Abstract
Aim:
To evaluate the structure–activity relationship of oxime ether lipids (OELs) containing modifications in the hydrophobic domains (chain length, degree of unsaturation) and hydrophilic head groups (polar domain hydroxyl groups) toward complex formation with siRNA molecules and siRNA delivery efficiency of resulting complexes to a human breast cancer cell line (MDA-MB-231).
Materials & methods:
Ability of lipoplex formation between oxime ether lipids with nucleic acids were examined using biophysical techniques. The potential of OELs to deliver nucleic acids and silence green fluorescent protein (GFP) gene was analyzed using MDA-MB-231 and MDA-MB-231/GFP cells, respectively.
Results & conclusion:
Introduction of hydroxyl groups to the polar domain of the OELs and unsaturation into the hydrophobic domain favor higher transfection and gene silencing in a cell culture system.
Keywords: : breast cancer cells, lipoplexes, nonsymmetric hydrophobic domain, oxime ether lipids, RNA interference, structure–activity relationship
Small interfering RNA (siRNA)-based gene silencing is a promising approach for the treatment of diseases including cancer [1–5]. However, delivery of uncomplexed (naked) siRNA is challenging due to nuclease degradation and the overall negative charge of the siRNA [6,7]. Therefore, carriers have been developed for siRNA delivery. Cationic lipids are the most commonly used nonviral agents to deliver nucleic acids including plasmid DNA, antisense oligos, siRNA and small hairpin RNA (shRNA) [7,8–10]. The most popular cationic lipids, DOTAP and DOTMA associate with negatively charged DNA or siRNA via electrostatic interactions resulting in the formation of lipoplexes (lipid-nucleic acid nanoparticles). These lipoplexes have been demonstrated to provide high in vitro transfection efficiency [11,12]. Modifications in the chemical structure of DOTAP and DOTMA have been extensively studied to determine structure–function relationships for optimal transfection efficiency. Delivery of nucleic acids using cationic lipids as carriers is reliant on various factors such as the type of cationic head group [13], length and degree of unsaturation of the hydrophobic domain chains [14], and lipid backbone functionality [15]. The nature of the anionic counterion [16,17], and the nucleic acid-to-cationic lipid charge ratio [18] also play a role in the overall transfection efficiency [14,19].
The optimal traits of nucleic acid carriers can be classified into two major categories, high transfection efficiency and minimal nonspecific cytotoxicity. Although DOTAP and DOTMA have been examined in detail as nucleic acid transfection agents, one of the most widely used commercially available nucleic acid transfection agents is Lipofectamine 2000 (L2K). Although this formulation is highly efficacious, it has considerable nonspecific cytotoxic effects presumably due to its multivalent cationic nature [20]. This limitation has prompted investigators to explore monovalent cationic lipids as alternate transfection agents [21,9,20] Interestingly, the linking functionality, and to a lesser extent the nature of the cationic head group of the lipids have been demonstrated to contribute toward the observed nonspecific toxic effects [22]. Therefore, in addition to ether and ester linking domains in transfection lipids, such as in DOTMA and DOTAP, respectively, other linking functionalities, such as orthoester [23], carbamate [24], amide [25,26] and phosphoramidate [27] moieties have been studied. Besides the linking functionalities described above, lipid hydrocarbon chains lengths are also likely to contribute to the cytotoxicity of cationic lipids [14].
With an aim to develop lipid transfection agents with low toxicity and high transfection efficiency, we pursued the synthesis of oxime ether lipids (OELs). Oxime linkages are readily assembled using click chemistry [28] and are present in US FDA approved pharmaceuticals, such as fluvoxamine [29]. Moreover, the resulting oxime linkages are robust at pH 7 and prone to hydrolysis only under strongly acidic conditions. Nonetheless, their use in pro-drug formulations suggests suitable metabolism after cellular incorporation [30,31]. Therefore, it can be anticipated that oxime ether lipids are not likely to bear long-term in vivo toxicity issues.
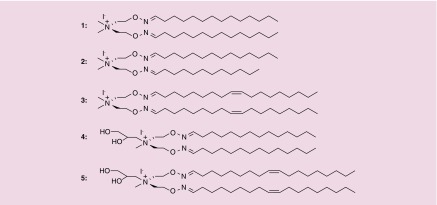
Synthesis of oxime ether lipids (containing the quaternary ammonium head groups) by utilizing the oximation approach has been reported previously by Biswas et al. [9]. These lipids when examined as tools to deliver nucleic acids (pDNA and siRNA) to cultured cells under in vitro conditions exhibited lower toxicity [9]. In the present investigation, we have further examined the suitability of OEL as feasible transfection agents by introducing hydroxyl groups in the hydrophilic region of these lipid molecules; as well as modification in hydrophobic core by varying the chain lengths and/or degree of unsaturation. The lipids used in the study shown in Figure 1 use an oxime ether group to conjugate the hydrophobic domain to the positively charged quaternary ammonium ion head group. OELs 1–3 were varied in the carbon chain length from C12-C18 and the degree of unsaturation as previously reported [9]. Hydroxylated transfection agents have been noted previously to be better-suited agents for nucleic acid delivery [32]. Consequently, in addition to the above-mentioned variations; we prepared OELs that contain hydroxyl groups attached to the head group region (lipids 4 and 5). Herein, we disclose the ability of this panel of OELs to deliver siRNA for RNAi in human breast cancer cells. We describe their delivery efficiency on the basis of their differences in chain length, degree of unsaturation and the presence of hydroxylated head groups.
Figure 1. . Panel of oxime ether lipids.
Materials & methods
Materials
1,2-Dioleoyl phosphatidylethanolamine (DOPE) was procured from Avanti polar lipids, Inc. (AL, USA). Human breast cancer cell lines MDA-MB-231 and green fluorescent protein expressing MDA-MB-231/GFP cells were procured from ATCC (VA, USA) and Cell Biolabs, Inc. (CA, USA), respectively. Nuclease protease free water was purchased from Quality Biological, Inc. (MD, USA). Early endosomal marker (EEA1) and secondary fluorescein antibody were procured from Cell Signaling, Inc. Cell titer blue reagent and RQ1 RNase free DNase buffer and DNAse were obtained from Promega Corp. (WI, USA). Cell culture reagents and media were purchased from Invitrogen (NY, USA). The sequences for the sense and anti-sense strands for the Dicer substrate of RNAs (DS RNAs), Alexa-488 and Alexa-546 labeled RNA/DNA hybrid duplexes, Alexa-488 and Iowa black quencher labeled DNA duplexes were purchased from Integrated DNA Technologies, Inc. (IA, USA):
RNA sequences for DS RNA: sense: 5′–pACCCUGAAGUUCAUCUGCACCACCG; antisense: 5′–CGGUGGUGCAGAUGAACUUCAGGGUCA, 5′ side of sense strand is phosphorylated;
Fluorescently labeled DNA: DNA antisense labeled with Alexa488: 5′GGAGACCGTGACCGGTGGTGCAGATGAACTTCAGGGTCAtt/3AlexF488N/;
DNA sense labeled with Alexa546: 5′–/5AlexF546N/aa TGACCCTGAAGTTCATCTGCACCACCGGTCACGGTCTCC;
DNA sense with Iowa Black Quencher (IBQ): 5′–/IBQ/aa TGACCCTGAAGTTCATCTGCACCACCGGTCACGGTCTCC.
