Abstract
Spatial and temporal dissection of the genomic changes occurring during the evolution of human non–small cell lung cancer (NSCLC) may help elucidate the basis for its dismal prognosis. We sequenced 25 spatially distinct regions from seven operable NSCLCs and found evidence of branched evolution, with driver mutations arising before and after subclonal diversification. There was pronounced intratumor heterogeneity in copy number alterations, translocations, and mutations associated with APOBEC cytidine deaminase activity. Despite maintained carcinogen exposure, tumors from smokers showed a relative decrease in smoking-related mutations over time, accompanied by an increase in APOBEC-associated mutations. In tumors from former smokers, genome-doubling occurred within a smoking-signature context before subclonal diversification, which suggested that a long period of tumor latency had preceded clinical detection. The regionally separated driver mutations, coupled with the relentless and heterogeneous nature of the genome instability processes, are likely to confound treatment success in NSCLC.
Lung cancer is the leading cause of cancer-related mortality (1, 2). Understanding the pathogenesis and evolution of lung cancer may lead to greater insight into tumor initiation and maintenance and may guide therapeutic interventions. Previous work characterizing the genome of non–small cell lung cancer (NSCLC) has demonstrated that NSCLC genomes exhibit hundreds of nonsilent mutations together with copy number aberrations and genome doublings (3-9). Although subclonal populations have been identified within single biopsies (9), the extent of genomic diversity within primary NSCLCs remains unclear. Moreover, although both exogenous mutational processes, such as smoking (10-12), and endogenous processes, such as up-regulation of APOBEC cytidine deaminases (13-15), have been found to contribute to the large mutational burden in NSCLC, the temporal dynamics of these processes and their contribution to driver somatic aberrations over time remain unknown.
To investigate lung cancer evolution, we performed multiregion whole-exome and/or whole-genome sequencing (M-seq WES/WGS) on a total of 25 tumor regions, collected from seven NSCLC patients who underwent surgical resection before receiving adjuvant therapy. The major NSCLC histological subtypes, including adenocarcinoma (LUAD) and squamous cell carcinoma (LUSC), were represented (table S1). Sequencing of tumor and normal DNA to mean coverage depths of 107× and 54× for M-seq WES and M-seq WGS, respectively (table S2), identified 1884 nonsilent and 76,129 silent mutations (16).
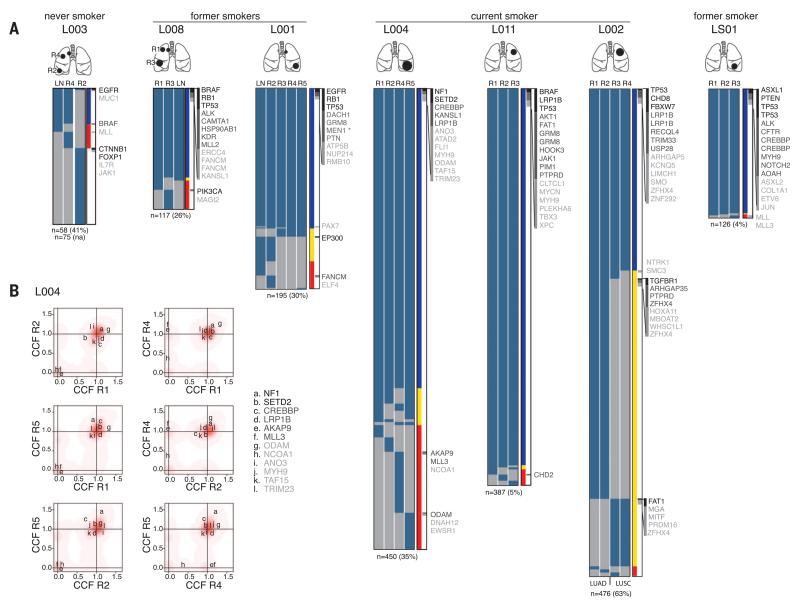
To evaluate the intratumor heterogeneity of nonsilent mutations, we classified each mutation as ubiquitous (present in all tumor regions) or heterogeneous (present in at least one, but not all, regions). Spatial intratumor heterogeneity was identified in all seven NSCLCs, with a median of 30% heterogeneous mutations (range 4 to 63%) (Fig. 1A and fig. S1). In the adenosquamous tumor from patient L002, heterogeneous mutations separated concordant with LUAD (regions R1 and R2) or LUSC (regions R3 and R4) histopathologies (fig. S2). Patients L003 and L008 each presented with two tumors in separate lobes of the lung. M-seq WES revealed 74% ubiquitous mutations in the tumors from L008, which indicated a clonal origin. However, in L003, only a single mutation (EGFRL858R, the epidermal growth factor receptor in which Leu858 is replaced with Arg) was detected in both tumors (Fig. 1A). Given that EGFRL858R is a highly recurrent mutation (17) and also that no silent mutations were shared, we concluded that the tumors in L003 were of independent clonal origin, with the evolution of identical oncogenic events in parallel.
