Abstract
Endonucleolytic double-strand DNA break production requires separate strand cleavage events. Although catalytic mechanisms for simple dimeric endonucleases are available, there are many complex nuclease machines which are poorly understood in comparison. Here we studied the single polypeptide Type ISP restriction-modification (RM) enzymes, which cleave random DNA between distant target sites when two enzymes collide following convergent ATP-driven translocation. We report the 2.7 Angstroms resolution X-ray crystal structure of a Type ISP enzyme-DNA complex, revealing that both the helicase-like ATPase and nuclease are unexpectedly located upstream of the direction of translocation, inconsistent with simple nuclease domain-dimerization. Using single-molecule and biochemical techniques, we demonstrate that each ATPase remodels its DNA-protein complex and translocates along DNA without looping it, leading to a collision complex where the nuclease domains are distal. Sequencing of single cleavage events suggests a previously undescribed endonuclease model, where multiple, stochastic strand nicking events combine to produce DNA scission.
Introduction
The prokaryotic ATP-dependent restriction-modification (RM) enzymes provide a potent defence against infection by foreign and bacteriophage DNA, and accordingly have a widespread distribution.1,2 Whilst recognition of specific sequences (targets) in the foreign DNA leads to nucleolytic cleavage (restriction), cleavage of self-DNA is prevented by the methylation (modification) of the target by the same enzyme/enzyme complex. Since the isolation of the first such enzymes in 1968,3,4 which helped launch the molecular biology revolution, many ATP-dependent RM enzymes have been characterized, including the classical heteropentameric Type I systems5 and the closely related but monomeric Single Polypeptide Type ISP systems.5,6 These enzymes are a paradigm for understanding modular, multifunctional protein machines,7 particularly in formulating concepts of protein-DNA recognition, DNA methylation and base flipping, nuclease activity,6,8 dsDNA translocation by superfamily 2 (SF2) helicases,9,10 and long-range communication by enzymes.11,12 The first insights into the molecular organisation of ATP-dependent enzymes came from structural analysis of the Type I RM enzymes EcoKI and EcoR124I using a combination of negative stain electron microscopy, neutron scattering and structural modelling.13 However, despite over 40 years of research, the molecular details of their actions are unclear due to the lack of high-resolution structures.
To address the lack of high-resolution structural data, we have undertaken structure-function studies of Type ISP enzymes LlaGI and LlaBIII from Lactococcus lactis. The Type ISP enzymes perform the disparate functions of restriction and modification by coordinating four functional domains within a single polypeptide: a target recognition domain (TRD) that recognises a 6 or 7 bp asymmetric sequence (Supplementary Results, Supplementary Fig. 1a); an N6-adenine methyltransferase (MTase) which modifies one strand of the sequence using S-adenosyl methionine (AdoMet) stimulated by ATP; an SF2 helicase-like ATPase motor; and a nuclease.6 Unlike the well-studied SF1 and SF2 helicases that translocate ssDNA and ssRNA, much less is known about the mechanism of dsDNA translocation by SF2 ATPases which is important in many biological phenomena such as nucleosome remodelling, removal of stalled RNA polymerases, or modulation of gene expression.14,15,16,17 A structure-function analysis of Type ISP enzymes would further our understanding of dsDNA translocation, particularly in the context of a fully functioning molecular machine.
We also sought to understand how the Type ISP nuclease domains interact to produce a dsDNA break. Previous biochemical analysis pointed to a loop translocation mechanism leading to DNA cleavage, most similar to classical Type I RM enzymes (Supplementary Fig. 1b).6,18,19,20 Following Type ISP target recognition, extensive ATP hydrolysis (~1-2 ATP per bp) produces unidirectional DNA translocation downstream of the target.18,19,20 Because of the directional motion, DNA cleavage only occurs when there is at least one pair of head-to-head targets on the same DNA – DNA with isolated sites, or pairs of sites in other orientations are, at best, only nicked.6 dsDNA breaks are produced at non-specific loci between the head-to-head targets consistent with convergent enzyme translocation and collision.18,19,20
The prevailing Type ISP model predicts that the ATPase domain will be sited downstream of the MTase-TRD and that, despite the complexity of communication, cleavage will result from direct dimerization of nuclease domains within a collision complex. However, the structure presented here shows that the nuclease-ATPase domains of the Type ISP enzymes are located upstream of the MTase-TRD domains relative to the bound directional target sequence. This unexpected arrangement has important implications for both DNA translocation and cleavage. Guided by the structure and based on single-molecule biophysical and biochemical studies, we propose a model for dsDNA break production resulting from multiple, random strand nicks.
Results
Architecture of Type ISP RM enzymes
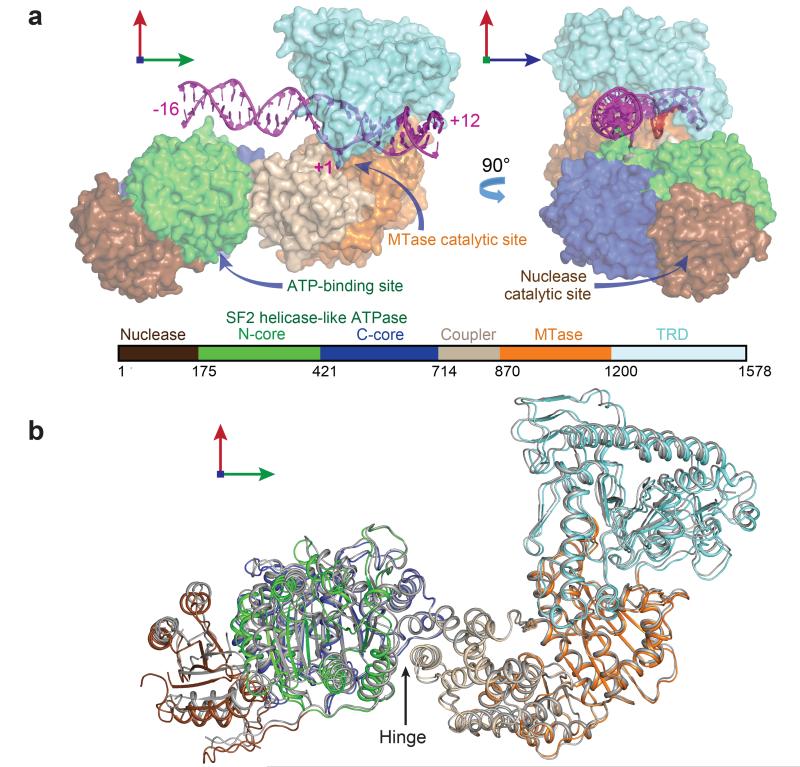
X-ray crystallographic studies initiated using full-length LlaGI yielded crystals that diffracted X-rays poorly. As an alternate strategy, studies were carried out on the close homologue LlaBIII which shares a very high sequence identity of ~80% (the first 1020 amino acids have ~98% sequence identity, while the remainder have 48% identity). Correspondingly, the two proteins can functionally cooperate to cleave DNA.20 The structure of full-length LlaBIII bound to a 28 bp DNA substrate mimic (Supplementary Fig. 2) was determined to a resolution of 2.7 Å (Supplementary Table 1).
The asymmetric unit contained two LlaBIII-DNA molecules related by a two-fold rotational symmetry resulting in a pseudo-continuous DNA of 56 bp (Supplementary Fig. 3a). This assembly appeared to be a result of crystal packing and may not be functionally relevant as the targets are oriented tail-to-tail (which does not support cleavage). The remaining crystal contacts arose from protein-protein and protein-DNA interactions between symmetry-related molecules (Supplementary Fig. 3b). Characterisation by size-exclusion chromatography (SEC) coupled to multi-angle light scattering (MALS) confirmed that the protein is monomeric (Supplementary Fig. 3c).
The structure revealed six structural domains with an unexpected arrangement: the N-terminal Mrr-family nuclease followed by the N-core and C-core RecA folds of the ATPase located upstream of the target; and an all α-helical domain (referred hereafter as the coupler) that links the ATPase to the MTase and the C-terminal TRD (Fig. 1a, Supplementary Fig. 4). The DNA interacted extensively with the TRD and MTase, while making fewer contacts with the ATPase as the DNA length limited the interaction to only a part of the N-core (Supplementary Fig. 2, Fig. 1a ). Amongst the six domains, quality of electron density for the nuclease was comparatively poor, possibly due to intra- and inter-domain conformational mobility.
Figure 1. Architecture of the modular Type ISP RM enzymes.
a, Surface representation of LlaBIII-DNA. b, Superposition of the two chains of LlaBIII reveals conformational plasticity. The coupler domain (residues 715 to 870) of Chain B (grey) was superimposed onto the coupler domain of chain A using the Secondary-structure matching method in Coot.52 The structural domains of Chain A are coloured as in Fig. 1b. The hinge about which the nuclease-ATPase domains move with respect to the coupler is indicated (Supplementary Table 1).
