Abstract
Recently we have presented data supporting the notion that PIKfyve not only produces the majority of constitutive phosphatidylinositiol 5-phosphate (PtdIns5P) in mammalian cells but that it does so through direct synthesis from PtdIns. Another group, albeit obtaining similar data, suggests an alternative pathway whereby the low-abundance PtdIns(3,5)P2 undergoes hydrolysis by unidentified 3-phosphatases, thereby serving as a precursor for most of PtdIns5P. Here, we review the experimental evidence supporting constitutive synthesis of PtdIns5P from PtdIns by PIKfyve. We further emphasize that the experiments presented in support of the alternative pathway are also compatible with a direct mechanism for PIKfyve-catalyzed synthesis of PtdIns5P. While agreeing with the authors that constitutive PtdIns5P could theoretically be produced from PtdIns(3,5)P2 by 3-dephosphorylation, we argue that until direct evidence for such an alternative pathway is obtained, we should adhere to the existing experimental evidence and quantitative considerations, which favor direct PIKfyve-catalyzed synthesis for most constitutive PtdIns5P.
Keywords: 3-phosphatases; direct PtdIns5P synthesis; phosphoinositides; PIKfyve; PtdIns(3,5)P2; PtdIns3P; PtdIns(3,5)P2-PtdIns3P conversion
Introduction
All in the family: PI intercommunications
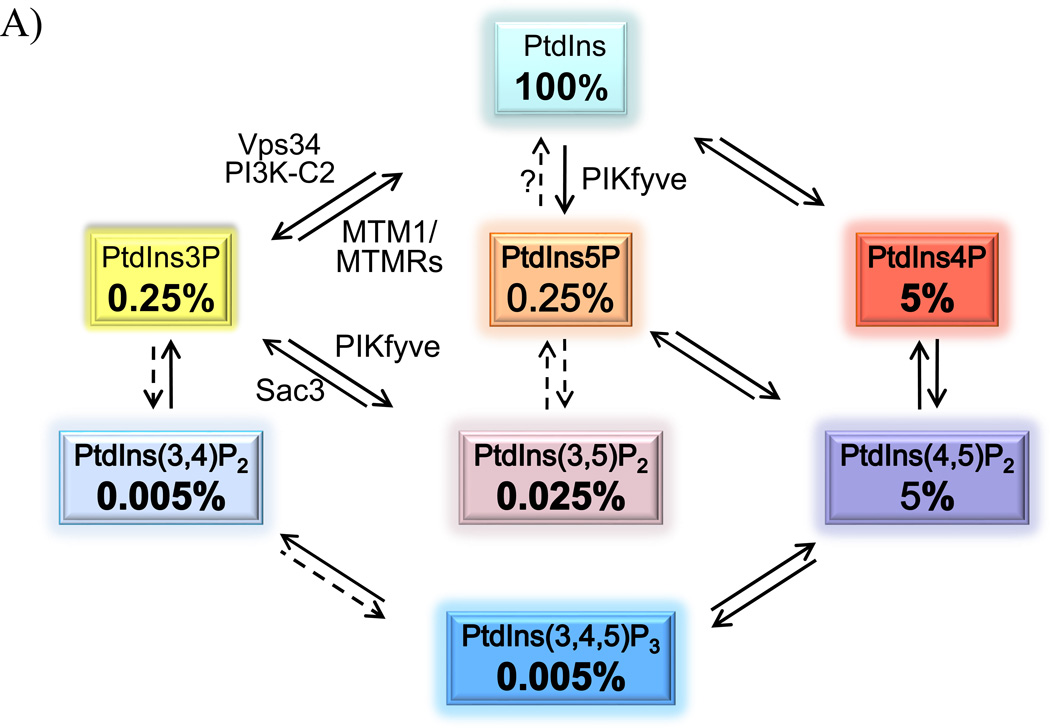
The seven phosphoinositides (PIs) that are detected in mammalian cells are produced directly or indirectly from phosphatidylinositol (PtdIns) through enzymatic reactions involving kinases and phosphatases (Fig. 1A). Because the PIs are subjected to dynamic interconversion by one or more enzymatic activities, it is often challenging to pinpoint the underlying physiological pathways that generate, maintain and curtail the constitutive or stimulated cellular levels of a given PI. Furthermore, with the exception of PtdIns(4,5)P2 and PtdIns4P, in quiescent cells PIs are present in minute amounts (Fig. 1A). This, together with the persistent technical challenges for simultaneous PI detection and quantification, as in the case of PtdIns(3,5)P2 and PtdIns5P, poses further roadblocks preventing firm conclusions about the immediate PI precursor. To further complicate the matter, the substrate specificity of a given PI-metabolizing enzyme determined in a test tube (in vitro) may not necessarily mirror the one the enzyme would have in intact cells (in vivo). Thus, variables such as substrate availability, amounts or mode of presentation, as well as the presence of regulatory factors and stimuli modifying the enzymatic activity, potentially affect the substrate specificity of in vivo enzymatic reactions. Moreover, some PI metabolizing enzymes are promiscuous and act on more than one substrate, adding to the complexity. Understanding of what a PI enzyme does or does not do in mammalian cells is essential from a basic standpoint. More importantly, in light of the accumulated evidence implicating the PI metabolizing enzymes and their products in serious human diseases, including diabetes, cancer and neurological disorders [1–8], precise and accurate determination of the enzyme substrate specificity in vivo has taken a central stage. Such knowledge will undoubtedly help in diversifying and improving treatment strategies.
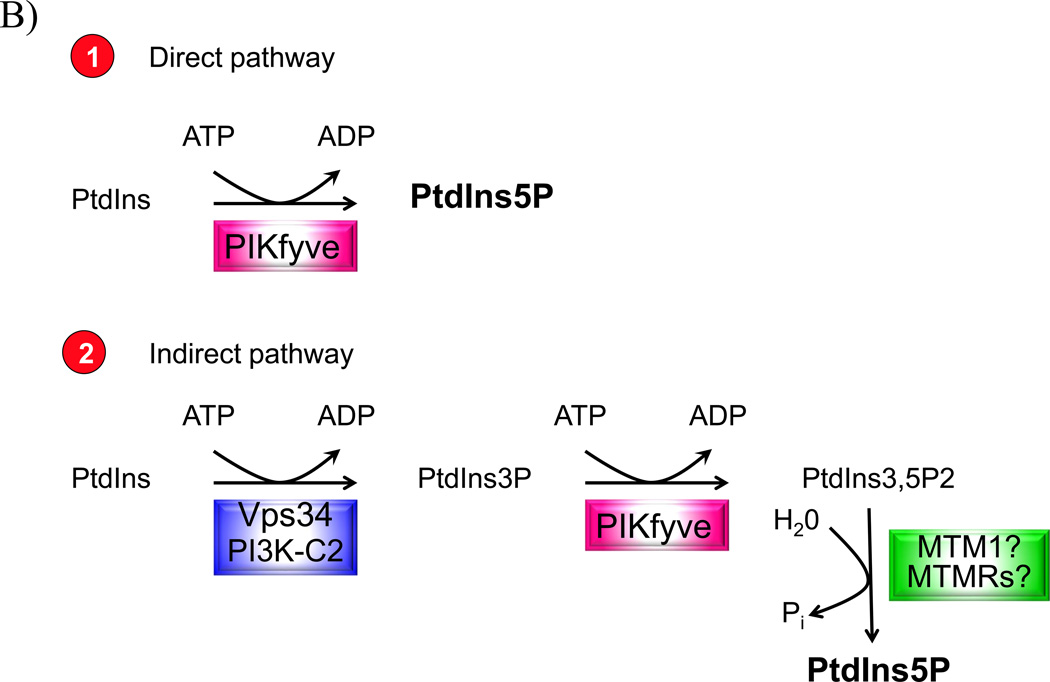
Figure 1.
A: The seven PIs and their relative abundance in resting mammalian cells. The approximate relative abundance is derived from our work [23, 87] and that of others [88,89] in various resting mammalian cells. Indicated are experimentally confirmed cellular pathways involved in PI production (solid arrows). In vitro established conversions are indicated by dashed arrows. Only the enzymes relevant for this discussion are denoted. B: The two pathways for PIKfyve-catalyzed synthesis of constitutive PtdIns5P in resting mammalian cells. The direct pathway (1), for which evidence is detailed in this paper, consumes one mol ATP and uses PtdIns substrate that is in huge excess. The indirect pathway (2), proposed by the authors [32, 36], consumes 2 PIKfymol ATP and incorporates a third reaction of 3-dephosphorylation of the PtdIns(3,5)P2 substrate that in quiescent cells constitutes ~1/10 of the product PtdIns5P. The enzyme(s) involved in the 3-dephosphorylation reaction of PtdIns(3,5)P2 is(are) still unidentified.
PIKfyve making both PtdIns(3,5)P2 and PtdIns5P in a cell context
Redefining and refining the substrate specificity of a PI-metabolizing enzyme is a continuing process going hand in hand with improvement of the detection approaches and development of new biological tools. A distinctive example in this regard is the substrate specificity of the Type II PI 5-Kinases, which – owing to improved PtdIns5P separation from the bulky PtdIns4P by Rameh et al. – was clarified to be for PtdIns5P rather than PtdIns4P, as initially thought [9]. Substrate specificities of Class II PI 3-kinases for PtdIns, PtdIns4P or both under constitutive and stimulated conditions are also in a process of clarification [10–14].
One PI metabolizing enzyme whose substrate specificity is still debated is PIKfyve, an evolutionarily conserved PI5-kinase (EC 2.7.1.150), found in animals, plants and fungi [2]. Following cloning of mouse PIKfyve in 1999 [15], we determined in vitro that both the endogenous and recombinant PIKfyve made PtdIns5P from a PtdIns substrate and PtdIns(3,5)P2 from a PtdIns3P substrate [16, 17]. The identity of the two products generated under these in vitro conditions as well as in cellular contexts was verified by HPLC inositol-head-group analysis [16, 18, 19], the gold standard in PI confirmation. Usage of PIKfyve point mutants, combined with exogenous delivery of individual lipids, allowed the implication of PIKfyve-catalyzed PtdIns(3,5)P2 synthesis in regulating endosomal membrane homeostasis [19, 20]. PIKfyve-catalyzed production of PtdIns5P in the cellular context was also confirmed by the PtdIns5P-mass assay [18], an approach developed in Irvine’s laboratory to circumvent the challenging separation of PtdIns5P from the bulky PtdIns4P by HPLC [21]. These data corroborated our functional observations that PtdIns5P-dependent F-actin breakdown was simulated by ectopic expression of PIKfyve in mammalian cells [22]. Hence, we proposed early on that in mammalian cells PIKfyve is responsible for direct synthesis of both PtdIns(3,5)P2 and PtdIns5P [23], a view that has continued to be supported by subsequent data from our lab and that of others.