Methods
Synthesis of oxime ether lipids
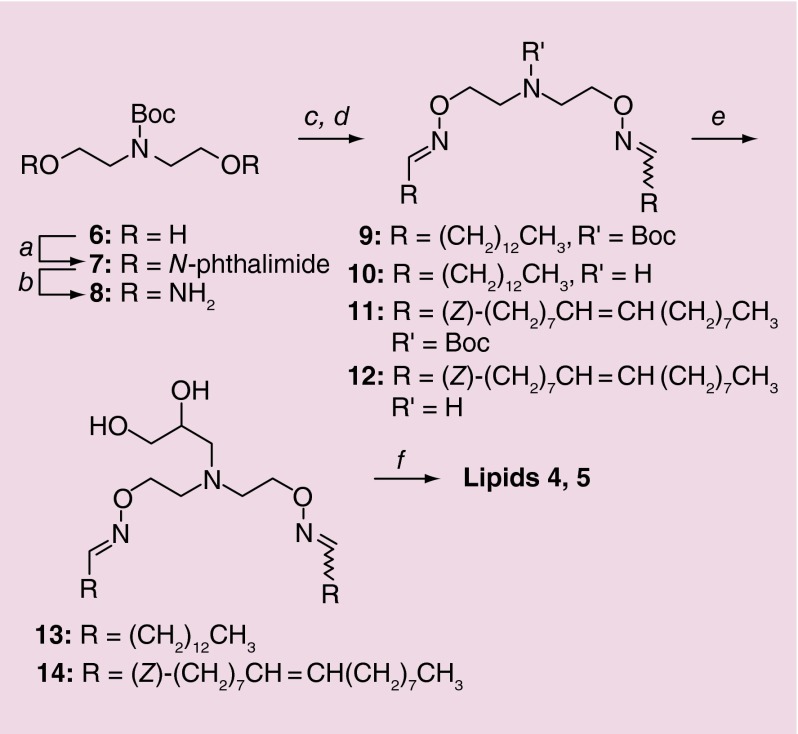
Lipids 1–3 have been reported previously [9]. Full experimental details for synthesis of hydroxylated OELs 4 and 5 are described in the Supplementary Material. The approach to prepare these lipids is given in Figure 2.
Figure 2. . Synthetic scheme for synthesis of lipids 4 and 5.
Conditions: a. N-hydroxyphthalimide, PPh3, DIAD, 0 °C to rt, 12h; b. N2H4•H2O, CH2Cl2, rt, 2.5h (16%, 2 steps); c. tetradecyl aldehyde (for 9) or oleyl aldehyde (for 11), CH2Cl2, rt, 10h (47–71%); d. TFA, CH2Cl2, 0 °C to rt, 3h (91–100%); e. glycidol, EtOH, rt, ca. 2.5d (38–41%); f. CH3I, CH2Cl2 (sealed tube), 55 °C, 18h (63–64%).
Nucleic acid duplex assembly
Nucleic acids used in this study were DS RNA duplexes [33], fluorescent duplexes of Alexa-488 RNA/DNA hybrids, DNA duplexes and quenched DNA duplexes containing Al488 and Iowa black fluorescence quencher (FQ), Alexa 546 labeled RNA/DNA hybrids [34,35] designed against green fluorescent protein (GFP). The specific sense and antisense strands of all the designed duplexes were heated at 90°C for 1 min and then annealed at room temperature before further use as described [34].
Preparation of oxime ether liposomes
Lipids were solubilized in CHCl3 at a concentration of 10 mg/ml. OELs (1 mg) were placed in glass vials and then mixed with an equimolar amount of the matrix lipid DOPE. Lipid films were prepared using a rotary evaporator under reduced pressure, and kept overnight in desiccator at room temperature. Lipids films were hydrated with 3 ml of enzyme-free water under vortexing and were probe sonicated for 10 min on ice bath with 1-min cycle of sonication and 1-min rest to obtain 0.33 mg/ml of OEL in the liposomes [9]. The resulting OEL liposome preparations were either used for further complexation with nucleic acids (described below), or alternatively, the OEL liposomes were stored at 4°C and used within 2 weeks of their preparation. Although the liposomes contain DOPE as a helper lipid, the liposomes used in this study will be referred as OEL liposomes.
Formation of oxime ether liposomes/nucleic acid complexes
To prepare oxime ether liposome/nucleic acid complexes, the lipids at different concentrations were introduced to a fixed concentration of nucleic acids (DS RNAs or RNA/DNA hybrids or DNA duplexes) and incubated for 30 min at room temperature. (We also conducted silencing studies to examine the effects of varying concentrations of DS RNA for a range of lipid 5 (see Supplementary Material). After incubation, the mixture was diluted tenfold by using either enzyme-free water/buffer or serum media at room temperature and the resulting mixture was used immediately for different experiments. The final concentration of nucleic acids and the lipid concentrations are mentioned in the respective experimental section and figure legends.
Size & zeta-potential measurements
The liposomes before or after complexation with DS RNAs were analyzed for their hydrodynamic size and zeta potential using Zeta Sizer Nano ZS (Malvern Instruments, MA, USA). OEL alone and at different concentrations in complexation with DS RNAs (50 nM) were run in triplicate in an automatic mode at 25°C equipped with a 633 nm laser and a back-scattering detector. However, only data for 10 µM for each OEL in complexation with 50 nM DS RNA are shown. Hydrodynamic size of the samples was measured using the Stokes–Einstein equation. The zeta potential of the complexes was calculated from the electrophoretic mobility using Smoluchowski approximation [36].
Cryo-electron microscopy
Cryo-electron microscopy (Cryo-EM) was performed on two representative oxime ether lipids, lipid 2 and lipid 5. Liposomes without nucleic acids were visualized using 1000 µM OEL. Similarly, liposome/nucleic acid complexes were examined at concentrations of 1000 µM OEL and 10 µM DS RNA were used. Four microliters of sample were blotted onto freshly glow-discharged holey carbon grids (Quantifoil R2/2, SPI, PA, USA) and vitrified in a Vitrobot plunge freezer (FEI, OR, USA). Images were recorded with a T20 microscope (FEI) at 200 kV on an Eagle CCD camera (FEI).
Fluorescent anisotropy/polarization measurements
Binding affinities of OEL with nucleic acids were determined by conducting fluorescent anisotropy/polarization measurements using Tecan Infinite M1000 (Tecan, USA). Fluorescent Alexa-488 labeled RNA/DNA hybrid duplexes (100 nM) were incubated with various concentrations of different OELs ranging from 1 to 40 µM per well in the 96-well plate. Changes in fluorescent (anisotropy/polarization) values of hybrid duplexes upon binding to OEL with respect to the hybrids alone was measured at λex 470 and λem 517 nm.
Protection of nucleic acids
The protection of nucleic acid duplexes by OEL upon digestion by nucleases was determined by fluorescence resonance energy transfer (FRET) experiments [37]. In brief, quenched DNA duplexes in which the 3′ antisense strand labeled with fluorescent Alexa-488 and the 5′ sense strand labeled with fluorescence quencher Iowa Black FQ were used. DNA duplexes (50 nM) alone or complexed with OEL (5 and 10 µM) in 1× RQ1 RNase free DNase buffer were incubated and fluorescence was measured at 37°C for 5 min in a fluorimeter (Horiba Jobin Yvon, NJ, USA) to get a stable baseline at λex460 and λem520 nm for Alexa 488 (slit width at 2 nm). After 5 min, RQ1 RNase free DNase was added according to the manufacturer's protocol to the samples while at 37ºC and the degradation of the duplexes was monitored following the dequenching of Alexa 488 for 3 h.
Cell culture studies
MDA-MB-231 (human breast cancer) cells either stably expressing enhanced green fluorescent protein (GFP) or non-GFP cells were maintained in a Dulbecco's modified Eagle's medium (DMEM) 10% (v/v) heat-inactivated FBS (fetal bovine serum), 100 iu./ml penicillin and 100 μg/ml streptomycin under a humidified 5% CO2 incubator at 37°C. On the basis of the experimental requirements, either GFP-expressing or non-GFP MDA-MB-231 cells were used.
Uptake & silencing measurement by flow cytometry
One day before the experiments, 60,000 cells/well of MDA-MB-231 cells and 30,000 cells/well of MDA-MB-231/GFP in the serum containing media were plated in two separate 24-well plates for uptake and silencing experiments. On the day of transfection, OEL (1–5) at different concentrations from 1–20 µM complexed with 50 nM of either Alexa-488 RNA/DNA hybrid duplexes or dicer substrates of RNA duplexes (DS RNAs) were added to the cells plated in each well of the plate in serum containing media. The cells were then incubated for 4 h at 37°C. After 4 h, the media was replaced with the fresh serum media. Uptake efficiency of RNA/DNA hybrid duplexes and silencing of GFP was quantified using Cell Quest software after 1 day and 3 days of transfections respectively by fluorescence-activated cell sorting (FACS).
