Abstract
Cancer chromosomal instability (CIN) results in an elevated rate of change of chromosome number and structure and generates intratumour heterogeneity1,2. CIN is observed in the majority of solid tumours and is associated with both poor prognosis and drug resistance3,4. Therefore, understanding a mechanistic basis for CIN is paramount. Here we find evidence for impaired replication fork progression and elevated DNA replication stress in CIN+ colorectal cancer (CRC) cells relative to CIN− CRC cells, with structural chromosome abnormalities precipitating chromosome missegregation in mitosis. We identify three novel CIN-suppressor genes (PIGN (MCD4), RKHD2 (MEX3C) and ZNF516 (KIAA0222)) encoded on chromosome 18q, which is subject to frequent copy number loss in CIN+ CRC. 18q loss was temporally associated with aneuploidy onset at the adenoma-carcinoma transition. CIN-suppressor gene silencing leads to DNA replication stress, structural chromosome abnormalities and chromosome missegregation. Supplementing cells with nucleosides, to alleviate replication-associated damage5, reduces the frequency of chromosome segregation errors following CIN-suppressor gene silencing and attenuates segregation errors and DNA damage in CIN+ cells. These data implicate a central role for replication stress in the generation of structural and numerical CIN, which may inform new therapeutic approaches to limit intratumour heterogeneity.
Structural and numerical chromosomal instability are commonly observed together in solid tumours (Supplementary Fig. 1a-c)6. This co-occurrence can be experimentally induced via defective mitotic checkpoint function or chromosome attachment to the mitotic spindle, or through pre-mitotic defects affecting chromosome structure, such as faulty DNA repair and replication6-10. However, the mechanisms underlying CIN in cancer remain unclear.
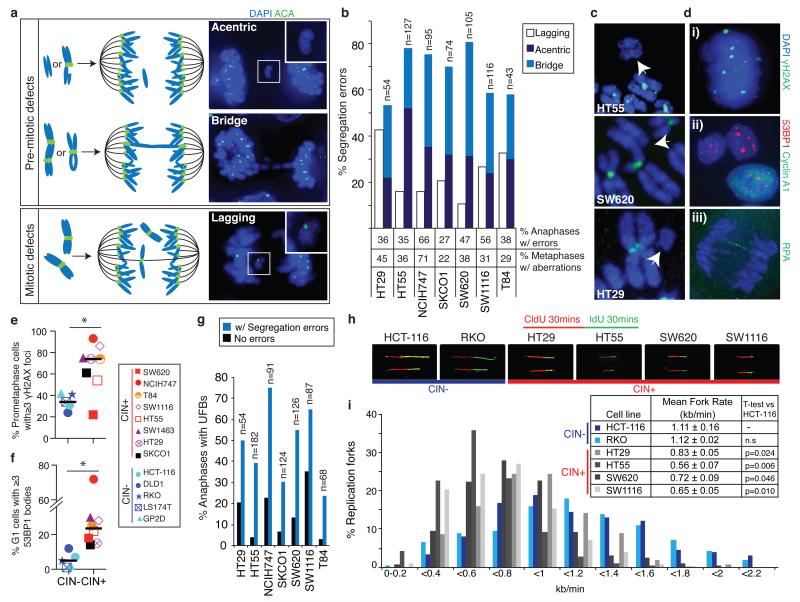
Colorectal cancers can be broadly classified as CIN+/aneuploid or CIN−/microsatellite unstable3. CIN+ cells displayed an increased frequency of chromosome segregation errors compared to CIN− cells (median = 38% versus 18%, p=0.0025, Mann-Whitney test, Supplementary Fig. 2a)11. To address whether mitotic or pre-mitotic mechanisms are responsible for these segregation errors, we analysed high resolution images of anaphases in a panel of CIN+ CRC cells. The majority of segregation errors consisted of chromosome fragments without centromeres (acentrics, Figure 1a) and anaphase bridges (54-81%, median 70%, Figure 1a,b), indicative of structural chromosome aberrations arising through pre-mitotic defects12. In contrast, only 10-43% of segregation errors were lagging chromosomes with centromeres (median 20%, Figure 1a,b), suggesting that mitotic dysfunction resulting in improper chromosome attachments6 cannot explain the majority of segregation errors in CIN+ CRC cells (Supplementary Fig. 2b). Furthermore, kinetochore distortion of lagging chromosomes (reflecting merotelic attachments6) was rare (0-12%, median 8% of segregation errors, Supplementary Fig. 2c). We did not observe differences in mitotic timing, mitotic checkpoint function, sister chromatid cohesion or supernumerary centrioles between CIN+ and CIN− cells, and multipolar spindles were infrequent (0-18%, median 8%, Supplementary Fig. 2d-i). These data suggest that mitotic dysfunction occurs at low frequency in CIN+ CRC cells and that the majority of observed chromosome segregation errors result from structural chromosome aberrations. Accordingly, 22-71% (median 36%) of CIN+ cell metaphases displayed structurally abnormal chromosomes, including acentric chromosomes, dicentric chromosomes and DNA double strand breaks (Figure 1b,c and Supplementary Fig. 2j).
Figure 1. Replication stress generates chromosome segregation errors in CIN+ cells.
a) Schematic illustrating pre-mitotic and mitotic origins of chromosome segregation errors. Right panels: example images in SW1116 (CIN+) cells stained with DAPI and anti-centromere antibodies (ACA). b) % Segregation errors in CIN+ cell lines classified into lagging chromosomes, acentrics and anaphase bridges. Bridges extend fully between DNA masses; acentrics and lagging chromosomes were distinguished using ACA staining. Segregation errors not classifiable as bridges, lagging chromosomes or acentrics (<15%) are omitted for clarity. % Anaphases displaying segregation errors (n>100/cell line), and % metaphases displaying structural aberrations (n=38-84/cell line) shown below graph. c) Examples of structurally abnormal chromosomes identified on metaphase chromosome spreads, hybridised to an all-centromere probe (green) and stained with DAPI. d) Examples of replication stress-associated cellular phenotypes in NCIH747 (CIN+) cells stained with DAPI and antibodies as indicated; (i) γH2AX foci in prometaphase; (ii) 53BP1 bodies in G1 (cyclin A1-negative) cells; (iii) anaphase UFBs, detected with antibodies for the single-stranded DNA binding protein RPA27. e) % Prometaphase DNA damage in CIN+ versus CIN− cells (n>100 cells/cell line, *p=0.033). f) % G1 53BP1 bodies in CIN+ versus CIN− cells (n>250 cells/cell line, *p=0.028). g) % Anaphases with UFBs. h,i) 4 CIN+ and 2 CIN− cell lines were incubated sequentially with 5-chlorodeoxyuridine (CldU) and 5-iododeoxyuridine (IdU) for 30 minutes each. DNA fibre assays were performed and replication rates at individual replication forks were assessed. Representative fibres from each cell line are shown (h). i) Distribution of replication fork rates (CldU, n>300 forks in total/cell line from 3 experiments), with mean replication fork rates (CldU, n>60 forks/experiment, mean±s.e.m of 3 experiments) shown in the key (inset). Two-tailed t-test relative to HCT-116 cells.