Despite this compelling in vitro and in vivo evidence, the majority view about PIKfyve specificity has been centered on PtdIns(3,5)P2 production from PtdIns3P substrate, mainly due to two observations in yeast, made in the late 90s: 1) expression of mouse PIKfyve in yeast mutant (S. cerevisiae) with deleted fab1 (Δfab1), the yeast PIKfyve ortholog, did not yield PtdIns5P yet PtdIns(3,5)P2 was restored as assayed by HPLC [24]; 2) absence of constitutive PtdIns5P occurrence in wild-type yeast [25–27]. Due to this evidence the prevalent opinion in the field was that, like fab1 in yeast, PIKfyve does not produce PtdIns5P in mammalian cells. The source of the relatively large constitutive PtdIns5P pool in mammalian cells remained unresolved, though dephosphorylation of bisphosphorylated PIs, such as PtdIns(4,5)P2 and PtdIns(3,5)P2, gained credibility [28–30]. However, note that steady-state levels of PtdIns(3,5)P2 are ~10 fold lower than those of PtdIns5P (Fig. 1A). Together with the lack of experimental evidence for the quantitative conversion of PtdIns(3,5)P2 to PtdIns5P in resting cells (discussed below), the notion of a PtdIns(3,5)P2 turnover pathway as a main source for constitutive PtdIns5P would seem improbable.
Two sides of the same story
After 2010, new tools for PIKfyve research, such as knockout mouse models and chemical inhibitors, bore the promise of finally clarifying the question of whether constitutive PtdIns5P is synthesized by PIKfyve in mammals. Thus, using our KO mice, the first genetically modified PIKFYVE mouse model, we obtained data unequivocally supporting the conclusion that PIKfyve is indeed responsible for production of both PtdIns(3,5)P2 and PtdIns5P in vivo. Specifically, our observation for similar decreases in steady-state levels of PtdIns(3,5)P2 and PtdIns5P in embryonic fibroblasts derived from heterozygous mice with Pikfyve gene disruption indicated that PIKfyve makes both lipids, as we published in Ikonomov et al. 2011 [31]. Similarly, Zolov et al. 2012 [32] found a simultaneous reduction in PtdIns(3,5)P2 and PtdIns5P in fibroblasts derived from an independent hypomorphic mouse model with a Pikfyve gene trap. Thus, they reached the same conclusion, that is, PIKfyve is responsible for both PtdIns(3,5)P2 and PtdIns5P production. It should be noted that related to PIKfyve research, our team [18, 31, 33, 34] and that of Weisman [32, 35] are the only groups that simultaneously detect and quantify steady-state levels of PtdIns5P and PtdIns(3,5)P2 in mammalian cells by a single HPLC run, due to various technical limitations discussed elsewhere [5, 18, 34]. Quantitative details of these data obtained by the two groups are summarized in Table 1.
Table 1.
Levels of PtdIns(3,5)P2 and PtdIns5P concurrently measured and quantified by HPLC analyses
| Cell Type | Model | PthIns5P % of control |
PthIns(3,5)P2 % of control |
Steady-State PthIns5P:PthIns(3,5)P2 in control |
Protein Levels % of control |
Ref |
|---|---|---|---|---|---|---|
| Genetic manipulation# | ||||||
| MEFs | PikfyveWT/KO | 64 | 61 | 7 | 50 | [31] |
| MFs | Pikfyve−/− | 47a (49b) | 54a (58b) | 11 | 10 | [32] |
| MFs | Pikfyve+/− | 70 | 100 | 8 | 50 | [32] |
| MFs | Vac14−/−/Arpikfyve−/− | 33 | 50 | 7.5 | 100 | [35] |
| Pharmacological Inhibition* | ||||||
| 3T3L1 adipocytes | YM201636, 160 nM | 37 | 71 | 6.8 | 100 | [34] |
| HEK-293 cells | YM201636, 160 nM | 29 | 53 | 5.8 | 100 | [34] |
| CHO-T cells | YM201636, 160 nM | 38 | 70 | 38 | 100 | [34] |
| 3T3L1 adipocytes | L41, 1–20 µM | 82–63 | 100 | 6.7 | 100 | [51] |
In the indicated references, steady-stale levels of PIs are measured under similar cell labeling conditions and eluted by HPLC under acomparable shallow gradient.
Relative lipid levels subsequent to 40 min treatment with indicated PIKfyve inhibitors. MEFs, mouse embryonic fibroblasts; MFs, mouse fibroblasts;
data from Fig. S7 and Fig. 3, respectively.
Furthermore, using these new tools, our laboratory has made additional observations allowing a second important conclusion, i.e., that PIKfyve not only produces PtdIns5P but that it does so by direct synthesis from PtdIns (Fig. 1B). For example, we demonstrated that acute PIKfyve inhibition at low doses of the YM201636 compound rendered intracellular PtdIns5P more severely reduced than PtdIns(3,5)P2 (by ~ 2-fold) in several mammalian cells ([34] and Table 1). Importantly, under these conditions the decrease of PIKfyve-inhibitable PtdIns5P production was quite substantial in all of the tested cell types (71% – 62%), suggesting a significant portion of the basal PtdIns5P being made by PIKfyve [34]. Notably, these observations in cell systems were supported by in vitro kinase assays, demonstrating similar preferential reduction of PtdIns5P vs. PtdIns(3,5)P2 at low concentrations of the inhibitor [34]. These results coupled with: i. profoundly greater steady-state levels of PtdIns5P compared to that of PtdIns(3,5)P2 (Table 1); ii. direct PIKfyve-catalyzed synthesis in vitro of both products [16, 17]; iii. distinct roles of two lysines from the PIKfyve catalytic domain in PtdIns(3,5)P2 and PtdIns5P production [20] (detailed below), strongly support direct PIKfyve-catalyzed synthesis of PtdIns5P from PtdIns as the principal physiological pathway for constitutive PtdIns5P [34] (Fig. 1B).
Work by Weisman’s team also provided experimental evidence in support of this conclusion. For example, steady-state levels of PtdIns5P decreased preferentially to PtdIns(3,5)P2 in fibroblasts derived from their Pikfyve gene trap mouse model [32] (Table 1). Their data in fibroblasts derived from mice with disruption of the vac14 gene, whose protein product ArPIKfyve increases PIKfyve activity [2], also show preferentially reduced steady-state levels of PtdIns5P vs. PtdIns(3,5)P2 [35].
However, despite the data and quantitative considerations outlined above, a recent review article in BioEssays by McCartney et al. [36] promotes an alternative pathway whereby the low-abundance PtdIns(3,5)P2 serves as a precursor for synthesis of an-order-of-magnitude-greater amounts of PtdIns5P (subtitle: “PI(3,5)P2 is a precursor for PI5P synthesis”) (Fig. 1B). This conclusion rests on data from their original study published under a similarly emphatic title “In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P” [32]. Below, we review specific experimental approaches both by us and others in support of the direct route, which, together with data in Table 1, must be taken into consideration. We also critically assess recent data provided in support of PtdIns(3,5)P2 as a precursor for most of constitutive PtdIns5P [32, 36] and demonstrate that this evidence is also completely compatible with direct synthesis of PtdIns5P by PIKfyve.
PtdIns3P-PtdIns(3,5)P2 cycling: Handcuffed by a kinase-phosphatase complex
The natural occurrence of PtdIns(3,5)P2 in quiescent mammalian cells and the direction of PtdIns(3,5)P2 synthesis and turnover was first reported by Ulug’s team in 1997. This study [37] not only formally identified the predicted PtdIns(3,5)P2 species in resting mammalian cells [38] but also determined the metabolic route of PtdIns(3,5)P2 synthesis from PtdIns3P. By monitoring the time-course of the specific radioactivity associated with the individual 3 and 5 positions in PtdIns(3,5)P2 in pulse-labeled mouse fibroblasts, the authors convincingly demonstrated that the phosphate addition at position 5 occurs subsequent to phosphorylation of position 3. In the same year, this pathway was also shown to operate in yeast [39]. Importantly, Ulug’s team went a step further to clarify that PtdIns(3,5)P2 is precipitously converted to PtdIns3P rather than to PtdIns5P. Note that in this study the authors standardized their reaction products to a PtdIns5P standard, well separated from the PtdIns4P peak by HPLC [37]. Based on the robustness of the PtdIns3P-PtdIns(3,5)P2 cycling, Ulug’s group envisioned a concerted regulation of this interconversion by a 5-kinase and a 5-phosphatase [37]. Both mammalian enzymes were identified 2 and 10 years later, respectively [15, 40]. Time has also proven correct the rapid PtdIns3P-PtdIns(3,5)P2 cycling predicted in the study [37]. Thus, subsequent to our identification of the PI 5-kinase PIKfyve and the PI 5-phosphatase Sac3 (EC 3.1.3) as the responsible mammalian kinase and phosphatase, respectively, we found that the two enzymes, aided by a scaffolding protein ArPIKfyve, associate in a stable signaling complex [40] (Fig. 2). A similar arrangement is confirmed with the yeast orthologous proteins [41, 42]. The association of two enzymes with opposing activities in a single complex underscores PtdIns(3,5)P2 turnover to PtdIns3P and the robustness of the PtdIns(3,5)P2-PtdIns3P interconversion.