Comparison of the LlaBIII-DNA molecules in the asymmetric unit highlighted inter-domain conformational plasticity (Fig. 1b; Supplementary Table 2). In particular, the nuclease-ATPase domains moved as a unit with respect to the coupler about a hinge-like interface formed by two laterally packed α-helices, one from the C-core of the ATPase (E649-N656) and one from the coupler (P823-I835) (Fig. 1b, Supplementary Fig. 5). Interestingly, the HsdM subunit of classical Type I enzymes (PDB code: 2AR0, 3UFB21) and the multi-domain Type IIG enzyme BpuSI22 contain a similar all α-helical domain of unknown function N-terminal to the MTase domain. The domain has been postulated to be involved in switching between different conformational and functional states.22
Target recognition and adenine base flipping
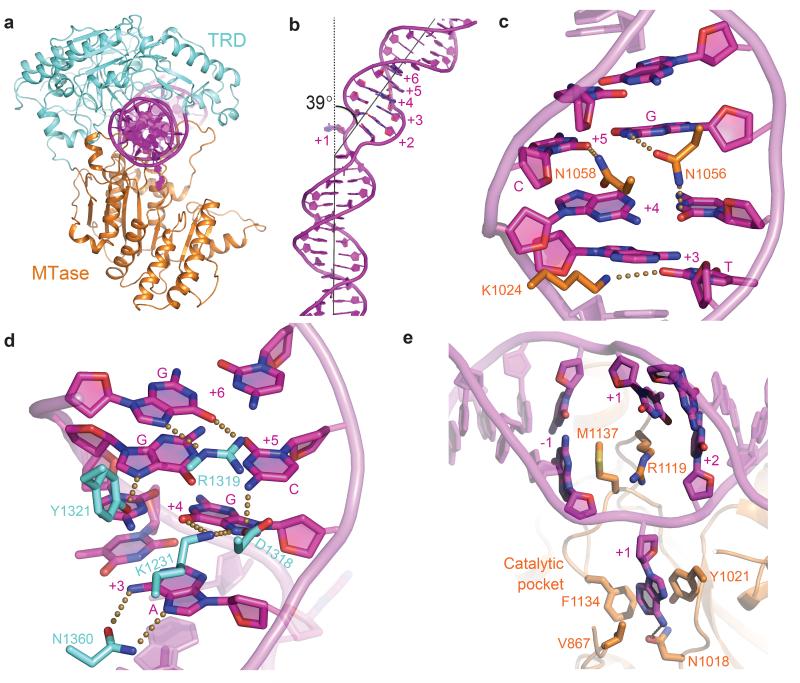
The target was clamped by the MTase and TRD (Fig. 2a). The MTase of LlaBIII had a structural fold similar to the prototype γ-class N6-adenine MTase, M.TaqI,23 and also to the MTase domains of the classical Type I enzymes (PDB ID: 2AR0, 2Y7C,13 3UFB21) and the Type IIG enzyme BpuSI.22 The core region of the C-terminal TRD of LlaBIII was structurally homologous to the TRD of M.TaqI,23 TRD1 and TRD2 of a Type I HsdS subunit,24,25 and the TRD of BpuSI.22
Figure 2. DNA target recognition by LlaBIII.
a, Ribbon diagram of the MTase-TRD clamp encircling the target. b, Extent of DNA bending. c, Minor groove readout by MTase. d, Major groove readout by TRD. e, MTase-DNA contacts stabilising the flipped adenine. Dotted lines represent interactions within hydrogen bonding distance (less than or equal to 3.5 Å).
The MTase-TRD clamp deformed the DNA leading to altered geometrical parameters at the target (Supplementary Fig. 6). The grip bent the DNA (Fig. 2b) and increased the width and decreased the depth of the major and minor grooves (Fig. 2b,c,d, Supplementary Fig. 6). The changes in the groove geometry facilitated sequence specific protein-DNA interactions. The MTase read the sequence via the minor groove (Fig. 2c), while the TRD read it via the major groove (Fig. 2d). Despite the absence of AdoMet in the crystals, the target adenine for methylation (position +1, Supplementary Fig. 2) was flipped out into the MTase catalytic pocket (Fig. 2e). The adenine stacked against Y1021 and hydrogen bonded with the catalytic N1018. F1134 made an energetically favourable T-shaped aromatic-aromatic interaction with the adenine and would sterically block a guanine from entering the pocket. The intercalating R1119 and M1137 plugged the resulting DNA cavity (Fig. 2e).
Mechanism of dsDNA translocation
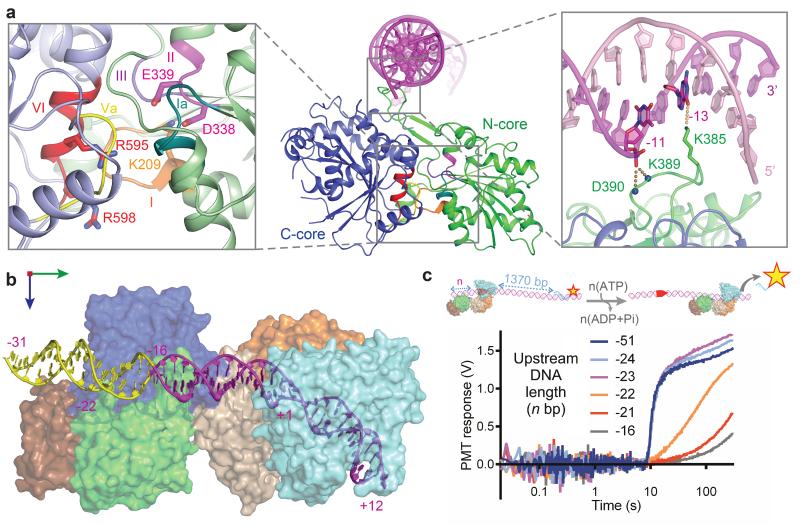
Sequence specific target binding by the MTase-TRD is essential for initiation of DNA translocation.18,19 The structure revealed that upon target recognition, the MTase-TRD steered the upstream DNA towards the ATPase thereby engaging the N-core and C-core (Fig. 1a, 3a, 3b), illustrating a mechanism of coupling of target recognition and DNA translocation, mediated by DNA conformational change. Structural comparison with other SF1 and SF2 ATPases revealed that the disposition of the RecA cores bound to dsDNA (Fig. 3a) was similar to that seen amongst ssDNA-bound helicases oriented to bind ATP26,27,28,29,30 and to the apo structure of the Swi2/Snf2 ATPase from zebrafish Rad5414 (Supplementary Fig. 7). In contrast, the orientation of the cores in the only other structure of a helicase-like ATPase bound to dsDNA, i.e. the Swi2/Snf2 ATPase from Sulfolobus solfataricus, differs by almost 180°.31 The relevance of this conformation is unclear.9,31
Figure 3. Architecture and upstream positioning of the ATPase domains.
a, ATPase architecture highlighting: (left inset) conserved SF2 helicase-like motifs, with ATP-interacting residues as sticks; (Right inset) DNA interaction of loop S383-D390, with side-chain of K385 as sticks and main chain nitrogens of K389 and D390 as spheres. b, Model of nuclease-ATPase interactions with an extended DNA (in yellow). c, The effect of upstream DNA length on triplex displacement. (cartoon) LlaGI motor activity initiated with ATP was monitored on DNA with varying upstream DNA.
The Type ISP N-core-DNA interaction was mediated by a β-hairpin loop (S383-D390) that interacted with the major groove and griped the 3′-terminated strand. The main chain amides of K389 and D390 interacted with phosphate −11, whilst in one of the monomers of LlaBIII, K385 inserted into the major groove and contacted base −13 (Fig. 3a). In the other monomer, the lysine side chain was disordered, suggesting that the interactions involving the main chain atoms were the primary contributors to the grip between the DNA and the hairpin loop. It is, however, likely that new interactions are formed between the hairpin loop and the DNA in other conformational states that may occur during a complete ATPase cycle. Assuming 3′-5′ motion, the LlaBIII ATPase will thus move downstream, consistent with the requirement for head-to-head targets for cleavage (Supplementary Fig. 1). A model of a longer DNA bound to the enzyme, suggested that the ATPase becomes fully engaged when the DNA extends to –22 (Fig. 3b). Accordingly, effective downstream DNA translocation (Fig. 3c) and cleavage (Supplementary Fig. 8) required at least 23 bp upstream of the target.