Next we sought a putative cause for these structural chromosome alterations. Activation of the DNA damage response has been observed in both colorectal adenomas and carcinomas13,14 and is thought to reflect DNA replication stress13,15,16. Pharmacological induction of replication stress in HCT-116 (CIN−) cells resulted in structural chromosome aberrations and segregation errors, 82% of which were bridges or acentric chromosomes (Supplementary Fig. 3a-f). Importantly, numerical chromosome changes were also induced (Supplementary Fig. 3g,h), demonstrating that replication stress can result in both structural and numerical chromosomal instability17.
DNA replication stress results in a number of cellular phenotypes including DNA damage in prometaphase10,15,16, ultra-fine anaphase DNA bridges (UFBs)16, and 53BP1-positive nuclear bodies in G1 cells15,18 (Figure 1d and Supplementary Fig. 3i-m). CIN+ cells displayed elevated prometaphase DNA damage (median 74% CIN+ versus 34% CIN− cells with ≥3 γH2AX foci, Figure 1d,e (p=0.033, Mann-Whitney test)) in the absence of increased oxidative DNA damage (Supplementary Fig. 4a-c). γH2AX foci were not confined to telomeres (Supplementary Fig. 4d,e). CIN+ cells also displayed more 53BP1 nuclear bodies in G1 cells (Figure 1d,f, p=0.028, Mann-Whitney test). Consistent with the hypothesis that replication stress may drive chromosome segregation errors in CIN+ cells, UFBs were enriched in anaphases with segregation errors compared to anaphases without segregation errors (Figure 1d,g, p=0.00018, paired t-test).
To assess DNA replication directly, DNA fibre assays were performed for two CIN− and four CIN+ cell lines, to measure progression of individual replication forks, fork stalling and asymmetry between sister replication forks. CIN+ cells displayed significantly slower fork rates relative to CIN− cell lines (0.56-0.83 kb/min compared to 1.11-1.12 kb/min, p<0.05, Figure 1h,i, Supplementary Fig. 5a,b. Additionally, there was evidence of increased replication fork stalling and asymmetric sister fork progression in several CIN+ cell lines (Supplementary Fig. 5c-e), consistent with impaired replication fork progression. These data indicate that elevated replication stress, as demonstrated using established hallmarks and direct measurement of replication fork progression, may contribute substantially to chromosome missegregation in CIN+ CRC cells.
We sought a genetic basis for the elevated replication stress and chromosome segregation errors in CIN+ cells. Examining whole-exome sequencing data in colorectal tumours from The Cancer Genome Atlas (TCGA)19, and mutation status for 64 genes in CRC cell lines (COSMIC), revealed that the only gene mutated at significantly higher frequencies in CIN+ tumours or cell lines was TP53, which is also mutated in 13-33% of CIN− tumours and cell lines (Supplementary Fig. 6a,b). Mutations in TP53 are thought to be permissive for CIN rather than causative20. These data suggest that whilst mutations in known oncogenes or tumour suppressors may contribute to replication stress in cancer cells14,21, these are unlikely to exclusively account for the elevated replication stress in CIN+ CRC cells. We therefore addressed whether additional genetic aberrations may contribute to CIN.
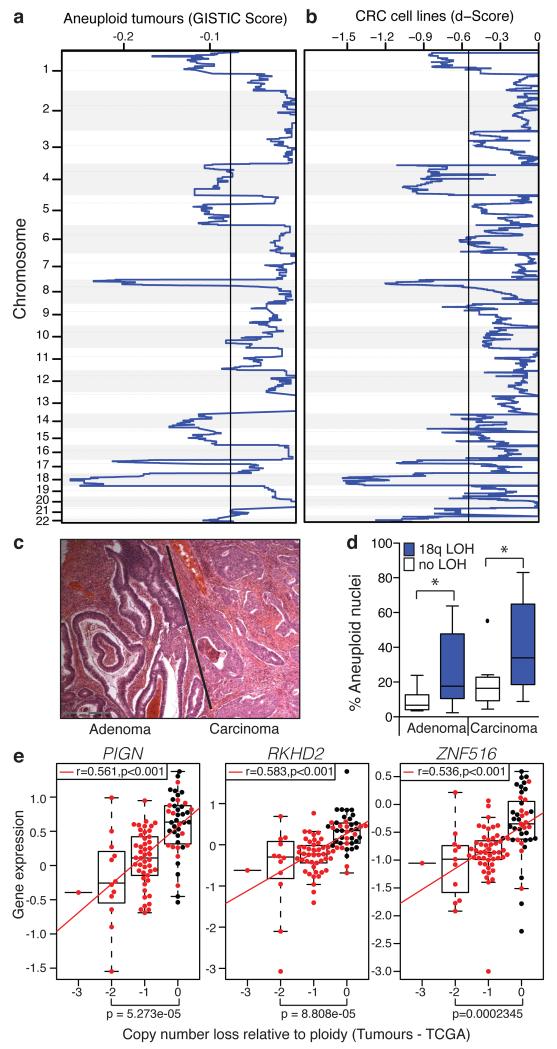
We hypothesised that regions of consistent somatic copy number loss in CIN+ CRC might encode CIN-suppressor genes, loss of which could contribute to the induction of chromosome missegregation. To identify CIN-specific regions of loss, comparative genomic hybridisation data (CGH) was analysed for a cohort of 26 aneuploid colorectal tumours and 20 CIN+ cell lines. Chromosome 18q was most frequently subject to copy number loss, observed in 88% of aneuploid tumours and 80% of CIN+ cell lines, (Figure 2a,b and Supplementary Table 1) consistent with published studies in CRC and other tumour types22-24. Copy number losses in CIN+ tumours and cell lines were highly concordant (p<0.001, Fisher’s exact test, Supplementary Table 2). Next we examined the temporal sequence and consequences of 18q copy number loss in tumours. In a cohort of 28 adenomas (preinvasive tumour) with carcinoma in the same specimen (Figure 2c), 18q loss of heterozygosity (LOH - indicative of copy number loss) was observed in 21/28 (75%) carcinomas, compared to 10/28 (35.7%) adenomas, implicating 18q loss in the adenoma-carcinoma transition. 18q LOH was significantly associated with aneuploidy in both adenomas and carcinomas (Figure 2d, Supplementary Fig. 6c).