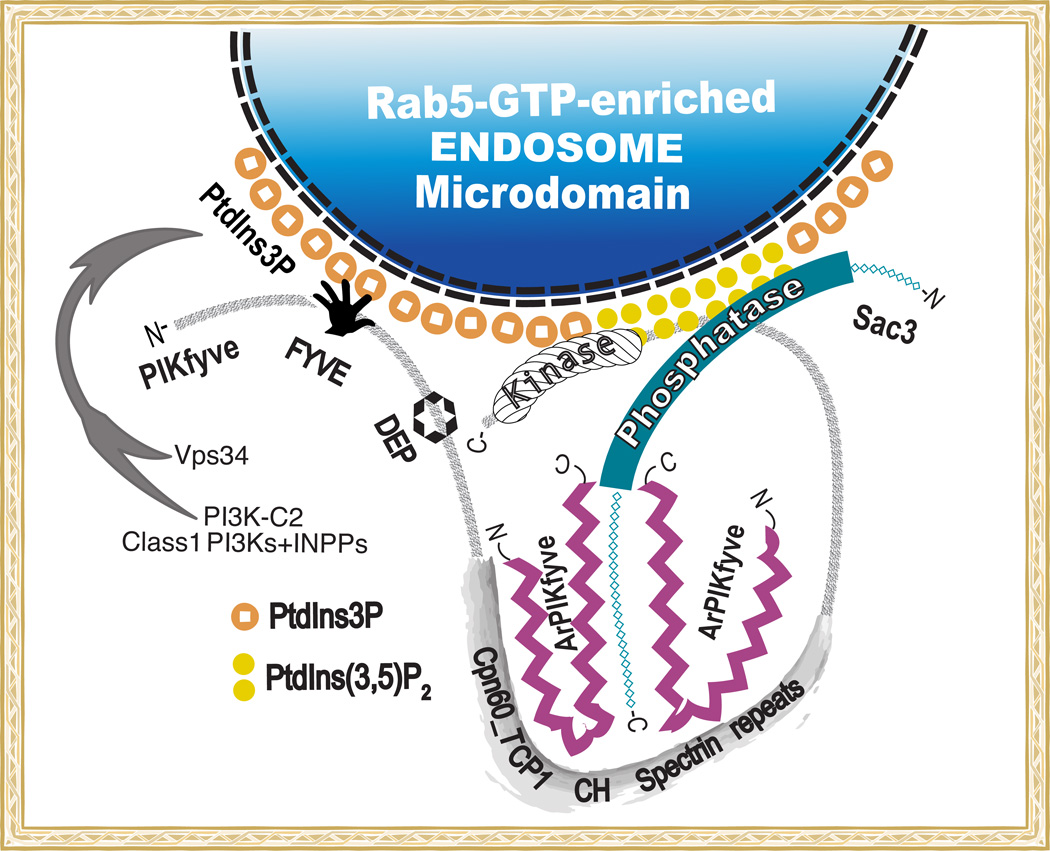
Figure 2.
Schematic model of PIKfyve association with ArPIKfyve and Sac3 in the PAS complex. The PtdIns3P - PtdIns(3,5)P2 cycling on endosomal membrane microdomains is achieved through a stable protein complex comprising PI 5-kinase PIKfyve, PI 5-phosphatase Sac3 and dimerized scaffolding regulator ArPIKfyve [40]. The complex attaches to RabGTP-early endosome platforms, enriched in PtdIns3P [90], presumed to be generated mainly by Vps34, with a possible contribution by the other two PI3K classes [11, 83, 84]. Indicated are the conserved domains in PIKfyve: FYVE, DEP, Cpn60_TCP1, CH homology/Spectrin repeats and lipid kinase homology. The two antagonistic enzymes act in concert to regulate PtdIns3P-PtdIns(3,5)P2 cycling. Whereas ArPIKfyve and Sac3 are clearly responsible for PIKfyve-dependent synthesis of PtdIns5P from PtdIns, whether this occurs in parallel with PtdIns(3,5)P2 synthesis on endosomal membranes is unknown. Interaction mapping details are from [49, 50].
Purified PIKfyve: Makes two products, both reduced by PIKfyve inhibition
Challenging the occurrence of direct synthesis of constitutive PtdIns5P by PIKfyve in a cellular context, the authors questioned our measurements of direct PtdIns5P synthesis by PIKfyve in in vitro assays [36]. Their criticism is centered on possible hydrolysis of PtdIns(3,5)P2 to PtdIns5P by 3-phosphatases that could co-immunoprecipitate with PIKfyve from mammalian cell lysates. They also discussed a presumed controversy of our in vitro data with that by another group.
Related to the first argument, a GST-purified recombinant PIKfyve produced by a baculovirus expression system and used in parallel with immunopurified PIKfyve from mammalian cell lysates yields, similarly, production of both PtdIns(3,5)P2 and PtdIns5P in vitro [17, 43]. Whereas in either expression system one cannot rule out a putative 3-phosphatase pulldown, note that none has been reported to date. Moreover, washing the PIKfyve immunoprecipates derived from mammalian cell lysates with SDS (0.05%), a step that strips even the stably associated proteins in the PAS complex (Fig. 2), also yields equal generation of the two products [44]. Perhaps more importantly, if 3-phosphatases were co-immunopurified with PIKfyve from mammalian cell lysates, hydrolysis to PtdIns5P ought to also occur when PtdIns(3,5)P2 is made from the PtdIns3P substrate. As this is repeatedly not the case [16, 43, 45], the claim by the authors for presumed PtdIns5P in vitro production by 3-phosphatases, pulled down from mammalian cell lysates [36], lacks experimental support. Clearly, these and other similar in vitro data by our lab [16, 17, 20, 31, 43–50] demonstrably show that the two products PtdIns(3,5)P2 and PtdIns5P are made in parallel by PIKfyve, unrelated to whether the endogenous or recombinant enzyme is purified from insect or various mammalian cell lysates.
Purified recombinant HA-PIKfyve produced by an adenoviral expression system in mammalian cells [18, 19, 47] also makes both PtdIns5P and PtdIns(3,5)P2 in vitro (Fig. 3A). Under these conditions the expressed protein is in vastly greater amounts vs. endogenous PIKfyve (>30-fold) [47], making the claimed PtdIns(3,5)P2 hydrolysis by putative pulled down endogenous 3-phosphatases [32, 36], also improbable. Most importantly, acute chemical inhibition of such adenovirally expressed PIKfyve by YM201636 or L41 results in arrested in vitro synthesis of both PtdIns(3,5)P2 and PtdIns5P, as we have reported for this [51] or another enzyme source [34, 52] (Fig. 3A). Thus, our data unambiguously demonstrate a YM201636- or L41-dependent arrest of both PtdIns5P and PtdIns(3,5)P2 syntheses in vitro due to direct PIKfyve inhibition, calling attention to YM201636 usage in support of the indirect pathway [32, 36] (discussed further).
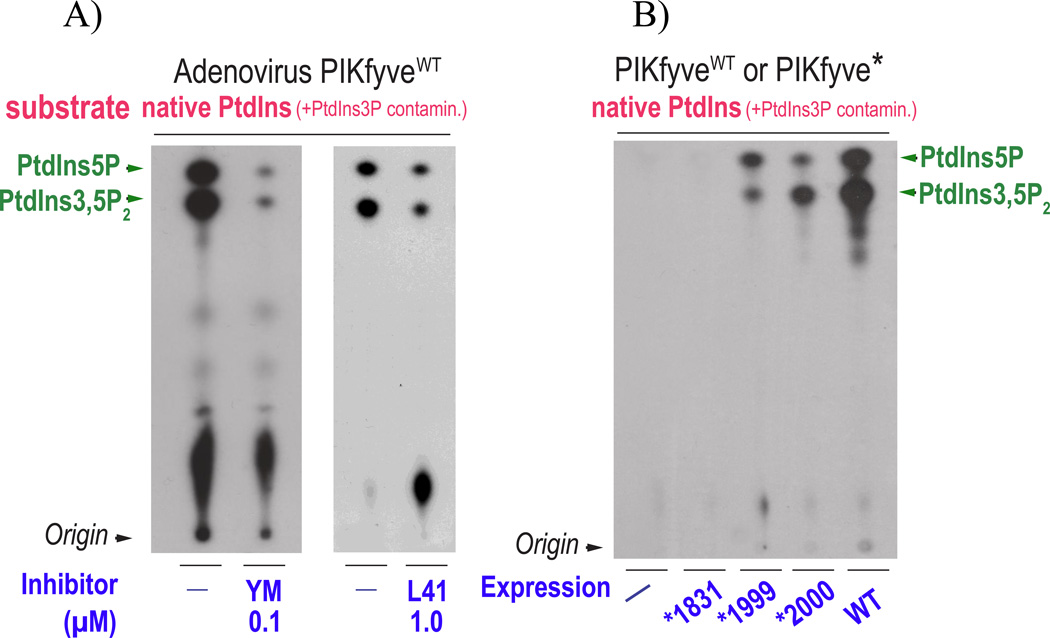
Figure 3.
In vitro production of PtdIns(3,5)P2 and PtdIns5P is equally inhibited by PIKfyve inhibitors but differentially affected by PIKfyve mutagenesis. A: HEK293 cells transduced with adenoviral vector encoding HA-PIKfyveWT were lysed in RIPA buffer (containing two detergents, 1% NP-40 and 0.5% Na deoxycholate) and immunoprecipitated with PIKfyve antibodies. PIKfyve protein expressed from adenoviral vector is >30-fold higher than the endogenous, making specific co-immunoprecipitation of potentially PIKfyve-associated myotubularins unlikely. Immunoprecipitates adsorbed onto Protein A Sepharose beads, extensively washed (>10 times) with RIPA buffer and high-salt buffers, were preincubated for 15 min with a DMSO solvent (−) or with DMSO-dissolved inhibitors at concentrations 0.1 µM of YM201636 or 1 µM of L41 (a novel PIKfyve inhibitor, [51]). The in vitro lipid kinase assay was carried out for 15 min at 30° C in the presence of 12.5 µCi of [γ-32P]ATP (50 µM), 100 µM freshly sonicated PtdIns substrate (natural form soybean, Avanti Polar Lipids, containing small amounts of natural PtdIns3P as a contaminant) and an assay buffer [25 mM Hepes, pH 7.5, 120 mM NaCl, 2.5 mM MnCl2, 2.5 mM MgCl2, 5 mM β–glycerophosphate (phosphatase inhibitor) and 1mM dithiothreitol [20]. This ionic composition prevents any generation of PtdIns3P during the assay as we described elsewhere [46, 52]. Lipids were resolved by TLC using an acidic solvent system [65:35 (v/v) 1-propanol:2 M acetic acid]. Indicated are PtdIns(3,5)P2 and PtdIns5P products confirmed by HPLC analyses of the radioactive spots. Apparent is similar reduction of the two products consistent with their direct production by and inhibition of PIKfyve during the in vitro reaction. Data are reported in Refs. [34, 51, 52]. B: Parallel dishes of COS cells, transiently transfected with wild-type (WT) and point mutants (*) of HA-PIKfyve in pCMV5 expression vector, or with an empty vector (−), were lysed in RIPA buffer, followed by immunoprecipitation with HA antibodies and lipid separation by TLC, as described in A. Whereas production of both lipids is perturbed by mutagenesis within the predicted PIKfyve substrate-binding activation loop, apparent is greater reduction of PtdIns5P compared to PtdIns(3,5)P2 with PIKfyveK2000E and, vice versa, greater reduction of PtdIns(3,5)P2 compared to PtdIns5P, with PIKfyveK1999E. Note that under identical immunoprecipitation conditions, synthesis of the two lipids is selectively affected, nullifying hydrolysis by hypothetical 3-phosphatases underlying the observed differences. These data support direct synthesis of both PtdIns(3,5)P2 and PtdIns5P by PIKfyve. No production of either lipid is seen by the PIKfyveK1831E mutant that harbors mutation in the predicted ATP-binding lysine [17]. Modified from [20].