Fluorescent microscopy
Approximately 3 days or approximately 72 h after the transfection, MDA-MB-231/GFP were imaged with a Nikon 200 TE inverted microscope (NJ, USA) using Meta Morph software (Universal Imaging Co., PA, USA) to determine the silencing efficiency of GFP by OEL/DS RNAs complexes. PanFluor 20X, ELWD, NA = 0.45 objective and a Nikon B-2E/C, 465–495/505/515–555 cube (Chroma Technology Corp., VT, USA) were used for the GFP imaging.
Endosomal colocalization
To observe the colocalization of fluorescently labeled Alexa-546 RNA/DNA hybrids (50 nM) in MDA-MB-231 cells, the early endosomal marker (EEA1) was used. A 30,000 cells/quadrant of a culture dish were seeded 1 day before the transfection. The next day, cells were transfected with fluorescently labeled hybrids (50 nM) complexed with respective OEL (10 µM) in serum-containing media in separate well quadrants. After 4 h of incubation, 1% BSA in 1× PBS (washing solution) was used to wash the cells three-times and the cells were fixed with 4% paraformaldehyde in 1× PBS for 10 min at room temperature. The cells were then washed again three-times and permeabilized with 0.2% Triton X-100 in 1× PBS for 10 min. After washing the cells three-times, the cells were treated with primary antibody EEA1 in 1% BSA in 1× PBS for 1 h. The cells were then washed three-times and were stained with anti-EEA1 secondary antibody labeled with Alexa-488. Thereafter, the cells were fixed again with 4% paraformaldehyde in 1× PBS as mentioned above and were imaged with a pinhole adjusted to 1 airy unit and analyzed for endosomal colocalization using an LSM 710 confocal microscope (Carl Zeiss) with a 63×, 1.4 NA magnification lens.
Cell viability assay
Viability of the MDA-MB-231 cells was determined on treatment with OEL alone and in complexation with DS RNAs. Typically, 20,000 cells/well were seeded in 96-well plates in serum containing media 24 h prior to experiments. OEL at different concentrations (1–20 µM) with DS RNAs (50 nM) were added to the cells in triplicate in serum-containing media in the plate and incubated for 4 h at 37°C. After incubation, the media were replaced with fresh serum-containing media. The cells were then incubated for another 24 h at 37°C. At the end of incubation, cell titer blue reagent was added to each well according to the manufacturer's protocol and the cells were further incubated for 4 h at 37°C. The fluorescence of the resofurin (converted from resazurin by viable cells) was measured at λex 560 and λem 590 nm with an auto cut-off in a fluorescent ELISA plate reader (Spectra MAX, Molecular Devices, CA, USA).
Results
Hydrodynamic diameter analysis
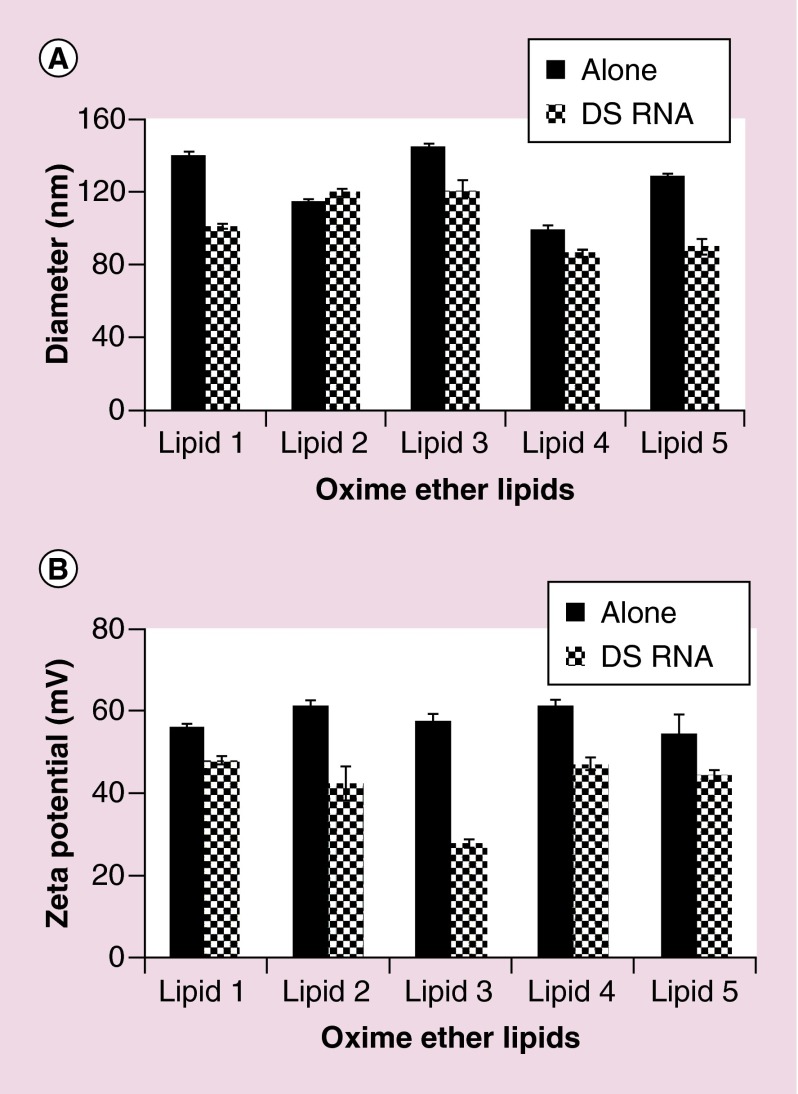
Formulations of OEL liposomes alone or in complexation with DS RNA (50 nM) were measured for their hydrodynamic sizes. All the OEL preparations showed particle diameters in the range of 80–140 nm. On complexation with DS RNA, the diameters of the resultant lipoplexes were smaller in comparison to the OEL formulations alone. The data for samples containing OEL at 10 µM and DS RNA at 50 nM are shown in Figure 3A.
Figure 3. . Size and zeta potential analysis of oxime ether lipid liposomes with or without addition of Dicer substrate of RNA.
Liposomes (10 µM) alone and in complexation with DS RNA (50 nM) were analyzed for their average diameter and zeta potential as described in the Methods section. (A) Diameter (nm) for various lipids (black bars) or in complexation with DS RNA (crossed bars). (B) Zeta potential analyses of oxime-ether lipids liposomes alone (black bars) and in complexation with DS RNA (crossed bars) are expressed in mV. The error bars in (A & B) represent standard deviation (n = 3).
DS RNA: Dicer substrate of RNA.
Zeta-potential measurements
Zeta potentials for OEL liposomes alone or in complexation with DS RNA (50 nM) were also measured (Figure 3B). All the OEL liposomes alone showed positive Zeta potential as expected due to their positively charged head groups. The zeta potential measured was in the range of 50–60 mV. In contrast, the zeta potential decreased in the presence of DS RNA due to the electrostatic interaction between the OEL and DS RNA as expected. The data for samples containing OEL at 10 µM and DS RNA at 50 nM are shown in Figure 3B.
Morphology
Although all the OELs used in this study differ in certain aspects, such as chain length, degree of unsaturation, presence of hydroxylated head group, they are all dual fatty acyl chain lipids. Therefore, we selected the two most diverse examples, lipid 2 (alkyl polar domain, nonsymmetric saturated hydrophobic domain, C12/C14) and lipid 5 (hydroxylated polar domain, symmetric unsaturated hydrophobic domain, C18:1/C18:1) and examined their morphology before and after complexation with DS RNA (Figure 4).
Figure 4. . Cryo-electron microscopy images.
OEL alone, and OEL complexed with DS RNA (OEL/DS RNA) for (A) lipid 2, and (B) lipid 5.
DS RNA: Dicer substrate of RNA; OEL: Oxime ether lipid.