Figure 2. Somatic copy number loss of chromosome 18q in CIN+ CRC.
a) GISTIC analysis for somatic copy number loss in 26 aneuploid colorectal tumours. Q=0.25 determines significance (black line). b) Copy number losses in 20 CIN+ CRC cell lines. Significant regions were defined relative to 9 CIN− cell lines, (Q=0.25, black line). c) Haematoxylin and eosin-stained tumour specimen, showing adenoma with adjacent carcinoma. d) % Aneuploid nuclei, measured by DNA image cytometry, in paired adenomas and carcinomas (n=20) with/without 18q LOH (Tukey box plot with outliers displayed, two-tailed t-test, *p<0.05). e) Spearman’s rank correlation between mRNA expression and DNA copy number for PIGN, RKHD2 and ZNF516 in CIN− (n=28, black dots) versus CIN+ (n=74, red dots) tumours (TCGA). Tumours were defined as CIN+ based on a weighted genome integrity index >0.2 (see Methods). Statistic: Wilcoxon Mann-Whitney test.
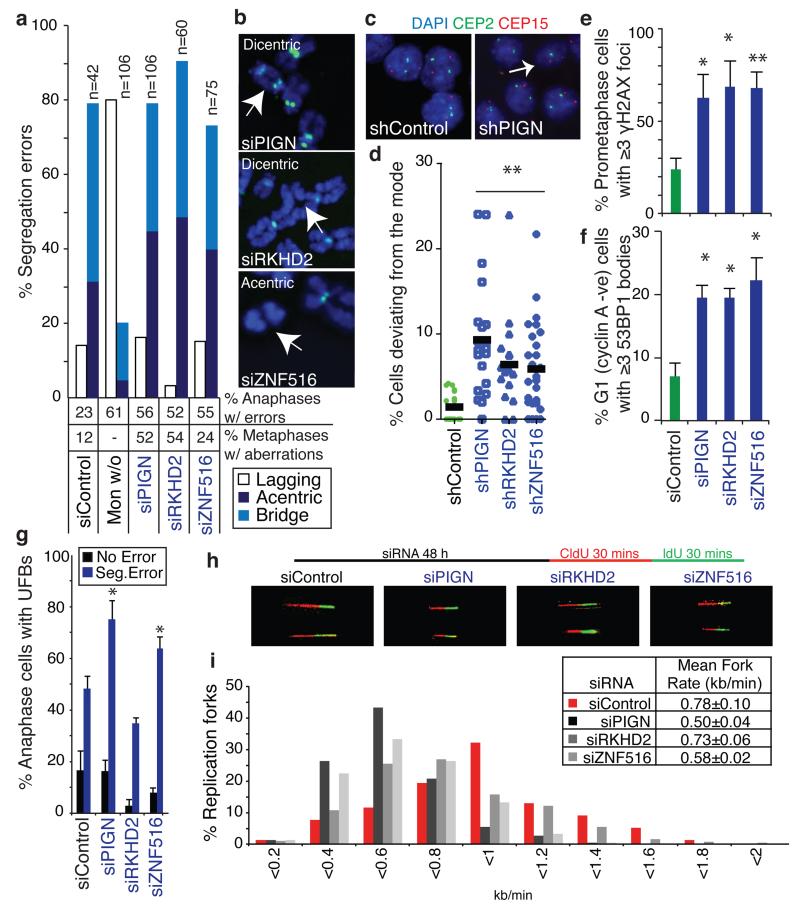
To identify candidate CIN-suppressor genes encoded within regions of recurrent copy number loss, HCT-116 cells were transfected with pools of four siRNAs targeting the most frequently lost genes, present at ≤1 copy in at least 30% of CIN+ cell lines (94 genes encoded on chromosome 18q, Supplementary Table 3). After 48 hours anaphase segregation errors were quantified (Supplementary Fig. 7a). Importantly, this approach would identify both mitotic and pre-mitotic defects. SiRNA pools inducing segregation errors to ≥3 standard deviations above the frequency in control-transfected cells were assessed in validation assays. All sequences targeting PIGN, RKHD2 and ZNF516 induced segregation errors and efficiently depleted mRNA levels (Supplementary Fig. 7b-d). Additional independent siRNA sequences targeting each of the three genes also induced segregation errors (Supplementary Fig. 7e) and these genes were prioritised for further analysis, although we cannot exclude a contribution of other genes encoded on 18q to CIN. Of note, RKHD2 is the only protein-coding gene located between two genes implicated in CRC carcinogenesis, DCC and SMAD4. Copy number loss of PIGN, RKHD2 or ZNF516 is observed in 85% of the 20 CIN+ cell lines and 84% of aneuploid tumours (n=103) in the independent TCGA cohort19, with loss of all three genes in 70% of CIN+ cell lines and 79% of aneuploid tumours (Supplementary Table 4). Importantly, reduced copy number correlated significantly with reduced mRNA expression for all three genes both in the TCGA tumour cohort and CRC cell lines (Figure 2e, Supplementary Fig. 8a and Supplementary Table 5).
We next ensured off-target siRNA effects reported against MAD2 (mitotic checkpoint protein) and RAD51 (DNA repair protein), were not causing chromosome missegregation (Supplementary Fig. 8b-e)25. Two RKHD2 sequences partially depleted MAD2 protein, and were excluded from further analysis. Additionally, expression of exogenous RKHD2-GFP rescued segregation errors induced upon silencing endogenous RKHD2 using a 3′ UTR-targeted siRNA, and expression of siRNA-resistant PIGN-GFP and ZNF516-GFP reduced segregation errors induced by depletion of endogenous PIGN or ZNF516 (Supplementary Fig. 8f). Silencing each CIN-suppressor also induced segregation errors in two additional CIN− CRC cell lines, DLD1 and RKO, and in 18q-normal CIN+ NCIH508 cells (Supplementary Fig. 9a-c). Induction of segregation errors was independent of oncogenic KRAS signalling in HCT-116 cells, as segregation errors were also induced following CIN-suppressor gene silencing in a KRAS wild-type isogenic cell line (Supplementary Fig. 9d).
CIN-suppressor gene silencing in HCT-116 cells primarily induced acentric chromosomes and anaphase bridges (Figure 3a) in the absence of gross mitotic defects (data not shown), similar to observations in CIN+ CRC cell lines with 18q loss (Figure 1b). This suggested a pre-mitotic origin for these chromosome segregation errors and accordingly, we observed an increased frequency of structurally abnormal chromosomes (Figure 3a,b, Supplementary Fig. 10a). To assess chromosome non-disjunction, HCT-116 cell lines stably expressing small-hairpin RNAs (shRNAs) were constructed. shRNA-mediated silencing of each CIN-suppressor gene increased segregation error frequency (Supplementary Fig. 10b), and single cell clones grown from each cell line displayed significantly increased intra-colony deviation for chromosomes 2 and 15 (Figure 3c,d and Supplementary Fig. 10c), indicating that CIN-suppressor gene silencing induces both structural and numerical instability.