Regarding the potential discrepancy of our data with the in vitro activity of PIKfyve expressed from a yeast plasmid - as documented in [24], under these conditions PIKfyve also produces in vitro mono-phosphorylated PI from a PtdIns substrate. Whereas this product has migration characteristics similar to PtdIns5P by TLC resolution, validation by HPLC has not been performed [24].
Two distinct lysines in the PIKfyve catalytic core define divergent enzymatic activity
If PIKfyve makes directly both PtdIns(3,5)P2 and PtdIns5P, a legitimate question then is the mechanism for this dual lipid substrate specificity. In the absence of structural information for the PIKfyve catalytic domain and facing the obstacle that even large peptides (~370 residues) encompassing the entire catalytic domain are devoid of enzymatic activity [17, 50, 53], the molecular basis of the two distinct PIKfyve lipid kinase activities is difficult to discern. We propose a potential mechanism whereby different residues in the PIKfyve catalytic domain define distinct substrate binding by selectively ligating the 3-phosphate and/or 1-phosphate of the inositol-head-groups in PtdIns3P and PtdIns, respectively.
Corroborating this prediction, targeted mutagenesis in full-length PIKfyve reveals that Lys2000 within the predicted PIKfyve substrate-binding pocket supports preferentially PtdIns5P vs. PtdIns(3,5)P2 synthesis in vitro, whereas Lys1999 shows preferences in the opposite direction [20] (Fig. 3B). The two point mutants also exhibit different morphological phenotypes upon cell expression (Fig. 4). Thus, only the PIKfyveK1999E mutant that exhibits greatly perturbed PtdIns(3,5)P2 synthesis provokes the characteristic cell vacuolation phenotype, readily rescued upon exogenous delivery of PtdIns(3,5)P2 but not of PtdIns5P [20]. Accordingly, Lys1999 but not Lys2000 specifically interacts with PtdIns3P-containing liposomes in in vitro binding assays utilizing PIKfyve peptides of the entire catalytic domain [54]. Our data showing lack of measurable specific binding between the PIKfyve catalytic domain peptide and PtdIns-containing liposome [54] are consistent with higher Km of PIKfyve (low affinity) for the PtdIns substrate compared to the PtdIns3P substrate. This is corroborated by multiple in vitro activity assays with either endogenous or wild-type PIKfyve [16, 17, 20, 31, 43–50], documenting equal or even preferential synthesis of PtdIns(3,5)P2 vs. PtdIns5P. Note that this is despite the greater amounts of PtdIns over PtdIns3P, present as a contaminant in PtdIns substrate preparations (Fig. 3). The ample cellular amounts of PtdIns substrate (Fig. 1A) together with relatively weaker affinity of PIKfyve for PtdIns compared to PtdIns3P (Fig. 3) are consistent with a more gradual rate of PtdIns5P appearance compared to PtdIns(3,5)P2, as observed under nutrient refeeding of starved cells [32, 36] (detailed further).
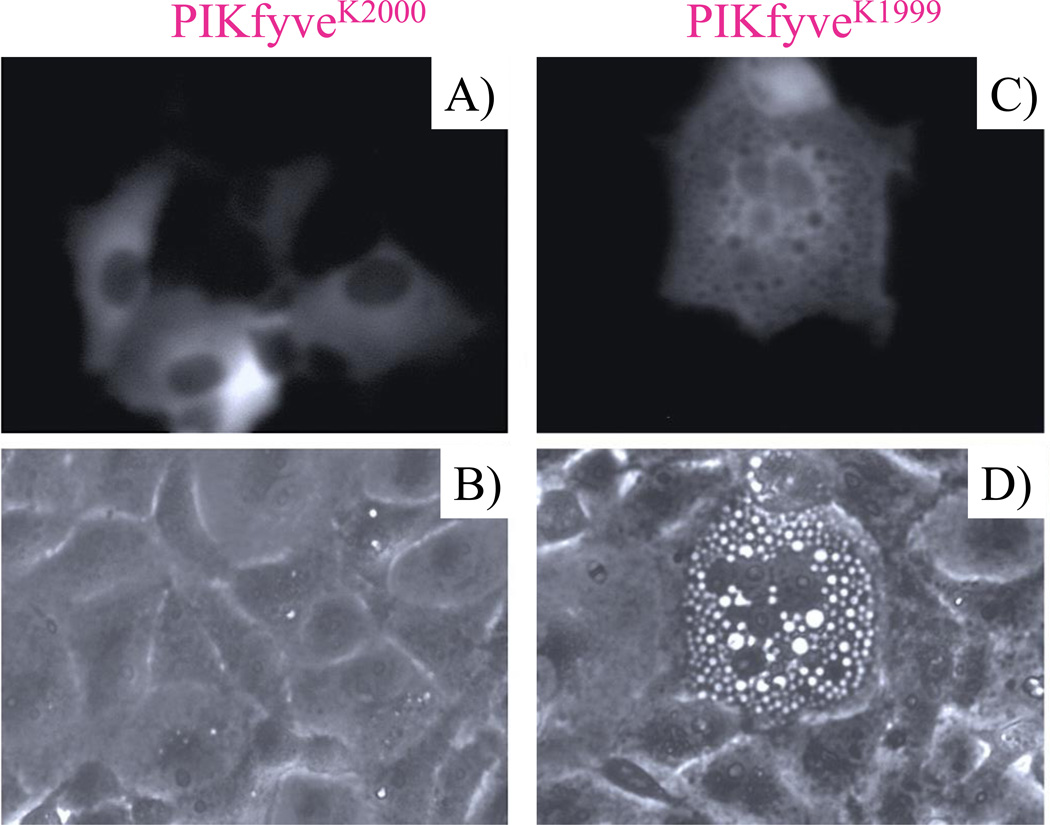
Figure 4.
Differential effect between PIKfyveK1999 and PIKfyveK2000 on cell morphology. A,C: COS cells transiently transfected with pEGFP-PIKfyveK2000E and pEGFP-PIKfyveK1999E were observed by fluorescence microscopy based on GFP fluorescence signals. PIKfyveK1999E mutant, exhibiting greatly perturbed PtdIns(3,5)P2 synthesis (shown in Fig. 3B) and binding to PtdIns3P-enriched liposomes vs. wild-type, examined by in vitro lipid kinase activity and liposome-binding assays [54], respectively, provokes the characteristic aberrant phenotype in the form of cytoplasmic vacuolation. The PIKfyveK2000E mutant, with preferentially impaired PtdIns5P synthesis vs. PtdIns(3,5)P2 (shown in Fig. 3B) and intact binding to PtdIns3P-enriched liposomes [54], does not trigger an aberrant phenotype. B,D: phase-contrast images of A and C, respectively. Modified from [20].
Myotubularins in constitutive PtdIns5P from PtdIns(3,5)P2 – Where is the in vivo evidence?
The direct and indirect models are not mutually exclusive. It is thus theoretically possible that they may be concurrent depending on the cell status. However, the claim that the indirect route is responsible for most of the constitutive PtdIns5P in quiescent cells [36] lacks experimental support. Specifically, the enzyme(s) performing the hydrolysis of the 3-phosphate in PtdIns(3,5)P2 to produce PtdIns5P (Fig. 1B) under basal conditions is/are unknown. The authors [36] assume that one or more members of the family of myotubularin 3-phosphatases (MTM1/MTMRs) account for this step (Fig. 1B). Indeed, in in vitro assays or when heterologously expressed in osmotically stressed yeast cells, most MTM1/MTMRs hydrolyze elevated amounts of PtdIns(3,5)P2 along with PtdIns3P (reviewed in [43, 55]). However, any proof of the physiological occurrence of PtdIns(3,5)P2-to-PtdIns5P hydrolysis by MTM1/MTMRs in resting mammalian cells is thus far unavailable. In fact, the vast majority of research efforts in different species (yeast, C. elegans, Drosophila and mammals) are in favor of PtdIns3P being the physiological substrate for myotubularin-dependent hydrolysis [27, 56–69]. A recent study in human cells [70] is cited by the authors [36] in support of their claim for MTMR-dependent hydrolysis of PtdIns(3,5)P2-to-PtdIns5P. This study, however, shows unaltered constitutive amounts of PtdIns5P upon protein depletion of MTMR3, the 3-phosphatase proposed to functionally crosstalk with PIKfyve (Suppl. Fig. S5 in [70]). Concordantly, MTMR3 protein depletion slightly abrogates the fibroblast-growth-factor1-stimulated increment in PtdIns5P, but again, steady-state levels of PtdIns5P are unaltered [70]). Thus, the reduced PtdIns5P levels in growth-factor-stimulated cells could be extrapolated to neither constitutive PtdIns5P nor the precursor function of PtdIns(3,5)P2, as the latter remained below detectable levels in both quiescent and stimulated cells [70]. It is likely that the PtdIns(3,5)P2 pool, when elevated due to cell stimulation, is turned over to PtdIns5P by MTMRs as reviewed in [43]. However, levels of constitutive PtdIns5P are most likely to be maintained through direct synthesis by PIKfyve (Fig. 1B).
