Abstract
Deciphering whether actionable driver mutations are found in all or a subset of tumor cells will likely be required to improve drug development and precision medicine strategies. We analyzed nine cancer types to determine the subclonal frequencies of driver events, to time mutational processes during cancer evolution, and to identify drivers of subclonal expansions. Although mutations in known driver genes typically occurred early in cancer evolution, we also identified later subclonal “actionable” mutations, including BRAF(V600E), IDH1(R132H), PIK3CA(E545K), EGFR(L858R), and KRAS(G12D), which may compromise the efficacy of targeted therapy approaches. More than 20% of IDH1 mutations in glioblastomas, and 15% of mutations in genes in the PI3K(phosphatidylinositol 3-kinase)–AKT–mTOR (mammalian target of rapamycin) signaling axis across all tumor types were subclonal. Mutations in the RAS–MEK (mitogen-activated protein kinase kinase) signaling axis were less likely to be subclonal than mutations in genes associated with PI3K-AKT-mTORsignaling. Analysis of late mutations revealed a link between APOBEC-mediated mutagenesis and the acquisition of subclonal driver mutations and uncovered putative cancer genes involved in subclonal expansions, including CTNNA2 and ATXN1. Our results provide a pan-cancer census of driver events within the context of intratumor heterogeneity and reveal patterns of tumor evolution across cancers. The frequent presence of subclonal driver mutations suggests the need to stratify targeted therapy response according to the proportion of tumor cells in which the driver is identified.
INTRODUCTION
The advent of large-scale genome characterization efforts has led to an explosion in the availability of next-generation sequencing data, with more than 5000 tumors sequenced by The Cancer Genome Atlas (TCGA) alone. These efforts have contributed to the identification of hundreds of cancer genes (1-3) and also shed light on the many mutational processes molding the cancer genome (4). For instance, across 30 different cancer classes, more than 20 distinct mutational processes have been identified, including those derived from exogenous sources, exemplified by tobacco carcinogens and ultraviolet (UV) light, as well as those resulting from endogenous mutational processes, such as mismatch repair deficiency or up-regulation of APOBEC cytosine deaminases (4, 5).
Accumulating evidence suggests that tumors often evolve through a process of branched evolution, involving genetically distinct subclones (6, 7). Therefore, drug development and precision medicine strategies will likely require not only an understanding of cancer genes and mutational processes but also an appreciation of the clonal status of driver events and the timing of mutational processes. The presence of subclonal mutations may reduce the clinical benefit of cancer therapies. For instance, in colorectal cancer, subclonal RAS mutations have been shown to precipitate resistance to cetuximab (8), and in glioblastomas, individual driver events can be present in distinct populations of cancer cells or subclones (9, 10). Furthermore, the use of targeted therapy against a subclonal driver present in a subset of cells within a tumor may lead to stimulation of wild-type subclones lacking the targeted mutation (11).
Targeting clonally dominant truncal somatic events or adopting multiple targeted therapies in a combination approach may therefore be necessary for optimal tumor control (12). However, although the clonal status of driver mutations has received attention in certain cancers (11, 13-17), a broad understanding of the heterogeneity of driver genes, deciphering the clonal and subclonal frequencies, and the timing of mutational processes involved in tumor evolution is lacking.
Here, we use TCGA data from nine major cancer types—urothelial bladder cancer (BLCA), breast cancer (BRCA), colon adenocarcinoma (COAD), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), clear cell kidney carcinoma (KIRC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and cutaneous melanoma (SKCM). Our results provide a pan-cancer census of driver events within the context of intratumor heterogeneity, determining the extent to which “actionable” mutations are subclonal. We also reveal how mutational processes defining cancer evolution vary over time during the course of a tumor’s development, in some cases fueling the acquisition of subclonal driver mutations, and uncover drivers of subclonal expansions.
RESULTS
A pan-cancer data set allows temporal dissection of mutations
Mutations and copy number variants from TCGA were processed and filtered to obtain a pan-cancer data set representing 2694 tumors from nine major cancer types with both copy number and mutation data. A total of 516,672 somatic mutations were used for downstream analysis, consisting of 326,918 missense, 145,009 silent, 24,236 nonsense, 9867 RNA[5′ untranslated region (UTR) or 3′UTR], 9828 splice-site, 341 non-stop, and 473 translation start-site mutations.
Clonal and subclonal mutations can be defined within single tumor samples
To explore the relative timing of mutations, we first estimated the mutation copy number (the number of alleles harboring the mutation) and cancer cell fraction (the fraction of tumor cells with the mutation) of each single nucleotide variant. Both measures were determined using single-nucleotide polymorphism (SNP) arrays and exome sequencing data, integrating the variant allele frequencies with the local copy number and normal cell contamination estimates (Fig. 1). Mutations were classified as clonal (present in all tumor cells sequenced) if the upper band of the 95% cancer cell fraction confidence interval was ≥ 1, and subclonal otherwise. We adopted this stringent classification to ensure that the number of subclonal mutations was not overestimated.
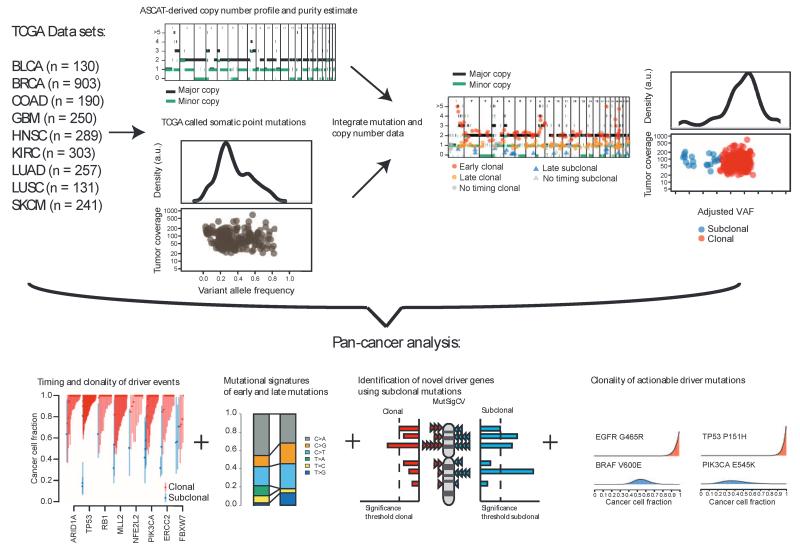
Fig. 1.
Temporal dissection of mutations and mutational processes in TCGA samples. Integration of copy number, purity estimates, and VAF of each somatic mutation permits calculation of the cancer cell fraction, describing the fraction of cancer cells with an alteration. The mutation copy number can also be estimated, allowing further timing of mutations in the case of a genome-doubling or amplification event. These data can reveal the clonality of driver events and shifts in mutational spectra and mutational signatures over time, as well as permitting identification of driver genes using subclonal mutations. a.u., arbitrary unit.