Due to technical limitations, we performed the cryo-electron microscopy (Cryo-EM) at 100-fold higher concentration than the concentrations used for other experiments. However, the OEL:DS RNA ratio of 100:1 corresponds to the one used for the transfection experiments (5 µM OEL/50 nM DS RNA). Both the OEL only samples formed nice spherical liposomes whereas upon addition of DS RNA, clumps of multilamellar or concentric liposomes were observed, consistent with previously reported observations [38]. The apparent visual discrepancy on the results obtained by the overall size distribution of OEL–siRNA complexes by cryo-EM in contrast to the DLS measurements can be reconciled based on utilization of 100-fold excess of samples used for cryo-EM.
Binding affinity of oxime ether lipid/nucleic acid complexes
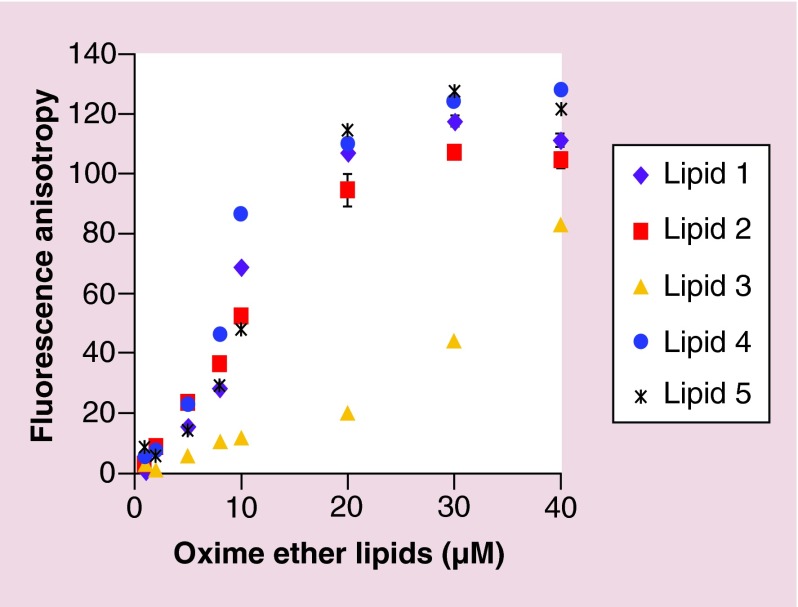
The binding affinity of OEL to nucleic acids was determined by fluorescence polarization measurements. This technique depends on the tumbling of a fluorescent molecule and exploits the relative change in the polarization/anisotropy values upon binding with a larger molecule. The slower the tumbling, the higher the polarization/anisotropy values [39]. These measurements were done using fluorescent Alexa488 labeled RNA/DNA hybrid duplexes. OELs at different concentrations from 1 to 40 µM were mixed with a constant concentration of fluorescent duplexes (100 nM). All the lipids showed an increase in anisotropy values with respect to the control duplexes even at the lowest concentration of 1 µM. Similar binding affinities for lipid 1, lipid 2, lipid 4 and lipid 5 to fluorescent duplexes were observed almost at all the concentrations of lipids tested. However, lipid 3 demonstrated significantly lower binding affinity to duplexes at all the concentrations (Figure 5).
Figure 5. . Nucleic acid binding affinity of oxime ether lipid liposomes.
Liposomes prepared from OEL were diluted to a range of lipid concentrations (1–40 µM) and 100 nM of Alexa 488 RNA/DNA hybrid was added to the samples. Fluorescence anisotropy was determined (see Methods section) as a measure of binding affinity. Lipid 1 (closed purple rhombus), lipid 2 (closed red square), lipid 3 (closed yellow triangle), lipid 4 (closed deep blue circle) and lipid 5 (black asterisk). The error bars represent standard error of the mean (±SEM) calculated from the values of two independent experiments (n = 2).
Protection of nucleic acids by OEL
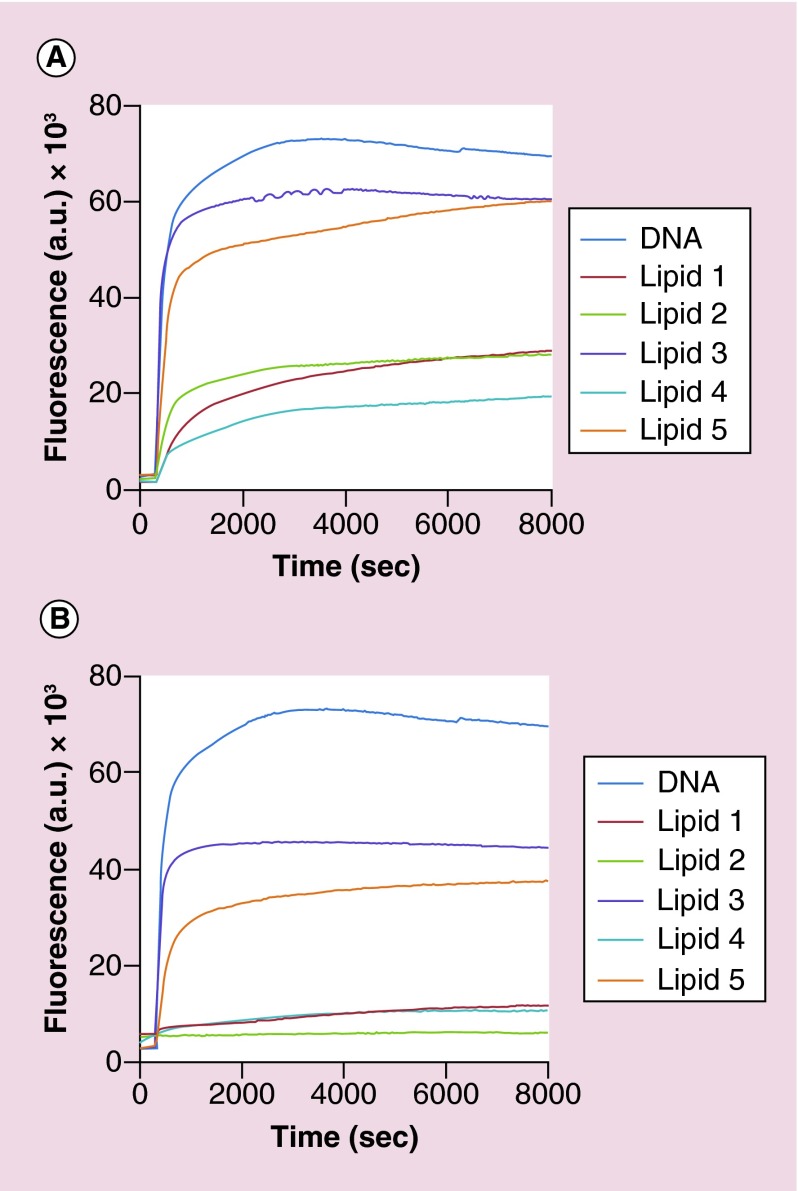
Here the protection of quenched DNA duplexes interacting with a DNase enzyme was evaluated. The fluorescence of Alexa488 upon DNase digestion was taken as the measurement of nucleic acid degradation and then correlated as protection by respective oxime ether lipids. We performed these experiments using two different concentrations of OELs: 5 and 10 µM. The data presented are representative of at least three independent experiments. All OEL lipoplexes at both concentrations tested showed protection of DNA duplexes against enzyme DNase. However, the unsaturated lipids (lipid 3 and lipid 5) did not protect the nucleic acids quite as well when compared with the other saturated lipids. In fact, lipid 3 showed the least protection which was followed by lipid 5 (Figure 6).
Figure 6. . Kinetics of DNA degradation by DNase.
Ability of oxime ether lipid to protect quenched DNA duplexes was determined at two lipid concentrations complexed with 50 nM DNA, as described in the Methods section. Fluorescence increase as a function of time represents dequenching of Alexa488 upon degradation from quenched duplex. DNA (dark blue line), lipid 1 (red line), lipid 2 (light green line), lipid 3 (purple line), lipid 4 (light blue line) and lipid 5 (light brown line). The data presented are representative of at least three independent experiments. (A) Oxime ether lipid: 5 µM, (B) oxime ether lipid: 10 µM.