In support of the claim for MTMR-dependent hydrolysis of PtdIns(3,5)P2, the authors [36] point out another evidence in fibroblasts from MTMR2 KO mice [71]. However, PtdIns(3,5)P2 - PtdIns5P conversion is not quantitatively supported in this study as the two lipids are assayed by different experimental approaches and in different cell types [71]. Moreover, PtdIns(3,5)P2 levels remained reduced in fibroblasts from mice with double KO of FIG4/Sac3 and MTMR2 [71], whereas elevation would be expected if MTMR2 dephosphorylates PtdIns(3,5)P2. Furthermore, it should also be considered that the PtdIns3P rise, observed in models with perturbed MTM1/MTMRs and ymr1 3-phosphatase activities [8, 27, 56, 57, 59, 61–63, 65, 67, 69] might result, by default, in elevated PtdIns(3,5)P2 through new PIKfyve-dependent synthesis from the increased PtdIns3P substrate. Finally, absence of statistical correlation between steady-state levels of PtdIns(3,5)P2 [or PtdIns(4,5)P2] and PtdIns5P in several cell types also implies PtdIns5P synthesis from PtdIns, as suggested by us and others [34, 72, 73]. Clearly, to date there exists no compelling in vivo evidence in mammalian cells for the role of myotubularin 3-phosphatase activity in producing most of constitutive PtdIns5P by PtdIns(3,5)P2 hydrolysis, hence questioning the proposed precursor-product relationship.
Considering limitations to free diffusion of the membrane-embedded PIs, McCartney et al. [36] further postulated that MTMRs reside within the PAS complex. However, no experimental evidence in support of plausible association between any member of the myotubularin family and PIKfyve (alone or associated in the PAS complex) has been provided to date. The PtdIns3P-PtdIns(3,5)P2 cycling channeled within the PAS complex (Fig. 2) might also pose limitations on the accessibility of PtdIns(3,5)P2 for myotubulrarin-dependent hydrolyses. Reportedly, many myotubularins are localized away from endosomes [74], providing further limitations. Strikingly, even when myotubularins localize to the endosomal system, as shown in A431 cells for MTM1 and MTMR2, their protein depletion does not affect the PtdIns(3,5)P2 pool yet PtdIns3P is greatly increased [59]. Together, these data and considerations indicate very significant uncertainties in assigning a physiological role of MTM1/MTMRs in constitutive PtdIns5P production from PtdIns(3,5)P2.
Evidence for direct synthesis of constitutive PtdIns5P
The above discussion indicates that PIKfyve’s implication in direct synthesis of constitutive PtdIns5P (Fig. 1A) is based on numerous observations by various in vivo and in vitro approaches. For the sake of clarity, we summarize the above-discussed evidence supporting this conclusion:
In vitro synthesis of PtdIns(3,5)P2 and PtdIns5P by purified PIKfyve preparations [16,17,20,31,43–50].
In vivo synthesis of both PtdIns(3,5)P2 and PtdIns5P upon heterologous expression of PIKfyve in several cell types [18, 19].
In vitro production of PtdIns(3,5)P2 and PtdIns5P is equally reduced upon PIKfyve inhibition at inhibitor doses above the in vitro IC50 (Fig. 3A and [34, 52].
MEFs derived from heterozygous PIKfyve KO mice show similar reduction of PtdIns(3,5)P2 and PtdIns5P despite ~10-fold lower steady-state levels of PtdIns(3,5)P2 compared to PtdIns5P (Table 1 and [31]).
Preferential reduction of PtdIns5P vs. PtdIns(3,5)P2 in different cell types upon PIKfyve inhibition at inhibitor doses below the in vivo IC50 (Table 1 and [34]). In vitro assays show similar preferential reduction of PtdIns5P vs. PtdIns(3,5)P2 at inhibitor doses below the in vitro IC50 [34].
Preferential diminution in steady-state levels of PtdIns5P vs. PtdIns(3,5)P2 in fibroblasts from mice with Pikfyve or Arpikfyve/Vac14 gene trap (Table 1 and [32, 35]).
Distinct lysines from the predicted PIKfyve substrate-binding activation loop are engaged in in vitro production of PtdIns(3,5)P2 and PtdIns5P (Fig. 3B and [20]). Concordantly, only the mutant with markedly defective PtdIns(3,5)P2 synthesis triggers the typical aberrant cell vacuolation (Fig. 4 and [20]).
Functional correlation of PIKfyve ectopic expression with PtdIns5P-dependent F-actin breakdown and, conversely, abrogation of induced F-actin disassembly by PIKfyve activity inhibition in CHO cells [22, 34].
Established direction of PtdIns(3,5)P2 turnover to PtdIns3P rather than to PtdIns5P [37]. Concordantly, PIKfyve couples to a 5-phosphatase rather than a 3-phosphatase (Fig. 2 and [40, 49]).
Unproven physiological role of myotubularins in maintaining constitutive PtdIns5P levels by hydrolysis of PtdIns(3,5)P2 in mammalian cells.
The combined evidence indicates that in resting mammalian cells PIKfyve directly generates PtdIns5P from PtdIns, along with PtdIns(3,5)P2 from PtdIns3P substrates, being responsible for most of constitutive PtdIns5P.
Evidence supporting the indirect pathway is also compatible with direct synthesis of PtdIns5P by PIKfyve
The review article [36] did not consider the observations discussed above. Instead, the authors [36] support their claim for a PtdIns(3,5)P2 precursor-PtdIns5P product relationship by new data from three sets of experiments in yeast or mammalian cells. Because the authors state that observations from heterologous expression of PIKfyve in yeast serves “as the strongest evidence for this hypothesis” [36], it is important that this evidence be carefully examined.
From yeast
The authors [36] assert that if direct synthesis of PtdIns5P from PtdIns by PIKfyve occurs, then the combined total of PtdIns3P, Ptdins5P and PtdIns(3,5)P2 upon heterologous expression of PIKfyve in yeast would rise over the control due to new conversion of PtdIns to PtdIns5P. In fact, this is the authors’ observation (Fig. 4C in the original study [32]). Specifically, the sum of individual numbers for the three lipids is greater in the condition with PIKfyve expression by an amount that is commensurate with the newly generated PtdIns5P (Box 1). Whereas we consider that the small statistically non-significant difference is due to variation of samples, we would like to point out that the one or the other model could be explained by only ~2% difference (the amount of newly produced PtdIns5P as % from the sum of the three lipids, Box 1). Given the sample size and the variation of samples, the study [32] lacks sufficient statistical power necessary to discern such a small difference. Clearly, the yeast study cannot selectively support the hypothesis of PtdIns(3,5)P2 serving as a precursor for PtdIns5P and, thus, does not disprove the direct pathway.
Box 1. Data in support of the indirect pathway are also compatible with direct synthesis of PtdIns5P by PIKfyve.
Yeast study:
Human PIKfyve expression in yeast*
| Condition | PtdIns3P % of total PI |
PtdIns(3,5)P2 % of total PI |
PtdIns5P % of total PI |
Σ [3P + (3,5)P2 + 5P] % of total PI |
|---|---|---|---|---|
| Control (Con) | 2.93 | 0.15 | 0.05 | 3.13 |
| PIKfyve | 2.35 | 0.75 | 0.11 | 3.21 |
| Difference, Δ | (−) 0.58 | (+) 0.6 | (+) 0.06 | (+) 0.08 |
| Δ(3,5)P2 + Δ5P = 0.66 | ||||
Data are derived from Fig. 4C and D in [32].
If PtdIns5P is made from PtdIns(3,5)P2, Eq. 1 and Eq. 2 are to be seen:
[3P + (3,5)P2 + 5P]PIKfyve = [3P + (3,5)P2 + 5P]con
[(3,5)P2 PIKfyve − (3,5)P2 con] + (5PPIKfyve − 5Pcon) = [(3Pcon) − (3PPIKfyve)]
Data from [32] show that in both cases there is inequality with a difference of ~0.08% (non-significant).
[3P + (3,5)P2 + 5P]PIKfyve > [3P + (3,5)P2 + 5P]con; (3.21%>3.13%);
[(3,5)P2 PIKfyve − (3,5)P2 con] + (5PPIKfyve − 5Pcon) > [(3Pcon) − (3PPIKfyve)]; 0.66%>0.58%
Note that this difference is commensurate to the newly formed PtdIns5P (~0.06%) due to PIKfyve expression. Therefore, this evidence remains compatible with PIKfyve direct synthesis of PtdIns5P.
Mammalian cell study: As rates of turnover and replenishment of PtdIns5P are unknown under the conditions of PIKfyve inhibition with YM203616 [32], one could simplify the system and evaluate the absolute molar changes of PtdIns(3,5)P2 and PtdIns5P during the initial time interval 0–2.5 min. Even if we assume that PtdIns(3,5)P2 is fully turned over to only PtdIns5P (not to PtdIns3P; Ref. 37), and PIKfyve does not directly inhibit PtdIns5P production [34,52], the molar reduction in PtdIns(3,5)P2 is 4-fold less than that of PtdIns5P during the initial time interval 0–2.5 min* and, thus, insufficient to account for the molar difference in PtdIns5P during this time interval.
In this simplified system, if PtdIns(3,5)P2 is a precursor for PtdIns5P as claimed by the authors [32,36], the following case should be seen:
([At0] − [At2.5]) ≥ ([Bt0] − [Bt2.5]), where A is PtdIns(3,5)P2 and B is PtdIns5P.
Data in Ref. 32 show:
([At0] − [At2.5]) << ([Bt0] − [Bt2.5]). t0 and t2.5 are the 0 min and the 2.5 min time, respectively.
The yeast experiment by the authors and reports by others allow important inferences about this model system related to PIKfyve research. Thus, the data that heterologous expression of PIKfyve in yeast yields a ~6-fold increase of PtdIns(3,5)P2, whereas PtdIns5P rises only ~2-fold [32], could suggest that yeast is a poor model system for the ATP-dependent anabolic route of PtdIns5P production from PtdIns by PIKfyve as well as by Fab1. The inability of wild-type yeast to make any, or negligible, amounts of PtdIns5P [25, 27, 32, 71, 75] may indicate that crucial protein adaptor(s)/regulator(s) are absent in this system. This conclusion is actually in agreement with complementation analysis of PIKfyve in yeast Δfab1 mutant, where no detectable PtdIns5P could be observed while PtdIns(3,5)P2 was elevated back to normal [24]. It is indeed surprising that overexpression of PIKfyve could even make PtdIns(3,5)P2 in wild-type yeast [36] given that overexpression of its own Fab1 was ineffective [25], evidently, due to the unavailability of regulatory proteins for Fab1-complex formation. This also underscores the critical requirement for the presence of regulatory protein machinery at yet to-be-determined stoichiometric ratios of the constituents in order to form a productive complex. The observations by the authors in yeast [32, 36] corroborate the notion that PtdIns5P synthesis by heterologously expressed PIKfyve is more sensitive to the absence of such regulatory signals than PtdIns(3,5)P2 synthesis. Absence of increased PtdIns(3,5)P2 upon Fab1 overexpression in yeast [25] does not refute PtdIns(3,5)P2 synthesis by Fab1. In the same vein, weak PtdIns5P synthesis by PIKfyve expression in yeast [32, 36] cannot refute PIKfyve’s ability to make PtdIns5P in mammalian cells directly. Moreover, several laboratories employing different approaches affecting PIKfyve protein levels and/or activity have seen changes in constitutive PtdIns5P levels in mammalian cells, as discussed above (Table 1 and [18, 70, 76, 77]). This analysis further reinforces the fact that the two orthologous kinases PIKfyve and Fab1, in addition to similarities, have many differences including substrate specificity, requirement for regulatory partners, behavior to inhibitors etc. [2, 78]. Together, these data and considerations suggest that the small PtdIns5P elevation seen in yeast is compatible with direct synthesis from PtdIns by heterologously expressed PIKfyve.