Mutations were then classified as “early” or “late” based on the cancer cell fraction and the mutation copy number. We reasoned that clonal mutations represent relatively early events in tumor evolution, occurring before or at the time of the most recent clonal expansion, whereas subclonal mutations represent later events. In the case of genome-doubling or amplification events, the timing of mutations was further refined; a mutation occurring before doubling would be expected to be present at multiple copies, whereas a mutation occurring after a doubling will likely only be present at one copy. Thus, mutations were defined as early if they were clonal and did not occur after amplification or a genome-doubling event. Conversely, we defined late mutations as those that were subclonal or occurred after genome-doubling or amplification events.
Our analysis, by necessity, was restricted to single tumor samples and was not powered to detect regional variation in driver events or subclonal copy number events (18-21). Our power to detect low-frequency subclonal events depended on sample sequencing depth (fig. S1A) and purity (fig. S1, B and C). At a sequencing depth of 70, the probability of detecting mutations at a variant allele frequency (VAF) of 0.1 is estimated to be 0.98; however, the probability is reduced to 0.69 when attempting to detect a variant with a VAF of 0.05 (22). Thus, in our analysis, we may have underestimated the number of low-frequency subclonal mutations.
Established driver mutations typically occur early in tumor evolution
To explore the clonal status of mutations in established cancer genes, we first identified all nonsilent mutations that occurred in known cancer driver genes based on the recent pan-cancer analysis by Lawrence et al. (1) combined with the manually curated COSMIC gene census (23) (table S1).
In general, we observed a clear tendency for mutations in driver genes to be clonal compared to mutations in noncancer genes (Fig. 2A). In every cancer type, with the exception of KIRC, mutations in driver genes (considered in aggregate) were enriched to a statistically significant degree for clonal mutations, compared to mutations in nondriver genes (Fig. 2A). In KIRC, we found that VHL was the only cancer gene that had a significantly higher proportion of clonal mutations than the background rate representing all nonsilent mutations, consistent with multiregion sequencing results in KIRC (19) and supporting our approach to distinguish clonal from subclonal somatic events (P= 0.0147; fig. S2). Our results also remained robust to the choice of cutoff to define subclonal mutations (Supplementary Materials and Methods and fig. S3).
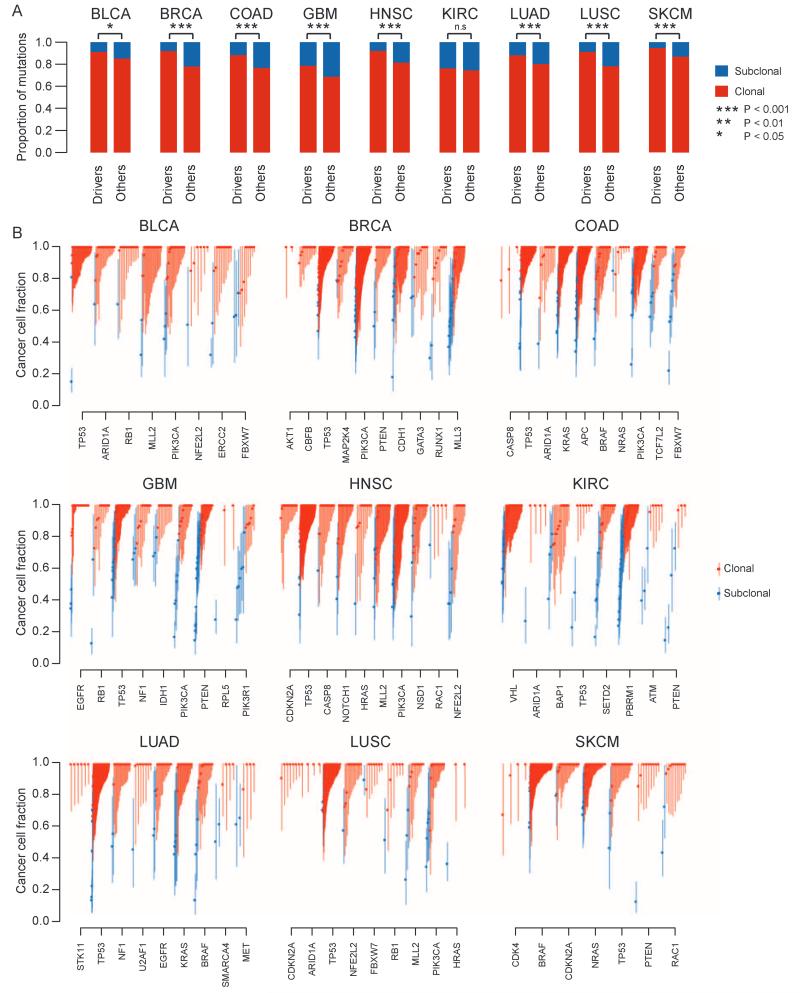
Fig. 2.
Clonal and subclonal mutations in nine cancer types. (A) The proportion of aggregated driver mutations versus other mutations that are clonal/subclonal is indicated for each cancer type. Red represents clonal mutations, and blue represents subclonal mutations. Notably, there is a higher proportion of clonal driver mutations compared to other clonal mutations. Significance from Fisher’s exact test is indicated. Exact P values are as follows: BLCA, P= 0.0292; BRCA, P= 8.19× 10−21; COAD, P= 1.45× 10−9; GBM, P= 0.000791; HNSC, P= 2.58× 10−8; KIRC, P= 0.619; LUAD, P= 0.00311; LUSC, P= 1.89× 10−5; SKCM, P= 2.71 × 10−5. (B) The cancer cell fraction of mutations in driver genes within each cancer type is depicted. Each symbol represents a somatic mutation in an individual tumor. On the basis of the probability distributions of the cancer cell fraction, mutations were determined to be either clonal (red circles, upper bound of confidence interval ≥1) or subclonal (blue circles, upper band of confidence interval <1). Error bars represent the 95% confidence interval.
Within other cancer types, we found that mutations in specific cancer genes showed a tendency to be clonal (Fig. 2B). For instance, in BRCA, mutations in CBFB were exclusively clonal, as were mutations in CDKN2A in LUSC and HNSC, and a similar trend was observed for mutations in ARID1A in BLCA. In the pan-cancer data set, somatic mutations in TP53 were significantly more often clonal than the background rate (fig. S2; P < 0.0001). We also observed a significant enrichment of TP53 mutations in genome-doubled tumors (fig. S4; P < 0.001, Fisher’s exact test), and almost invariably (>90% cases), these mutations occurred before doubling, consistent with TP53 playing an important role in tolerance of genome doubling (24).
Together, these results suggest that many known cancer genes have a tendency to be clonal within single samples and corroborate the “driver” capabilities of these genes. However, these findings may also reflect the fact that current methods for detecting cancer genes are likely biased to detect clonal drivers that occur at high frequency within single cancer samples.
Notably, we also identified nonsilent subclonal mutations in known cancer genes in every cancer type. In KIRC, more than 50% of nonsilent mutations in PTEN were found to be subclonal (significantly more than the background rate, P= 0.01; fig. S2), consistent with observations from a smaller cohort of KIRC samples subject to multiregion sequencing (19), and supporting the role PTEN may play later in tumor evolution in this cancer type. Mutations in PIK3R1 and MLL3 were frequently subclonal across cancer types, such that a higher proportion of subclonal mutations were observed in these genes compared to the background rate (fig. S2).