Cell viability
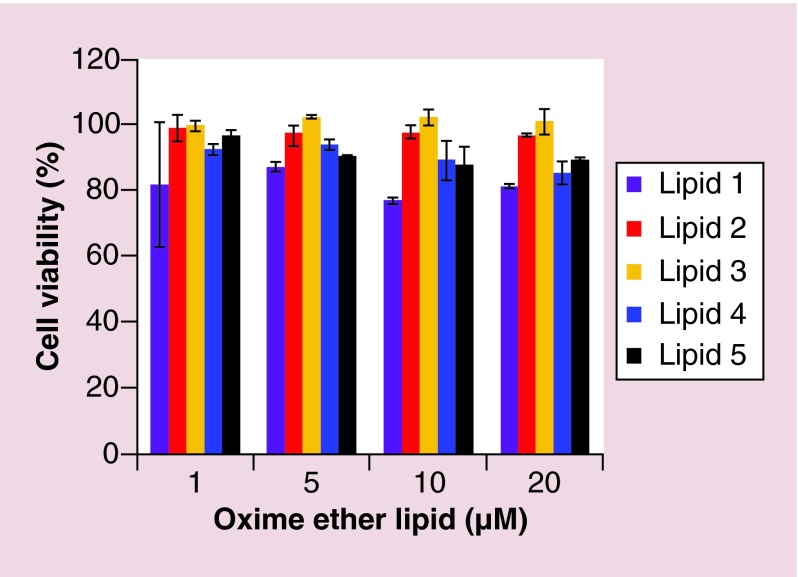
Impairment of cell viability upon addition of OEL liposomes alone or in complexation with DS RNA was assessed using a cell-titre blue reagent. The dye resazurin present in the reagent is converted to resorufin upon enzymatic reaction in viable cells, and the fluorescence of resofurin is then measured (λex 560 nm, λem 590 nm) to assay cell viability. MDA-MB-231 cells did not show any sign of impairment in their viability on treatment with the OEL (1–20 µM)/DS RNA (50 nM) lipoplexes after 24 h (Figure 7).
Figure 7. . Effect of addition of oxime ether lipid liposome/Dicer substrate of RNA complexes on viability of MDA-MB-231 cells.
Liposomes containing various lipid concentrations (1–20 µM) were complexed with 50 nM Dicer substrate of RNA and the resulting lipoplexes were added to MDA-MB-231 cells plated in 96-well clusters (see Methods section). Cell viability was monitored 24 h post-addition. The values are presented as percentage cell viability. Values of cells without any addition of samples were taken as 100%. Lipid 1 (purple bars), lipid 2 (red bars), lipid 3 (light brown bars), lipid 4 (dark blue bars) and lipid 5 (black bars). The error bars represent standard error of the mean (±SEM) calculated from the values of two independent experiments (n = 2).
Uptake efficiency of nucleic acids by OELs
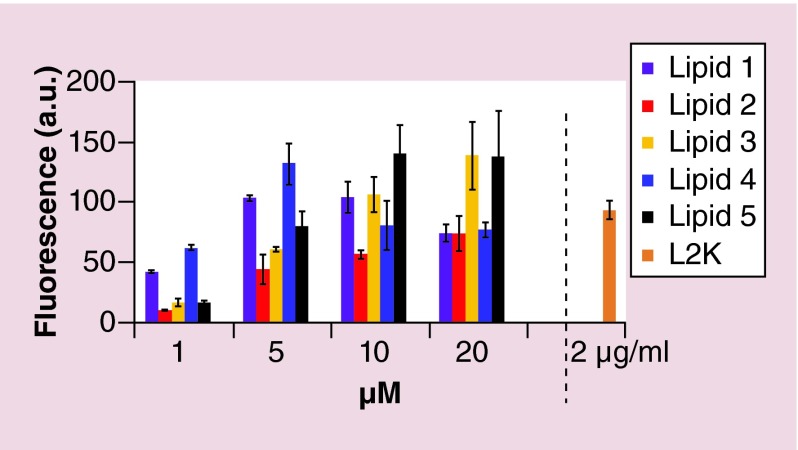
The uptake efficiency of Alexa488 RNA/DNA hybrid duplexes was measured using the breast cancer cell line, MDA-MB-231. FACS analysis showed that all OELs were capable of delivering the nucleic acids in the presence of serum (10% FBS) (Figure 8). Lipids 3 and 5, with hydrophobic domain unsaturation, showed better uptake at higher concentrations whereas lipids 1 and 4 (C14 unsaturated tails) showed better uptake at lower concentrations. Asymmetric lipid 2 showed generally poorer uptake overall. The uptake efficiency of lipids was also compared with the commercial transfection agent lipofectamine (L2K). Lipids 3, 4 and 5 demonstrated higher uptake than L2K at 20, 5 and 10 µM, respectively.
Figure 8. . Cellular uptake of oxime ether lipid liposomes/Alexa488 RNA/DNA hybrid complexes.
Various concentrations of OEL (1–20 µM) were complexed with Alexa488 RNA/DNA hybrid (50 nM) and added to MDA-MB-213 cells (non-GFP) plated on 24-well clusters and cellular uptake was monitored post 24 h as described in the Methods section. Lipofectamine (L2K) was used as a reference standard at a concentration of 2 µg/ml per well. The values are expressed as relative fluorescence arbitrary units (a.u.) as a function of increasing concentration of liposomes. Lipid 1 (purple bars), lipid 2 (red bars), lipid 3 (light brown bars), lipid 4 (dark blue bars), lipid 5 (black bars) and L2K (brown bar). The error bars represent standard error of the mean (±SEM) calculated from the values of two independent experiments (n = 2).
Silencing of green fluorescent protein gene
The release of nucleic acids such as siRNA from the delivery vectors is crucial to cause RNA interference (RNAi) that corresponds to the silencing of a particular gene. It was observed that all the OELs were capable of releasing DS RNA and were further processed by the RNAi pathway to cause silencing of the GFP gene in the presence of serum (10% FBS). Hydroxylated lipids 4 and 5 performed well with GFP gene silencing in human breast cancer cells (Figure 9). We observed that lipid 5, which has a hydrophobic domain comprised of C18:1/C18:1 chains and contains hydroxyl groups in the polar domain, consistently showed superior GFP silencing at all the doses tested and was also a better performer than the commercial transfection agent L2K and 10 and 20 µM concentration of OEL (see also Supplementary Figure 1). Lipid 2, which contains additional dissymmetry in the hydrophobic domain (C14/C12 chains) but does not contain OH groups in the polar domain consistently showed lower silencing activity in comparison to other lipids tested. The only exception was the silencing activity mediated by lipid 2 at 5 µM concentration. A representative fluorescent microscopic image (Figure 9B) demonstrates the silencing of GFP by lipid 5/DS RNA complexes.
Figure 9. . Green fluorescent protein gene silencing by oxime ether lipid liposomes/Dicer substrate of RNA complexes.
(A) Dose-dependent silencing activity of GFP gene mediated by OEL (1–20 µM)/DS RNA (50 nM) complexes is shown and compared with L2K (2 µg/ml)/DS RNA (50 nM) complexes. Lipid 1 (purple bars), lipid 2 (red bars), lipid 3 (light brown bars), lipid 4 (dark blue bars), lipid 5 (black bars) and L2K (brown bar). The error bars represent standard error of the mean (±SEM) calculated from the values of two independent experiments (n = 2). (B) Representative fluorescent microscopic images of MDA-MB-231/GFP cells showing silencing of the GFP gene mediated by lipid 5 (10 µM)/DS RNA (50 nM) complexes.
DS RNA: Dicer substrate of RNA.