The structure revealed an enzyme that was competent to carry out either methylation or translocation/cleavage in the presence of the appropriate co-factor – AdoMet or ATP. It appears that the tussle between the two activities is avoided by the apparent rate of methylation being slower than ATP hydrolysis (Supplementary Fig. 9). Consequently, ATPase activity would more often initiate translocation on unmodified foreign DNA, leading to cleavage rather than methylation. On the host DNA, the absolute requirement for two unmodified head-to-head targets for cleavage6,19,20 ensures that the newly-replicated host DNA, with one strand methylated and the other unmethylated, would not be cleaved. Following multiple redundant translocation events, maintenance methylation would eventually occur.
Mechanism of long-range communication along DNA
In contrast to expectations of the prevailing model, the location of the Type ISP ATPase upstream of the MTase-TRD-target complex appeared completely inconsistent with Type I-like loop translocation, since downstream ATPase motion would push the MTase-TRD off the target (Supplementary Fig. 10a). In addition, collision of two converging Type ISP enzymes at the MTase-TRD interface will leave the upstream nucleases at locations apparently too distant to either interact directly or produce a dsDNA break (Supplementary Fig. 10a).
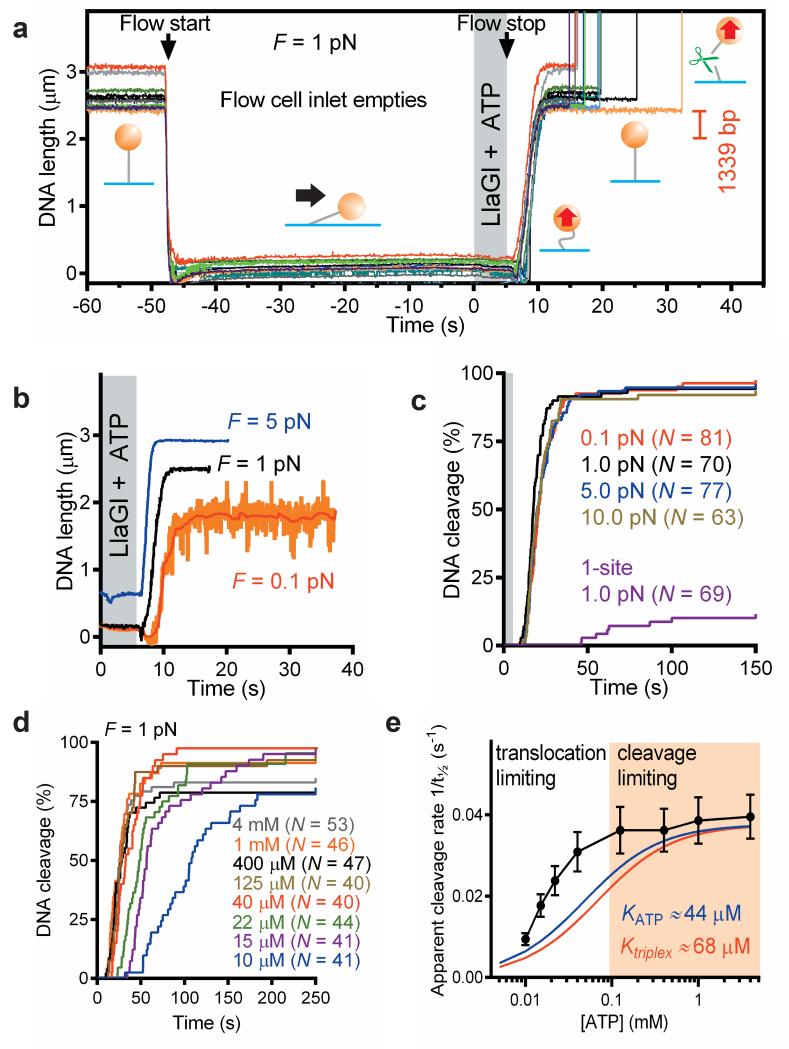
Magnetic tweezers microscope (MTM) assays have been used to distinguish between loop-dependent and –independent translocation models for ATP-dependent RM enzymes.32,33 We utilised a similar approach here, in which cleavage of DNA with two head-to-head Type ISP targets would be observed as loss of magnetic bead tracking, whilst loop translocation would be observed as a uniform downward bead movement (Supplementary Fig. 10b). Cleavage occurred rapidly following addition of LlaGI and ATP, independent of the DNA stretching force and without any evidence for DNA shortening and hence loop formation (Fig. 4a,b,c). The cleavage rate was nonetheless dependent on ATP concentration (Fig. 4d,e), consistent with translocation-driven communication and cleavage. Above the previously measured KM,app values for ATP hydrolysis and translocation,18 the cleavage rate was invariant, most likely because events after collision (e.g. nuclease activity) are rate-limiting. Below the KM,app values, the cleavage rate decreased rapidly, most likely due to a combination of slower translocation, and a concurrent decrease in processivity (as the translocation rate approaches the DNA dissociation rate), which would necessitate multiple translocation events before successful collision.
Figure 4. Loop-independent translocation.
a, DNA cleavage events (upward spikes) following transient introduction of LlaGI and ATP into the flow cell. DNA length change expected of looping is indicated. Lines are raw data (60 Hz). Flow is started as indicated to empty the inlet. Bead positions change because of flow and cell distortion due to hydrodynamic pressure. The LlaGI + ATP solution is added and the flow stopped after ~5 s. The DNA relaxes back to full extension slowly because of hydrostatic re-equilibration of the cell. b, examples of loop-independent cleavage at a range of stretching forces. Lines are raw data (31 Hz) except the red line (1 s filter). c, Cumulative dsDNA cleavage for N events on head-to-head DNA at 0.1-10 pN and on a one-site DNA (Supplementary Fig. 12) at 1.0 pN. d, Cumulative dsDNA cleavage for N events on head-to-head DNA at 1 pN at a range of ATP concentrations. e, ATP dependence of the apparent cleavage rate (taken from t½, the time to reach 50% cleavage in f). Blue and red lines are simulations of a hyperbolic relationship for the previously-determined apparent KM,app values for ATP hydrolysis (KATP) and DNA translocation (Ktriplex).18 Error bars were calculated from the average values divided by the square root of number of points (N) in the bin.
Evidence for changes in DNA topology during translocation
The previous suggestion that these enzymes translocate DNA loops was based on an indirect topoisomerase assay that measured changes in DNA topology during translocation of a nuclease mutant.18 Wild type enzyme could not be used as it would have rapidly cleaved the DNA substrate. The principal of the assay is that a motor complex that forms a DNA loop and translocates around the DNA helical pitch will trap changes in DNA supercoiling; the twin domain supercoiling model.11,34 The upstream DNA loop expands in size and has reduced twist (leading to negative supercoiling) whilst the downstream DNA has increased twist (leading to positive supercoiling). Using a one-site DNA, a LlaGI nuclease mutant and E. coli topoisomerase I, we were able to convert a relaxed DNA to a positively-supercoiled one, consistent with twin domain supercoiling.18
We re-examined the effect of translocation by a LlaGI nuclease mutant on DNA topology using the MTM assay (Supplementary Fig. 11a). On negatively-supercoiled one-site DNA at low force, we observed transient increases in DNA height consistent with loop translocation (the plectonemes being released by the increased twist downstream of the motor). In theory, upon reaching relaxed DNA the motor could continue to translocate leading to positive supercoil formation and a reduction in bead height. However, we never observed reductions in bead position below the resting height of the negatively-supercoiled DNA. This suggests that upon reaching relaxed DNA (or soon before or after), the loop is released allowing re-equilibration to negatively-supercoiled DNA. On positively-supercoiled DNA at low force the bead height transiently reduced, also consistent with loop translocation (the increased twist producing additional plectonemes). However, the events were longer lived and more frequent.
If loop translocation on a negatively-supercoiled DNA produced relaxed DNA, the difference in bead height compared to relaxed DNA in the absence of looping would indicate the length of DNA trapped in the loop.35 The differences we observed were larger than expected given a model where the motor initiates looping immediately next to the target (Supplementary Fig. 1). In Supplementary Fig. 11b, relaxed DNA should have formed after translocation of ~168 bp (+16 turns assuming 10.5 bp per turn), corresponding to a height difference of ~57 nm. The larger difference observed (e.g. 136 nm) could be explained if LlaGI trapped a larger loop initially. However, we cannot explicitly state that the relaxed DNA state had been reached (i.e. looping may be short-lived and always collapse before reaching 168 bp).
On topologically-unconstrained one-site DNA, neither the nuclease mutant nor wild type LlaGI produced loop translocation events (Supplementary Fig. 11c,d and 12a). Wild type LlaGI produced a much lower frequency of loop translocation events on positively-supercoiled DNA (Supplementary Fig. 11d). Using the wild type enzyme we also tested DNA cleavage on two-site DNA (Supplementary Fig. 11e). Individual strand cleavage events were observed; the first event relaxed the supercoils, lengthening the DNA, before a subsequent event generated a dsDNA break. However, reductions in bead height consistent with twin-domain supercoiling were not observed. Therefore it seems that although LlaGI can occasionally trap loops during translocation, these are not pre-requisites for cleavage of either supercoiled (Supplementary Fig. 11e) or relaxed DNA (Fig. 4a,b).