In more than 30 cases, we identified multiple subclonal mutations occurring in the same cancer gene or in separate cancer genes whose function might be expected to be redundant (fig. S5). For example, in KIRC-B0-5399, we identified two distinct mutations in SETD2, one occurring in 40% of cancer cells and the other in 19%. In GBM-28-2513, we identified two subclonal mutations in genes involved in the PI3K (phosphatidylinositol 3-kinase) signaling axis, with one mutation in PIK3R1 present in 54% of cancer cells and a mutation in PTEN present in only 36% of cancer cells. These observations are consistent with evidence for parallel tumor evolution, whereby the same genetic pathway is disrupted independently in distinct tumor subpopulations within one tumor (19). In further support of parallel evolution, in general, we found that when multiple nonsilent mutations were identified in the same cancer gene within one tumor sample, these mutations exhibited a significantly lower cancer cell fraction compared to mutations in cancer genes occurring only once in a cancer sample (fig. S5C; P< 0.001, Wilcoxon rank sum test).
Mutational processes vary dynamically during tumor evolution
Temporal analysis also allowed us to explore whether mutational processes vary during tumor evolution and whether the dynamics of genome instability processes can be elucidated. In the nine cancer types, we identified 10 robust mutational signatures, most of which were found to change in prevalence during the course of a tumor’s development (Fig. 3).
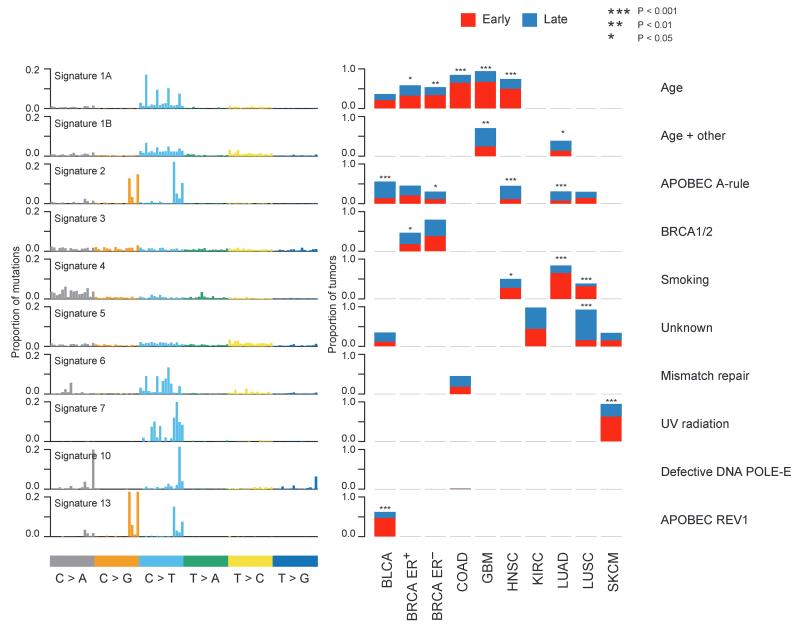
Fig. 3.
Temporal dissection of mutational signatures. For each mutational signature identified in at least one cancer type, the proportion of patients with either a higher fraction of early (red) or late (blue) mutations corresponding to a signature is indicated. A sample is classified as harboring a mutational signature if more than 100 mutations or more than 25%of mutations in that sample correspond to the signature. The factors responsible for the mutational processes are indicated in the right panel. The names of the signatures correspond to those used by Alexandrov et al. (4).
Signature 1A, reflecting a preponderance of C>T transitions at CpG sites, was identified in five cancer types (BLCA, BRCA, COAD, GBM, and HNSC). This signature has been linked to spontaneous deamination of methylated cytosines and has been found to correlate with patient age at diagnosis (4). In four cancer types, signature 1A was significantly more prevalent in early compared to late mutations [BRCA estrogen receptor (ER)–positive, P= 0.0359; BRCA ER-negative, P= 0.000629; COAD, P= 7.59 × 10−15; GBM, P= 3.55 × 10−10; and HNSC, P= 2.26 × 10−9]. These data suggest that a large proportion of early mutations represent non–cancer-specific mutational processes.
Consistent with additional cancer-specific mutational processes increasing in prevalence later in tumor evolution, signature 2, which has been linked with up-regulation of APOBEC cytosine deaminases, was found to increase over time in most of the tumor types in which it was detected. A significant increase in signature 2 was observed in HNSC (P= 5.57 × 10−7) and BLCA (P= 4.39 × 10−6), and a similar pattern was observed in LUAD (P= 3.33 × 10−6), as previously reported (20). We also found a tendency for signature 2 to increase in frequency in later mutations in ER-negative BRCA (P= 0.0126), but not in ER-positive breast cancers (P= 0.597), highlighting the differences between these two subtypes of breast cancer. These data suggest that the APOBEC mutational process may foster subclonal expansions across several tumor types. Notably, signature 2 was detected in both early and late mutations, albeit to a lesser degree in early mutations, suggesting that APOBEC activity is more than a transient event and does not simply represent a historical relic within the tumor genome, active at only one point in time during the disease course (25).
In contrast, signature 13, also linked to APOBEC cytosine deaminases, was found to be significantly more prevalent in early compared to late mutations in BLCA (P= 6.67 × 10−7). Signature 13 has been postulated to be associated with APOBEC coupled with excess activity of DNA repair protein REV1. These data suggest that signatures 2 and 13 are distinct mutational processes that can be separated temporally in BLCA (5).
In terms of exogenous mutational processes, we found that signature 7, related to mutations caused by exposure to UV, was elevated in early compared to late mutations in SKCM (P= 5.67 × 10−11). Signature 4, associated with smoking-induced mutations, was significantly more prevalent in early compared to late mutations in LUAD (P= 4.56 × 10−17) and LUSC (P= 8.1 × 10−7) (20), and a similar pattern was observed in HNSC (P= 0.0338). These data likely reflect the fact that despite the mutagenic effects of tobacco smoke and UV light, additional mutational processes occur during tumor development. In SKCM, it is also worth noting that most samples were derived from metastatic sites; thus, many of the later mutations will likely have been acquired when tumor cells were no longer exposed to UV light.
Mutational processes fuel the acquisition of somatic events in cancer genes
Next, we explored whether mutational processes could explain the temporal acquisition of nonsilent mutations in known cancer genes (fig. S6 and table S2). In both LUAD and LUSC, we found that more than 30% of clonal nonsilent mutations in cancer genes were C>A transversions, which can be attributed to tobacco smoke; for example, more than 40% of clonal mutations in TP53 were C>A transversions. In BLCA, by contrast, more than 40% of clonal mutations in cancer genes were found to occur in an APOBEC context, consistent with APOBEC-mediated mutagenesis shaping the early evolutionary trajectory of many BLCA tumors.