Endosomal co-localization
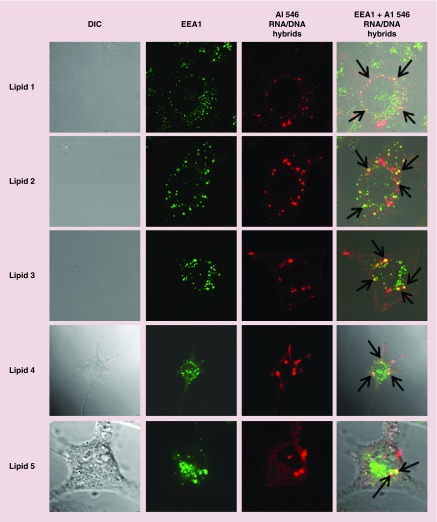
To determine whether the OEL/nucleic acid complexes enter the cells via endosomes or not, we used the early endosomal marker (EEA1, primary antibody) to co-localize the uptake of Alexa 546 RNA/DNA hybrid duplexes. We stained the EEA1 with the fluorescein tagged anti-EEA1 antibody and observed the colocalization of Alexa546 RNA/DNA hybrids (red) with EEA1 (green) as punctate yellow fluorescence. It was evident from the Figure 10 panel EEA1 + Alexa 546 RNA/DNA hybrids that RNA/DNA hybrid duplexes delivered by all the OEL tested were co-localized with EEA1 suggesting that the nucleic acids delivered by OELs were internalized by endosomes.
Figure 10. . Endosomal uptake of Alexa546 RNA/DNA hybrids delivered by oxime ether lipid liposomes.
Confocal microscopic images showing the colocalization of Alexa546 RNA/DNA hybrids (50 nM) delivered by oxime ether lipids 1–5 (10 µM) with early endosomal marker, EEA1. Black arrows show endosomal colocalization.
Discussion
Our investigations of oxime ether-based cationic lipids have established them as promising candidates for nonviral nucleic acid delivery in serum conditions. In our examination of the relationship between OEL structure and siRNA transfer efficacy, some trends emerged. Polar domain hydroxylation had the greatest influence on release properties, where hydroxylated lipids 4 and 5 outperformed their nonhydroxylated counterparts in regard to nucleic acid release upon cellular internalization, as indicated by GFP silencing.
Although tight lipid complexation with nucleic acids is desirable for protection against enzymatic degradation, the nucleic acid phosphate–lipid ammonium interactions [40] in concert with hydrophobic domain packing and any polar domain hydrogen bonding interactions must be readily overcome at a later stage for lipoplex disassembly to occur.
In this regard, the OELs appear well structured for effective siRNA protection and release. Hydrophobic domain asymmetry has been suggested as a structural factor that is beneficial for nucleic acid release, thus ultimately enhancing transfection activity [27,41]. Each of the OELs in our panel were used as a mixture of (E,E)- and (E,Z)-oxime ether isomers, meaning a nontrivial amount of nonsymmetric (E,Z)-lipid was present. Nonsymmetric hydrophobic domains likely benefit transfection due to better nucleic acid release through looser lipid packing in the lipoplex formulation [27,42–44]. A second level of asymmetry was examined in this study in the use of lipid 2 having the mixed fatty acyl lengths, C12 and C14 chains in the hydrophobic domain. We found by comparing lipid 1 (C14/C14) and 2, that the added asymmetry in the alkyl tails was not beneficial for transfection. Indeed, the poorest performing lipid by most criteria studied was the nonhydroxylated, nonsymmetric lipid 2.
Lipoplex uptake and activity was also facilitated by lipid hydroxylation, and this observation agrees well with the assessment of a recent review [45] citing greater transfection efficiencies for lipids possessing hydroxylated head groups. This effect is thought to be mediated by the decrease in head group hydration that results from the incorporation of functionality capable of hydrogen bonding. Neighboring lipid–lipid interactions in the lipoplex bilayers lead to exclusion of interstitial water, and the resulting high positive charge/low hydration motif promotes electrostatic interactions not only with the polynucleotide but also with cell membranes [46].
Unsaturation in the OEL lipid hydrophobic domain also strongly impacted cellular uptake and nucleic acid protection of the resulting complexes. Uptake of lipid–siRNA complexes by the cells showed concentration dependence wherein unsaturated-tail lipids outperformed at higher concentrations of the added complexes whereas saturated-tail lipids were optimal for mid-range concentrations. Prior studies designed to determine optimal lipid chain length and unsaturation have generated conflicting results [45]; the concentration dependence we have observed for our lipids may partially explain this inconsistency. Nucleic acid protection was also superior for the saturated lipids (1, 2, 4); however, despite weaker protection from the longer-chain unsaturated lipids, silencing of GFP was not compromised. Transfection efficiency has been known to be enhanced by the effects of unsaturation as unsaturated lipids are thought to fluidize endosomal bilayers and thus facilitate the lipoplex escape [47]. Similar effects might have occurred in the present studies by unsaturated lipids to fluidize endosomes and help in lipoplex escape to release siRNA to promote efficient silencing.
Conclusion
Over the past decade, Nantz and colleagues (co-authors in this study) have focused on the design and synthesis of oxime ether lipids as superior transfection agents [9]. The inspiration to use an oxime ether as a linking functionality follows from observations that oxime ether drugs such as Fluvoxamine are in wide clinical use [29], and initial studies indicate that oxime ether lipids can be developed as suitable transfection agents [9]. This study was designed to carefully examine the structure–function relationship of oxime ether lipids (OELs)/siRNA lipoplexes with their biological activity.
The average size distribution of the OEL/DS RNA (Dicer substrate of RNA) lipoplexes ranged from 80 to 120 nm in diameter. Binding experiments revealed that lipids 1, 2, 4 and 5 bind DS RNA with similar affinity. In contrast, binding of lipid 3 was drastically reduced. Using alexa488-labeled duplexes, the transfection efficiency of various OELs was determined in MDA-MB-231 cells. In general, the efficiency of GFP silencing mediated by the oxime ether lipid containing lipoplexes was dependent on the concentrations of the lipids used in our study, that is, addition of higher amounts of lipoplexes to a known number of cells yielded an increase in GFP silencing. Lipid 5 in particular, exhibited superior transfection efficiency at higher concentrations of the lipid. We also noted that lipid 2 containing the nonsymmetric fatty acid chains consistently showed reduced transfection efficiency at all concentrations tested. Experiments with an endosomal marker indicated that the lipid–siRNA complexes enter the cells via the endocytic pathway.
Interestingly, the higher binding affinity of lipid 5 to nucleic acids did not correlate well with protection against DNase. We hypothesize that the higher binding affinities of these lipids are mediated by lower fluidity (ordered lipid packing due to saturated fatty acyl chains). In addition, the presence of the hydroxyl head group further strengthens the association with the nucleic acids via the formation of hydrogen bonds. We predict that the presence of unsaturation in the fatty acyl chains (such as in lipid 3) reduces the effects imparted by the hydroxyl groups that contribute to hydrogen bonding (such as in lipid 5). Therefore, lipoplexes containing lipid 5, although exhibiting strong affinity with the nucleic acids, fail to provide maximum protection against DNAse degradation. This, however, may facilitate dissociation of the nucleic acid from the lipid resulting in a higher degree of silencing (as observed in the case of lipid 5). In addition, fatty acyl chain symmetry/asymmetry as well as chain length is likely to contribute toward efficient gene delivery. Further experiments in this area will be needed to assess the nucleic acid–lipid interactions in vitro and in vivo.
The results of this study led us to conclude that the lipids containing hydroxylated head groups are superior carriers for siRNA delivery to cells and the fatty acyl chain length and degree of unsaturation also contributes toward enhancement of transfection and silencing activity in vitro. To our knowledge, a systematic examination of OEL structure–function activity with respect to their ability to carry siRNA to the cells interior has not been reported earlier. Therefore, we believe that this is the first report to this end. Further examination of our OEL liposomes in animal studies will validate their suitability as in vivo nucleic acid carriers.
Future perspective
RNAi-based therapeutics has become an important field of research since the first clinical trial studies led by Davis and colleagues in year 2010 [48]. The future of this technology bears merit and is believed to play an important role for personalized medicine in the future. However, one of the bottlenecks in success of clinically suitable RNAi agents calls for development of suitable delivery platforms [10]. Lipids (including phospholipids) have been long-sought as suitable carriers for drugs and/or nucleic acids for chemotherapy and gene delivery including vaccines [7]. Application of lipids as carriers of therapeutics is based on their inherent biological properties and potentially relatively nontoxic effects [9,20,21]. However, nucleic acid delivery using the lipids as carriers is predominantly reliant on the positive charge of the latter. It is discernable that positively charged lipid molecules with user-friendly chemical synthesis routes, desired biodegradability and the ability to complex with nucleic acids may lead to successful RNAi-based therapies [9]. Structure-function studies indicate that overall charge [18], size, fatty acyl composition [14,41] and the polarity of head groups of the lipid carriers [13,45] or surface properties will likely dictate the suitability of resulting nucleic acid–lipid complexes for patient care in the future. In addition inclusion of molecules (such as PEG lipids) that will allow longer circulation times as well as molecules (such as pH-sensitive lipids) that will facilitate endosomal escape in the formulations would aid in designing superior lipid-based delivery systems. This study provides a first step to accomplish these objectives.