We suggest an alternative model to account for supercoiling changes observed here and previously18 (Supplementary Fig. 11f). An individual enzyme cannot trap a DNA loop. However, where a translocating motor interacts with a static motor, continued translocation leads to twin domain supercoiling. These interactions are promoted and maintained by DNA supercoils, particularly positive DNA nodes. However, they are facile and are not necessary for DNA translocation or cleavage. The higher event frequency observed with nuclease mutants may reflect changes in motor properties; for the classical Type I RM enzymes, nuclease domain mutations caused decreased translocation and ATPase rates, enzyme population heterogeneity, failure to initiate and altered turnover dynamics.36
Evidence for release of the target during translocation
Molecular motors have a finite chance of dissociating during each ATP-dependent step along DNA. The forward stepping rate divided by the sum of the stepping rate and the dissociation rate defines the processivity. During loop-independent translocation, the enzyme must leave its target site and subsequent dissociation will release the complete enzyme. This contrasts with translocation termination by the classical Type I RM enzymes where the motor dissociates from the core MTase which remains at the site.33 Translocation termination can be quantified using a triplex displacement assay. We initiated translocation by mixing ATP with a pre-formed LlaGI-DNA triplex complex in a stopped flow fluorimeter. The termination pathway was probed by including “traps” along with the ATP (Supplementary Fig. 12b). We trapped dissociated enzymes using an excess of DNA carrying the target sequence or, as a control, a random, non-specific DNA (Supplementary Fig. 12c). Alternatively, we blocked the exposed DNA target by adding a helicase mutant (LlaGI K210A) which can bind the site but not translocate.20
Both traps caused a reduction in the amplitude of triplex displacement which scaled with the translocation distance (Supplementary Fig. 12d). The burst amplitude represented the proportion of pre-bound LlaGI proteins which initiated translocation and reached the triplex without being trapped. The specific DNA was an efficient trap over short time scales, but protein dissociation from the trap allowed re-association with the triplex substrate, leading to a background triplex displacement rate. The non-specific oligoduplex had a minor trapping effect but turnover was more rapid. The helicase mutant was more efficient in sequestering the site and preventing re-association by dissociated enzymes (i.e. the background turnover rate was slower). But for technical reasons we could only add an equimolar concentration of mutant to wild type protein. Therefore some wild type protein could rebind the site and the apparent amplitude was higher than that seen with the DNA trap. Nonetheless, the distance-dependent changes in displacement amplitude seen by trapping either the site or the dissociated enzyme are consistent with release of the target during loop-independent translocation.
Regulation of the nuclease activity
DNA cleavage does not occur during translocation, but is activated upon convergent protein collision.6,18,19,20 The key catalytic residues of the Mrr-family nuclease37 were clustered into a putative active site cleft (Fig. 5a), which pointed away from the DNA path (Fig. 1a). The loop containing the essential D7437 was unstructured (Fig. 5a), suggesting a catalytically impaired conformation that becomes activated on engaging with the DNA upon collision. The loop could also be unstructured due to the absence of divalent cation in the buffer, which is thought to interact with the catalytic triad of D74, D78 and K94.37 We note that the side chains of D78 and K94 were also disordered in the structure.
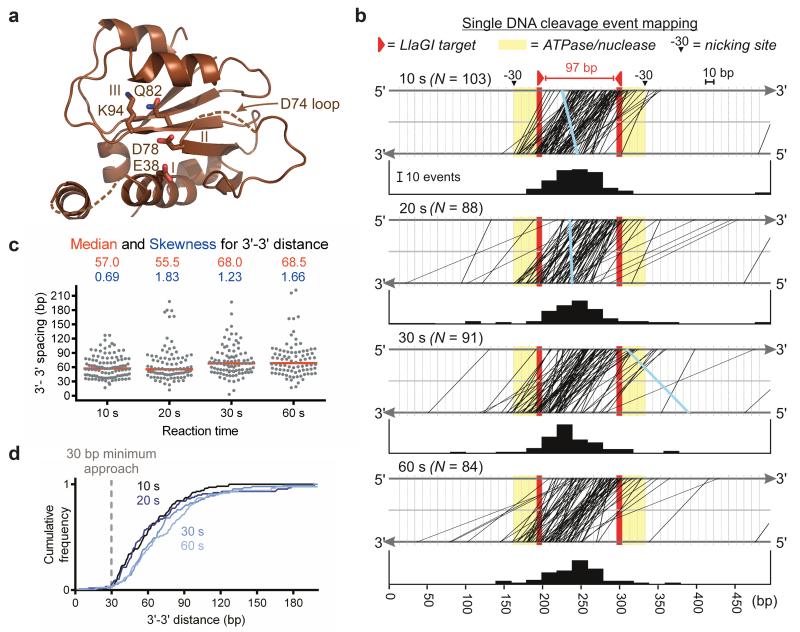
Figure 5. Single cleavage event mapping.
a, Mrr-family nuclease, highlighting catalytic motifs and residues. b, DNA cleavage by LlaGI at each time point is plotted as top/bottom strand locations joined by a line (rare 5′-5′ overhangs in blue). Bar graphs are the collision centroids (20 bp bins). c, Scatter plot of the outermost 3′-3′ cleavage distances, with median (red line) and skewness values (the latter showing a time-dependent increase in data asymmetry). d, Cumulative frequency (normalised for N events) of outermost 3′-3′ cleavage distances.
Nuclease activation, along with structuring of the D74 loop, may additionally arise due to ATP-dependent or ATP-independent nucleoprotein rearrangements that engage the active site with the DNA. A simple modelling based on the structures of the SF2 helicase NS3 from hepatitis virus C bound to ssDNA and an ATP analogue30 suggested that ATP-induced domain closure would move the nuclease closer to the DNA path. However, in this orientation the nuclease active site still pointed away from the DNA, and the active orientation is possibly only attained upon collision. In addition, pivoting of the nuclease-ATPase unit about the hinge between the ATPase and the coupler (Fig. 1b, Supplementary Fig.5) could help engage the nuclease with the DNA. Activation of the upstream nuclease effected by interdomain plasticity was supported by the mapping of a slow ATP-independent nicking activity, which exclusively targeted the bottom 3′-5′ strand at −30 (Supplementary Fig. 13a, b).
Mechanism of dsDNA break formation
Although activation must occur upon collision, the enzyme architecture suggested that the nuclease domains in a collision complex will be located too far apart to interact directly and, additionally, that the DNA nicks introduced would be too far apart to form a dsDNA break (Supplementary Fig. 10a). To investigate the locations of the strand breaks resulting from stochastic translocation and randomised collision, we mapped single DNA cleavage events at different time points during the reaction using a DNA with two head-to-head sites spaced 97 bp apart (Fig. 5b). The assay (Supplementary Fig. 14) reports either on the outermost cleavage loci for 3′ overhangs or the innermost cleavage loci for 5′ overhangs. The majority (>98%) of collision events produced 3′ overhangs, with an average centroid of collision midway between the sites, as predicted18 (Fig. 5b). The Mrr-family nuclease had an unexpected dinucleotide preference (Supplementary Fig.13c), although this did not influence the distributions observed on the DNA used here.
Early (10s) cleavage events clustered between the sites and in the immediate upstream regions (as defined by Fig. 5b). The 3′-3′ distances between the outermost nicks on the two strands revealed a distinct minimum of 30 bp but also a wide range of longer values up to 120 bp, with a median value of 57 bp (Fig. 5c,d). The minimum represents the closest approach of two nuclease-ATPase units during collision. The wide range of values observed supports a distant arrangement of nuclease domains. As the reaction progressed, the 3′-3′ distance increased, consistent with DNA end processing as observed in gel assays6 (Supplementary Fig. 8), raising the median value to ~68 bp. In addition, cleavage events began to be observed further upstream than −30, consistent with an ability of the collision complex to move from its primary location.
Discussion
The data presented here indicates that the upstream ATPase of a Type ISP enzyme remodels its downstream MTase-TRD-target complex to allow target release (Fig. 6). Upon binding of ATP, Type ISP domain movement and ensuing rearrangements in the ATPase-DNA interactions, including those involving the β-hairpin loop, would pull the DNA in an upstream direction leading to movement downstream of the complete enzyme without a necessity for DNA loop formation. This model is quite distinct to that of the classical Type I RM enzymes. We suggest that the coupler will play a key role in transferring conformational strain during initiation. The flipped base seen in the MTase active site would also act as an “anchor” to prevent target release; this must be returned to its base-paired location in the DNA before initiation can proceed and this may be accelerated by ATP-induced strain relayed by the DNA and/or coupler.