From mammalian cells
To further support the claim for precursor-product relationship between PtdIns(3,5)P2 and PtdIns5P, the authors provided additional evidence from two experiments in mammalian cells [32,36]. However, whereas these data may not contradict their hypothesis they do not prove it either. In fact, as with the study in yeast, the evidence derived from these indirect experimental approaches remains compatible with direct synthesis of PtdIns5P and PtdIns(3,5)P2 by PIKfyve.
Specifically, in the first approach, the authors employed the PIKfyve inhibitor YM201636, which not only reduces PtdIns(3,5)P2 as originally reported [78] but also inhibits synthesis of PtdIns5P as observed both in vitro (Fig. 3A) and in cellular contexts [34, 52, 77]. The authors also observe reduction of both products at a high dose of YM201636, and show that the disappearance of PtdIns(3,5)P2 and PtdIns5P occurs at a different rate [32]. The straightforward inference from this experiment is that the two lipids have different turnover rates subsequent to concurrent synthesis arrest by PIKfyve inhibition. As each PI has distinct enzymes catalyzing different directions of conversion (Fig. 1A), the different turnover rates are expected. Because the authors dispute PIKfyve making directly PtdIns5P [32, 36] and, hence, direct inhibition of PtdIns5P synthesis, the slow attenuation of PtdIns5P vs. PtdIns(3,5)P2 is interpreted in support of a PtdIns(3,5)P2 precursor - PtdIns5P product relationship. However, note that the slower rate of PtdIns5P disappearance does not disprove that PtdIns5P is, in part, reduced due to direct PIKfyve inhibition by the YM201636 compound. In fact, due to continuous interconversion of the PIs, the rates of which remain unknown, and direct inhibition of PtdIns5P production by PIKfyve, this experiment does not provide support for the indirect pathway. Even if we make assumptions as those made by the authors, and analyze the initial 0–2.5 min time period of inhibition [32], it would appear that the absolute decrease in the molar concentration of PtdIns(3,5)P2 is too small to account for the change of PtdIns5P mass, and thus, would be inconsistent with the indirect model (Box 1).
Likewise, nutrient refeeding of starved cells, an approach affecting multiple enzyme activities in various signaling pathways (not solely “activation of endogenous PIKfyve”, as stated in [36]), fails to lend selective support in favor of the claim that “PtdIns(3,5)P2 is a precursor for PtdIns5P synthesis”. In fact, the observation that PtdIns(3,5)P2 reaches equilibrium faster than PtdIns5P [32] by nutrient refeeding is also compatible with PIKfyve directly catalyzing both reactions. Thus, based on Michaelis-Menten kinetics, at constant enzyme amounts, the initial reaction rate is dependent on the substrate concentration and enzyme affinity for the substrate (Km), with equilibrium reached faster at low substrate concentrations and low Km [79]. Accordingly, the PtdIns3P substrate for PtdIns(3,5)P2 synthesis is greatly less than the PtdIns substrate for PtdIns5P synthesis (Fig. 1A) and the PIKfyve affinity for PtdIns3P is higher than that for PtdIns, as discussed above. In conclusion, the two approaches in mammalian cells provide limited evidence in support of the claim that constitutive PtdIns5P is produced from PtdIns(3,5)P2 [36]. More importantly, they remain consistent with direct synthesis of PtdIns(3,5)P2 and PtdIns5P by PIKfyve.
Conclusions and outlook
The claim for PtdIns(3,5)P2 serving as a precursor for most of the cellular PtdIns5P pool [32, 36] faces many challenges, first and foremost being the lack of direct and convincing experimental evidence in support. Given its inconsistency with experimental evidence, corroborating direct synthesis of constitutive PtdIns5P by PIKfyve, together with quantitative considerations and lack of in vivo evidence about PtdIns(3,5)P2-PtdIns5P enzymatic conversion, this claim remains speculative. While we consider that some speculations might be proven as science continuously advances, we believe that in the absence of critical experimental support, this claim should currently be viewed as a hypothesis.
We wish to propose several direct approaches that might empower future investigation in seeking direct validation and coherent explanation of constitutive PtdIns5P occurrence. First, [3H]myo-inositol with substituted 3-hydroxyl that is most likely synthesizable [80], will be a key tool in cell labeling experiments, unambiguously defining the contribution of PIKfyve vs. MTM/MTMRs to PtdIns5P production under different conditions. Second, PIKfyve1999 and PIKfyve2000 knock-in mouse models, where the mutations differentially affect production of the two lipids (Fig. 3) [20], will provide unequivocal evidence for synthesis of each of the two lipids, in addition to enlightening their distinct physiological functions. Third, new PIKfyve chemical inhibitors [51, 81, 82] together with other yet-to-be-designed compounds hold promise for a greater selectivity window than that observed with YM201636 [34] in inhibiting synthesis of one lipid over the other. Such inhibitors will also help sort through the distinct functions regulated by the PIKfyve-catalyzed PtdIns5P vs. PtdIns(3,5)P2. Finally, PtdIns3P ablation through perturbation of the 3-kinases feeding into the PtdIns3P pool [11, 83–85] might appear as a potential approach. However, it should be considered that PIKfyve requires PtdIns3P not only as a substrate but also as membrane targeting signal that promotes its functionality [19, 54, 86]. Therefore, PtdIns3P ablation is likely to compromise not only PtdIns(3,5)P2 but also PtdIns5P production.
Acknowledgement
The authors thank Drs. Brian Edwards, Dave Evans, Sorin Draghici, Nicolas Kostoff, Ellen Tisdale and Steve Cala for helpful discussions. The first author is grateful to the late Violeta Shisheva for her many years of support. The work described from our laboratory was supported by ADA (1-05-RA-120; 7-13-BS-161), JDRF, NIH (DK58058) and WSU/WSU School of Medicine Research Offices
Abbreviations
- HPLC
high-pressure liquid chromatography
- KO
knockout
- PAS complex
PIKfyve-ArPIKfyve-Sac3 complex
- PIs
phosphoinositides
- PtdIns
phosphatidylinositol
- TLC
thin-layer chromatography
Footnotes
Conflict of Interest: The authors have no conflict of interest.
References
- 1.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 2.Shisheva A. PIKfyve and its lipid products in health and in sickness. Curr Top Microbiol Immunol. 2012;362:127–162. doi: 10.1007/978-94-007-5025-8_7. [DOI] [PubMed] [Google Scholar]
- 3.Hakim S, Bertucci MC, Conduit SE, Vuong DL, et al. Inositol polyphosphate phosphatases in human disease. Curr Top Microbiol Immunol. 2012;362:247–314. doi: 10.1007/978-94-007-5025-8_12. [DOI] [PubMed] [Google Scholar]
- 4.Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Viaud J, Lagarrigue F, Ramel D, Allart S, et al. Phosphatidylinositol 5-phosphate regulates invasion through binding and activation of Tiam1. Nat Commun. 2014;5:4080. doi: 10.1038/ncomms5080. [DOI] [PubMed] [Google Scholar]
- 6.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayinger P. Phosphoinositides and vesicular membrane traffic. Biochim Biophys Acta. 2012;1821:1104–1113. doi: 10.1016/j.bbalip.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun Y, Thapa N, Hedman AC, Anderson RA. Phosphatidylinositol 4,5-bisphosphate: targeted production and signaling. BioEssays. 2013;35:513–522. doi: 10.1002/bies.201200171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rameh LE, Tolias KF, Duckworth BC, Cantley LC. A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature. 1997;390:192–196. doi: 10.1038/36621. [DOI] [PubMed] [Google Scholar]
- 10.Leibiger B, Moede T, Uhles S, Barker CJ, et al. Insulin-feedback via PI3K-C2alpha activated PKBalpha/Akt1 is required for glucose-stimulated insulin secretion. FASEB J. 2010;24:1824–1837. doi: 10.1096/fj.09-148072. [DOI] [PubMed] [Google Scholar]
- 11.Falasca M, Maffucci T. Regulation and cellular functions of class II phosphoinositide 3-kinases. Biochem J. 2012;443:587–601. doi: 10.1042/BJ20120008. [DOI] [PubMed] [Google Scholar]
- 12.Bridges D, Ma JT, Park S, Inoki K, et al. Phosphatidylinositol 3,5-bisphosphate plays a role in the activation and subcellular localization of mechanistic target of rapamycin 1. Mol Biol Cell. 2012;23:2955–2962. doi: 10.1091/mbc.E11-12-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Posor Y, Eichhorn-Gruenig M, Puchkov D, Schoneberg J, et al. Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature. 2013;499:233–237. doi: 10.1038/nature12360. [DOI] [PubMed] [Google Scholar]
- 14.Franco I, Gulluni F, Campa CC, Costa C, et al. PI3K class II alpha controls spatially restricted endosomal PtdIns3P and Rab11 activation to promote primary cilium function. Dev Cell. 2014;28:647–658. doi: 10.1016/j.devcel.2014.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shisheva A, Sbrissa D, Ikonomov O. Cloning, characterization, and expression of a novel Zn2+-binding FYVE finger-containing phosphoinositide kinase in insulin-sensitive cells. Mol Cell Biol. 1999;19:623–634. doi: 10.1128/mcb.19.1.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sbrissa D, Ikonomov OC, Shisheva A. PIKfyve, a mammalian ortholog of yeast Fab1p lipid kinase, synthesizes 5-phosphoinositides. Effect of insulin. J Biol Chem. 1999;274:21589–21597. doi: 10.1074/jbc.274.31.21589. [DOI] [PubMed] [Google Scholar]