Fig. 1. Intratumor heterogeneity of somatic mutations in human NSCLC.
(A) Heat maps show the regional distribution of all nonsilent mutations; presence (blue) or absence (gray) of each mutation is indicated for every tumor region. Cartoons depict the location of each tumor. Column next to heat map shows the intratumor heterogeneity; mutation present in all regions (blue), in more than one but not all (yellow), or in one region (red). Mutations are ordered on tumor driver category with categories 1 to 3 indicated in the right column in black, dark gray, and light gray, respectively (details in table S3). Total number of nonsilent mutations is provided below each tumor with percentage of heterogeneous mutations in brackets. In L001, the mutation marked by an asterisk (*) is additional to the germline MEN1 mutation. LN, lymph node; R, region. (B) Two-dimensional Dirichlet plots show the cancer cell fraction (CCF) of the mutations in all regions of tumors L004; increasing intensity of red indicates the location of a high posterior probability of a cluster. In region R5, the majority of heterogeneous mutations are subclonal, and a cluster of mutations with a CCF below 1 can be observed.
To resolve the extent of genomic diversity in NSCLC and to infer the ancestral relations between tumor regions, we estimated the fraction of tumor cells within each region harboring each mutation (16, 18). Almost all ubiquitous mutations (>99%) were classified as fully clonal within each region. Moreover, in most regions, the majority of heterogeneous mutations was clonal and, thus, present in all cells within the region (Fig. 1B and fig. S3). However, certain regions displayed considerable subclonal diversity. For example, >75% of heterogeneous mutations present in L004 R5 were subclonal (Fig. 1B), and this region consisted of two distinct subclonal populations. The subclonal structure of each tumor region was then used to construct phylogenetic trees, by using both maximum parsimony and unweighted pair-group methods. We also took into account regional copy number losses that resulted in shared truncal mutations becoming heterogeneous later in tumor evolution (16), such as a segment of chromosome 6 in LS01 (fig. S4) and the PAX7 mutation in the lymph node of L001 (Fig. 1A). Notably, all seven NSCLCs showed evidence of branched tumor evolution (fig. S5).
We next evaluated the regional heterogeneity of potential NSCLC driver mutations, classified into three categories on the basis of current evidence supporting driver mutation status (16). Every tumor showed evidence for ubiquitous, as well as heterogeneous, driver mutations, many of which were clonally dominant in a subset of tumor regions and entirely absent in others (Fig. 1B, fig. S3, and table S3). Note that the probability of missing a category 1 “high-confidence” driver gene by analyzing a single region for each tumor was on average 42% (range 0 to 67%) and 83% (range 67 to 100%) for all potential driver genes (categories 1 to 3), which highlights the potential limitations of assessing single tumor regions. Nevertheless, category 1 and 2 driver mutations were significantly more often truncal compared with mutations in nondriver genes in our M-seq analysis (P = 0.04). Consistent with these data, in The Cancer Genome Atlas (TCGA) cohort, previously reported driver genes (5, 19, 20) were significantly enriched for clonal mutations (P < 0.001) (fig. S6). These data indicate that, in NSCLC, most known driver mutations occur early in tumor evolution.