Focusing on subclonal mutations in known cancer driver genes, APOBEC represented a dominant mutational process in five cancer types: BLCA, BRCA, HNSC, LUAD, and LUSC. In LUAD, LUSC, and HNSC samples showing evidence of APOBEC-mediated mutagenesis, on average only 21% (range, 19 to 24%) of clonal mutations in cancer driver genes occurred in an APOBEC context, but more than 45% (range, 35 to 59%) of subclonal mutations in cancer genes could be explained by APOBEC-mediated mutagenesis. Strikingly, in these cancers, more than 90% of subclonal mutations in PIK3CA occurred in an APOBEC context, and we also identified subclonal APOBEC mutations in multiple other cancer driver genes, including PTEN, EGFR, and TP53. Similarly, in APOBEC-associated BLCA, more than 45% of subclonal mutations in driver genes occurred in an APOBEC context. These data suggest that APOBEC cytosine deaminases may play a key role in driving subclonal diversification in these cancer types.
Together, these data highlight the extent to which endogenous and exogenous mutational processes can fuel the acquisition of somatic events in cancer genes. Moreover, the dynamics of specific mutational processes may alter the subclonal architecture of a tumor by providing the mutational fuel upon which selection can act.
Subclonal analysis identifies drivers of subclonal expansions
Given the tendency for mutations in many known cancer genes to be clonal and the observation that many clonal mutations likely occur before tumorigenesis, we reasoned that it might be possible to identify drivers of subclonal expansions by focusing exclusively on late or subclonal mutations. We therefore applied the MutSigCV algorithm (1), a statistical analysis that takes into account nucleotide context, gene expression, replication timing, and the somatic background mutation rate to identify cancer genes, to our temporally and clonally dissected mutations.
In total, we identified 32 late putative driver genes across the nine cancer types (q < 0.05). Of these, 12 would have been missed in at least one cancer type without temporally dissecting mutations (table S3). Temporal dissection was required to uncover the cancer gene PIK3CA in HNSC, suggesting that mutations in this gene may often lead to, or be permissive for, subclonal expansions in this cancer type.
A number of putative cancer genes that we identified can be linked to tumor development, maintenance, and progression. A notable example is cell adhesion gene CTNNA2, catenin (cadherin-associated protein) a2, identified in LUAD. CTNNA2 has previously been implicated in laryngeal cancer as a tumor suppressor, and its inactivation in HNSC cells is associated with migration and invasion advantages, consistent with it playing a role at later stages of tumor development (26). In LUAD, we also identified mutations in NRXN3 as a putative subclonal driver event. A polymorphic site of this gene (rs10146997) has been associated with higher risk of breast cancer development (27), and low expression of NRXN3 is associated with poorer survival in lung cancer (fig. S7).
Another putative cancer gene, identified in COAD, is ATXN1. The ATXN1 protein family plays an important role in transcriptional control of extracellular matrix remodeling, and mutations in ATXN1 have been putatively linked to cancer metastasis, consistent with its occurrence as a later event in COAD (28). In LUSC, we identified LRP1B, which encodes a member of the low-density lipoprotein receptor gene family. It has been suggested that LRP1B acts as a tumor suppressor gene (29), and in KIRC, its depletion leads to increased anchorage-independent growth, cell migration, and invasion in vitro (30).
By temporally separating mutations, we also identified driver genes that may be missed by the inclusion of late or subclonal mutations. For example, when focusing exclusively on clonal mutations, we identified BRAF in COAD, consistent with BRAF playing an important role early in tumor evolution and the clonal nature of this event in published studies (31). Likewise, in LUSC, BRAF, KRAS, and EGFR were only identified as drivers by focusing on early mutations.
Mutations in genes with therapeutic relevance can be subclonal within single tumor samples
We next considered all nonsilent mutations in genes and gene pathways for which therapeutics have been developed or are in development (table S4). For instance, the presence of mutations that activate the PI3K–AKT–mTOR (mammalian target of rapamycin) pathway and contribute to carcinogenesis has engendered much interest in inhibitors of this signaling axis (32).
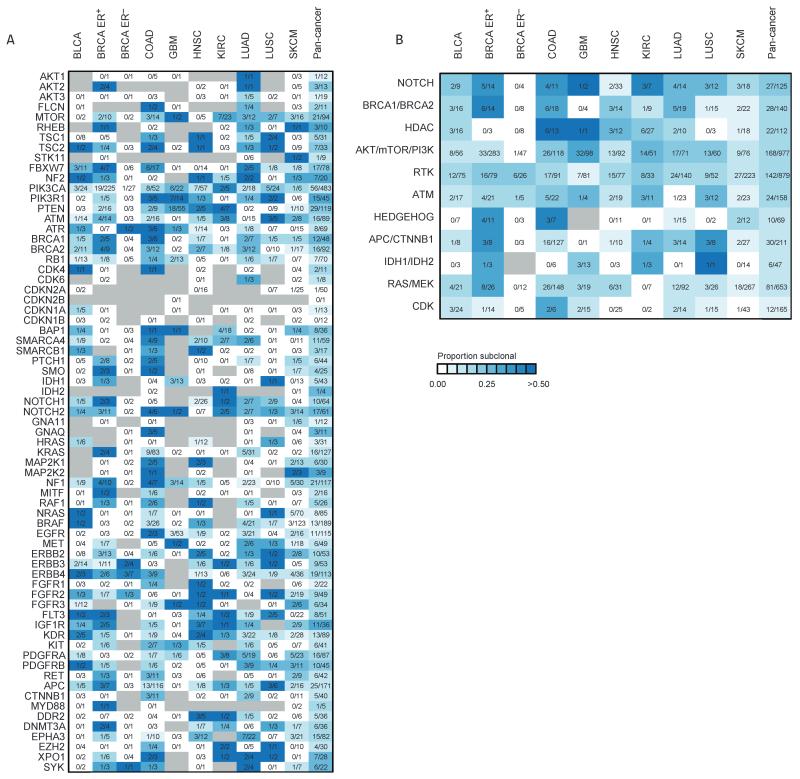
It is therefore notable that, with the exception of CDKN2B and CDKN1B, for every gene that has been linked with a targeted therapy approach, a subclonal mutation was identified in at least one tumor in the pan-cancer cohort (Fig. 4A). More than 10%of all nonsilent PIK3CA mutations and more than 20% of all nonsilent mutations in PTEN were found to be subclonal, and more than 15% of mutations in genes in the PI3K-AKT-mTOR pathway overall were subclonal (Fig. 4B). In GBM, the use of IDH-targeted therapies has been proposed for tumors with IDH1 or IDH2 mutations (33), and yet, we observed that more than 20% of IDH1 mutations are subclonal in GBM (Fig. 4). On the other hand, all IDH1 mutations detected in SKCM were clonal. In KIRC, mTOR inhibition is a common therapeutic option; however, we found that more than 30% of mutations in mTOR were subclonal within this disease type within single tumor samples.
Fig. 4.
Clonal heterogeneity of mutations in genes linked to therapies. (A) Heatmap showing the proportion of nonsilent mutations that are subclonal for each potentially actionable gene across nine cancer types. For each mutation, the number of subclonal mutations identified is indicated for each cancer type and the combined pan-cancer data set. Gray indicates the absence of a mutation. (B) The clonality of actionable pathways is depicted for each cancer type and the combined pan-cancer data set. Pathways are ordered according to subclonality in the pan-cancer data set, with pathways that have a higher proportion of subclonal mutations at the top of the heatmap. Genes related to CDKs (cyclin-dependent kinases) have very few subclonal mutations. RTK, receptor tyrosine kinases. For details of all the genes within each pathway, see table S4.