Figure 3. Candidate suppressors of replication stress and CIN encoded on chromosome 18q.
a,b) CIN-suppressor genes were silenced in HCT-116 (CIN−) cells for 48 h. a) % Segregation errors accounted for by lagging chromosomes, acentrics and anaphase bridges. Other segregation errors (<15%) are omitted for clarity. For comparison, segregation errors arising via improper chromosome attachments were induced by monastrol treatment (100 μM, 1 h, 75 min release). % Anaphases displaying segregation errors and % metaphases displaying ≥1 structurally abnormal chromosome (n>100) shown below graph. b) Examples of structurally abnormal chromosomes as indicated. c,d) Cell lines stably expressing shRNAs as indicated were seeded at low density on glass slides to allow colony formation. Slides were fixed and hybridised to DNA probes for centromeres 2 and 15. c) Example images of control and shPIGN cells. d) % Deviation from the modal centromere copy number per colony (mean of two probes (CEP2 and CEP15)). Lines are median values, statistic: Dunn’s Multiple Comparison test, p<0.01. e-g) HCT-116 cells were scored for replication stress-associated phenotypes following siRNA-mediated CIN− suppressor gene silencing: e) % Prometaphases exhibiting ≥3 γH2AX foci (mean±s.e.m, 3 experiments, n>100/experiment); f) % G1 cells with ≥3 53BP1 bodies (mean±s.e.m, 3 experiments, n>150/experiment); g) % Anaphases with UFBs (mean±s.e.m, 3 experiments, n=100/experiment). Statistical tests for e-g) were two-tailed t-tests, *p<0.05, **p<0.01. h,i) DNA fibre assays were performed following siRNA transfection as indicated. Representative fibre images for siRNA transfections as indicated as shown (h). i) Distribution of replication fork rates (n>200 forks in total per siRNA transfection from 2 experiments) with mean fork rates (n>70 forks/experiment, mean±s.d of 2 experiments) shown in the key (inset).
We next examined replication stress following CIN-suppressor gene silencing. Elevated prometaphase DNA damage was observed (Figure 3e), and this increased concomitantly with, rather than after, the rise in segregation error frequency (Supplementary Fig. 10d), supporting the hypothesis that the observed DNA damage reflects a cause, rather than a consequence, of segregation errors. Silencing each CIN-suppressor gene also resulted in elevated 53BP1 bodies in G1 cells (Figure 3f), which were unaffected by cytokinesis-inhibition, and therefore unlikely to reflect cytokinesis-induced chromosome damage7,15 (Supplementary Fig. 10e-g). Silencing PIGN and ZNF516 also significantly increased the frequency of UFBs (Figure 3g). Consistent with loss of CIN-suppressor genes contributing to replication stress in CIN+ cells, transient co-expression of PIGN, RKHD2 and ZNF516 resulted in a partial reduction in G1 53BP1 bodies in 3 CIN+ cell lines with 18q loss (Supplementary Fig. 11a-d). DNA fibre assays revealed a shift in distribution towards reduced replication fork speeds upon silencing each of the CIN-suppressor genes in HCT-116 cells, with reduced average fork rates after silencing PIGN and ZNF516 (Figure 3h,i and Supplementary Fig. 12a,b). Furthermore, we observed increased sister fork asymmetry after silencing each of the three genes (Supplementary Fig. 12c) consistent with impaired replication fork progression.
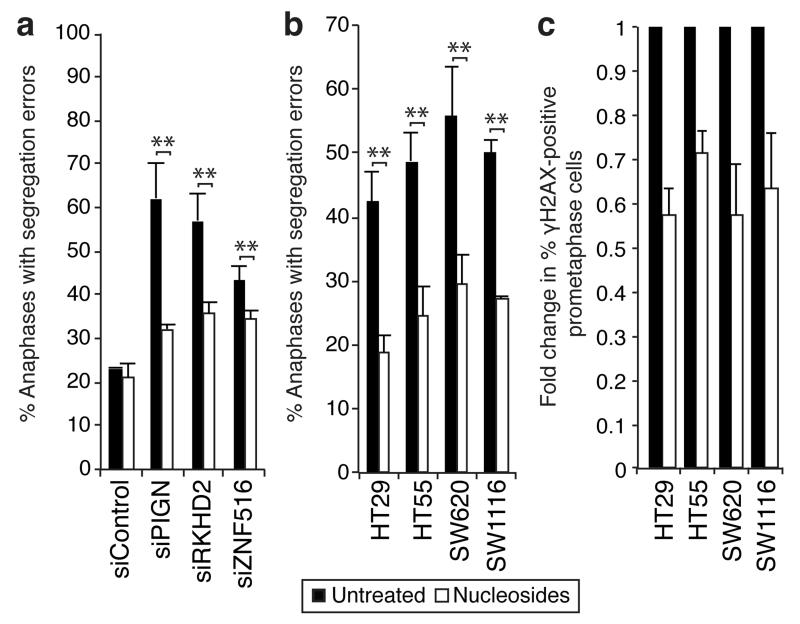
These data suggest that segregation errors resulting from CIN-suppressor gene silencing are driven by replication stress. To further test this hypothesis, HCT-116 cells transfected with siRNAs against CIN-suppressor genes were supplemented with nucleosides, previously shown to reduce replication-induced DNA damage5. Nucleosides significantly reduced the induction of segregation errors following silencing of PIGN (62% to 32%), RKHD2 (57% to 36%) and ZNF516 (43% to 34%) whilst control segregation errors were unaffected (Figure 4a). We then tested whether nucleoside supplementation could attenuate chromosome missegregation in CIN+ cell lines with 18q loss. Nucleoside supplementation significantly reduced segregation error frequency by 45-55% and attenuated prometaphase DNA damage by 28-43% in four CIN+ cell lines (Figure 4b,c, Supplementary Fig. 13a), indicating suppression of replication-induced DNA damage and subsequent chromosome missegregation. The extent of the nucleoside-mediated reduction in segregation errors implies that de novo generation of structural chromosome aberrations is responsible for a large proportion of chromosome missegregation events in CIN+ cells. In contrast, nucleoside supplementation did not affect segregation error frequency in four CIN− cell lines or 18q-normal CIN+ NCIH508 cells (Supplementary Fig. 13b,c). Nucleoside supplementation did not affect proliferation, cell cycle distribution, or cellular ATP levels (Supplementary Fig. 14a-f)5.