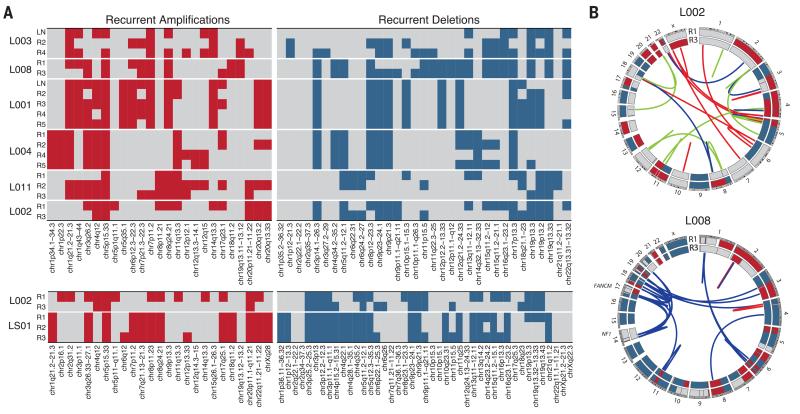
To determine the intratumor heterogeneity of copy number aberrations, we estimated integer DNA copy numbers for each tumor region (7, 16, 21, 22). A large fraction of the genome had undergone alterations in all tumors, and genomic profiles were more similar within tumors than between different tumors (fig. S7). To evaluate the spatial heterogeneity of potential tumor driver copy number aberrations, we explored the regional distribution of chromosomal segments identified as recurrently gained or lost in TCGA LUAD or LUSC tumors. Most segments were identified as aberrant in at least one tumor region, and many recurrent gains and losses were found to be heterogeneous in at least one tumor (Fig. 2A). For example, in L001, a focal EGFR amplification (chr7p11.2), as well as deletions of chromosomal segments harboring CDKN2A (chr9p21.3) and PTEN (chr10q23.31), was observed in all regions, whereas, in L008, we observed heterogeneous copy number losses involving CDKN2A and PTEN. In support of copy number aberrations occurring later in tumor development, we also identified subclonal copy number aberrations within tumor regions. For instance, more than 15% of the genome in region R1 of L008 was subject to subclonal copy number alterations (fig. S8). Consistent with evidence of subclonal copy number aberrations, centromeric fluorescence in situ hybridization analyses confirmed numerical chromosomal diversity within individual tumor regions (fig. S9), which suggested that chromosomal instability may provide a substrate for subclonal competition.
Fig. 2. Intratumor heterogeneity of chromosomal alterations in human NSCLC.
(A) Distribution of potential tumor driver copy number alterations is indicated for each tumor region. The upper heat maps show the regional distribution of recurrently amplified (left) or deleted (right) chromosomal segments based on TCGA LUAD data, and the lower heat maps show the regional distribution of recurrently amplified or deleted chromosomal segments based on TCGA LUSC data. For each region, gain (red) or loss (blue) was determined relative to the mean ploidy. (B) Circos plots depicting inter- and intrachromosomal translocations, as well as deletions and insertions for regions R1 and R3 for L002 (upper) and L008 (lower); shared events are indicated in blue, events private to region R1 are indicated in red, and private to region R3 in green. The outer circle represents the integer copy number data for R1 and the inner circle for R3 for each tumor sample; copy number segments with an integer value greater than mean ploidy are in red and those less than mean ploidy in blue.
The high-coverage M-seq WGS (mean 96×) for L002 and L008 enabled us to investigate the regional separation of large-scale genomic events in these samples. For the adenosquamous tumor L002, we identified 30 structural variants, most of which were found either in the LUAD region R1 or the LUSC region R3, but not both (table S4), which suggested that they occurred after subclonal diversification (Fig. 2B). By contrast, for L008, 48 of the 52 identified structural variants were shared between the two tumor regions from different lobes of the lung, which suggested that the majority of these variants occurred before tumor metastasis to the other lobe (Fig. 2B). Notably, in L008, “chains” of translocations with highly clustered breakpoints were found between chromosomes 14 and 17, as well as chromosomes 17 and 19 (fig. S10 and table S4), which disrupted the FANCM and NF1 tumor suppressor genes. Breakpoint homology profiling suggests involvement of either nonhomologous or alternative end-joining (23, 24), indicative of double-strand break events. This lesion pattern is consistent with chromoanagenesis (25) and indicates a punctuated evolution pattern where multiple oncogenic events may have occurred simultaneously (26).
Four tumors displayed evidence for whole-genome–doubling events (16). In three tumors (L001, L004, and L008), the genome-doubling event was shared across every tumor region; it occurred before diversification, with the majority of truncal mutations (84 to 88%) present at ploidy ≥2, indicative of a large mutational burden before genome doubling. In one tumor, L002, the majority of heterogeneous mutations were also present at ploidy ≥2, indicative of two independent genome-doubling events: one in the LUAD region and one in the LUSC region (fig. S11). Notably, every truncal driver mutation likely occurred before genome doubling.