Subclonal mutations were detected in every actionable cancer pathway explored (Fig. 4B and table S4). However, we also observed clear differences between different cancer pathways; significantly fewer subclonal mutations in genes associated with cyclin-dependent kinases (CDKN1A, CDKN1B, CDKN2B, RB1, CDKN2A, CDK6, and CDK5) and the RAS–MEK(mitogen-activated protein kinase kinase) pathway were observed compared to mutations in the AKT-mTOR-PI3K pathway (P< 0.05, Fisher’s exact test).
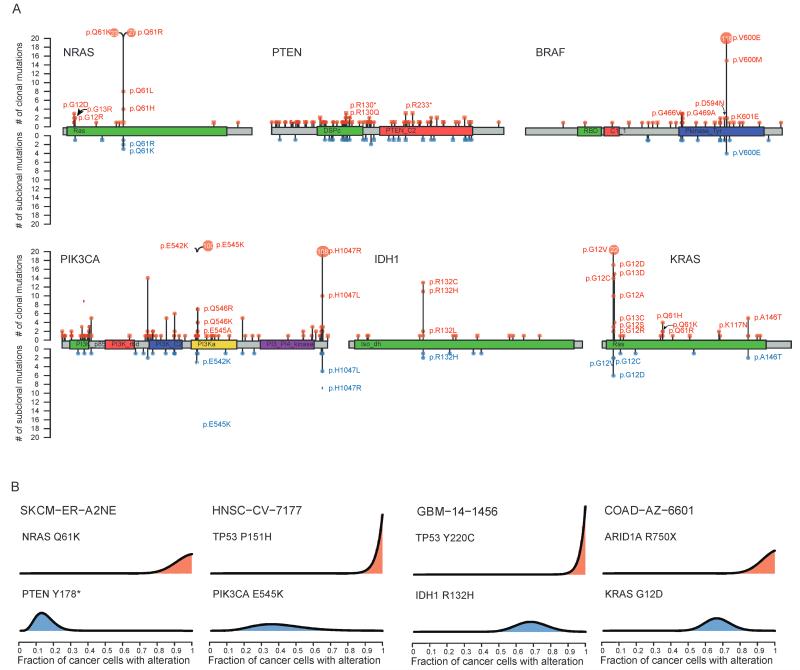
Finally, we restricted our analysis to well-characterized mutations occurring in the database of curated mutations (DoCM; docm.genome.wustl.edu) and in at least three tumor samples within the cohort. For most of these mutations, we identified both clonal and subclonal mutations in at least one cancer type (fig. S8). We identified tumor samples with subclonal mutations in known sites with therapeutic relevance such as NRAS(Q61K, Q61R, Q61L), BRAF (V600E), KRAS (G12C, G12D, G12V), PIK3CA (E542K, 545K, H1047R), and IDH1 (R132H), as well as many subclonal loss-of-function mutations in tumor suppressor genes such as PTEN (Fig. 5A and fig. S8). Identified subclonal driver mutations often occurred in tumors where clonal mutations in established cancer genes were also present [Fig. 5B; mean, 45% (15 to 67%)]. For example, in patient HNSC-CV-7177, a PIK3CA (E545K) mutation in the highly conserved helical domain was estimated to be present in only 36% of cancer cells, whereas a mutation in TP53in the same tumor was found to be present in all cancer cells. Similarly, patient SKCM-ER-A2NE exhibited a clonal mutation in NRAS(Q61K), whereas a PTEN mutation (Y178*) was present only in 13% of cells.
Fig. 5.
Clonal and subclonal actionable mutations in known cancer genes. (A) The distribution of nonsilent clonal and subclonal mutations in known cancer genes across the entire pan-cancer cohort. Red lollipops indicate clonal mutations, whereas blue lollipops represent subclonal mutations. Square lollipops indicate loss-of-function mutations (such as stop codon or frameshift). Hotspot sites harbor both clonal and subclonal mutations. For hotspot sites that harbor more than 20 mutations, the number of mutations is indicated inside the lollipop. (B) In many cases, mutations in known cancer genes occur in tumors that also harbor clonal driver mutations. Probability distributions over the cancer cell fraction for individual mutations are shown for specific tumors.
Together, these data highlight the need to understand the subclonal composition of tumors, which may be fueled by the temporal nature of mutational processes, to guide the use of targeted therapies. Moreover, these results demonstrate that known driver mutations do not only play a role in tumor initiation but also likely influence tumor behavior after tumor branching within distinct subclones.
DISCUSSION
Precision medicine will ultimately require not only a catalog of cancer genes and mutational processes but also an understanding of their spatial and temporal dynamics during a tumor’s evolution. Notwithstanding co-operative subclonal interactions (34, 35), targeting clonally dominant truncal somatic events may represent one therapeutic approach to optimize tumor control (36). Therefore, defining whether driver mutations are found in all or only a subset of cancer cells is likely to become increasingly relevant in cancer drug development. Moreover, identification of rules dictating the temporal acquisition of somatic events and mutational processes may inform the use of anticancer therapies and cancer detection strategies.
Our results demonstrate the presence of considerable intratumor heterogeneity in driver events, including known canonical hotspot mutations, such as IDH1 (R132H) and PIK3CA (E545K). Strikingly, we find that almost every gene linked with a targeted therapy harbors subclonal mutations in at least one tumor within the cohort. Moreover, we found that genes involved in the PI3K-AKT-mTOR pathway harbor a higher proportion of subclonal mutations compared to genes associated with cyclin-dependent kinases (CDKN1A, CDKN1B, CDKN2B, RB1, CDKN2A, CDK6, and CDK5) or the RAS-MEK pathway. We found further evidence for parallel evolution of subclones, with distinct somatic events affecting the same gene or pathway appearing to occur in distinct tumor subclones, suggesting constraints to tumor evolution that maybe therapeutically exploitable in the future.
Our census of subclonal driver events has important implications for targeted therapy strategies. Therapy targeting a mutation present in a fraction of tumor cells may only affect that subclone, resulting in limited clinical benefit. Conceivably, targeted therapy applied to cancer cells lacking the targeted mutation could have a paradoxical stimulatory effect, resulting in increased growth of wild-type subclones (11). Our results therefore suggest the need to stratify targeted therapy response according to the proportion of tumor cells in which the driver is identified and, moreover, indicate that certain pathways may be more actionable than others.
Nevertheless, despite extensive heterogeneity, we also demonstrate that mutations in established driver genes have a tendency to be clonal compared to mutations in nondriver genes, suggesting that these mutational events may often be required as early events in tumorigenesis and might represent suitable candidates for cancer screening approaches (37). The enrichment for clonal mutations in established driver genes is likely related to the fact that current methods for detecting driver genes are underpowered to detect subclonal drivers of tumor subclade expansions, present at low variant allele frequencies or spatially or temporally separated within tumors. Consistent with this, by focusing on later mutations, we demonstrate that it is possible to detect drivers that may be responsible for subclonal clade expansions. These genes may play roles in tumor maintenance and progression. Indeed, some of the cancer genes we identify, such as ATXN1 and CTNNA2, have been linked to tumor metastasis and cell migration (28). Although follow-up studies will be required to validate the functional impact of these genes, our results suggest that the catalog of cancer genes is far from complete.
