Abstract
The human BCL6 gene encodes a transcriptional repressor that is crucial for germinal center B cell development and T follicular helper cell differentiation. It is involved in the pathogenesis of certain human lymphomas. In an effort to identify targets of BCL6 repression, we used a previously described cell system in which BCL6 repressive effects are inhibited, followed by subtractive hybridization, and identified the integral membrane 2B gene (ITM2B, formerly BRI2) as a potential target. Here we show that BCL6 can bind to its preferential consensus binding site within the first intron of ITM2B and represses its transcription. Knockdown of endogenous BCL6 in a human B cell lymphoma line increases ITM2B expression. Further, there is an inverse relationship between the expression levels of BCL6 and ITM2B proteins in 16 human B- and T-cell lymphomas studied by immunohistochemistry. Both the BCL6 and ITM2B proteins are expressed ubiquitously. Similar to some other targets of BCL6, a short form of the ITM2B protein generated by alternative splicing induces apoptosis in hematopoietic cell lines. Molecular alterations in the ITM2B gene are associated with two neurodegenerative diseases, Familial British and Familial Danish dementia. ITM2B dysfunction also may be relevant for the development of Alzheimer’s disease. Our data confirm ITM2B as a target of BCL6 repression in lymphoma. A further understanding of the genes that function as regulators of the ITM2B protein may provide insights for the development of new molecular tools not only for targeted lymphoma therapy but also for the treatment of these dementias.
Keywords: ITM2B gene, BCL6 target, Familial British dementia, Familial Danish dementia, Alzheimer’s disease
1. Introduction
The BCL6 nuclear zinc finger protein is encoded by a gene located on chromosome 3, band q27, and functions as a transcriptional repressor [1–4]. It has long been known to play an important role in the pathogenesis of diffuse large B cell lymphomas, and, more recently, its additional role in T-cell biology has been appreciated [5,6]. BCL6 has been called a “master regulator” of germinal center formation and is believed to repress the transcription of hundreds of proteins [7]. In a study of germinal center B cells and diffuse large cell lymphomas, it was found to bind to the promoters of about 3,000 genes (enhancer and intronic elements were not studied). Less frequently, BCL6 has been implicated in the regulation of the growth of other cancers, e.g., colorectal and breast cancer, as well as in the control of other disease processes, e.g., myasthenia gravis [8–10].
In an effort to identify BCL6 target genes, we previously developed a dominant-negative cell system in which the BCL6 repressive effects are inhibited, enabling the detection of genes that are ordinarily repressed. By subtractive hybridization, we selectively amplified differentially expressed sequences, thus detecting upregulated messages [11]. With the use of this methodology, we now describe the identification of the integral membrane 2B gene (ITM2B, formerly called BRI2) as a novel target of BCL6 repression.
Like the BCL6 protein, the ITM2B protein is expressed ubiquitously [12]. A short form of the ITM2B protein, which is generated by alternative splicing, has been shown to induce apoptosis in hematopoietic cell lines [13], a function that is similar to some other targets of BCL6 [11,14]. However, ITM2B has not been studied in the context of human lymphomas. Interestingly, alterations in the ITM2B gene are associated with two neurodegenerative diseases, Familial British dementia (FBD) and Familial Danish dementia (FDD), and data have been presented indicating that aberrant ITM2B function also may play a role in the development of Alzheimer’s disease (AD) [15–20]. Therefore, an understanding of the regulation of ITM2B expression by BCL6 is likely to have important implications for new therapeutic tools.
2. Materials and methods
2.1. Differential expression by Northern blotting
Preparation of constructs, cDNA subtraction, and amplification of differentially expressed sequences have been described previously [11]. Briefly, through the use of a cell system in which the BCL6 repressive effects were inhibited, we identified upregulated genes that are ordinarily its targets of repression. We converted the BCL6 zinc fingers (BCL6ZF), which bind DNA but lack repressive effects, into a transcriptional activator as described and used this construct to compete with wild-type endogenous BCL6 in a cell line (BJAB, an Epstein–Barr virus-negative Burkitt lymphoma cell line) expressing BCL6 at high levels [11]. The cells were transiently transfected with this construct or the vector in which it had been cloned. Subtractive hybridization was performed, and the inserts of the cDNA clones obtained after PCR were sequenced. Potential targets of the BCL6 repressive effects were 32P-labeled and hybridized to Northern blots prepared from BJAB cells that had been transfected with the BCL6ZF construct or the vector. The blots were washed under stringent conditions, exposed to film, then the probes were stripped, and the blots were rehybridized with a 32P-labeled human β-actin cDNA control probe (Clontech Laboratories, Inc, Mountain View, CA), washed under stringent conditions, and reautoradiographed. Quantitation of relative band intensity was normalized to β-actin by scanning densitometry.