To further explore the dynamics of the mutational processes shaping lung cancer genomes over time, the spectra of point mutations in each tumor were temporally dissected. Early (truncal) mutations likely reflect processes involved before and during tumor initiation and early development, whereas late (branched) mutations reveal mutational processes shaping the genome during tumor maintenance and progression, including those contributing to intratumor heterogeneity. For L002, we analyzed regions R1 and R3 separately to allow comparisons of LUAD and LUSC histologies within the same tumor.
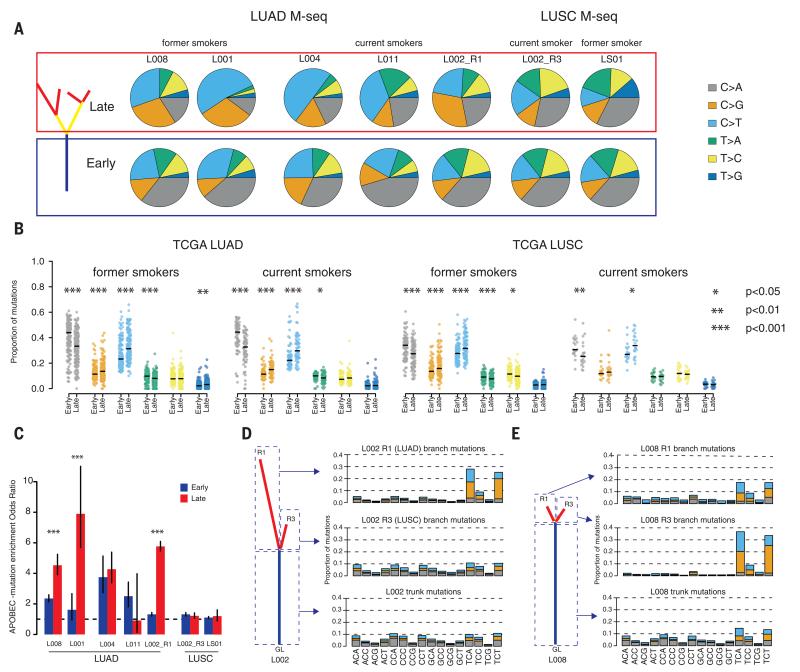
In all tumors, we observed statistically significant shifts in the mutation spectra over time (P < 0.05 all cases) (Fig. 3A). Furthermore, every tumor exhibited a statistically significant decrease in the proportion of C>A transversions in late compared with early mutations (P < 0.05) (Fig. 3A), although this was more pronounced in the LUAD cases [mean odds ratio: LUAD 3.13 (range 2.07 to 5.55) and LUSC 1.34 (range 1.21 to 1.46)]. Because C>A transversions are associated with the mutagenic effects of tobacco smoke (12), a decrease in the proportion of C>A transversions indicates a relative decrease in the mutational burden attributable to smoking during LUAD development, in both former smokers and current smokers.
Fig. 3. Temporal and spatial dissection of mutation spectra in LUAD and LUSC samples.
(A) Fraction of early mutations (trunk) and late mutations (branch) accounted for by each of the six mutation types in all M-seq samples. (B) Beeswarm plots showing the fraction of early mutations and late mutations accounted for by each of the six mutation types in every TCGA former smoker or current smoker with both early and late mutations. Significance is indicated. (C) APOBEC mutation enrichment odds ratio for early (trunk, blue bars) and late (branch, red bars) mutations for M-seq samples. The APOBEC signature encompasses C>T and C>G mutations in a TpC context (16).The 95% confidence intervals for Fisher’s exact test are indicated. (D and E) Three mutation types (C>A; C>G and C>T) at all 16 possible trinucleotide contexts for L002 (D) and L008 (E). For both samples, trunk mutations as well as branch mutations from two regions are depicted.
To validate these observations in a larger NSCLC cohort, mutations in TCGA LUAD and LUSC samples were temporally dissected (16). Consistent with our M-seq analyses, both TCGA LUAD and LUSC smokers and former smokers exhibited a decrease in the proportion of C>A transversions in late mutations (LUAD current smokers, P < 0.0001; former smokers, P < 0.0001; never-smokers, P = 0.147; LUSC current smokers, P = 0.003; former smokers, P < 0.0001; and never-smokers, P = 0.673) (Fig. 3B). Similarly, the least-pronounced decrease was observed in LUSC current smokers; 25% of LUSC displayed no decrease in C>A transversions, compared with less than 10% in LUAD. The mutational footprint of smoking exhibits a strand bias with C>A transversions accumulating preferentially on the transcribed strand (10, 12). Both LUAD and LUSC former smokers revealed a statistically significant decrease in strand bias in late, compared with early, C>A transversions (LUAD, P = 0.00354; LUSC, P = 0.046), consistent with an ancestral footprint of smoking on these genomes. Conversely, no statistically significant difference was observed between early and late mutations in current smokers (LUAD, P = 0.23; LUSC, P = 0.22).