Finally, on the basis of our results, we can also conclude that patterns for the temporal acquisition of mutations can be deciphered. We found an age-related signature that decreases over the course of a tumor’s life history in COAD, GBM, and HNSC, and a similar trend is observed in BRCA (16, 38). By contrast, cancer-specific signatures reflecting endogenous processes gone awry, such as APOBEC-mediated mutagenesis, were found to increase later on in tumor evolution (20). These data suggest that a mutator phenotype may be prevalent across cancers. We also found evidence linking APOBEC-mediated mutagenesis with the acquisition of subclonal driver events, highlighting how this mutational process can alter tumor evolutionary trajectories.
Our strategy for subclonal stratification has limitations. Given that we used single tumor samples taken at one point in the disease course, we likely underestimated the true extent of heterogeneity within tumors. As recently demonstrated in KIRC as well as in LUAD and LUSC, mutations from single biopsies may give the illusion of clonality, that is, although a mutation appears to be present in every cell from one tumor biopsy, it can be entirely absent from other regions of the same tumor (19, 20). Moreover, our ability to identify subclonal mutations was restricted by the sequencing depth and purity of samples. Comprehensive longitudinal studies similar to TRACERx, involving deep multiregion sequencing and the analysis of copy number events, coupled with single-cell sequencing, will be needed to obtain a more complete understanding of the prevalence of subclonal driver events over space and time (39). Moreover, further studies will be required to gain a deeper understanding of the impact of subclonal mutations on the efficacy of targeted therapies.
In conclusion, our study highlights the importance of viewing tumor development as a dynamic evolutionary process (40). Despite our emerging understanding of the catalog of cancer aberrations and mutational processes involved in tumor progression, we are only beginning to decipher the rules that constrain the evolution of tumors and to identify the later events that might guide or precipitate subclonal expansions (41,42). Ultimately, an understanding of these rules and an appreciation of the subclonal nature of many actionable driver events may be required to improve drug development strategies in cancer medicine.
MATERIALS AND METHODS
Study design
This was an observational study using publicly available mutation and copy number data from nine cancer types to explore the clonal status of driver mutations (including those linked to targeted therapies), the dynamics of mutational processes over time, and whether we could identify additional cancer genes through temporal and clonal dissection (Fig. 1). All data were obtained from TCGA (see below), and we did not consider clinical outcome endpoints.
Mutation data
All mutation data were downloaded from Broad Institute MAF dashboard (https://confluence.broadinsttitute.org/display/GDAC/MAF+Dashboard). Mutations were filtered to ensure that each variant had at least five tumor reads and coverage of ≥ 30. Only single nucleotide variants were used for analysis.
Classification of known driver genes
Driver mutations were classified as those residing in genes representing the intersection of genes identified by the recent Lawrence analysis (1) and those residing in the cancer gene census (23). For a full list of driver genes, see table S1.
Copy number and loss-of-heterozygosity analysis
Affymetrix SNP 6.0 arrays for TCGA tumors from nine cancer types—BLCA, BRCA, COAD, GBM, HNSC, KIRC, LUAD, LUSC, and SKCM—were obtained from TCGA (https://tcga-data.nci.nih.gov/tcga). Allele-specific integer copy numbers, as well as ploidy and purity estimates, were obtained using ASCAT (for further details, see Supplementary Materials and Methods)
Estimating the cancer cell fraction and temporal dissection of mutations
Supplementary Materials and Methods provide details regarding estimation of the cancer cell fraction and temporal dissection of mutations.
Assessing clonal enrichment of mutations in known cancer genes
To assess enrichment for clonal mutations in cancer genes versus other genes, we implemented a two-sided Fisher’s exact test. To explore the significance of specific cancer genes, we implemented a permutation test (for further details, see Supplementary Materials and Methods).
Identification of cancer genes
To identify cancer genes, MutSigCV was applied separately to temporally and clonally dissected mutations within each cancer cohort (for details, see Supplementary Materials and Methods).
Classification of genes relevant for genomics-driven therapy
To identify genes relevant for genomics-driven therapy, we used version 2 of TARGET (tumor alterations relevant for genomics-driver therapy) database (www.broadinstitute.org/cancer/cga/target). We restricted our analysis to genes that are predicted to result in sensitivity to specific therapies when somatically altered in cancers. The classification of genes into related modules can be seen in table S4.
Identification of specific mutations with therapeutic relevance
The DoCM was used to identify mutations with clinical evidence (drug targets associated with a mutation; diagnostic or prognostic markers associated with a mutation) or functional evidence (disease function described in cell lines; disease function described in animal models). The database is available online at docm.genome.wustl.edu.
Identification and temporal dissection of mutational signatures
Mutational signature analysis was performed using the Wellcome Trust Sanger Institute’s signatures framework (4) (for details, see Supplementary Materials and Methods).
Supplementary Material
Acknowledgments
We thank E. Mardis, B. Ainscough, O. L. Griffith, and M. Griffith for sharing the DoCM. We also thank R. Harris for critical reading of the manuscript. The results published here are in part based on data generated by TCGA pilot project established by the National Cancer Institute and National Human Genome Research Institute. The data were retrieved through dbGaP (Database of Genotypes and Phenotypes) authorization (accession no. phs000178.v5.p5). Information about TCGA and the investigators and institutions that constitute the TCGA research network can be found at http://cancergenome.nih.gov/.
Funding: C.S. is a senior Cancer Research UK clinical research fellow and is funded by Cancer Research UK, the Rosetrees Trust, EU FP7 (projects PREDICT and RESPONSIFY, ID: 259303), the Prostate Cancer Foundation, the Breast Cancer Research Foundation, and the European Research Council (THESEUS). This research is supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre.
Footnotes
www.sciencetranslationalmedicine.org/cgi/content/full/7/283/283ra54/DC1
Materials and Methods
Fig. S1. Tumor coverage and purity estimates.
Fig. S2. Clonal heterogeneity of mutations in specific genes.
Fig. S3. High-confidence clonal and subclonal mutations in nine cancer types.
Fig. S4. TP53 and genome doubling.
Fig. S5. Examples of parallel evolution.
Fig. S6. Mutational spectra of clonal and subclonal mutations in cancer genes.
Fig. S7. Overall survival and expression of NRXN3.
Fig. S8. Clonality of actionable mutations.
Table S1. Driver genes within each cancer type (provided as a separate Excel file).
Table S2. Mutational spectra of cancer genes (provided as a separate Excel file).
Table S3. Cancer genes identified through clonality and temporal dissection analysis (provided as a separate Excel file).
Table S4. Genes linked with targeted therapeutics (provided as a separate Excel file).
References (43–45)
Competing interests: C.S. sits on the Roche/Genentech clinical trial steering committee. All other authors declare that they have no competing interests.