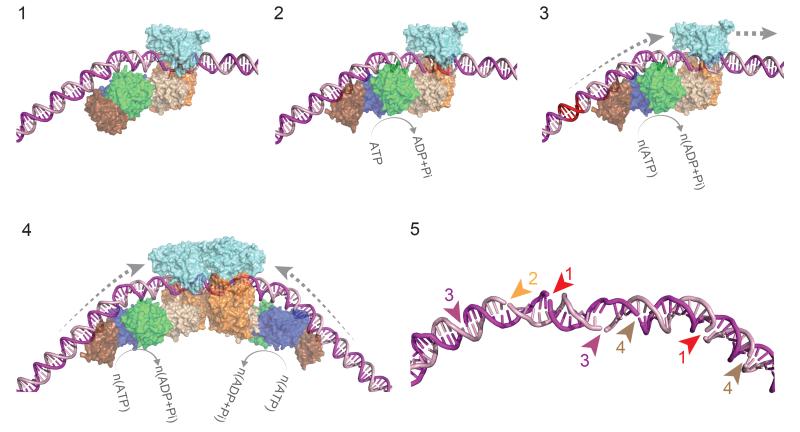
Figure 6. Model for loop-independent DNA translocation and extensive nucleolytic DNA processing.
Model for loop-independent DNA translocation and extensive nucleolytic DNA processing. (1) The pre-initiation complex. The nuclease is in an inactive conformation. (2) The ATPase cycle loosens the MTase-TRD grip on the DNA. (3) dsDNA translocation downstream of the target (red). (4) Convergence of two enzymes initially brings the nucleases approximately 75 bp apart. (5) Example of stochastic “DNA shredding” by a collision complex. Numbers are the order of the nicking events.
Remodelling of the MTase-TRD must involve loss of target interactions. However, the remodelled MTase-TRD may remain associated with the DNA to act as a sliding clamp, enhancing processivity. Alternatively, the clamp may open fully, allowing the coupler-MTase-TRD units to swing completely off the DNA. This motion may help explain the ~30 bp closest approach of the nuclease domains (Fig. 5c,d).
The Type ISP remodelling activity is somewhat similar to the role of the ATPase subunit of the Type III RM enzymes.12 However, whilst those enzymes use a burst of ATPase activity to break the MTase-target interactions, subsequent bidirectional movement along DNA is thermally-driven without the need for ATP hydrolysis. In contrast, the Type ISP enzymes continue to consume ATP during unidirectional translocation.18 It remains to be seen whether the Type III enzymes also move towards their cognate MTase subunits, “pushing” them off the target as in the Type ISP scheme (Fig. 6), or move in the opposite direction to “pull” the MTase subunits off the target.
The Type ISP structure also provides a simple, single polypeptide framework to understand other remodelling processes that may involve active disruption of protein-nucleic acid complexes by an ATPase motor, such as the DExH/D RNA helicase,38 SF1 DNA helicase PcrA,39,40 the Swi2/Snf2 family ATPase Mot1,15,17 or nucleosome remodellers.15,16 The above structural comparison suggests that the ATPase cycle proposed for bona fide helicases, involving domain closure and opening of N- and C-cores upon ATP binding and hydrolysis26,27,29,30 is conserved in dsDNA translocating ATPases. In turn, the Type ISP ATPase would therefore use an equivalent inchworm mechanism for both remodelling and translocation. Interestingly, the β-hairpin loop is unique to Type ISP ATPases (Supplementary Fig. 7), indicating variation in the translocation mechanisms amongst SF2 ATPases despite conservation of canonical motifs and overall structures.
The wide spacing of the DNA strand breaks produced (Fig. 5) is consistent with the upstream locations of the nuclease domains as predicted in a head-on collision complex (Supplementary Fig. 10a). Since the nucleases cannot interact directly, activation upon collision is most likely due to conformational strain from the ATPases transferred via the couplers. Nonetheless, additional remodelling and movement of the collision complex must be required to further process the DNA to produce a dsDNA break. We propose a “DNA Shredder” model in which cumulative nicking events eventually result in a dsDNA break (Fig. 6). This contrasts with the precise DNA cleavage produced by two closely-spaced nucleases as suggested for Type I and III enzymes (Supplementary Fig. 1) and observed in a great many other, simpler nucleases.
The collision complex would initially result in formation of one or two strand breaks. The location of this initial cleavage will be random, dictated by the preceding stochastic translocation events. Rearrangements of the complex due to interdomain plasticity together with the nuclease’s sequence preference (Supplementary Fig. 13c), can result in varied loci of cleavage and hence a range of initial 3′-3′ spacings. The observed 30 bp minimum could reflect a collision complex where both coupler-MTase-TRD units have swung off the DNA, allowing the closest approach of two helicase-nuclease units that still allows for stress-activation. Continued ATPase activity can then remodel the collision complex, leading to further up- and downstream movement and nicking events, causing compound damage culminating in a dsDNA break (Fig. 6). The increases in the median and skewness values (Fig. 5) are thus due to the longer incubation times during which collision complex motion leads to further cleavage events at other sites. Notably, post-collision mobility can also account for cleavage further upstream than −30 and for rare 5′ overhangs (Fig. 5b), where presumably the collision complex nicks one strand and immediately moves in a 3′ direction before nicking the opposite strand.
The loading of multiple translocating enzymes on the DNA that is a potential consequence of target release could result in a pile-up of enzymes on both sides of the primary collision complex. If these enzymes were activated they could cause a time-dependent increase in 3′-3′ cleavage distance. However, post-collision mobility can more readily account for cleavage events upstream of −30 and the 5′ overhangs. Nonetheless, rear-end collisions may play a supplementary role in DNA processing. We discounted an alternative model where strand nicking by multiple, separate collision events at random sites generates the widely-spaced overhangs since this would generate 5′ overhangs with equal frequency and would not show a time-dependent increase in distance.
We speculate that, depending on the cellular host, broken DNA with variable 3′-overhangs produced by Type ISP enzymes would not be a substrate for the corresponding dsDNA break repair enzymes (RecBCD or AddAB), which have a preference for blunt ends.41,42,43 Consequently, even if the foreign DNA were to have the regulatory sequences required for homologous recombination, they would not enter that repair pathway. dsDNA break formation by multiple nicking events would additionally prevent simple religation of the DNA ends. Where the cell also encodes a CRISPR-Cas system, the small DNA fragments generated during cleavage by Type ISP enzymes (or during post-cleavage processing by classical Type I enzymes44) may feed into the spacer acquisition pathway (adaptation), as suggested for the DNA fragments generated by RecBCD.45
In conclusion, the distinct nucleolytic activity of the Type ISP enzymes contrasts with mechanisms employed by dimeric nucleases and further illustrates yet another strategy evolved towards resistance in the perpetual bacteriophage-host arms race.2
Online Methods
Generation of a LlaGIΔN mutant
pRSFLlaGI6 was used as template in PCR protocol using Phusion polymerase (Finnzymes Oy, Finland) as per manufacturer’s guidelines. The phosphorylated primers 5′-CATGGTATATCTCCTTATTAAAGT-3′ and 5′-CGTCCAGAAAATGTAGTCGT-3′ were designed to delete the region of the gene corresponding to amino acids 2-165. Template DNA was removed by digestion with DpnI and clones selected and sequenced following transformation of Escherichia coli Top10 cells (Invitrogen).
Purification of LlaGI, LlaGIΔN and LlaBIII for crystallisation
LlaGI, LlaGIΔN, selenomethionine containing LlaGIΔN (LlaGIΔNSe) and LlaBIII were purified using the same strategy. The protein was overexpressed in 10L of Escherichia coli BL21 (DE3) from recombinant clones of the genes in pRSF vector. LlaGIΔNSe was overexpressed in Escherichia coli B834 (DE3) grown in LeMaster media.46 A higher amount of soluble protein in the crude lysate was observed upon inducing the culture at 25°C with 0.5 mM IPTG at OD600 = 0.6. The incubation temperature was lowered from 37°C to 25°C before addition of IPTG. Induced cells were harvested after 5 hours further incubation. To minimise proteolytic degradation of the protein, protease inhibitors (Roche) were added to the lysis buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 10 mM MgCl2, 1mM EDTA and 1 mM DTT), all the steps of purification were carried out at 4°C, and the protein was purified to homogeneity within 24 hours of lysis. The cells were lysed by sonication. The protein was salted-out using 70% w/v ammonium sulphate. The re-suspended protein pellet was further purified using heparin and MonoQ columns (GE Healthcare), respectively. Finally, size exclusion chromatography using Superdex 200 10/300 (GE Healthcare) ensured homogenous monomeric protein. Purified LlaGI and LlaGIΔN were stored in a buffer containing 10 mM Tris-HCl pH 7.4, 100 mM NaCl and 1 mM DTT, and LlaBIII was stored in 10 mM Tris-HCl pH 7.4, 100 mM KCl and 1 mM DTT. Purified samples stored at –80°C remained intact and could be thawed and used for crystallisation later.