Figure 4. Nucleoside supplementation reduces segregation error frequency and prometaphase DNA damage.
a) % Anaphases with segregation errors ± 48 h nucleoside supplementation 24 h after siRNA transfection as indicated (mean±s.e.m, 3 experiments, Two-tailed t-test, **p<0.01). b) % Anaphases with segregation errors ± 48 h nucleoside supplementation in CIN+ cell lines as indicated (mean±s.e.m, 3 experiments, Two-tailed t-test, **p<0.01). c) Fold change in % prometaphases exhibiting ≥3 γH2AX foci ± 48 h nucleoside supplementation in CIN+ cell lines as indicated (n≥100 cells per condition per cell line, mean±s.e.m of 3 experiments). Un-normalised data is shown in Supplementary 13a.
In conclusion, our findings implicate replication stress as a major driver of chromosomal instability in CRC. In addition to impaired replication fork progression, CIN+ cells exhibit replication stress-associated DNA damage and structurally abnormal chromosomes that missegregate during mitosis, linking structural and numerical instability. Supplementing CIN+ cells with nucleosides reduced both DNA damage and segregation errors, supporting a direct role for replication stress in driving CIN. Given the complex nature of replication stress and CIN, it is likely that there are multiple genetic aberrations contributing to these phenotypes within an individual tumour. Here, we suggest a putative genetic mechanism that may contribute to CIN in CRC, through the recurrent loss of a region on chromosome 18q, encoding three newly identified suppressors of replication stress and chromosome missegregation. The loss of chromosome 18q in many solid tumour types suggests the potential contribution of this locus to CIN in tumours beyond CRC. Furthermore, reports of DNA replication stress across multiple tumour types suggest replication stress may be a common route to CIN and intratumour heterogeneity13,26. Efforts to target or restrain replication stress may therefore provide a rational approach to limit tumour diversity, genome evolution and adaptation.
Materials and Methods
Cell lines
Cells were maintained at 37 °C with 5% CO2 in DMEM with L-Glutamine (Gibco) or RPMI 1640 media (Gibco – NCIH747 cells), supplemented with 10% FBS, and 1/10000 units of Penicillin-Streptomycin (Sigma). Cell line CIN status was defined as described previously28.
Cell treatments
All compounds were from Sigma. Nocodazole: 50 or 100 ng/ml. Aphidicolin: 0.2 μM, 24 h. Monastrol wash-out: 100 μM, 1 or 16 h, washed 3x in fresh medium before 75 min recovery. Blebbistatin: 100 μM, 4 h. Nucleosides: Adenosine, cytidine, guanosine and uridine were freshly prepared for each experiment, filter sterilized, and used at 0.3 or 30 μM. H2O2: 350 μM, 4 h, before 16 h recovery.
Metaphase spreads and clonal FISH preparation
Metaphase spreads: Cells were harvested after 1 h treatment with 10 mM colcemid (Gibco), swelled with KCl (0.4%, 37°C, 7 min) before fixation in 3:1 methanol:acetic acid. Cells were dropped onto glass slides and aged for ~2 weeks. All-human centromere probe (Poseidon) was used according to the manufacturer’s instructions. Clonal FISH: 500 cells were expanded into colonies of 30-60 cells on glass slides before KCl treatment and fixation in 3:1 methanol:acetic acid. Slides were denatured (70°C in 2X sodium citrate (2XSSC)/75% formamide, 2 min, quenched in ice-cold 70% ethanol) and dehydrated through an ethanol series. Probes (CEP2 (D2Z1 Spectrum orange), CEP15 (D15Z1 Spectrum green, Abbott Molecular probes), were denatured (90°C, 6 min) and hybridized to slides (16 h, 37°C), then washed (50% formamide/2XSSC and 2XSSC at 42°C, followed by 4XSSC and PBS washes at room temperature). Slides were dehydrated and mounted in Vectashield hardset plus DAPI mounting medium (H-1500).
Immunofluorescence
Cells grown on coverslips were fixed in: 10% Triton X-100, 1 M PIPES, 0.5M EGTA, 1 M MgCl2, 4% formaldehyde. Mouse primary antibodies: α-tubulin (1:1000 Sigma T6074), NDC80 (1:800 Abcam Ab3613), centrin3 (1:1000 Abcam Ab54531) cyclin A1 (1:350 Santa Cruz sc-56299), phospho-histone H2A.X Ser 139 (1:500 Millipore 05-636), RPA (1:500 Neomarkers MS-691-P0) Rabbit primary antibodies: 53BP1 (1:500 sc-22760), β-tubulin (1:1000, Abcam), and human anti-centromere antibodies (ACA) (1:250; Antibodies Incorporated). Secondary antibodies (Molecular probes, used at 1:500): Goat anti-mouse conjugated to AlexaFluor 488 (A11017), goat anti-rabbit AF594 (A11012), goat anti-human AF647 (A21445). DNA was stained with DAPI (Roche) and coverslips mounted in Vectashield (Vector H-1000). Preparation and immunostaining of metaphase spreads was performed as described previously29. Most images were acquired using an Olympus DeltaVision RT microscope (Applied Precision, LLC) equipped with a Coolsnap HQ camera. 3D-image stacks were acquired in 0.2 μm steps, using an Olympus 100x or 60x 1.4NA UPlanSApo oil immersion objective. Deconvolution of image stacks and quantitative measurements was performed with SoftWorx Explorer (Applied Precision, LLC).
DNA Fibre Assays
Cells were plated (±siRNA transfection) 48 h prior to sequential pulse labelling with CldU and IdU (Sigma Aldrich, 30 min each) and harvesting for DNA fibre assays as described previously30.
RNA interference
siRNA transfections were performed at 40 nM by reverse transfection with Lipofectamine RNAiMax (Invitrogen). See Supplementary Table 6 for sequences used. The screen was performed in 12-well plates (0.5 × 105 HCT-116 cells/well on coverslips) using siGENOME SMARTpools (Dharmacon) with one control well per plate (Dharmacon control no.2). After 48 h cells were fixed and stained for α-tubulin. 30 anaphases/siRNA pool were scored manually for segregation errors.