Data and materials availability: Data and code are available online at https://bitbucket.org/nmcgranahan/pancancerclonality/downloads/McGranahan_data.zip.
REFERENCES AND NOTES
- 1.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, Leiserson MD, Miller CA, Welch JS, Walter MJ, Wendl MC, Ley TJ, Wilson RK, Raphael BJ, Ding L. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tamborero D, Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Kandoth C, Reimand J, Lawrence MS, Getz G, Bader GD, Ding L, Lopez-Bigas N. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci. Rep. 2013;3:2650. doi: 10.1038/srep02650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, VanBuuren MM, van ’t Veer L, Vincent-Salomon A, Waddell N, Yates LR, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat. Rev. Genet. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 7.Yates LR, Campbell PJ. Evolution of the cancer genome. Nat. Rev. Genet. 2012;13:795–806. doi: 10.1038/nrg3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M, Siravegna G, Bencardino K, Cercek A, Chen CT, Veronese S, Zanon C, Sartore-Bianchi A, Gambacorta M, Gallicchio M, Vakiani E, Boscaro V, Medico E, Weiser M, Siena S, Di Nicolantonio F, Solit D, Bardelli A. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, Akhavanfard S, Cahill DP, Aldape KD, Betensky RA, Louis DN, Iafrate AJ. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20:810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, Ozawa T, Holland EC, Huse JT, Jhanwar S, Leversha MA, Mikkelsen T, Brennan CW. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc. Natl. Acad. Sci. U.S.A. 2012;109:3041–3046. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, Sougnez C, Knoechel B, Gould J, Saksena G, Cibulskis K, McKenna A, Chapman MA, Straussman R, Levy J, Perkins LM, Keats JJ, Schumacher SE, Rosenberg M, Multiple Myeloma Research Consortium. Getz G, Golub TR. Widespread genetic heterogeneity in multiple myeloma: Implications for targeted therapy. Cancer Cell. 2014;25:91–101. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C. Intratumor heterogeneity: Seeing the wood for the trees. Sci. Transl. Med. 2012;4:127ps10. doi: 10.1126/scitranslmed.3003854. [DOI] [PubMed] [Google Scholar]
- 13.Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, Yoon CJ, Ellis P, Wedge DC, Pellagatti A, Shlien A, Groves MJ, Forbes SA, Raine K, Hinton J, Mudie LJ, McLaren S, Hardy C, Latimer C, Della Porta MG, O’Meara S, Ambaglio I, Galli A, Butler AP, Walldin G, Teague JW, Quek L, Sternberg A, Gambacorti-Passerini C, Cross NC, Green AR, Boultwood J, Vyas P, Hellstrom-Lindberg E, Bowen D, Cazzola M, Stratton MR, Campbell PJ, Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, Dawson KJ, Iorio F, Nik-Zainal S, Bignell GR, Hinton JW, Li Y, Tubio JM, McLaren S, Butler AP, Teague JW, Mudie L, Anderson E, Rashid N, Tai YT, Shammas MA, Sperling AS, Fulciniti M, Richardson PG, Parmigiani G, Magrangeas F, Minvielle S, Moreau P, Attal M, Facon T, Futreal PA, Anderson KC, Campbell PJ, Munshi NC. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat. Commun. 2014;5:2997. doi: 10.1038/ncomms3997. O.M.S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, Bashashati A, Prentice LM, Khattra J, Burleigh A, Yap D, Bernard V, McPherson A, Shumansky K, Crisan A, Giuliany R, Heravi-Moussavi A, Rosner J, Lai D, Birol I, Varhol R, Tam A, Dhalla N, Zeng T, Ma K, Chan SK, Griffith M, Moradian A, Cheng SW, Morin GB, Watson P, Gelmon K, Chia S, Chin SF, Curtis C, Rueda OM, Pharoah PD, Damaraju S, Mackey J, Hoon K, Harkins T, Tadigotla V, Sigaroudinia M, Gascard P, Tlsty T, Costello JF, Meyer IM, Eaves CJ, Wasserman WW, Jones S, Huntsman D, Hirst M, Caldas C, Marra MA, Aparicio S. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, Raine K, Jones D, Marshall J, Ramakrishna M, Shlien A, Cooke SL, Hinton J, Menzies A, Stebbings LA, Leroy C, Jia M, Rance R, Mudie LJ, Gamble SJ, Stephens PJ, McLaren S, Tarpey PS, Papaemmanuil E, Davies HR, Varela I, McBride DJ, Bignell GR, Leung K, Butler AP, Teague JW, Martin S, Jönsson G, Mariani O, Boyault S, Miron P, Fatima A, Langerød A, Aparicio SA, Tutt A, Sieuwerts AM, Borg A, Thomas G, Salomon AV, Richardson AL, Borresen-Dale AL, Futreal PA, Stratton MR, Campbell PJ, Breast Cancer Working Group of the International Cancer Genome Consortium The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, Wan Y, Zhang W, Shukla SA, Vartanov A, Fernandes SM, Saksena G, Cibulskis K, Tesar B, Gabriel S, Hacohen N, Meyerson M, Lander ES, Neuberg D, Brown JR, Getz G, Wu CJ. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos CR, Martinez P, Phillimore B, Begum S, Rabinowitz A, Spencer-Dene B, Gulati S, Bates PA, Stamp G, Pickering L, Gore M, Nicol DL, Hazell S, Futreal PA, Stewart A, Swanton C. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Gen. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, Jamal-Hanjani M, Shafi S, Murugaesu N, Rowan AJ, Gronroos E, Muhammad MA, Horswell S, Gerlinger M, Varela I, Jones D, Marshall J, Voet T, Van Loo P, Rassl DM, Rintoul RC, Janes SM, Lee SM, Forster M, Ahmad T, Lawrence D, Falzon M, Capitanio A, Harkins TT, Lee CC, Tom W, Teefe E, Chen SC, Begum S, Rabinowitz A, Phillimore B, Spencer-Dene B, Stamp G, Szallasi Z, Matthews N, Stewart A, Campbell P, Swanton C. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bashashati A, Ha G, Tone A, Ding J, Prentice LM, Roth A, Rosner J, Shumansky K, Kalloger S, Senz J, Yang W, McConechy M, Melnyk N, Anglesio M, Luk MT, Tse K, Zeng T, Moore R, Zhao Y, Marra MA, Gilks B, Yip S, Huntsman DG, McAlpine JN, Shah SP. Distinct evolutionary trajectories of primary high-grade serous ovarian cancers revealed through spatial mutational profiling. J. Pathol. 2013;231:21–34. doi: 10.1002/path.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aylon Y, Oren M. p53: Guardian of ploidy. Mol. Oncol. 2011;5:315–323. doi: 10.1016/j.molonc.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts SA, Gordenin DA. Clustered and genome-wide transient mutagenesis in human cancers: Hypermutation without permanent mutators or loss of fitness. BioEssays. 2014;36:382–393. doi: 10.1002/bies.201300140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fanjul-Fernández M, Quesada V, Cabanillas R, Cadinanos J, Fontanil T, Obaya A, Ramsay AJ, Llorente JL, Astudillo A, Cal S, López-Otin C. Cell-cell adhesion genes CTNNA2 and CTNNA3 are tumour suppressors frequently mutated in laryngeal carcinomas. Nat. Commun. 2013;4:2531. doi: 10.1038/ncomms3531. [DOI] [PubMed] [Google Scholar]