Purification of DNA substrates for crystallisation
The dsDNA substrates used for co-crystallisation were obtained by annealing two chemically synthesized, PAGE-purified, complementary ssDNA purchased from Integrated DNA Technologies, USA. The dsDNA was purified to remove ssDNA using a MonoQ column. The pure duplex DNA was concentrated and stored in sterile water at −20°C.
Crystallisation of LlaBIII-DNA and data collection
LlaBIII-DNA complex was crystallised in 200 nanolitre drops (1:1 of protein : reservoir buffer) by vapour diffusion at 291 K using sitting-drop method (Supplementary Fig. 15). The reservoir buffer contained 200 mM KCl and 20% w/v PEG 3350. This crystal diffracted to 2.7 Å at 100 K with ethylene glycol as a cryoprotectant. Data was collected at the I02 beamline, Diamond Light Source, at a wavelength of 0.9793 Å. The data collection statistics are provided in Supplementary Table 1.
Crystallisation of LlaGIΔNSe-DNA and data collection
A complex of the purified protein and DNA was formed by mixing the two in 1:1.3 molar ratio at 4°C. A protein concentration of 5 mg/ml and a crystallisation drop size of 200 nL were used for all the initial crystallisation trials by sitting drop vapour diffusion method. Conditions of varying buffers, additives, precipitants, DNA substrates and temperatures were screened for crystallisation of LlaGI-DNA and LlaGIΔN-DNA complex. Crystals of LlaGI-DNA diffracted poorly. However, a crystal form of LlaGIΔN bound to a DNA substrate mimic (Supplementary Fig. 15) grown in 4 μL (1:1 of protein : reservoir buffer) hanging drops at 291 K by the vapour diffusion method diffracted better. The reservoir buffer contained 100 mM Tris-HCl pH 7.4, 20% w/v PEG 20,000, 4% w/v PEG 550 MME, and 150 to 250 mM sodium acetate. This condition was used to grow crystals of LlaGIΔNSe-DNA. A 3.2 Å multi-wavelength anomalous data set was collected from one of these crystals at 100 K. The data was collected at I03 beamline, Diamond Light Source.
Structure solution of LlaGIΔNSe-DNA
The data set was processed using XDS47 and the intensities were scaled and merged using XSCALE,47 and converted to structure factors using TRUNCATE48 (Supplementary Table 2). LlaGIΔNSe-DNA crystallised in the space group P21 and Matthews coefficient suggested two molecules in the asymmetric unit. Experimental phases to a resolution of 3.2 Å were determined by single-wavelength anomalous diffraction (SAD) method from the data collected at the peak wavelength of 0.9797 Å (Supplementary Table 2). The quality of the anomalous signal was good enough to obtain the substructure of 46 of the 58 selenomethionines in the asymmetric unit using ShelxD as implemented in the program PHASER-EP.49 Phases were calculated by PHASER-EP. Density modification with solvent flattening and histogram matching was carried out using DM.50 The quality of the map, though of medium resolution, was sufficiently good to visually identify the correct hand (Supplementary Fig. 15). The maps were visualized and model building carried out using COOT.51 Initial cycles of structure refinements were carried out by REFMAC552 and subsequently by phenix.refine.53 This map was used to manually build most of the structure of LlaGIΔN-DNA. This structure was used as a model to solve the structure of LlaBIII-DNA by molecular replacement.
Structure determination of LlaBIII-DNA
The data was processed using XDS47 and the intensities were scaled and merged using XSCALE,47 and converted to structure factors using TRUNCATE48. LlaBIII-DNA crystallised in the space group P1. The structure solution for LlaBIII-DNA was obtained by molecular replacement with MOLREP54 using the coordinates of an N-terminal deletion mutant of LlaGI. Alternate cycles of model building using Coot51 and refinement of coordinates and individual B-factors using phenix.refine53 were carried out to build the structure of the full-length LlaBIII-DNA complex. Hydrogens were automatically added to the model during refinement. The refinement statistics for the structures are provided in Supplementary Table 1. The final model had 96% of the residues in the favoured, 4% in the allowed and 0% in the disallowed region of the Ramachandran plot. PYMOL (http://www.pymol.org) was used to generate figures.
SEC-MALS
The Type ISP enzymes (0.5-1.0 mg/ml) were run in reaction buffer (for LlaGI, 50 mM Tris-Cl, pH 8.0, 10 mM MgCl2, 1 mM DTT; for LlaBIII, 50 mM Tris-Cl, pH 8.0, 10 mM MgCl2, 150 mM KCl, 1 mM DTT) at 1 ml/min on a Superdex 200 analytical size exclusion column (GE Healthcare), attached to a light scattering diode array and a differential refractive index detector (Wyatt Technology). Chromatograms were analysed using the ASTRA software (v6.0.5.3, Wyatt Technology). Absolute molecular masses of elutes were calculated from the ratio of light scattering and the differential refractive index. Reproducibility of the column chromatography was checked using bovine serum albumin as in Butterer et al.55
Modelling studies
The models of LlaBIII bound to longer DNA in Fig. 3b and the collision complex in Supplementary Fig. 10a were generated using Coot.51 The former was generated by addition of nucleotides upstream to the co-crystal DNA. The conformation of the nucleotides was approximated to that of a B-form DNA. A model of the collision complex was generated by placing the structure of two molecules of LlaBIII-DNA side-by-side such that the two TRDs and the downstream ends of the DNA were contiguous. The pseudo-continuous DNA thus formed has two molecules of LlaBIII oriented head-to-head. The upstream ends were extended by addition of nucleotides having the conformation of B-form DNA.
DNA cleavage assay
To produce the linear substrates used in Supplementary Fig. 8, pHH-318 was used as a substrate for PCR with the downstream primer FOli LlaGI-529 and the following upstream primers (Supplementary Table 3) : ROli LlaGI-16 (for n =16); ROli LlaGI-21 (for n =21); ROli LlaGI-22 (for n =22); ROli LlaGI-23 (for n =23); ROli LlaGI-24 (for n =24); and, ROli LlaGI-51 (for n =51). All substrates were purified by phenol-chloroform extraction and ethanol precipitation. Assays contained 2 nM DNA, 4 mM ATP and 200 nM LlaGI in TMD buffer. Reactions were started by adding ATP, and incubated at 25 °C for the times indicated. Samples were stopped by adding ½ volume of STEB [0.1 M Tris (pH 7.5), 0.2 M EDTA, 40% (w/v) sucrose, 0.4 mg/ml bromophenol blue] and analysed by agarose gel electrophoresis. Gels were stained for 30 minutes in 0.5 μg/ml ethidium bromide and destained for at least two hours. The gels were quantified from 8-bit images using ImageQuant TL (GEHealthcare), correcting band intensity for DNA length. Average values and standard deviations were calculated in GraphPad Prism.