We foresee that lipid-based delivery systems will have a crucial role in systemic application of siRNA in the clinic. However, the lipid-based delivery systems for siRNA are mostly targeted to the liver/spleen. Therefore, more extensive studies are needed at the in vivo level to make them available to other tissues based on their pharmacokinetic profile, distribution in organs and diseased tissues, interaction with serum proteins, degradation of siRNA and lipid delivery system by enzymes, immune-stimulatory effects, targeting diseased tissue/cells selectivity versus nonspecific normal tissue/cells uptake, escape from endosomes and release profile of siRNA in the cytoplasm for efficient gene silencing.
Executive summary.
Lipid-based nonviral delivery systems for RNAi or siRNA delivery
Lipid-based delivery agents has been widely used for the delivery of nucleic acids including siRNAs and several of such formulations are under clinical trials.
Oxime ether lipids for RNAi or siRNA delivery
We have been developing oxime ether lipids as transfection agents. Oxime ether lipids (OELs) 1, 2, 3, 4 and 5 containing variations in the hydrophobic domain chain length, degree of unsaturation and/or with hydroxyl groups incorporated polar domain were synthesized and compared for siRNA delivery.
Structure–activity relationship between designed OELs & siRNA delivery or key observations of the study
Lipids 1, 2, 4 & 5 bind DS RNA with similar affinity whereas binding of lipid 3 was drastically reduced.
All the lipids mediated transfection under serum conditions and were nontoxic in human breast cancer cells. Lipid 5 exhibited superior transfection efficiency at higher concentrations of the lipid. Whereas lipid 2 (nonsymmetric) consistently showed reduced transfection efficiency at all concentrations tested.
Incorporation of hydroxyl groups to the polar domain and unsaturation into the hydrophobic domain of the OELs favor higher transfection and gene silencing (˜70%) activity.
Potential of OELs as siRNA delivery agents
Results led us to confidently say that the lipid 5 mediated delivery of siRNA will show promising results in vivo and we hope to take it to clinical trials.
Supplementary Material
Acknowledgements
The authors thank Ulrich Baxa (Leidos Biomedical Research, Inc., Frederick National Laboratory for Cancer Research) for electron microscopy analysis of the liposomes.
Footnotes
Disclaimer
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
Financial & competing interests disclosure
This research has been funded in whole or in part with Federal funds from the Frederick National Laboratory for Cancer Research, NIH, under contract HHSN261200800001E. This research was supported (in part) by the Intramural Research Program of NIH, Center for Cancer Research. RJ Knipp thanks the University of Louisville School of Interdisciplinary and Graduate Studies for fellowship support. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.de FA, Manoharan M, Meyers R, Vornlocher HP. RNA interference in vivo: toward synthetic small inhibitory RNA-based therapeutics. Methods Enzymol. 2005;392:278–296. doi: 10.1016/S0076-6879(04)92016-2. [DOI] [PubMed] [Google Scholar]
- 2.Zimmermann TS, Lee AC, Akinc A, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441(7089):111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]; • First systemic study to show the potential of RNAi in nonhuman primates.
- 3.Querbes W, Ge P, Zhang W, et al. Direct CNS delivery of siRNA mediates robust silencing in oligodendrocytes. Oligonucleotides. 2009;19(1):23–29. doi: 10.1089/oli.2008.0165. [DOI] [PubMed] [Google Scholar]
- 4.Alvarez R, Elbashir S, Borland T, et al. RNA interference-mediated silencing of the respiratory syncytial virus nucleocapsid defines a potent antiviral strategy. Antimicrob. Agents Chemother. 2009;53(9):3952–3962. doi: 10.1128/AAC.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Novobrantseva TI, Borodovsky A, Wong J, et al. Systemic RNAi-mediated gene silencing in nonhuman primate and rodent myeloid cells. Mol. Ther. Nucleic Acids. 2012;1:e4. doi: 10.1038/mtna.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akinc A, Querbes W, De S, et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18(7):1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ozpolat B, Sood AK, Lopez-Berestein G. Liposomal siRNA nanocarriers for cancer therapy. Adv. Drug Deliv. Rev. 2014;66:110–116. doi: 10.1016/j.addr.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Interesting review that overviews the lipid-based approaches with preclinical application in cancer therapy.
- 8.Akinc A, Goldberg M, Qin J, et al. Development of lipidoid-siRNA formulations for systemic delivery to the liver. Mol. Ther. 2009;17(5):872–879. doi: 10.1038/mt.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biswas S, Knipp RJ, Gordon LE, et al. Hydrophobic oxime ethers: a versatile class of pDNA and siRNA transfection lipids. ChemMedChem. 2011;6(11):2063–2069. doi: 10.1002/cmdc.201100259. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Previous study carried out by Nantz group, which underlies the basis of the current work.
- 10.Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat. Mater. 2013;12(11):967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]; •• Excellent review describes various delivery materials for siRNA delivery.
- 11.Landen CN, Jr, Chavez-Reyes A, Bucana C, et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65(15):6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- 12.Miller CR, Bondurant B, McLean SD, McGovern KA, O'Brien DF. Liposome–cell interactions in vitro: effect of liposome surface charge on the binding and endocytosis of conventional and sterically stabilized liposomes. Biochemistry. 1998;37(37):12875–12883. doi: 10.1021/bi980096y. [DOI] [PubMed] [Google Scholar]
- 13.Berchel M, Le GT, Couthon-Gourves H, et al. Lipophosphonate/lipophosphoramidates: a family of synthetic vectors efficient for gene delivery. Biochimie. 2012;94(1):33–41. doi: 10.1016/j.biochi.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 14.Balasubramaniam RP, Bennett MJ, Aberle AM, Malone JG, Nantz MH, Malone RW. Structural and functional analysis of cationic transfection lipids: the hydrophobic domain. Gene Ther. 1996;3(2):163–172. [PubMed] [Google Scholar]
- 15.Rajesh M, Sen J, Srujan M, Mukherjee K, Sreedhar B, Chaudhuri A. Dramatic influence of the orientation of linker between hydrophilic and hydrophobic lipid moiety in liposomal gene delivery. J. Am. Chem. Soc. 2007;129(37):11408–11420. doi: 10.1021/ja0704683. [DOI] [PubMed] [Google Scholar]
- 16.Aberle AM, Bennett MJ, Malone RW, Nantz MH. The counterion influence on cationic lipid-mediated transfection of plasmid DNA. Biochim. Biophys. Acta. 1996;1299(3):281–283. doi: 10.1016/0005-2760(95)00230-8. [DOI] [PubMed] [Google Scholar]
- 17.Reynier P, Briane D, Coudert R, et al. Modifications in the head group and in the spacer of cholesterol-based cationic lipids promote transfection in melanoma B16-F10 cells and tumours. J. Drug Target. 2004;12(1):25–38. doi: 10.1080/10611860410001683040. [DOI] [PubMed] [Google Scholar]