- 27.Kusinska R, Górniak P, Pastorczak A, Fendler W, Potemski P, Mlynarski W, Kordek R. Influence of genomic variation in FTO at 16q12.2, MC4Rat 18q22 and NRXN3 at 14q31 genes on breast cancer risk. Mol. Biol. Rep. 2012;39:2915–2919. doi: 10.1007/s11033-011-1053-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee Y, Fryer JD, Kang H, Crespo-Barreto J, Bowman AB, Gao Y, Kahle JJ, Hong JS, Kheradmand F, Orr HT, Finegold MJ, Zoghbi HY. ATXN1 protein family and CIC regulate extracellular matrix remodeling and lung alveolarization. Dev. Cell. 2011;21:746–757. doi: 10.1016/j.devcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varela I, Klijn C, Stephens PJ, Mudie LJ, Stebbings L, Galappaththige D, van der Gulden H, Schut E, Klarenbeek S, Campbell PJ, Wessels LF, Stratton MR, Jonkers J, Futreal PA, Adams DJ. Somatic structural rearrangements in genetically engineered mouse mammary tumors. Genome Biol. 2010;11:R100. doi: 10.1186/gb-2010-11-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ni S, Hu J, Duan Y, Shi S, Li R, Wu H, Qu Y, Li Y. Down expression of LRP1Bpromotes cell migration via RhoA/Cdc42 pathway and actin cytoskeleton remodeling in renal cell cancer. Cancer Sci. 2013;104:817–825. doi: 10.1111/cas.12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brannon A, Vakiani E, Sylvester BE, Scott SN, McDermott G, Shah RH, Kania K, Viale A, Oschwald DM, Vacic V, Emde AK, Cercek A, Yaeger R, Kemeny NE, Saltz LB, Shia J, MI DA, Weiser MR, Solit DB, Berger MF. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol. 2014;15:454. doi: 10.1186/s13059-014-0454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janku F, Tsimberidou AM, Garrido-Laguna I, Wang X, Luthra R, Hong DS, Naing A, Falchook GS, Moroney JW, Piha-Paul SA, Wheler JJ, Moulder SL, Fu S, Kurzrock R. PIK3CA mutations in patients with advanced cancers treated with PI3K/AKT/mTOR axis inhibitors. Mol. Cancer Ther. 2011;10:558–565. doi: 10.1158/1535-7163.MCT-10-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, Tsoi J, Clark O, Oldrini B, Komisopoulou E, Kunii K, Pedraza A, Schalm S, Silverman L, Miller A, Wang F, Yang H, Chen Y, Kernytsky A, Rosenblum MK, Liu W, Biller SA, Su SM, Brennan CW, Chan TA, Graeber TG, Yen KE, Mellinghoff IK. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inda MM, Bonavia R, Mukasa A, Narita Y, Sah DW, Vandenberg S, Brennan C, Johns TG, Bachoo R, Hadwiger P, Tan P, Depinho RA, Cavenee W, Furnari F. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24:1731–1745. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marusyk A, Tabassum DP, Altrock PM, Almendro V, Michor F, Polyak K. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature. 2014;514:54–58. doi: 10.1038/nature13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C. Intratumor heterogeneity: Seeing the wood for the trees. Sci. Transl. Med. 2012;4:127ps110. doi: 10.1126/scitranslmed.3003854. [DOI] [PubMed] [Google Scholar]
- 37.Martinez P, McGranahan N, Birkbak NJ, Gerlinger M, Swanton C. Computational optimisation of targeted DNA sequencing for cancer detection. Sci. Rep. 2013;3:3309. doi: 10.1038/srep03309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, Menzies A, Martin S, Leung K, Chen L, Leroy C, Ramakrishna M, Rance R, Lau KW, Mudie LJ, Varela I, McBride DJ, Bignell GR, Cooke SL, Shlien A, Gamble J, Whitmore I, Maddison M, Tarpey PS, Davies HR, Papaemmanuil E, Stephens PJ, McLaren S, Butler AP, Teague JW, Jönsson G, Garber JE, Silver D, Miron P, Fatima A, Boyault S, Langerød A, Tutt A, Martens JW, Aparicio SA, Borg A, Salomon AV, Thomas G, Borresen-Dale AL, Richardson AL, Neuberger MS, Futreal PA, Campbell PJ, Stratton MR, Breast Cancer Working Group of the International Cancer Genome Consortium Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jamal-Hanjani M, Hackshaw A, Ngai Y, Shaw J, Dive C, Quezada S, Middleton G, de Bruin E, Le Quesne J, Shafi S, Falzon M, Horswell S, Blackhall F, Khan I, Janes S, Nicolson M, Lawrence D, Forster M, Fennell D, Lee SM, Lester J, Kerr K, Muller S, Iles N, Smith S, Murugaesu N, Mitter R, Salm M, Stuart A, Matthews N, Adams H, Ahmad T, Attanoos R, Bennett J, Birkbak NJ, Booton R, Brady G, Buchan K, Capitano A, Chetty M, Cobbold M, Crosbie P, Davies H, Denison A, Djearman M, Goldman J, Haswell T, Joseph L, Kornaszewska M, Krebs M, Langman G, MacKenzie M, Millar J, Morgan B, Naidu B, Nonaka D, Peggs K, Pritchard C, Remmen H, Rowan A, Shah R, Smith E, Summers Y, Taylor M, Veeriah S, Waller D, Wilcox B, Wilcox M, Woolhouse I, McGranahan N, Swanton C. Tracking genomic cancer evolution for precision medicine: The lung TRACERx study. PLOS Biol. 2014;12:e1001906. doi: 10.1371/journal.pbio.1001906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat. Rev. Cancer. 2006;6:924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 41.Hiley C, de Bruin EC, McGranahan N, Swanton C. Deciphering intratumor heterogeneity and temporal acquisition of driver events to refine precision medicine. Genome Biol. 2014;15:453. doi: 10.1186/s13059-014-0453-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 2015;27:15–26. doi: 10.1016/j.ccell.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Bengtsson H, Wirapati P, Speed TP. A single-array preprocessing method for estimating full-resolution raw copy numbers from all Affymetrix genotyping arrays including GenomeWideSNP 5 & 6. Bioinformatics. 2009;25:2149–2156. doi: 10.1093/bioinformatics/btp371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ortiz-Estevez M, Aramburu A, Bengtsson H, Neuvial P, Rubio A. CalMaTe: A method and software to improve allele-specific copy number of SNP arrays for downstream segmentation. Bioinformatics. 2012;28:1793–1794. doi: 10.1093/bioinformatics/bts248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Loo P, Nordgard SH, Lingjaerde OC, Russnes HG, Rye IH, Sun W, Weigman VJ, Marynen P, Zetterberg A, Naume B, Perou CM, Borresen-Dale AL, Kristensen VN. Allele-specific copy number analysis of tumors. Proc. Natl. Acad. Sci. U.S.A. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.