocytes)
Count isolated cardiomyocytes using a hemocytometer and inverted fluorescent microscope and resuspend 10–30,000 GFP-positive cells in 300 μL heat medium containing 15% FBS.
Plate cell solution on the upper surface of fibronectin-coated insert and add 1 mL of heart medium containing 15% FBS into the lower chamber. Place transwell in a 37°C incubator with an atmosphere of 5% CO2. Do not disturb for 24 h.
After 24 h, gently rinse the top and bottom chambers three times with PBS. Add 300 μL of serum-free heart media to the top chamber and add 1 mL of heart medium containing 15% FBS to the bottom chamber (to provide a chemo-attractant gradient). Prior to returning to the incubator, count the number of GFP-positive cardiomyocytes attached to the insert using an inverted fluorescent microscope with a 10× objective (see Note 3).
Place transwell in a 37°C incubator in an atmosphere of 5% CO2 for 24 h.
Aspirate media from both chambers, rinse three times with PBS and fix cells by adding 1 mL of 4% paraformaldehyde to the lower chamber for 20 min at room temperature.
Remove nonmigrated cells from the upper surface of the insert with the cotton-tipped applicator (swab in circular fashion while applying pressure for approximately 15 s/insert, rinse with PBS and repeat).
Count the number of GFP-positive cardiomyocytes attached to the bottom of the insert using an inverted fluorescent microscope with a 10× objective.
Normalize the number of migrating cells to the number attached prior to the induction of chemotaxis.
3.3. Single-Cell Tracking Analysis from Cardiac Explants
While the Boyden transwell system is useful for population studies, the value of this approach is limited with respect to defining molecular mechanisms that drive chemotaxis, since primary cultured cardiomyocytes are particularly difficult to transfect. To overcome this limitation, we have recently developed a single-cell tracking assay from cardiac explants in which myocyte chemotaxis can be tracked via time-lapse imaging. The aforementioned β-GFP mice are pivotal to the analysis of migration in these explants as tracking of nuclear GFP in real time provides a direct measure of cardiomyocyte migration without the confounding effects of proliferation that could not be ruled out in past studies.
The wind-rose plots in Fig. 2 depict the change in individual cell tracks of wild-type GFP-tagged cardiomyocytes from explant cultures over a 13-h period. The high power images in panels d–g reveal that motile myocytes (marked by nuclear GFP and positive for cardiac troponin T) exhibit a polarized cell phenotype with identifiable leading and trailing edges. Subsequent kymographic analysis (described in detail below) was used to define the parameters of cardiomyocyte chemokinesis (i.e., the speed and directional persistence of these cells). For these experiments, data obtained from individual cardiomyocytes at the periphery of the explants were chosen. These studies revealed that in the presence of serum, embryonic myocytes move at a rate of 11.4 ± 1.2 nm/s. This speed is slightly lower than the “basal” speed of approximately 15–20 nm/s that we and others have found for mammalian fibroblasts or smooth muscle cells, which contain a less rigid cyto-architecture. As is apparent from the highlighted cell tracks, the outward movement of myocytes from the periphery of the explants is quite persistent (i.e., relatively few changes in direction are observed). Indeed, on a scale of 0–1 (with 1 being a straight line), the directional persistence for these cells was 0.76. Note, the cell explants can be transfected with various m-Cherry-tagged cDNA constructs using the Amaxa Nucleofector electroporation system and/or treated with pharmacological inhibitors (see Note 4) prior to imaging to identify molecules that regulate myocyte speed and directional persistence.
As noted above, an inverted fluorescent microscope with temperature, humidity, and CO2 ontrol that is equipped with a motorized programmable x–y stage, a digital CCD camera, and cell tracking image analysis software are all necessary for this technique. We use an Olympus IX70 inverted microscope encased in Plexiglas housing to control the internal environment (37°C, 5% CO2, and a relative humidity of 60%) that is equipped with a programmable DeltaT motorized x–y stage. Our image collection is performed using OpenLab software, and our image analysis is performed using Imaris (described below in Subheading 3.4).
The following protocol enables simultaneous quantification of the speed and directional persistence of large numbers of migrating cells (typically >400 cells corresponding to approximately 50 cells/ condition).
Continuing from step 14 in Subheading 3.1, secure 12-well plate containing attached explants in place on the microscope stage, and initiate CO2 at a rate of 80–90 PCO2.
Use the joystick to identify the center of each well. In the X–Y stage click on: Add → New Point. Label Well 1, Well 2, etc. Once this is completed, it is simpler to go through and tell the program to “go to” Well 1, Well 2, etc., without having to remove the covering of the chamber each time.