DNA transfections and GFP-tagged construct mutagenesis
H2B-mRFP cells: Cells were transfected with pH2B-mRFP (gift from A. Straube) using Fugene 6.0 (Promega) and selected in 1 mg/ml G418 before flow-sorting for mRFP expression. Cells were maintained in 500 μg/ml G418. ShRNA cell line synthesis: HCT-116 cells were transfected with shRNA plasmids (Open Biosystems, see Supplementary Table 7) using Fugene 6.0 (Promega) and selected in 0.5 μM puromycin for 2-3 weeks. 3 colonies per shRNA were amplified, RNA extracted and silencing assessed by qPCR. Colonies with efficient silencing were selected. Cell lines were maintained in 0.5 μM puromycin. PIGN/RKHD2/ZNF516-GFP transfections: For siRNA rescue experiment in Supplementary Fig. 8f, 1 μg (PIGNins-GFP, RKHD2-GFP) or 0.5 μg (ZNF516ins-GFP) DNA was either co-transfected with siRNA using Lipofectamine RNAiMax for 48 h (RKHD2) or transfected 24 h following siRNA transfection using Lipofectamine 2000 (Invitrogen) for 24 h (PIGN and ZNF516). For co-transfection into CIN+ cell lines, 2 μg total DNA was transfected using Lipofectamine LTX Plus (Invitrogen), according to the manufacturer’s instructions. PIGN- and RKHD2-GFP were from Geneocopoeia, ZNF516-GFP was from Origene. siRNA insensitive mutagenesis: Quikchange XL site-directed mutagenesis kit (Stratagene) was used to create three base mismatches in siRNA target sequences for PIGN oligo 3 and ZNF516 oligo 3 in PIGN-GFP and ZNF516-GFP respectively. Mutagenesis was confirmed by sequencing.
RNA extraction and RT-qPCR
RNA was extracted using the Qiagen RNeasy Kit. Reverse transcription was performed using the First strand cDNA synthesis kit (Amersham). qPCR was performed in 96-well plates using pre-designed TaqMan® probe/primers on a ABI 7900HT system (Applied Biosystems). All reactions were performed in duplicate. The relative amount of mRNA was calculated using the comparative CT method after normalisation to GAPDH.
Western blotting
Cellular protein extracts were separated on NuPAGE 4-12% Bis-Tris gels (Invitrogen) then transferred to poly-vinylidene fluoride membrane (Millipore). Membranes were incubated with antibodies: MAD2 (mouse 1:1,000 BD-Biosciences 610678), RAD51 (mouse 1:1,000 Abcam ab213), GFP (mouse 1:1000, sc-9996), Turbo-GFP (rabbit 1:1000, Evrogen) in 5% milk in Tris-buffered saline, and detected with HRP-conjugated secondary antibody (1:10,000 Dako) and chemiluminescence (ECL, Amersham Biosciences). Loading was quantified with HRP-conjugated anti-β-actin (1:100,000, Sigma).
Time-lapse microscopy
H2B-mRFP-labelled cells were grown in 8-well chamber slides (LabTek). 14 μm z-stacks (7 images) were acquired using an Olympus 40x 1.3 NA UPlanSApo oil immersion objective every 3 min for 6 h using a DeltaVision microscope in a temperature and CO2-controlled chamber. Analysis was performed using Softworx Explorer.
Flow cytometry
Mitotic index: Cells were fixed in 70% ethanol and stained with mouse anti-MPM2 antibody (3:500 Millipore 05-368, overnight 4°C) then goat anti-mouse AF647 (Molecular probes A21463) and DAPI. 8-oxo-guanine: cells (±H2O2 treatment) were fixed in 4% formaldehyde, DNA denatured in 2 M HCl for 20 min, and stained with mouse anti-8-oxoguanine (1:200, Abcam ab62623). Data were processed using FlowJo software.
Immunohistochemistry
Paraffin-embedded cell pellets (±H2O2 treatment), were sectioned at 4 μm, de-waxed in xylene, then rehydrated through ethanol series to water. Endogenous peroxidase was blocked (1.6% H2O2, 10 min), followed by incubation with 10% normal horse serum (30 min). Sections were incubated with mouse anti-8-oxoguanine (1:1000, 1 h), washed 3x in PBS, then incubated with biotinylated horse anti-mouse antibodies (Vector labs 1:400, 35 min). After washing, peroxidase substrate (DAB) was added (2 min), slides were washed in water and counterstained with haematoxylin. Slides were washed, dehydrated and mounted in DPX-type mountant.
Proliferation assays
Plates were imaged using an IncuCyte Long-term in-situ Cell Imaging System, within an incubator. Phase contrast images were acquired every 2 h for 72 h and % cell monolayer confluence determined automatically. Outlying wells were excluded, and growth curves constructed. Growth rates were calculated by measuring the gradient of the linear growth phase.
ATP measurement
Following nucleoside supplementation, cells were treated with Cell Titer Glo reagent (Promega). Control measurements were taken from wells with media only±nucleosides. ATP levels were normalised to the biomass/well (In Vitro Toxicology assay kit Sulforhodamine B solution – Sigma).
SNP 6.0 array processing
Cell lines: Affymetrix SNP 6.0 data was obtained for 20 CIN+ and 9 CIN− cell lines (Wellcome Trust Sanger Institute). Integer copy numbers were estimated for each SNP probe using the PICNIC algorithm31. TCGA: Affymetrix SNP 6.0 data was downloaded for 365 CRC samples and logR ratios and allelic differences were estimated by the Affymetrix Genotyping Console™. Samples that failed the Affymetrix quality control parameters were excluded. All tumours with <60% tumour nuclei (based on pathological estimates of adjacent sections) were excluded. To estimate copy number, the GAP algorithm was used32. Ploidy was estimated by calculating the weighted median copy number across all copy number segments, with weights equal to the segment length. Copy number segments of loss and gain were defined relative to the ploidy status of each sample by subtracting the ploidy estimate from the estimated copy number of the segment.
Karyotypic Complexity Scores
Structural Complexity Score (SCS): defined as the sum of all structurally aberrant regions. Regions of intra-chromosomal gain and loss were defined relative to the modal copy number of the chromosome, and each region counted as one structural aberration. To avoid over-estimation, aberrant regions <1 MB were excluded. Numerical complexity score (NCS): the sum of all whole chromosome gains and losses (chromosomes with >75% of SNP copy number values higher or lower than the ploidy of the sample were counted as whole chromosome gains or losses respectively). Multiple copy number events affecting the same chromosome were scored separately (e.g. −2 copies = 2 chromosome losses). NCS and SCS scores were divided by 1.5 for triploid cell lines, and by 2 for tetraploid cell lines, to account for the increased likelihood of karyotypic abnormalities in polyploid genomes. Weighted genome instability index: As FACS-based DNA index measures were not available for the TCGA tumours, and information about MSI status was unavailable for a sufficient number of tumours, an alternative means of classification was required. The genome instability index (GII)33 is the percentage of SNPs across the genome present at an aberrant copy number, relative to the baseline ploidy of the sample. We adapted the GII in order to account for variation in chromosome size, so that large chromosomes do not have a greater effect on the score than small chromosomes: % aberrant SNPs for each chromosome was calculated separately and mean % aberration then calculated across all 22 chromosomes. To define a threshold for CIN− versus CIN+, the weighted GII (wGII) was calculated for the cell lines. A threshold of 0.2 accurately distinguished CIN+ from CIN−, as previously defined28. The same threshold was then applied to the TCGA cohort of tumours.