2.2. Consensus binding site
To clone the sequences surrounding the preferential consensus binding site for BCL6, 0.697 kb of human genomic DNA was amplified by PCR (forward primer, 5’-GGACACTGCTGTAGCATATTGG-3’; reverse primer, 5’-CTTTCATTGTGAAGCACAGCTC-3’); 1 μl of the PCR product was blunted and ligated to 50 μg of the pJET 1.2/Blunt Cloning Vector (Thermo Scientific, Waltham, MA) per the manufacturer’s guidelines. The appropriate insert was isolated from this vector with BglII restriction enzyme digests and ligated to the BglII site of the pGL3 basic luciferase reporter vector (Promega, Madison, WI) (PGL3ITM2B).
2.3. ChIP assay
Two ChIP assays were performed with an EZ-ChIP™ Chromatin Immunoprecipitation Kit (#17-371, EMD Millipore Corp, Billerica, MA). BJAB cells (Burkitt lymphoma line rich in endogenous BCL6, described above), 4.7 × 106 per sample, were cross-linked with 1% formaldehyde for 5 min at room temperature. The reaction was terminated with an excess of glycine. Cross-linked chromatin was sonicated to ~200–1000 bp with a Microson™ XL 2000 sonicator (Qsonica, LLC, Newtown, CT); cells were vortexed then sonicated four times on ice, on average, at 9 W, 7 W, and 5 W twice, for 10 s each, respectively, with a 50 s cooling period on ice between sonications, followed by immunoprecipitation with antibodies to BCL6 (sc-858 X, Santa Cruz Biotechnology, Dallas, TX) and anti-rabbit IgG as a negative control. ChIP DNA was amplified by PCR. One set of primers (described in the Consensus Binding Site section) amplified the BCL6 consensus site in the first intron of the ITM2B gene, and another set of primers amplified a coding region of ITM2B, which did not contain any putative BCL6 binding sites. These primers amplified 100 bp within the second exon of ITM2B: forward primer, 5’-GGTACCAG TTGGCCAAAGAA-3’; reverse primer, 5’-TTTGTACAAGTATGCTCCTCCT AGA-3’. Bands were detected on 0.8% and 0.9% agarose gels, respectively.
2.4. Transfection assays/functional analysis
NIH3T3 cells (murine fibroblasts with minimal expression of both endogenous BCL6 [21] and ITM2B [22], which we have previously used for similar studies [4,23], were grown under standard conditions and plated at 1.5 to 4 × 105 cells per well in a six-well dish. They were transfected the next day by calcium phosphate DNA-coprecipitation or by TurboFect™ Transfection Reagent (Thermo Scientific) with PGL3ITM2B (0.6 to 0.65 μg)—a construct containing the exact BCL6 consensus binding site identified in the first intron of the ITM2B gene, and either full-length BCL6 cDNA (1.25 to 1.46 μg) subcloned in the pCGN expression vector (pCGNBCL6) or an equivalent amount of a truncated BCL6 expression construct (control), also subcloned in the pCGN vector. We have used this truncated construct in prior studies as a control [4,23] because it lacks the zinc finger DNA-binding region of BCL6 and, therefore, cannot bind DNA. A CMV-driven β-galactosidase expression construct was cotransfected. Cells were harvested at 47.5 h to 70 h. Luciferase levels were normalized for transfection efficiency by using the values of β-galactosidase assays as previously described [4].
Relative luciferase activity was defined as the luciferase levels obtained with the BCL6 construct divided by the luciferase levels obtained with the truncated control. A paired t test was used to evaluate whether the mean relative luciferase activity of the ITM2B consensus binding site in the cells transfected with BCL6 vs. those transfected with the truncated control from three independent experiments (triplicate wells in each) was significantly different from 1.