View plate with a 10× fluorescent objective (using a suitable FITC/EGFP filter cube) and scan the wells, using the joystick, to identify the locations of up to 24 well-spread colonies containing 25–100 GFP positive cardiomyocytes. Record an appropriate exposure time, gain, and offset to visualize individual cells within each colony. Also, record the focus value in the corresponding MoveTo [ ] variable array position (the focus value will appear inside the brackets). This is located in the Variables Window under the View menu. This is important to do before running the automation because it records the focus value you choose for each colony.
After appropriate adjustments are made and the image is as clear as possible, click on Add → New Point, and number colonies sequentially from 1 to 24. This is done for each well, moving to Well 2, Well 3, etc. Once all the points are set and all the parameters are adjusted, Select “GoTo Well 1.” View each colony and readjust the exposure time, gain, offset, and focus as necessary.
Program automation: Set the delay between loops and total number of loops to be collected. For monitoring cell motility over a 24-h window a 5–10 min delay is best. For monitoring lamellipodial dynamics (usually performed at 40×), a 15–30 s delay is more appropriate. However, it is important to note that the cycle time cannot be shorter than the time it takes the stage to visit each preidentified locale. Thus, the total number of cells or colonies imaged may need to be limited to accommodate the chosen interval.
Run the automation: Initiate a test run and review images after completion of one full cycle. If the images need additional focusing and adjustment, stop the automation, close the image windows, and adjust the parameters that need correcting. When satisfied with the program constraints, proceed with automation.
After completing the time-lapse imaging, close the image windows, and save files. When using IMARIS software for subsequent analysis, save images as “256 grays” Liff files (note that Tiff files are too large to process in IMARIS but may be suitable for other image analysis software). For viewing purposes, save the files as QuickTime movies.
3.4. Quantification of Cell Speed and Persistence Using IMARIS Programming
Open the IMARIS program. Navigate to the folder where all images are saved. Select your first point of interest and click Open. This may take a few minutes; especially if the interval chosen between loops was short.
Select Edit → Image Properties. A window will appear with the Voxel Size, Min, and Max, for each X, Y, and Z variable. Since the Live Cell Imaging was performed in 2-dimensions, the Z variable will be used for the Time dimension (see step 3). The Voxel size should be entered in each dimension (this is a standard value, use 0.9803 for a 10× objective). Enter 0 for all viable minimums. Pressing tab will fill in the rest of the information, as the maximum coordinates are specific for each point and determined by each file.
Next, change the Z variable to Time. To this end, first go to the Main Menu and Select→Image Processing, then Select→Swap Time and Z. This is important because the IMARIS program defaults to a 3-dimension program and this step changes the reference frame to a 2-dimensional image.
Record the image interval used for the Live Cell Imaging (i.e., 5 or 10 min). In the main menu, Select Edit → Image Properties and chose All Equidistant. This will open a window denoted Set Equidistant Time Points. There will be four boxes where information can be entered. For the purpose of these experiments, it is only necessary to enter the appropriate value in the Time Interval box (see Note 5).
Select color channel and adjust contrast. In the main menu, select File→Open. Choose the first file you wish to process and click Open. Select the Display Adjustment Box and check the color channel (i.e., green for GFP). The background, Spot and image intensity can be adjusted accordingly in this window. Note the image intensity (the far right toggle) should remain as dim as possible so that individual nuclei can be appropriately identified.
-
In the Main Menu Select fourth box from the left in the nine-box series (the orange-spotted box) to initiate a Wizard that includes a series of six steps for focusing, aligning, and tracking cell migration. It is recommended that data be saved after each step. To move between steps, click on the forward or reverse blue arrow below the work area.
Step 1: Algorithm: There are three settings to choose from:
Segment Only a Region of Interest: this allows you to choose a particular region of the image to analyze. For these experiments, leave unchecked. A box will appear below this choice stating that the entire image will be processed.
Different Spot Sizes (Region Growing): this will implement spheres over each fluorescent cell in the image. There is a separate process for adjusting the parameters of these spheres if necessary. Note: the size of the sphere will be defined below.
Track Spots (over Time): This box needs to be checked to perform single-tracking analysis.