In the majority of M-seq tumors, the decreased proportion of C>A mutations was accompanied by an increase in C>T and C>G mutations at TpC sites, indicative of APOBEC cytidine deaminase activity (13-15). Mutations consistent with APOBEC-mediated mutagenesis were more pronounced on the branches than the trunk in four out of five LUAD M-seq samples (Fig. 3C). On average 31% (8 to 41%) of nonsilent branch mutations occurred in an APOBEC-mutation context compared with 11% (7 to 16%) of truncal nonsilent mutations. Branched driver genes PIK3CA, EP300, TGFBR1, PTPRD, and AKAP9 harbored mutations in an APOBEC context, which indicated a possible functional impact of APOBEC activity on subclonal expansion. Likewise, TCGA LUAD tumors with detectable APOBEC mutational signatures showed significant enrichment in late, compared with early, APOBEC mutations (P < 0.001) (fig. S12), and 20% of subclonal driver mutations were found to occur in an APOBEC context, compared with 11% of clonal driver mutations. However, for TCGA LUSC tumors with detectable APOBEC mutational signatures, temporal dissection of APOBEC mutations did not reveal such a clear trend (fig. S12), which indicated potential differences in the temporal dynamics of APOBEC-mediated mutagenesis between histological subtypes. In addition to temporal heterogeneity, spatial heterogeneity in both the proportion of APOBEC-associated mutations (Fig. 3, D and E) and APOBEC mRNA expression was observed in the M-seq tumors (fig. S13).
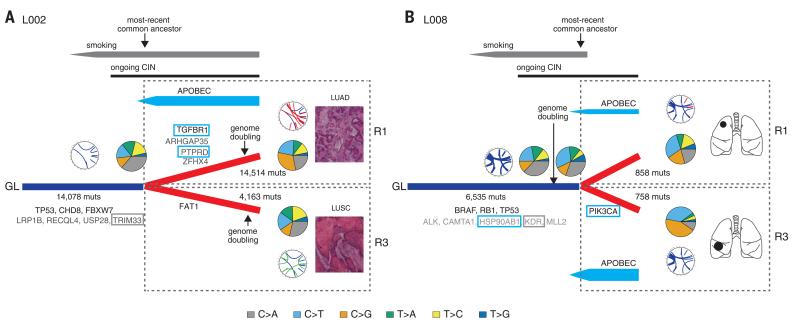
To gain a deeper understanding of NSCLC evolution, we focused on the two tumors with high-coverage M-seq WGS and temporally placed the genomic instability processes relative to the emergence of the most-recent common ancestor (Fig. 4). In patient L002, a current smoker, tobacco carcinogens played a significant role early in tumor development, with C>A transversions representing 39% of truncal mutations (Fig. 4A). Early mutations included multiple driver genes, such as TP53 and CHD8. Upon diversification into a LUAD subclone and a LUSC subclone, copy number alterations (fig. S7) and driver mutations were acquired independently in both subclones, such as a stop-gain mutation in the tumor suppressor gene FAT1 on the LUSC branch and mutations affecting TGFBR1, ZFHX4, ARHGAP35, and PTPRD in the LUAD region. APOBEC-associated mutations were elevated specifically in the LUAD region, which included the driver mutations in TGFBR1 and PTPRD, and the highest APOBEC3B mRNA expression was detected in this region (fig. S13).
Fig. 4. A model of the evolutionary history of NSCLC.
Evolutionary histories of tumors from patients L002 (A) and L008 (B) are depicted. Genomic instability processes defining NSCLC evolution have been placed on their phylogenetic trees. Driver mutations occurring in an APOBEC context are highlighted with a blue box, and those occurring in a smoking context with a gray box. In each case, the timing of genome-doubling events is indicated with an arrow. CIN, chromosomal instability; muts, mutations.