Triplex Displacement assays
To produce the linear substrate used in Fig. 3c, pRMA03F20 was used as a substrate for PCR with the downstream primer FOli LlaGITr-529 and the following upstream primers (Supplementary Table 3): ROli LlaGI Tr-16 (for n =16); ROli LlaGI Tr-21 (for n =21); ROli LlaGI Tr-22 (for n =22); ROli LlaGI Tr-23 (for n =23); ROli LlaGI Tr-24 (for n =24); and, ROli LlaGI Tr-51 (for n =51). To produce the linear substrates used in Supplementary Fig. 12, either pRMA03R20 was cut with BlpI (for the 4663 bp spacing) or pRMA03F was cut with EcoRI (for the 168 bp spacing). All substrates were purified by phenol-chloroform extraction and ethanol precipitation. Triplexes were formed overnight as described previously using a tetramethylrhodamine-labelled triplex forming oligonucleotide.18 Reactions were carried out using 1 nM DNA (0.5 nM triplex), 100 nM LlaGI, 4 mM ATP in TMD [50 mM Tris-Cl, pH 8.0, 10 mM MgCl2, 1 mM DTT] at 25 °C. Fluorescence intensity measurements were performed using an SF61-DX2 stopped-flow fluorimeter (TgK Scientific).18 DNA and enzyme were premixed and the reactions initiated by mixing with ATP. In Supplementary Fig. 12, translocation traps were added to the ATP syringe, and gave final concentrations of 1 μM oligoduplex or 100 nM LlaGI(K210A).20
ATP-independent nick mapping assay
To produce the linear substrate (Supplementary Fig. 14a), pHH-3 was used as a substrate for PCR with the primers FOli LlaGI +75 (32P-labelled for top strand substrates) and ROli LlaGI −75 (32P-labelled for bottom strand substrates) (Supplementary Table 3). Assays contained 2 nM DNA and 200 nM wild type LlaGI, nuclease mutant LlaGI(D74A) or nuclease domain deletion mutant LlaGIΔN in TMD buffer. Reactions were started by adding enzyme (or blank dilution buffer), and incubated at 25 °C for 60 minutes. Samples were stopped using 10 mM EDTA, 1 mM NaOH, 80% (v/v) formamide, 0.1% (w/v) bromophenol blue) and analysed by electrophoresis on a urea denaturing 8% (w/v) polyacrylamide gel. Sequencing lanes were produced using labelled primers following the USB T7 sequencing kit (Affymetrix) according to the manufacturer’s instructions. The pixel positions of the 16-bit scanned wild type lanes in Supplementary Fig. 13a were corrected for DNA length using an exponential function calculated based on the positions of DNA bands in the sequencing lanes, according to Šišáková et al.19
Magnetic Tweezers assay
pKA20 was generated by QuikChange multi-site directed mutagenesis of pSFV-1 (Life Technologies) using the following upstream primers (Supplementary Table 3): KA40F; KA41F; KA42F; KA43F; KA44F; KA45F; KA46F; KA47F to remove LlaGI sites at 131, 1539, 2120, 3637, 3863, 5619, 7252 and 9499 bp, respectively, to leave two head-to-head sites 1339 bp apart. pKA21 was generated by QuikChange mutagenesis of pKA20 to remove the LlaGI site at 2640 bp using primer pair KA69F and KA69R (Supplementary Table 3). The DNA were cleaved using BamHI and SpeI. Biotin- or digoxigenin-modified attachment handles were made using 1.0 kbp DNA fragments that where labelled with biotin- or digoxigenin-dUTP by PCR of pUC19,56 using primers ms293SpeF and primers ms293NotR (Supplementary Table 3), and which were digested with either BamHI or SpeI. The pKA20 or pKA21 fragment was ligated with the biotin/dig-labelled handles using T4 DNA ligase.56
Single-molecule experiments were carried out as previously described32,56 using a commercial magnetic tweezers instrument (Picotwist, Saint Romain de Popey, France, equipped with a Jai CM-140GE camera, image acquisition at 31 Hz).57 Fluidic cells were constructed from an uncoated 24×60 mm coverslip (Menzel-Gläser No. 1), double-sided adhesive tape (3M 467MP, 50 μm depth) and polyester film (Melinex 401, DuPont, 50 μm depth). Anti-digoxigenin (Roche) and BSA were adsorbed directly to the glass by incubation for >3 hours at room temperature. Each DNA construct was bound at its biotin-modified end to excess streptavidin-coated magnetic beads (1 μm diameter, MyOne T1, Invitogen) and added into the fluidic cell to allow the DNA to bind the surface via its digoxigenin-modified end. Beads attached directly to the coverslip were monitored to correct for instrument drift. The three-dimensional position of the magnetic bead and thus the orientation and length of the attached DNA molecule was determined from video images at 31 Hz using real-time 3D particle tracking with sub-nm accuracy.57 Topologically-unconstrained (nicked) and –constrained (supercoilable) DNA were identified from rotations curves. The reaction temperature within the cell was maintained by Peltier elements to 25±1 °C.
Experiments with LlaGI were carried out in TMD buffer. In all time trajectories depicted, raw DNA length data taken at the camera acquisition rate is shown, except for 0.1 pN trace in Fig. 4b where the data was also smoothed with a 1 Hz moving average. Flow was started by manually opening a valve and 10 nM LlaGI and ATP (4 mM except where stated) were added to the flow cell in-port. After 5±1 s, the flow was stopped by closing the valve and the beads thereafter relaxed back to their stretched lengths. Cleavage times were taken from the initiation of flow to the point where bead tracking was lost. Error were calculated from the average values from the square root of number of points (N) in the bin. For topologically-constrained DNA, supercoiling levels were set by rotating the magnets at low force where the rotation curves are symmetrical.
Single DNA cleavage event mapping
The method was adapted from that of Jindrova et al.,58 (Supplementary Fig. 14). pKA20.100 was made from pKA20 by reverse PCR using primers ROli LlaGI int 0 and FOli LlaGI int +100 (Supplementary Table 3), followed by recircularization of the linear product using T4 DNA ligase. PCR was used to amplify the chloramphenicol gene from pACYC18459 using primers KA082F and KA082R (Supplementary Table 3). The amplified product was purified following agarose gel electrophoresis using a QIAquick Gel Extraction Kit (Qiagen). Cleavage reactions contained 2 nM pKA20.100, 200 nM LlaGI, 4 mM ATP in TMD. Reactions were initiated by adding ATP, and the reactions at 25°C stopped at 10, 20, 30 or 60 s by adding ½ volume of 100 mM Tris-Cl, pH 8.0, 100 mM EDTA. Cleaved DNA was purified using a QIAquick PCR Purification Kit (Qiagen), and treated with 1.5 units T4 DNA polymerase (New England Biolabs) per nM DNA and 100 μM dNTPs in the supplied reaction buffer supplemented with 100 μg BSA at 12°C for 30 min. The reaction was stopped by heating to 75°C for 30 min. Blunt-ended DNAs were purified using a QIAquick PCR Purification Kit and ligated to the chloramphenicol gene insert using T4 DNA ligase (New England Biolabs) at 16°C for 16 hours. E. coli TOP10 cells (Life Technologies) were transformed with the ligations and single colonies selected by overnight growth on LB agar plates with 50 μg/ml ampicillin and 34 μg/ml chloramphenicol. Plasmid DNAs representing single cleavage events were purified from single colonies and sent for DNA sequencing (Dundee Sequencing Service, Dundee, Scotland) using the primer KA050F (Supplementary Table 3). Data in Fig. 5c,d and Supplementary Fig. 14d is from three independent repeat cleavage reactions (average values and standard deviations were calculated in GraphPad Prism). 6-12% of clones had deletions of >225 bp that could have resulted from illegitimate recombination,58 and were thus excluded from the analysis. Clones without a sequence deletion or duplication, characteristic of a blunt end (Supplementary Fig. 14c), were not found. Note, however, that our model (Fig. 6) does not exclude the possibility of strand breaks occurring immediately opposite one another in rare instances. Median and skewness values in Fig. 5c were calculated using Excel (Microsoft).