Step 2: Source Channel: This step indicates the Channel being used (i.e., fluorescent, phase, or both) and allows you to define the source of interest (termed “Spot”). For these experiments, the GFP-labeled nucleus is the Spot. Thus, the size of the nucleus needs to be defined in this window. To this end, click on the Slice function located below the Main Menu. On the left side of the screen under Measure, make sure Line is highlighted. Next, click on the far right edge of the nucleus. A plus sign will appear. This marks the right edge of the Spot. Click on the far left side of the nucleus. Another plus sign will appear that marks the left side, and a line will form connecting the two marks (which is a rough estimate of the diameter of the nucleus). Record the length of this line (displayed in the Measure box) and repeat this step on several nuclei to obtain an average diameter. Switch back to Surpass mode to define Spot size. In the box next to Estimated Diameter, enter in a rough average of the values you obtained in Slice mode (this is a rough estimate and it is best to error on the low end). When finished, save and proceed to the next page.
Step 3: Classify Spots: This step allows for filtering and classifying Spots based on their size, diameter, quality, etc. Several different filters can be used to modify the Spots identified via usage of the aforementioned parameters. Using the Spherical mode, we adjust the Quality to ensure that a reasonable number of nuclei are demarcated in each explant. Addition or/and deletion of specific nuclei (Spots) can be accomplished in the next step.
Step 4: Edit Spots: This step allows you to view the Spots chosen by the program (as per the classifications defined above) and to manually select those you want to track. To this end, under Create (on the left side of the screen), select Add/ Delete, and choose “All Visible Channels.” Next, switch from the Navigate mode to the Select mode (the cursor will now appear as a small square). To delete a Spot, move the cursor over the Spot of interest, hold down the shift key and left click on the mouse. To add a Spot, move the cursor over the fluorescing nucleus you wish to identify as a Spot, hold down the shift key and left click on the mouse.
Step 5: Tracking: This step allows you to identify the algorithm you wish to utilize to analyze cell movements. For these experiments Autoregressive Motion was chosen in order to obtain the speed and track straightness (i.e., directional persistence) of individual cells. Next set the tracking Parameters. Max Distance is the maximum distance each Spot is expected to travel. This number should be estimated high. Once again, Slice mode can be used to estimate the average distance traveled over the course of the experiment for a few Spots. Max Gap size is the separation desired between Spots. This value can be set to deselect cells from the analysis due to their close proximity with other cells (i.e., to avoid affects of cell–cell interaction on rates of motility).
Step 6: Classify Tracks: This feature allows the tracking of paths based on certain variables. These variables can be turned on and off in the Edit→Preferences tab. For these experiments, Total Displacement Length was chosen.
Clicking the double right green arrows will exit the Wizard and initiate the single-cell tracking analysis.
Data can next be exported to Excel for further analysis. Data can be exported for the entire experiment, or from individual points. To choose individual points, highlight the Graphing symbol icon and click Detailed to view all statistics collected from all points. Click on any of the listings in this view to export.
Acknowledgments
The authors would like to thank Matthew Medlin and the UNC Microscopy Services Laboratory (including Bob Bagnell and Steven Ray) for excellent technical assistance with image collection and utilization of the OpenLab and Imaris imaging software. This work was supported, in part, by National Heart, Lung, and Blood Institute grants HL-081844 (to J.M.T.) and HL-071054 (to J.M.T.), and American Heart Association Grants 0355776U (to J.M.T.).
Footnotes
The effect of extracellular matrix on cellular motility is a bell-shaped curve. Too little matrix will not support nascent attachments at the leading edge of cells, while too much matrix will impede lamellipodial dynamics. It is best to start with a range of 1–100 μg/mL to determine the most efficacious concentration. We have found that the optimal concentrations vary between cardiomyocytes isolated from different staged embryos.
Paraformaldehyde: This solution should be carefully prepared in a fume hood. For a 100 mL solution: heat 90 mL of ddH2O in a glass flask in the microwave for 1 min. Transfer to a preheated hot plate in the fume hood. Add 4 g of Paraformaldehyde and one drop of 1 M NaOH and bring to a boil. Remove and cool in hood. Add 10 mL of 10× PBS, qs to 100 mL with ddH2O and pH to 7.4.
Instead of counting all of the cells attached to the filter, we find that it is generally sufficient to count the number of cells in four random fields at 10× in both the upper and lower chambers (as long as this process gives a value of at least 200 cells).