- 18.Lin AJ, Slack NL, Ahmad A, George CX, Samuel CE, Safinya CR. Three-dimensional imaging of lipid gene-carriers: membrane charge density controls universal transfection behavior in lamellar cationic liposome-DNA complexes. Biophys. J. 2003;84(5):3307–3316. doi: 10.1016/S0006-3495(03)70055-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim HS, Moon J, Kim KS, et al. Gene-transferring efficiencies of novel diamino cationic lipids with varied hydrocarbon chains. Bioconjug. Chem. 2004;15(5):1095–1101. doi: 10.1021/bc049934t. [DOI] [PubMed] [Google Scholar]
- 20.Spagnou S, Miller AD, Keller M. Lipidic carriers of siRNA: differences in the formulation, cellular uptake, and delivery with plasmid DNA. Biochemistry. 2004;43(42):13348–13356. doi: 10.1021/bi048950a. [DOI] [PubMed] [Google Scholar]
- 21.Biswas S, Huang X, Badger WR, Nantz MH. Nucleophilic cationization reagents. Tetrahedron Lett. 2010;51(13):1727–1729. doi: 10.1016/j.tetlet.2010.01.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leventis R, Silvius JR. Interactions of mammalian cells with lipid dispersions containing novel metabolizable cationic amphiphiles. Biochim. Biophys. Acta. 1990;1023(1):124–132. doi: 10.1016/0005-2736(90)90017-i. [DOI] [PubMed] [Google Scholar]
- 23.Chen H, Zhang H, McCallum CM, Szoka FC, Guo X. Unsaturated cationic ortho esters for endosome permeation in gene delivery. J. Med. Chem. 2007;50(18):4269–4278. doi: 10.1021/jm060128c. [DOI] [PubMed] [Google Scholar]
- 24.Liu D, Qiao W, Li Z, et al. Structure-function relationship research of glycerol backbone-based cationic lipids for gene delivery. Chem. Biol. Drug Des. 2008;71(4):336–344. doi: 10.1111/j.1747-0285.2008.00644.x. [DOI] [PubMed] [Google Scholar]
- 25.Byk G, Dubertret C, Escriou V, et al. Synthesis, activity, and structure–activity relationship studies of novel cationic lipids for DNA transfer. J. Med. Chem. 1998;41(2):229–235. doi: 10.1021/jm9704964. [DOI] [PubMed] [Google Scholar]
- 26.Savva M, Chen P, Aljaberi A, Selvi B, Spelios M. In vitro lipofection with novel asymmetric series of 1,2-dialkoylamidopropane-based cytofectins containing single symmetric bis-(2-dimethylaminoethane) polar headgroups. Bioconjug. Chem. 2005;16(6):1411–1422. doi: 10.1021/bc050138c. [DOI] [PubMed] [Google Scholar]
- 27.Le Corre SS, Berchel M, Belmadi N, et al. Cationic lipophosphoramidates with two different lipid chains: synthesis and evaluation as gene carriers. Org. Biomol. Chem. 2014;12(9):1463–1474. doi: 10.1039/c3ob42270d. [DOI] [PubMed] [Google Scholar]
- 28.Kolb HC, Finn MG, Sharpless KB. Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. Engl. 2001;40(11):2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 29.Figgitt DP, McClellan KJ. Fluvoxamine. An updated review of its use in the management of adults with anxiety disorders. Drugs. 2000;60(4):925–954. doi: 10.2165/00003495-200060040-00006. [DOI] [PubMed] [Google Scholar]
- 30.Zhong D, Li X, Wang A, Xu Y, Wu S. Identification of the metabolites of roxithromycin in humans. Drug Metab. Dispos. 2000;28(5):552–559. [PubMed] [Google Scholar]
- 31.Carmona S, Jorgensen MR, Kolli S, et al. Controlling HBV replication in vivo by intravenous administration of triggered PEGylated siRNA-nanoparticles. Mol. Pharm. 2009;6(3):706–717. doi: 10.1021/mp800157x. [DOI] [PubMed] [Google Scholar]
- 32.Hattori Y, Ding WX, Maitani Y. Highly efficient cationic hydroxyethylated cholesterol-based nanoparticle-mediated gene transfer in vivo and in vitro in prostate carcinoma PC-3 cells. J. Control. Release. 2007;120(1–2):122–130. doi: 10.1016/j.jconrel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 33.Rose SD, Kim DH, Amarzguioui M, et al. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 2005;33(13):4140–4156. doi: 10.1093/nar/gki732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Afonin KA, Grabow WW, Walker FM, et al. Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nat. Protoc. 2011;6(12):2022–2034. doi: 10.1038/nprot.2011.418. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Shapiro and coworkers are developing RNA architectures of a different genre, which are anticipated to produce significant silencing effects in serum conditions if delivered by oxime-ether lipids.
- 35.Afonin KA, Viard M, Koyfman AY, et al. Multifunctional RNA nanoparticles. Nano. Lett. 2014;14(10):5662–5671. doi: 10.1021/nl502385k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta K, Jang H, Harlen K, et al. Mechanism of membrane permeation induced by synthetic beta-hairpin peptides. Biophys. J. 2013;105(9):2093–2103. doi: 10.1016/j.bpj.2013.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim T, Afonin KA, Viard M, et al. In silico, in vitro, and in vivo studies indicate the potential use of bolaamphiphiles for therapeutic siRNAs delivery. Mol. Ther. Nucleic Acids. 2013;2:e80. doi: 10.1038/mtna.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Molecular dynamic simulation studies from Shapiro and coworkers to understand the interaction between lipid-like materials and siRNA.
- 38.Dan N, Danino D. Structure and kinetics of lipid–nucleic acid complexes. Adv. Colloid Interface Sci. 2014;205:230–239. doi: 10.1016/j.cis.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 39.Zhou S, Anupam R, Hines JV. Fluorescence anisotropy: analysis of tRNA binding to the T box riboswitch antiterminator RNA. Methods Mol. Biol. 2015;1240:143–152. doi: 10.1007/978-1-4939-1896-6_11. [DOI] [PubMed] [Google Scholar]
- 40.Nantz MH, Li L, Zhu J, Aho-Sharon KL, Lim D, Erickson KL. Inductive electron-withdrawal from ammonium ion headgroups of cationic lipids and the influence on DNA transfection. Biochim. Biophys. Acta. 1998;1394(2–3):219–223. doi: 10.1016/s0005-2760(98)00114-3. [DOI] [PubMed] [Google Scholar]
- 41.Nantz MH, Dicus CW, Hilliard B, Yellayi S, Zou S, Hecker JG. The benefit of hydrophobic domain asymmetry on the efficacy of transfection as measured by in vivo imaging. Mol. Pharm. 2010;7(3):786–794. doi: 10.1021/mp900298f. [DOI] [PubMed] [Google Scholar]
- 42.Heyes JA, Niculescu-Duvaz D, Cooper RG, Springer CJ. Synthesis of novel cationic lipids: effect of structural modification on the efficiency of gene transfer. J. Med. Chem. 2002;45(1):99–114. doi: 10.1021/jm010918g. [DOI] [PubMed] [Google Scholar]
- 43.Santel A, Aleku M, Keil O, et al. A novel siRNA-lipoplex technology for RNA interference in the mouse vascular endothelium. Gene Ther. 2006;13(16):1222–1234. doi: 10.1038/sj.gt.3302777. [DOI] [PubMed] [Google Scholar]
- 44.Santel A, Aleku M, Keil O, et al. RNA interference in the mouse vascular endothelium by systemic administration of siRNA-lipoplexes for cancer therapy. Gene Ther. 2006;13(18):1360–1370. doi: 10.1038/sj.gt.3302778. [DOI] [PubMed] [Google Scholar]
- 45.Zhi D, Zhang S, Cui S, Zhao Y, Wang Y, Zhao D. The headgroup evolution of cationic lipids for gene delivery. Bioconjug. Chem. 2013;24(4):487–519. doi: 10.1021/bc300381s. [DOI] [PubMed] [Google Scholar]
- 46.Bennett MJ, Aberle AM, Balasubramaniam RP, Malone JG, Malone RW, Nantz MH. Cationic lipid-mediated gene delivery to murine lung: correlation of lipid hydration with in vivo transfection activity. J. Med. Chem. 1997;40(25):4069–4078. doi: 10.1021/jm970155q. [DOI] [PubMed] [Google Scholar]; • Interesting paper to understand how lipid hydration affects transfection.
- 47.Niculescu-Duvaz D, Heyes J, Springer CJ. Structure–activity relationship in cationic lipid mediated gene transfection. Curr. Med. Chem. 2003;10(14):1233–1261. doi: 10.2174/0929867033457476. [DOI] [PubMed] [Google Scholar]
- 48.Davis ME. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464(7291):1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.