Sequencing data (TCGA)
Preprocessed level 2 somatic mutation data was obtained from The Cancer Genome Atlas Research Network19 for 101 colorectal tumours for which SNP 6.0 copy number data was also available. All genes with ≤5 somatic mutations were excluded from the analysis. Colorectal tumours were classified as CIN+ using wGII > 0.2. Somatic mutation data for the 29 colorectal cancer cell lines were obtained from the COSMIC database (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/). The association of CIN status with somatic mutation status of TP53, APC, SMAD4 and KRAS was tested with one-sided Fisher’s exact tests and the p-values were adjusted for multiple testing with the Benjamini & Hochberg method34
Defining somatic copy number losses in CIN+ versus CIN− tumours and cell lines
Aneuploid tumours: BAC array-CGH data was obtained for 26 aneuploid tumour samples, and segmented by circular binary segmention (R package DNAcopy). The GISTIC algorithm35 was used to identify regions of consistent gain and loss, with thresholds of 0.1/-0.1 for gain/loss respectively, and a q-value threshold of 0.25. Aneuploidy was defined by flow cytometry (DNA index >1.2). Cell lines: Minimum consistent regions of genomic alteration across all cell lines were assessed for DNA copy number. Each region in each cell line was normalized to the ploidy baseline of the cell line to give Xnorm and was then defined as either lost (copy number < ploidy baseline), or not lost (copy number ≥ ploidy baseline) and set to 0. Each region was assessed for gain in the same manner. To test for statistical significance between CIN+ and CIN− cell lines a d-score for each lost region was computed by calculating the mean normalised copy number Xnorm across CIN+ (mean(Xnorm,C)) and CIN− (mean(Xnorm,M)) cell lines, thereby accounting for both amplitude and frequency of genomic aberrations. A Significance Analysis of Microarrays (SAM – R package siggenes) was then performed with a modified two-sample t-statistic:
The parameter s(i) defines the region-specific standard deviation36. In contrast to a standard two-sample t-statistic, SAM includes an additional parameter s0, which decreases the influence of high sample variance. This was empirically set to 0.5, resulting in a balanced weighting of frequency and amplitude. To detect significant regions, we randomly permuted (N = 10000) SNP probes for each sample separately. To save computation time, we randomly drew copy numbers for each sample, setting the probability for a given copy number to the percentage of SNP probes showing this copy number level across the genome. For each tested region, p-values were estimated by counting the percentage of permutation d-scores greater or equal than the observed d-score. To adjust for multiple testing, q-values were estimated with the R-package qvalue and genes with q<0.25 were called significant. To ensure selection of genes consistently altered across CIN+ cell lines, genomic changes not seen in ≥50% of cell lines were excluded from further analysis. Genes were then mapped to regions using the R package biomaRt37. All genes present at ≤1 copies in ≥30% of CIN+ cell lines, and no more than 1 CIN− cell line, were selected for functional investigation.
Carcinoma-in-adenoma samples
20 archival FFPE tumours showing adjacent but discrete colorectal carcinoma and adenoma were identified. Samples and records were used in accordance with UK research ethics approval (MREC06/Q1702/99). H&E slides of the samples were reviewed, regions marked as normal (if present)/adenoma/carcinoma. and used as a guide to take tissue from each region from unstained slides by needle microdissection. DNA was extracted with standard proteinase K digestion, and purified (DNeasy kit - Qiagen). Ploidy analysis was performed using automated image-based cytometry (Fairfield Imaging, Nottingham, UK) as previously described38,39. LOH analysis: SNP arrays: The Illumina Sentrix Beadarray human linkage mapping panel version IVb was used according to the Goldengate genotyping assay instructions (Illumina, San Diego, USA). DNA was amplified, fragmented and hybridized to the Beadchip, followed by single-base extension. Beadchips were stained, dried and scanned using a Beadarray reader (Illumina, San Diego, USA). Image data were visualised using Genomestudio (Illumina, San Diego, USA). All samples had call rates above 0.97. Adenomas and carcinomas were marked as having LOH or no LOH in 18q according to the absence or presence of heterozygous alleles respectively. LOH analysis using microsatellites: Standard PCR conditions were used with the forward primer fluorescently labelled with HEX or FAM. At each marker, LOH was considered present when a peak area in the adenoma or the carcinoma was reduced to 50% of the other allele, relative to the normal paired DNA. Up to 5 microsatellites in 18q21 (D18S46, D18S1110, D18S35, D18S69 and D18S1407) were analysed. All PCRs were performed twice and all samples analysed with SNP arrays had concordant results.
Gene expression analysis
TCGA: Agilent 244K custom gene expression (G4502A-07-3) data was downloaded for 154 CRC samples and was print-tip normalised with the R-package marray. Duplicated probes were averaged. Cell lines: Affymetrix HT-HGU133A microarray data for 20 CIN+ and 9 CIN− cell lines was obtained from the Wellcome Trust Sanger Institute. The data was RMA normalized with the R-package affy. Spearman’s rank correlation coefficient was used for the correlation of copy number and expression data.
Supplementary Material
Acknowledgements
We thank A. Straube for reagents. CS is a senior Medical Research Council clinical research fellow and is funded by Cancer Research UK (CRUK), Medical Research Council, EU FP7 (projects PREDICT and RESPONSIFY) Prostate Cancer Foundation and the Breast Cancer Research Foundation (BCRF). IPT is supported by the Oxford Biomedical Research Centre and CRUK. JB is funded by the Danish Cancer Society, the Lundbeck Foundation, and the European Commission (FP7 projects: DDResponse, Biomedreg and Infla-Care). TH is funded by the Swedish Cancer Society, the Swedish Research Council and the Torsten and Ragnar Söderberg Foundation.
Footnotes
The authors declare no competing interests.