Use of pharmacological inhibitors: Pathway inhibitors can be added to determine the effect of particular enzymes on the chemotactic response. For best results, let the automation run for at least 4 h before adding the inhibitor, so that initial rates can be compared to the rates obtained following treatment. To this end, dilute the appropriate amount of the inhibitor into 1 mL of warm heart media. Upon completion of a scanning cycle, carefully remove the Plexiglass lid and top of cell culture dish, add the media containing the inhibitor to the side of the well and gently resuspend, being careful not to move the plate. When finished, replace the cover and allow the automation to proceed. It is important to complete this step before initiation of the next cycle (i.e., within the 5–10 min delay).
It is very important to change the Image Properties and then change how the image is processed, in this order. If this is done in the reverse order, the IMARIS program will take on too much file information too fast and will stall.
References
- 1.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–9. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 2.van den Hoff MJ, Moorman AF, Ruijter JM, Lamers WH, Bennington RW, Markwald RR, Wessels A. Myocardialization of the cardiac outflow tract. Dev Biol. 1999;212:477–90. doi: 10.1006/dbio.1999.9366. [DOI] [PubMed] [Google Scholar]
- 3.Moralez I, Phelps A, Riley B, Raines M, Wirrig E, Snarr B, Jin JP, Van Den Hoff M, Hoffman S, Wessels A. Muscularizing tissues in the endocardial cushions of the avian heart are characterized by the expression of h1-calponin. Dev Dyn. 2006;235:1648–58. doi: 10.1002/dvdy.20738. [DOI] [PubMed] [Google Scholar]
- 4.Shai SY, Harpf AE, Babbitt CJ, Jordan MC, Fishbein MC, Chen J, Omura M, Leil TA, Becker KD, Jiang M, Smith DJ, Cherry SR, Loftus JC, Ross RS. Cardiac myocyte-specific excision of the beta1 integrin gene results in myocardial fibrosis and cardiac failure. Circ Res. 2002;90:458–64. doi: 10.1161/hh0402.105790. [DOI] [PubMed] [Google Scholar]
- 5.Schroeder JA, Jackson LF, Lee DC, Camenisch TD. Form and function of developing heart valves: coordination by extra-cellular matrix and growth factor signaling. J Mol Med. 2003;81:392–403. doi: 10.1007/s00109-003-0456-5. [DOI] [PubMed] [Google Scholar]
- 6.Shubeita HE, Thorburn J, Chien KR. Microinjection of antibodies and expression vectors into living myocardial cells. Development of a novel approach to identify candidate genes that regulate cardiac growth and hypertrophy. Circulation. 1992;85:2236–46. doi: 10.1161/01.cir.85.6.2236. [DOI] [PubMed] [Google Scholar]
- 7.Brancaccio M, Fratta L, Notte A, Hirsch E, Poulet R, Guazzone S, De Acetis M, Vecchione C, Marino G, Altruda F, Silengo L, Tarone G, Lembo G. Melusin, a muscle-specific integrin beta1-interacting protein, is required to prevent cardiac failure in response to chronic pressure overload. Nat Med. 2003;9:68–75. doi: 10.1038/nm805. [DOI] [PubMed] [Google Scholar]
- 8.Hescheler J, Fleischmann BK. Integrins and cell structure: powerful determinants of heart development and heart function. Cardiovasc Res. 2000;47:645–7. doi: 10.1016/s0008-6363(00)00164-4. [DOI] [PubMed] [Google Scholar]
- 9.Yang JT, Bader BL, Kreidberg JA, Ullman-Cullere M, Trevithick JE, Hynes RO. Overlapping and independent functions of fibronectin receptor integrins in early mesodermal development. Dev Biol. 1999;215:264–77. doi: 10.1006/dbio.1999.9451. [DOI] [PubMed] [Google Scholar]
- 10.Valencik ML, Keller RS, Loftus JC, McDonald JA. A lethal perinatal cardiac phenotype resulting from altered integrin function in cardiomyocytes. J Card Fail. 2002;8:262–72. doi: 10.1054/jcaf.2002.127335. [DOI] [PubMed] [Google Scholar]
- 11.Parsons JT. Focal adhesion kinase: the first ten years. J Cell Sci. 2003;116:1409–16. doi: 10.1242/jcs.00373. [DOI] [PubMed] [Google Scholar]
- 12.Hakim ZS, Dimichele LA, Doherty JT, Homeister JW, Beggs HE, Reichardt LF, Schwartz RJ, Brackhan J, Smithies O, Mack CP, Taylor JM. Conditional Deletion of Focal Adhesion Kinase Leads to Defects in Ventricular Septation and Outflow Tract Alignment. Mol Cell Biol. 2007;27:5352–64. doi: 10.1128/MCB.00068-07. [DOI] [PMC free article] [PubMed] [Google Scholar]