The tumors from patient L008 also displayed truncal C>A transversions and spatial heterogeneity in APOBEC enrichment, with a more pronounced APOBEC signature in the tumor of the middle lobe compared with the upper lobe (Fig. 4B). In L008, we gained further temporal resolution by exploring the mutations before and after the truncal genome-doubling event. All truncal driver mutations were found to occur before genome doubling. However, a tobacco smoke signature of C>A transversions was observed in more than 30% of truncal mutations both before and after doubling, and only in 21% and 9% of heterogeneous mutations in the two regions R1 and R3 from separate lobes of the lung. Because L008 ceased smoking more than 20 years before surgery (table S1), these data suggest that the genome-doubling event and truncal driver mutations occurred within a smoking carcinogenic context more than 20 years ago. Similarly, the genome-doubling event and truncal driver mutations in former smoker L001 also appeared to occur before smoking cessation more than 20 years before surgery (fig. S14). These data suggest a prolonged tumor latency period after genome doubling and before clinical detection in NSCLC.
Through sequencing multiple surgically resected tumor regions, we were able to unravel both the extent of genomic heterogeneity and the evolutionary history of seven NSCLCs. In contrast to the situation in clear cell renal cell carcinoma (ccRCC) (27, 28), known driver mutations typically occurred early in NSCLC development, and the majority of high-confidence driver events were fully clonal. Conceivably, this explains the progression-free survival benefits associated with NSCLC oncogenic driver targeting (29). However, like ccRCC (27, 28), all NSCLCs exhibited heterogeneous driver mutations and/or recurrent copy number aberrations and many heterogeneous mutations gave the “illusion of clonality,” as they are present in all cells from certain regions but undetectable within other regions. Notably, although our multiregional sampling approach allowed us to evaluate spatial heterogeneity, only a small part of the entire tumor was sampled (on average <5%), which indicates that we might be underestimating the full extent of heterogeneity in these tumors.
Conceivably, intratumor heterogeneity may compromise the ability of a single biopsy to define all driver events comprehensively for optimal tumor control. For instance, L008 presented with an activating BRAF (G469A) mutation (30) in all regions and an activating PIK3CA (E542K) mutation (31) only in region R3. Thus, a biopsy taken from R3 might suggest treatment with an inhibitor of the phosphatidylinositol 3-kinase–mammalian target of rapamycin (PI3K/mTOR) signaling axis and combination therapy. Conversely, a single biopsy from any other region would suggest treatment with a BRAF inhibitor, for which the tumor cells from R3 might be resistant because of the PIK3CA mutation (32).
Our study also sheds light on the divergent genomic instability processes involved in NSCLC evolution and their dynamics over time. Evidence for spatial diversity in genomic instability processes suggests that opportunities to exploit such mechanisms therapeutically may be limited in this disease (33). In three tumors, we detected genome-doubling events occurring before subclonal diversification but after acquisition of driver mutations, consistent with findings in colorectal cancer that genome doubling may accelerate cancer genome evolution (34). The relation of chromosomal instability with drug resistance and early tumor recurrence (35, 36) suggests that targeting truncal driver events may be compromised by the initiation of chromosomal instability later in tumor evolution. These results, coupled with the observation that NSCLC tumors may have prolonged latency periods, support continued efforts to optimize methods for earlier detection.
Unexpectedly, we found that despite continuous exposure to the mutagens in tobacco smoke, tumors from smokers showed evidence that an additional genomic instability process (APOBEC-associated mutagenesis) likely contributes to tumor progression. A large proportion of subclonal driver mutations were found to occur in an APOBEC context, which suggests that the differences in mutation spectra over time and space may reflect the activity of the process generating the mutations, as well as the selective advantage of the acquired mutations.
The presence of subclonal, regionally separated driver events, coupled with the relentless and dynamic nature of genomic instability processes observed in this study, highlight the therapeutic challenges associated with NSCLC. Engaging an adaptable immune system may present a tractable approach to manage the dynamic complexity in NSCLC (37). Longitudinal studies will be required to decipher drivers of subclonal expansion, identify the origins of subclones contributing to metastatic recurrence, and resolve the evolutionary principles that underpin the dismal outcome associated with this disease.