References
- 1.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. doi:10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding L, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. doi:10.1038/nature10738 nature10738 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. doi:10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 4.McGranahan N, Burrell RA, Endesfelder D, Novelli MR, Swanton C. Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep. 2012;13:528–538. doi: 10.1038/embor.2012.61. doi:10.1038/embor.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bester AC, et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. doi:S0092-8674(11)00378-3 [pii] 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson SL, Compton DA. Chromosomes and cancer cells. Chromosome Res. 2011;19:433–444. doi: 10.1007/s10577-010-9179-y. doi:10.1007/s10577-010-9179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–1898. doi: 10.1126/science.1210214. doi:10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- 8.Crasta K, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. doi:10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pampalona J, Soler D, Genesca A, Tusell L. Whole chromosome loss is promoted by telomere dysfunction in primary cells. Genes Chromosomes Cancer. 2010;49:368–378. doi: 10.1002/gcc.20749. doi:10.1002/gcc.20749. [DOI] [PubMed] [Google Scholar]
- 10.Ichijima Y, et al. DNA lesions induced by replication stress trigger mitotic aberration and tetraploidy development. PLoS One. 2010;5:e8821. doi: 10.1371/journal.pone.0008821. doi:10.1371/journal.pone.0008821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. doi:jcb.200712029 [pii] 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gisselsson D. Classification of chromosome segregation errors in cancer. Chromosoma. 2008;117:511–519. doi: 10.1007/s00412-008-0169-1. doi:10.1007/s00412-008-0169-1. [DOI] [PubMed] [Google Scholar]
- 13.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. doi:nature03482 [pii] 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 14.Tort F, et al. Retinoblastoma pathway defects show differential ability to activate the constitutive DNA damage response in human tumorigenesis. Cancer Res. 2006;66:10258–10263. doi: 10.1158/0008-5472.CAN-06-2178. doi:66/21/10258 [pii] 10.1158/0008-5472.CAN-06-2178. [DOI] [PubMed] [Google Scholar]
- 15.Lukas C, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13:243–253. doi: 10.1038/ncb2201. doi:ncb2201 [pii] 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- 16.Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11:753–760. doi: 10.1038/ncb1882. doi:ncb1882 [pii] 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- 17.Kawabata T, et al. Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol Cell. 2011;41:543–553. doi: 10.1016/j.molcel.2011.02.006. doi:10.1016/j.molcel.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrigan JA, et al. Replication stress induces 53BP1-containing OPT domains in G1 cells. J Cell Biol. 2011;193:97–108. doi: 10.1083/jcb.201011083. doi:jcb.201011083 [pii] 10.1083/jcb.201011083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Research Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. doi:10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bunz F, et al. Targeted inactivation of p53 in human cells does not result in aneuploidy. Cancer Res. 2002;62:1129–1133. [PubMed] [Google Scholar]
- 21.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. doi:319/5868/1352 [pii] 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 22.Rowan A, et al. Refining molecular analysis in the pathways of colorectal carcinogenesis. Clin Gastroenterol Hepatol. 2005;3:1115–1123. doi: 10.1016/s1542-3565(05)00618-x. doi:S1542-3565(05)00618-X [pii] [DOI] [PubMed] [Google Scholar]
- 23.Pasello G, et al. DNA copy number alterations correlate with survival of esophageal adenocarcinoma patients. Mod Pathol. 2009;22:58–65. doi: 10.1038/modpathol.2008.150. doi:10.1038/modpathol.2008.150. [DOI] [PubMed] [Google Scholar]
- 24.Yatsuoka T, et al. Association of poor prognosis with loss of 12q, 17p, and 18q, and concordant loss of 6q/17p and 12q/18q in human pancreatic ductal adenocarcinoma. Am J Gastroenterol. 2000;95:2080–2085. doi: 10.1111/j.1572-0241.2000.02171.x. doi:10.1111/j.1572-0241.2000.02171.x. [DOI] [PubMed] [Google Scholar]
- 25.Sigoillot FD, et al. A bioinformatics method identifies prominent off-targeted transcripts in RNAi screens. Nat Methods. 2012;9:363–366. doi: 10.1038/nmeth.1898. doi:10.1038/nmeth.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dereli-Oz A, Versini G, Halazonetis TD. Studies of genomic copy number changes in human cancers reveal signatures of DNA replication stress. Mol Oncol. 2011;5:308–314. doi: 10.1016/j.molonc.2011.05.002. doi:S1574-7891(11)00051-2 [pii] 10.1016/j.molonc.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan KL, Hickson ID. On the origins of ultra-fine anaphase bridges. Cell Cycle. 2009;8:3065–3066. doi: 10.4161/cc.8.19.9513. doi:9513 [pii] [DOI] [PubMed] [Google Scholar]
Additional references
- 28.Lee AJ, et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 2011;71:1858–1870. doi: 10.1158/0008-5472.CAN-10-3604. doi:71/5/1858 [pii] 10.1158/0008-5472.CAN-10-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cesare AJ, et al. Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat Struct Mol Biol. 2009;16:1244–1251. doi: 10.1038/nsmb.1725. doi:10.1038/nsmb.1725. [DOI] [PubMed] [Google Scholar]
- 30.Groth P, et al. Methylated DNA causes a physical block to replication forks independently of damage signalling, O(6)-methylguanine or DNA single-strand breaks and results in DNA damage. J Mol Biol. 2010;402:70–82. doi: 10.1016/j.jmb.2010.07.010. doi:S0022-2836(10)00763-1 [pii] 10.1016/j.jmb.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 31.Greenman CD, et al. PICNIC: an algorithm to predict absolute allelic copy number variation with microarray cancer data. Biostatistics. 2010;11:164–175. doi: 10.1093/biostatistics/kxp045. doi:kxp045 [pii] 10.1093/biostatistics/kxp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Popova T, et al. Genome Alteration Print (GAP): a tool to visualize and mine complex cancer genomic profiles obtained by SNP arrays. Genome Biol. 2009;10:R128. doi: 10.1186/gb-2009-10-11-r128. doi:gb-2009-10-11-r128 [pii] 10.1186/gb-2009-10-11-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chin SF, et al. High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol. 2007;8:R215. doi: 10.1186/gb-2007-8-10-r215. doi:gb-2007-8-10-r215 [pii] 10.1186/gb-2007-8-10-r215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 35.Beroukhim R, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. doi:0710052104 [pii] 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Storey JD, Siegmund D. Approximate p-values for local sequence alignments: numerical studies. J Comput Biol. 2001;8:549–556. doi: 10.1089/106652701753216530. doi:10.1089/106652701753216530. [DOI] [PubMed] [Google Scholar]
- 37.Durinck S, et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. doi:21/16/3439 [pii] 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 38.Thirlwell C, et al. Clonality assessment and clonal ordering of individual neoplastic crypts shows polyclonality of colorectal adenomas. Gastroenterology. 2010;138:1441–1454. 1454, e1441–1447. doi: 10.1053/j.gastro.2010.01.033. doi:S0016-5085(10)00097-1 [pii] 10.1053/j.gastro.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 39.Leedham SJ, et al. Clonality, founder mutations, and field cancerization in human ulcerative colitis-associated neoplasia. Gastroenterology. 2009;136:542–550. e546. doi: 10.1053/j.gastro.2008.10.086. doi:S0016-5085(08)02039-8 [pii] 10.1053/j.gastro.2008.10.086. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.