Supplementary Material
Acknowledgements
This work was funded by the Wellcome Trust-DBT India Alliance (500048-Z-09-Z to K.S.), Wellcome Trust (084086 to M.D.S., K.v.A, F.M.D, C.P.), DBT India (to M.K.C), and CSIR India (to N.N.). K.S. acknowledges Kiyoshi Nagai and his group members at MRC Laboratory of Molecular Biology, Cambridge, U.K., for hosting him as an India Alliance visiting scientist during the initial stage of this work. We are grateful to Kiyoshi Nagai, Dale Wigley, Lori Passmore and Radha Chauhan for their comments on the manuscript. We acknowledge Diamond Light Source (DLS), Oxfordshire, UK, European Synchrotron Radiation Facility (ESRF), Grenoble, France, for access to beamlines, and DBT India for funding the use of BM14 beamline at ESRF.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Accesssion Code
Protein Data Bank: 4XQK
Reference list
- 1.Dryden DTF, Murray N, Rao DN. Nucleoside triphosphate-dependent restriction enzymes. Nucleic Acids Res. 2001;29:3728–3741. doi: 10.1093/nar/29.18.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nature Rev. Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 3.Linn S, Arber W. Host specificity of DNA produced by Escherichia coli, X. In vitro restriction of phage fd replicative form. Proc. Natl. Acad. Sci. USA. 1968;59:1300–1306. doi: 10.1073/pnas.59.4.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meselson M, Yuan R. DNA restriction enzyme from E. coli. Nature. 1968;217:1110–1114. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- 5.Loenen WAM, Dryden DTF, Raleigh EA, Wilson GG. Type I restriction enzymes and their relatives. Nucleic Acids Res. 2014;42:20–44. doi: 10.1093/nar/gkt847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith RM, Diffin FM, Savery NJ, Josephsen J, Szczelkun MD. DNA cleavage and methylation specificity of the single polypeptide restriction-modification enzyme LlaGI. Nucleic Acids Res. 2009;37:7206–7218. doi: 10.1093/nar/gkp790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murray NE. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle) Microbiol. Mol. Biol. Rev. 2000;64:412–434. doi: 10.1128/mmbr.64.2.412-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rao DN, Dryden DT, Bheemanaik S. Type III restriction-modification enzymes: a historical perspective. Nucleic Acids Res. 2014;42:45–55. doi: 10.1093/nar/gkt616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dürr H, Flaus A, Owen-Hughes T, Hopfner K-P. Snf2 family ATPases and DExx box helicases: differences and unifying concepts from high-resolution crystal strucures. Nucleic Acids Res. 2006;34:4160–4167. doi: 10.1093/nar/gkl540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stanley LK, et al. When a helicase is not a helicase: dsDNA tracking by the motor protein EcoR124I. EMBO J. 2006;25:2230–2239. doi: 10.1038/sj.emboj.7601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halford SE, Welsh AJ, Szczelkun MD. Enzyme-mediated DNA looping. Annual Rev. Biophys. Biomol. Struct. 2004;33:1–24. doi: 10.1146/annurev.biophys.33.110502.132711. [DOI] [PubMed] [Google Scholar]
- 12.Schwarz FW, et al. The helicase-like domains of type III restriction enzymes trigger long-range diffusion along DNA. Science. 2013;340:353–356. doi: 10.1126/science.1231122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kennaway CK, et al. Structure and operation of the DNA-translocating type I DNA restriction enzymes. Genes Dev. 2012;26:92–104. doi: 10.1101/gad.179085.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thoma NH, et al. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nature_Struct. Mol. Biol. 2005;12:350–356. doi: 10.1038/nsmb919. [DOI] [PubMed] [Google Scholar]
- 15.Hopfner K-P, Gerhold C-B, Lakomek K, Wollmann P. Swi2/Snf2 remodelers: hybrid views on hybrid molecular machines. Current Opinion Struct. Biol. 2012;22:225–233. doi: 10.1016/j.sbi.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wollmann P, et al. Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP. Nature. 2011;475:403–407. doi: 10.1038/nature10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith RM, Josephsen J, Szczelkun MD. The single polypeptide restriction-modification enzyme LlaGI is a self-contained molecular motor that translocates DNA loops. Nucleic Acids Res. 2009;37:7219–7230. doi: 10.1093/nar/gkp794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Šišáková E, van Aelst K, Diffin FM, Szczelkun MD. The Type ISP Restriction-Modification enzymes LlaBIII and LlaGI use a translocation-collision mechanism to cleave non-specific DNA distant from their recognition sites. Nucleic Acids Res. 2013;41:1071–1080. doi: 10.1093/nar/gks1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Aelst K, Šišáková E, Szczelkun MD. DNA cleavage by Type ISP Restriction-Modification enzymes is initially targeted to the 3′-5′ strand. Nucleic Acids Res. 2013;41:1081–1090. doi: 10.1093/nar/gks1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park SY, et al. Structural characterization of a modification subunit of a putative type I restriction enzyme from Vibrio vulnificus YJ016. Acta Crystallogr. D Biol.Crystallogr. 2012;68:1570–1577. doi: 10.1107/S0907444912038826. [DOI] [PubMed] [Google Scholar]
- 22.Shen BW, et al. Characterization and crystal structure of the type IIG restriction endonuclease RM.BpuSI. Nucleic Acids Res. 2011;39:8223–8236. doi: 10.1093/nar/gkr543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goedecke K, Pignot M, Goody RS, Scheidig AJ, Weinhold E. Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. Nature Struct. Mol. Biol. 2001;8:121–125. doi: 10.1038/84104. [DOI] [PubMed] [Google Scholar]
- 24.Kim JS, et al. Crystal structure of DNA sequence specificity subunit of a type I restriction-modification enzyme and its functional implications. Proc. Natl. Acad. Sci. USA. 2005;102:3248–3253. doi: 10.1073/pnas.0409851102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calisto BM, et al. Crystal structure of a putative type I restriction-modification S subunit from Mycoplasma genitalium. J Mol. Biol. 2005;351:749–762. doi: 10.1016/j.jmb.2005.06.050. [DOI] [PubMed] [Google Scholar]
- 26.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 27.Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buttner K, Nehring S, Hopfner K-P. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nature Struct. Mol. Biol. 2007;14:647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 29.Saikrishnan K, Powell B, Cook NJ, Webb MR, Wigley DB. Mechanistic basis of 5′-3′ translocation in SF1B helicases. Cell. 2009;137:849–859. doi: 10.1016/j.cell.2009.03.036. [DOI] [PubMed] [Google Scholar]
- 30.Gu M, Rice CM. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc. Natl. Acad. Sci. USA. 2010;107:521–528. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dürr H, Körner C, Müller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 32.Ramanathan SP, et al. Type III restriction enzymes communicate in 1D without looping between their target sites. Proc Natl Acad Sci USA. 2009;106:1748–1753. doi: 10.1073/pnas.0807193106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seidel R, et al. Dynamics of initiation, termination and reinitiation of DNA translocation by the motor protein EcoR124I. EMBO J. 2005;24:188–197. doi: 10.1038/sj.emboj.7600881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seidel R, van Noort J, et al. Real-time observation of DNA translocation by the Type I RM enzyme EcoR124I. Nature Structural & Molecular Biol. 2004;11:838–843. doi: 10.1038/nsmb816. [DOI] [PubMed] [Google Scholar]
- 36.Šišáková E, Weiserova M, Dekker C, Seidel R, Szczelkun MD. The Interrelationship of Helicase and Nuclease Domains during DNA Translocation by the Molecular Motor EcoR124I. J. Mol. Biol. 2008;384:1273–1286. doi: 10.1016/j.jmb.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith RM, Josephsen L, Szczelkun MD. An Mrr-family nuclease motif in the single polypeptide restriction-modification enzyme LlaGI. Nucleic Acids Res. 2009;37:7231–7238. doi: 10.1093/nar/gkp795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jankowsky E, Gross CH, Shuman S, Pyle AM. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- 39.Park J, et al. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 2010;142:544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fagerburg MV, et al. PcrA-mediated disruption of RecA nucleoprotein filaments--essential role of the ATPase activity of RecA. Nucleic Acids Res. 2012;40:8416–8424. doi: 10.1093/nar/gks641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor AF, Smith GR. Substrate specificity of the DNA unwinding activity of the RecBC enzyme of Escherichia coli. J. Mol. Biol. 1985;185:431–443. doi: 10.1016/0022-2836(85)90414-0. [DOI] [PubMed] [Google Scholar]
- 42.Dillingham MS, Kowalczykowski SC. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 2008;72:642–671. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeeles JT, Cammack R, Dillingham MS. An iron-sulfur cluster is essential for the binding of broken DNA by AddAB-type helicase-nucleases. J. Biol Chem. 2009;284:7746–7755. doi: 10.1074/jbc.M808526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Endlich B, Linn S. The DNA restriction endonuclease of Escherichia coli B. II. Futher studies of the structure of DNA intermediates and products. J. Biol. Chem. 1985;260:5729–5738. [PubMed] [Google Scholar]
- 45.Levy A, et al. CRISPR adaptation biases explain preferences for acquisition of foreign DNA. Nature. 2015;520:505–510. doi: 10.1038/nature14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 46.LeMaster DM, Richards FM. NMR sequential assignment of Escherichia coli thioredoxin utilizing random fractional deuteriation. Biochemistry. 1988;27:142–150. doi: 10.1021/bi00401a022. [DOI] [PubMed] [Google Scholar]
- 47.Kabsch W. XDS. Acta Crystallogr. D Biol.Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.French S, Wilson K. On the treatment of negative intensity observations. Acta Crystallogr. A. 1978;34:517–525. [Google Scholar]
- 49.Read RJ, McCoy AJ. Using SAD data in Phaser. Acta Crystallogr. D Biol.Crystallogr. 2011;67:338–344. doi: 10.1107/S0907444910051371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cowtan KD, Zhang KYJ. Density modification for macromolecular phase improvement. Prog. Biophy. Mol. Biol. 1999;72:245–270. doi: 10.1016/s0079-6107(99)00008-5. [DOI] [PubMed] [Google Scholar]
- 51.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol.Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol.Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Afonine PV, et al. Joint X-ray and neutron refinement with phenix.refine. Acta Crystallogr. D Biol.Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444910026582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol.Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 55.Butterer A, et al. Type III restriction endonucleases are heterotrimeric: comprising one helicase-nuclease subunit and a dimeric methyltransferase that binds only one specific DNA. Nucleic Acids Res. 2014;42:5139–5150. doi: 10.1093/nar/gku122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Szczelkun MD, et al. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci USA. 2014;111:9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lionnet T, et al. Magnetic trap construction. Cold Spring Harb Protoc. 2012;2012:133–138. doi: 10.1101/pdb.prot067496. [DOI] [PubMed] [Google Scholar]
- 58.Jindrova E, Schmid-Nuoffer S, Hamburger F, Janscak P, Bickle TA. On the DNA cleavage mechanism of Type I restriction enzymes. Nucleic Acids Res. 2005;33:1760–1766. doi: 10.1093/nar/gki322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.