Supplementary Material
ACKNOWLEDGMENTS
E.B. is a Rosetrees Trust fellow; M.J.H. has a Cancer Research UK fellowship; N.Mu. received funding from the Rosetrees Trust; M.G. is funded by the UK Medical Research Council; I.V. is funded by Spanish Ministerio de Economía y Competitividad subprograma Ramón y Cajal; R.C.R. and D.M.R. are partly funded by the Cambridge Biomedical Research Centre and Cancer Research UK Cancer Centre; P.V.L. is a postdoctoral researcher of the Research Foundation—Flanders (FWO); S.M.J. is a Wellcome Senior Fellow in Clinical Science; and C.S. is a senior Cancer Research UK clinical research fellow and is funded by Cancer Research UK, the Rosetrees Trust, European Union Framework Programme 7 (projects PREDICT and RESPONSIFY, ID:259303), the Prostate Cancer Foundation, the European Research Council and the Breast Cancer Research Foundation. This research is supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre. We would like to thank Servicio Santander Supercomputación for their support. P.C. is a paid consultant for and holds equity in 14M Genomics Ltd. The results published here are in part based upon data generated by the Cancer Genome Atlas pilot project established by the National Cancer Institute and National Human Genome Research Institute, NIH. Information about TCGA and the investigators and institutions who constitute the TCGA research network can be found at http://cancergenome.nih.gov/. The data were retrieved through dbGaP authorization (accession no. phs000178.v5.p5). Sequence data have been deposited at the European Genome-Phenome Archive (EGA, www.ebi.ac.uk/ega/), which is hosted by the European Bionformatics Institute (EBI), under accession numbers EGAS00001000840 and EGAS00001000809.
Footnotes
www.sciencemag.org/content/346/6206/251/suppl/DC1
Materials and Methods
Figs. S1 to S14
Tables S1 to S4
References (38–64)
REFERENCES AND NOTES
- 1.World Health Organization 2013 www.who.int/cancer/en/
- 2.Siegel R, Naishadham D, Jemal A. CA Cancer J. Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka H, et al. Cancer Res. 2007;67:6007–6011. doi: 10.1158/0008-5472.CAN-06-4774. [DOI] [PubMed] [Google Scholar]
- 4.Weir BA, et al. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding L, et al. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kan Z, et al. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 7.Carter SL, et al. Nat. Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zack TI, et al. Nat. Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Govindan R, et al. Cell. 2012;150:1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pleasance ED, et al. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee W, et al. Nature. 2010;465:473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 12.Pfeifer GP, Hainaut P. Mutat. Res. 2003;526:39–43. doi: 10.1016/s0027-5107(03)00013-7. [DOI] [PubMed] [Google Scholar]
- 13.Roberts SA, et al. Nat. Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burns MB, Temiz NA, Harris RS. Nat. Genet. 2013;45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexandrov LB, et al. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Detailed information on methods is available on Science online.
- 17.Sharma SV, Bell DW, Settleman J, Haber DA. Nat. Rev. Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 18.Bolli N, et al. Nat. Commun. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imielinski M, et al. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawrence MS, et al. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Loo P, et al. Proc. Natl. Acad. Sci. U.S.A. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nik-Zainal S, et al. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malhotra A, et al. Genome Res. 2013;23:762–776. doi: 10.1101/gr.143677.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bunting SF, Nussenzweig A. Nat. Rev. Cancer. 2013;13:443–454. doi: 10.1038/nrc3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang CZ, Leibowitz ML, Pellman D. Genes Dev. 2013;27:2513–2530. doi: 10.1101/gad.229559.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baca SC, et al. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerlinger M, et al. Nat. Genet. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerlinger M, et al. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reck M, Heigener DF, Mok T, Soria JC, Rabe KF. Lancet. 2013;382:709–719. doi: 10.1016/S0140-6736(13)61502-0. [DOI] [PubMed] [Google Scholar]
- 30.Davies H, et al. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 31.Kang S, Bader AG, Vogt PK. Proc. Natl. Acad. Sci. U.S.A. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi H, et al. Cancer Discov. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burrell RA, McGranahan N, Bartek J, Swanton C. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 34.Dewhurst SM, et al. Cancer Discov. 2014;4:175–185. doi: 10.1158/2159-8290.CD-13-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sotillo R, Schvartzman JM, Socci ND, Benezra R. Nature. 2010;464:436–440. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee AJ, et al. Cancer Res. 2011;71:1858–1870. doi: 10.1158/0008-5472.CAN-10-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soria JC, et al. Eur. J. Cancer. 2013;49(suppl. 2):3408. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.