- 17.Sbrissa D, Ikonomov OC, Shisheva A. PIKfyve lipid kinase is a protein kinase: downregulation of 5'-phosphoinositide product formation by autophosphorylation. Biochemistry. 2000;39:15980–15989. doi: 10.1021/bi001897f. [DOI] [PubMed] [Google Scholar]
- 18.Sbrissa D, Ikonomov OC, Deeb R, Shisheva A. Phosphatidylinositol 5-phosphate biosynthesis is linked to PIKfyve and is involved in osmotic response pathway in mammalian cells. J Biol Chem. 2002;277:47276–47284. doi: 10.1074/jbc.M207576200. [DOI] [PubMed] [Google Scholar]
- 19.Ikonomov OC, Sbrissa D, Shisheva A. Mammalian cell morphology and endocytic membrane homeostasis require enzymatically active phosphoinositide 5-kinase PIKfyve. J Biol Chem. 2001;276:26141–26147. doi: 10.1074/jbc.M101722200. [DOI] [PubMed] [Google Scholar]
- 20.Ikonomov OC, Sbrissa D, Mlak K, Kanzaki M, et al. Functional dissection of lipid and protein kinase signals of PIKfyve reveals the role of PtdIns 3,5-P2 production for endomembrane integrity. J Biol Chem. 2002;277:9206–9211. doi: 10.1074/jbc.M108750200. [DOI] [PubMed] [Google Scholar]
- 21.Morris JB, Hinchliffe KA, Ciruela A, Letcher AJ, et al. Thrombin stimulation of platelets causes an increase in phosphatidylinositol 5-phosphate revealed by mass assay. FEBS Lett. 2000;475:57–60. doi: 10.1016/s0014-5793(00)01625-2. [DOI] [PubMed] [Google Scholar]
- 22.Sbrissa D, Ikonomov OC, Strakova J, Shisheva A. Role for a novel signaling intermediate, phosphatidylinositol 5-phosphate, in insulin-regulated F-actin stress fiber breakdown and GLUT4 translocation. Endocrinology. 2004;145:4853–4865. doi: 10.1210/en.2004-0489. [DOI] [PubMed] [Google Scholar]
- 23.Shisheva A. PIKfyve: the road to PtdIns 5-P and PtdIns 3,5-P(2) Cell Biol Int. 2001;25:1201–1206. doi: 10.1006/cbir.2001.0803. [DOI] [PubMed] [Google Scholar]
- 24.McEwen RK, Dove SK, Cooke FT, Painter GF, et al. Complementation analysis in PtdInsP kinase-deficient yeast mutants demonstrates that Schizosaccharomyces pombe and murine Fab1p homologues are phosphatidylinositol 3-phosphate 5-kinases. J Biol Chem. 1999;274:33905–33912. doi: 10.1074/jbc.274.48.33905. [DOI] [PubMed] [Google Scholar]
- 25.Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, et al. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J Cell Biol. 1998;143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooke FT, Dove SK, McEwen RK, Painter G, et al. The stress-activated phosphatidylinositol 3-phosphate 5-kinase Fab1p is essential for vacuole function in S. cerevisiae. Curr Biol. 1998;8:1219–1222. doi: 10.1016/s0960-9822(07)00513-1. [DOI] [PubMed] [Google Scholar]
- 27.Parrish WR, Stefan CJ, Emr SD. Essential role for the myotubularin-related phosphatase Ymr1p and the synaptojanin-like phosphatases Sjl2p and Sjl3p in regulation of phosphatidylinositol 3-phosphate in yeast. Mol Biol Cell. 2004;15:3567–3579. doi: 10.1091/mbc.E04-03-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts HF, Clarke JH, Letcher AJ, Irvine RF, et al. Effects of lipid kinase expression and cellular stimuli on phosphatidylinositol 5-phosphate levels in mammalian cell lines. FEBS Lett. 2005;579:2868–2872. doi: 10.1016/j.febslet.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 29.Coronas S, Ramel D, Pendaries C, Gaits-Iacovoni F, et al. PtdIns5P: a little phosphoinositide with big functions? Biochem Soc Symp. 2007:117–128. doi: 10.1042/BSS0740117. [DOI] [PubMed] [Google Scholar]
- 30.Lecompte O, Poch O, Laporte J. PtdIns5P regulation through evolution: roles in membrane trafficking? Trends Biochem Sci. 2008;33:453–460. doi: 10.1016/j.tibs.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 31.Ikonomov OC, Sbrissa D, Delvecchio K, Xie Y, et al. The phosphoinositide kinase PIKfyve is vital in early embryonic development: Preimplantation lethality of PIKfyve−/− embryos but normality of PIKfyve+/− mice. J Biol Chem. 2011;286:13404–13413. doi: 10.1074/jbc.M111.222364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zolov SN, Bridges D, Zhang Y, Lee WW, et al. In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc Natl Acad Sci USA. 2012;109:17472–17477. doi: 10.1073/pnas.1203106109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sbrissa D, Shisheva A. Acquisition of unprecedented phosphatidylinositol 3,5-bisphosphate rise in hyperosmotically stressed 3T3-L1 adipocytes, mediated by ArPIKfyve-PIKfyve pathway. J Biol Chem. 2005;280:7883–7889. doi: 10.1074/jbc.M412729200. [DOI] [PubMed] [Google Scholar]
- 34.Sbrissa D, Ikonomov OC, Filios C, Delvecchio K, et al. Functional dissociation between PIKfyve-synthesized PtdIns5P and PtdIns(3,5)P2 by means of the PIKfyve inhibitor YM201636. Am J Physiol Cell Physiol. 2012;303:C436–C446. doi: 10.1152/ajpcell.00105.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Zolov SN, Chow CY, Slutsky SG, et al. Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc Natl Acad Sci USA. 2007;104:17518–17523. doi: 10.1073/pnas.0702275104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCartney AJ, Zhang Y, Weisman LS. Phosphatidylinositol 3,5-bisphosphate: low abundance, high significance. BioEssays. 2014;36:52–64. doi: 10.1002/bies.201300012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whiteford CC, Brearley CA, Ulug ET. Phosphatidylinositol 3,5-bisphosphate defines a novel PI 3-kinase pathway in resting mouse fibroblasts. Biochem J. 1997;323(Pt 3):597–601. doi: 10.1042/bj3230597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cantley LC, Auger KR, Carpenter C, Duckworth B, et al. Oncogenes and signal transduction. Cell. 1991;64:281–302. doi: 10.1016/0092-8674(91)90639-g. [DOI] [PubMed] [Google Scholar]
- 39.Dove SK, Cooke FT, Douglas MR, Sayers LG, et al. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature. 1997;390:187–192. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- 40.Sbrissa D, Ikonomov OC, Fu Z, Ijuin T, et al. Core protein machinery for mammalian phosphatidylinositol 3,5-bisphosphate synthesis and turnover that regulates the progression of endosomal transport. Novel Sac phosphatase joins the ArPIKfyve-PIKfyve complex. J Biol Chem. 2007;282:23878–23891. doi: 10.1074/jbc.M611678200. [DOI] [PubMed] [Google Scholar]
- 41.Botelho RJ, Efe JA, Teis D, Emr SD. Assembly of a Fab1 phosphoinositide kinase signaling complex requires the Fig4 phosphoinositide phosphatase. Mol Biol Cell. 2008;19:4273–4286. doi: 10.1091/mbc.E08-04-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin N, Chow CY, Liu L, Zolov SN, et al. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J. 2008;27:3221–3234. doi: 10.1038/emboj.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shisheva A. PtdIns5P: news and views of its appearance, disappearance and deeds. Arch Biochem Biophys. 2013;538:171–180. doi: 10.1016/j.abb.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ikonomov OC, Sbrissa D, Dondapati R, Shisheva A. ArPIKfyve-PIKfyve interaction and role in insulin-regulated GLUT4 translocation and glucose transport in 3T3-L1 adipocytes. Exp Cell Res. 2007;313:2404–2416. doi: 10.1016/j.yexcr.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y, Lee SA, Kutateladze TG, Sbrissa D, et al. Chemical synthesis and molecular recognition of phosphatase-resistant analogues of phosphatidylinositol-3-phosphate. J Am Chem Soc. 2006;128:885–897. doi: 10.1021/ja0554716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sbrissa D, Ikonomov O, Shisheva A. Selective insulin-induced activation of class I(A) phosphoinositide 3-kinase in PIKfyve immune complexes from 3T3-L1 adipocytes. Mol Cell Endocrinol. 2001;181:35–46. doi: 10.1016/s0303-7207(01)00539-1. [DOI] [PubMed] [Google Scholar]
- 47.Ikonomov OC, Sbrissa D, Mlak K, Shisheva A. Requirement for PIKfyve enzymatic activity in acute and long-term insulin cellular effects. Endocrinology. 2002;143:4742–4754. doi: 10.1210/en.2002-220615. [DOI] [PubMed] [Google Scholar]
- 48.Sbrissa D, Ikonomov OC, Strakova J, Dondapati R, et al. A mammalian ortholog of accharomyces cerevisiae Vac14 that associates with and up-regulates PIKfyve phosphoinositide 5-kinase activity. Mol Cell Biol. 2004;24:10437–10447. doi: 10.1128/MCB.24.23.10437-10447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sbrissa D, Ikonomov OC, Fenner H, Shisheva A. ArPIKfyve homomeric and heteromeric interactions scaffold PIKfyve and Sac3 in a complex to promote PIKfyve activity and functionality. J Mol Biol. 2008;384:766–779. doi: 10.1016/j.jmb.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ikonomov OC, Sbrissa D, Fenner H, Shisheva A. PIKfyve-ArPIKfyve-Sac3 core complex: contact sites and their consequence for Sac3 phosphatase activity and endocytic membrane homeostasis. J Biol Chem. 2009;284:35794–35806. doi: 10.1074/jbc.M109.037515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fant X, Durieu E, Chicanne G, Payrastre B, et al. cdc-like/dual-specificity tyrosine phosphorylation-regulated kinases inhibitor leucettine L41 induces mTOR-dependent autophagy: implication for Alzheimer's disease. Mol Pharmacol. 2014;85:441–450. doi: 10.1124/mol.113.090837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ikonomov OC, Sbrissa D, Shisheva A. YM201636, an inhibitor of retroviral budding and PIKfyve-catalyzed PtdIns(3,5)P2 synthesis, halts glucose entry by insulin in adipocytes. Biochem Biophys Res Commun. 2009;382:566–570. doi: 10.1016/j.bbrc.2009.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shisheva A. PIKfyve: Partners, significance, debates and paradoxes. Cell Biol Int. 2008;32:591–604. doi: 10.1016/j.cellbi.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sbrissa D, Ikonomov OC, Shisheva A. Phosphatidylinositol 3-phosphate-interacting domains in PIKfyve. Binding specificity and role in PIKfyve. Endomenbrane localization. J Biol Chem. 2002;277:6073–6079. doi: 10.1074/jbc.M110194200. [DOI] [PubMed] [Google Scholar]
- 55.Viaud J, Boal F, Tronchere H, Gaits-Iacovoni F, et al. Phosphatidylinositol 5-phosphate: a nuclear stress lipid and a tuner of membranes and cytoskeleton dynamics. BioEssays. 2014;36:260–272. doi: 10.1002/bies.201300132. [DOI] [PubMed] [Google Scholar]
- 56.Taylor GS, Maehama T, Dixon JE. Myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc Natl Acad Sci USA. 2000;97:8910–8915. doi: 10.1073/pnas.160255697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chaussade C, Pirola L, Bonnafous S, Blondeau F, et al. Expression of myotubularin by an adenoviral vector demonstrates its function as a phosphatidylinositol 3-phosphate [PtdIns(3)P] phosphatase in muscle cell lines: involvement of PtdIns(3)P in insulin-stimulated glucose transport. Mol Endocrinol. 2003;17:2448–2460. doi: 10.1210/me.2003-0261. [DOI] [PubMed] [Google Scholar]
- 58.Srivastava S, Li Z, Lin L, Liu G, et al. The phosphatidylinositol 3-phosphate phosphatase myotubularin- related protein 6 (MTMR6) is a negative regulator of the Ca2+-activated K+ channel KCa3.1. Mol Cell Biol. 2005;25:3630–3638. doi: 10.1128/MCB.25.9.3630-3638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cao C, Backer JM, Laporte J, Bedrick EJ, et al. Sequential actions of myotubularin lipid phosphatases regulate endosomal PI(3)P and growth factor receptor trafficking. Mol Biol Cell. 2008;19:3334–3346. doi: 10.1091/mbc.E08-04-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vergne I, Roberts E, Elmaoued RA, Tosch V, et al. Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. EMBO J. 2009;28:2244–2258. doi: 10.1038/emboj.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Velichkova M, Juan J, Kadandale P, Jean S, et al. Drosophila Mtm and class II PI3K coregulate a PI(3)P pool with cortical and endolysosomal functions. J Cell Biol. 2010;190:407–425. doi: 10.1083/jcb.200911020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taguchi-Atarashi N, Hamasaki M, Matsunaga K, Omori H, et al. Modulation of local PtdIns3P levels by the PI phosphatase MTMR3 regulates constitutive autophagy. Traffic. 2010;11:468–478. doi: 10.1111/j.1600-0854.2010.01034.x. [DOI] [PubMed] [Google Scholar]
- 63.Razidlo GL, Katafiasz D, Taylor GS. Myotubularin regulates Akt-dependent survival signaling via phosphatidylinositol 3-phosphate. J Biol Chem. 2011;286:20005–20019. doi: 10.1074/jbc.M110.197749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davies E, Sheffield D, Tibarewal P, Fedele C, et al. The PTEN and Myotubularin Phosphoinositide 3-Phosphatases: Linking Lipid Signalling to Human Disease. In: Balla T, Wymann M, York J, editors. Phosphoinositides I. Enzymes of Synthesis and Degradation. Springer; 2012. pp. 281–336. [DOI] [PubMed] [Google Scholar]
- 65.Jean S, Cox S, Schmidt EJ, Robinson FL, et al. Sbf/MTMR13 coordinates PI(3)P and Rab21 regulation in endocytic control of cellular remodeling. Mol Biol Cell. 2012;23:2723–2740. doi: 10.1091/mbc.E12-05-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hnia K, Vaccari I, Bolino A, Laporte J. Myotubularin phosphoinositide phosphatases: cellular functions and disease pathophysiology. Trends Mol Med. 2012;18:317–327. doi: 10.1016/j.molmed.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 67.Lu N, Shen Q, Mahoney TR, Neukomm LJ, et al. Two PI 3-kinases and one PI 3-phosphatase together establish the cyclic waves of phagosomal PtdIns(3)P critical for the degradation of apoptotic cells. PLoS Biol. 2012;10:e1001245. doi: 10.1371/journal.pbio.1001245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pulido R, Stoker AW, Hendriks WJ. PTPs emerge as PIPs: protein tyrosine phosphatases with lipid-phosphatase activities in human disease. Hum Mol Genet. 2013;22:R66–R76. doi: 10.1093/hmg/ddt347. [DOI] [PubMed] [Google Scholar]
- 69.Amoasii L, Hnia K, Chicanne G, Brech A, et al. Myotubularin and PtdIns3P remodel the sarcoplasmic reticulum in muscle in vivo. J Cell Sci. 2013;123:1806–1819. doi: 10.1242/jcs.118505. [DOI] [PubMed] [Google Scholar]
- 70.Oppelt A, Lobert VH, Haglund K, Mackey AM, et al. Production of phosphatidylinositol 5-phosphate via PIKfyve and MTMR3 regulates cell migration. EMBO Rep. 2013;14:57–64. doi: 10.1038/embor.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vaccari I, Dina G, Tronchere H, Kaufman E, et al. Genetic interaction between MTMR2 and FIG4 phospholipid phosphatases involved in Charcot-Marie-Tooth neuropathies. PLoS Genet. 2011;7:e1002319. doi: 10.1371/journal.pgen.1002319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ikonomov OC, Filios C, Sbrissa D, Chen X, et al. The PIKfyve-ArPIKfyve-Sac3 triad in human breast cancer: Functional link between elevated Sac3 phosphatase and enhanced proliferation of triple negative cell lines. Biochem Biophys Res Commun. 2013;440:342–347. doi: 10.1016/j.bbrc.2013.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sarkes D, Rameh LE. A novel HPLC-based approach makes possible the spatial characterization of cellular PtdIns5P and other phosphoinositides. Biochem J. 2010;428:375–384. doi: 10.1042/BJ20100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Clague MJ, Lorenzo O. The myotubularin family of lipid phosphatases. Traffic. 2005;6:1063–1069. doi: 10.1111/j.1600-0854.2005.00338.x. [DOI] [PubMed] [Google Scholar]
- 75.Amoasii L, Bertazzi DL, Tronchere H, Hnia K, et al. Phosphatase-dead myotubularin ameliorates X-linked centronuclear myopathy phenotypes in mice. PLoS Genet. 2012;8:e1002965. doi: 10.1371/journal.pgen.1002965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Coronas S, Lagarrigue F, Ramel D, Chicanne G, et al. Elevated levels of PtdIns5P in NPM-ALK transformed cells: implication of PIKfyve. Biochem Biophys Res Commun. 2008;372:351–355. doi: 10.1016/j.bbrc.2008.05.062. [DOI] [PubMed] [Google Scholar]
- 77.Kawasaki T, Takemura N, Standley DM, Akira S, et al. The second messenger phosphatidylinositol-5-phosphate facilitates antiviral innate immune signaling. Cell Host Microbe. 2013;14:148–158. doi: 10.1016/j.chom.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 78.Jefferies HB, Cooke FT, Jat P, Boucheron C, et al. A selective PIKfyve inhibitor blocks PtdIns(3,5)P(2) production and disrupts endomembrane transport and retroviral budding. EMBO Rep. 2008;9:164–170. doi: 10.1038/sj.embor.7401155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Berg JM, Tymoszko JL, Stryer L. Biochemistry Freeman, W.H. 2011 [Google Scholar]
- 80.Best MD, Zhang H, Prestwich GD. Inositol polyphosphates, diphosphoinositol polyphosphates and phosphatidylinositol polyphosphate lipids: structure, synthesis, and development of probes for studying biological activity. Nat Prod Rep. 2010;27:1403–1430. doi: 10.1039/b923844c. [DOI] [PubMed] [Google Scholar]
- 81.Cai X, Xu Y, Cheung AK, Tomlinson RC, et al. PIKfyve, a class III PI kinase, is the target of the small molecular IL-12/IL-23 inhibitor apilimod and a player in Toll-like receptor signaling. Chem Biol. 2013;20:912–921. doi: 10.1016/j.chembiol.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hayakawa N, Noguchi M, Takeshita S, Eviryanti A, et al. Structure-activity relationship study, target identification, and pharmacological characterization of a small molecular IL-12/23 inhibitor, APY0201. Bioorg Med Chem. 2014;22:3021–3029. doi: 10.1016/j.bmc.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 83.Salamon RS, Backer JM. Phosphatidylinositol-3,4,5-trisphosphate: tool of choice for class I PI 3-kinases. BioEssays. 2013;35:602–611. doi: 10.1002/bies.201200176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- 85.Schink KO, Raiborg C, Stenmark H. Phosphatidylinositol 3-phosphate, a lipid that regulates membrane dynamics, protein sorting and cell signalling. Bioessays. 2013;35:900–912. doi: 10.1002/bies.201300064. [DOI] [PubMed] [Google Scholar]
- 86.Stuffers S, Malerod L, Schink KO, Corvera S, et al. Time-resolved ultrastructural detection of phosphatidylinositol 3-phosphate. J Histochem Cytochem. 2010;58:1025–1032. doi: 10.1369/jhc.2010.955815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shisheva A. Phosphoinositides in insulin action on GLUT4 dynamics: not just PtdIns(3,4,5)P3. Am J Physiol Endocrinol Metab. 2008;295:E536–E544. doi: 10.1152/ajpendo.90353.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rameh LE, Cantley LC. The role of phosphoinositide 3-kinase lipid products in cell function. J Biol Chem. 1999;274:8347–8350. doi: 10.1074/jbc.274.13.8347. [DOI] [PubMed] [Google Scholar]
- 89.Leslie NR, Downes CP. PTEN: The down side of PI 3-kinase signalling. Cell Signal. 2002;14:285–295. doi: 10.1016/s0898-6568(01)00234-0. [DOI] [PubMed] [Google Scholar]
- 90.Ikonomov OC, Sbrissa D, Shisheva A. Localized PtdIns 3,5-P2 synthesis to regulate early endosome dynamics and fusion. Am J Physiol Cell Physiol. 2006;291:C393–C404. doi: 10.1152/ajpcell.00019.2006. [DOI] [PubMed] [Google Scholar]