2.5. Transfection of BCL6 siRNAs and Western blotting
BJAB cells were transfected with human BCL6 siGENOME SMARTpool reagent or CONTROL nontargeting siRNA 1 (Dharmacon, LaFayette, CO) by electroporation as described previously [23]. Whole cell extracts were prepared from the transfected cells and subjected to Western blotting as described. The antibodies used included rabbit polyclonal antibodies to BCL6 (sc-858 or sc-368, Santa Cruz Bio-technology, Santa Cruz, CA), a validated affinity-isolated Prestige antibody to ITM2B produced in rabbit (Sigma-Aldrich Co. LLC, Saint Louis, MO, #HPA029292), and affinity-isolated actin antibody produced in rabbit (#A2066, Sigma-Aldrich). The membranes were washed and incubated with anti-rabbit IgG (Fc), alkaline phosphatase conjugate (Promega, Madison, WI), then washed again. Protein bands were detected with Western Blue Stabilized Substrate for Alkaline Phosphatase (Promega).
Calculations of relative band intensity on four Western blots were normalized to the intensity of β-actin expression by scanning densitometry. The paired t test was used to compare BCL6 and ITM2B protein levels, respectively, in the siRNA BCL6-transfected cells with the corresponding control cells.
2.6. Immunohistochemistry
Human paraffin-embedded lymphoma blocks were retrieved from the Surgical Pathology archives under an institutional review board-approved protocol. In each case, the sections that are stained with the different antibodies are from the same tissue block, but they are not necessarily consecutive sections. BCL6 staining was performed as described previously [24] with mouse monoclonal anti-human BCL6 (clone LN22; Novocastra). Staining for ITM2B was performed overnight at 4 °C with an affinity-isolated Prestige antibody produced in rabbit (Sigma-Aldrich, Saint Louis, MO, #HPA029292) that was diluted 1:20–1:25. Antigen–antibody binding was detected with DAB chromogen, and tissues were counterstained with hematoxylin. Images were taken with a BX41 microscope (Olympus), DP72 digital camera, and cellSens Standard imaging software (Olympus).
3. Results
3.1. Subtractive hybridization and amplification of differentially expressed cDNAs in BJAB cells transfected with the BCL6 zinc fingers or vector control followed by nucleotide sequencing
We used the BCL6 zinc fingers (BCL6ZF), which bind DNA but lack the BCL6 repressive effects, to compete with endogenous BCL6 in BJAB cells (Epstein–Barr virus-negative Burkitt lymphoma cell line) [11]. Cells also were transfected with the vector used for cloning, and subtractive hybridization was performed. Primary and secondary PCR products of the experimental subtracted sample revealed differentially expressed bands on an ethidium bromide-stained 2% agarose gel when compared with the unsubtracted control. As previously described [11], the finding that sequences from two of the clones analyzed were identical to overlapping regions of BCL6ZF (indicating identification of BCL6ZF mRNA that had been overexpressed in the study cells) implied that subtractive hybridization had been successful. The sequences from another clone matched the human integral membrane protein 2B (ITM2B) gene (formerly called BRI2).
3.2. Northern blotting confirms differential expression of ITM2B
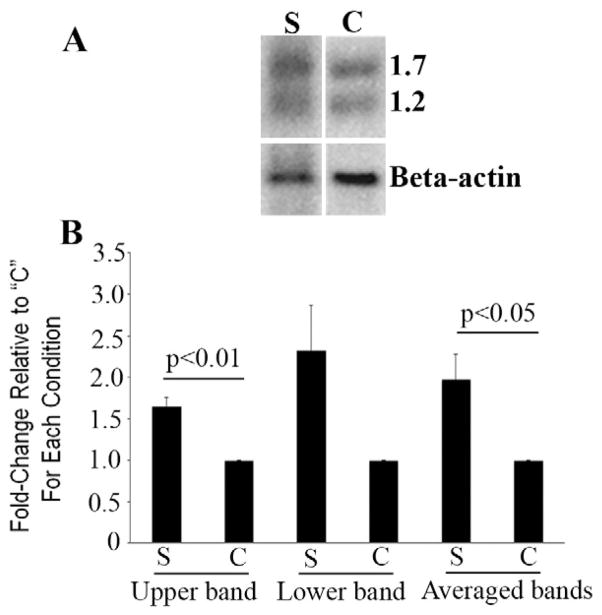
Four Northern blots were prepared from the total RNA of BJAB cells transiently transfected with the BCL6ZF study construct (S lane) or vector control (C lane) as previously described [11], and they were hybridized with the cDNA fragment of ITM2B obtained from cDNA subtraction/ amplification. A representative Northern blot is shown in Fig. 1A. The expected two transcripts [12] were noted (1.7 kb and 1.2 kb, respectively). Relative band intensity normalized to β-actin by scanning densitometry showed that, for the four blots, differential expression of ITM2B (S lane) as compared with the control (C) was, by paired t test, mean ± SEM = 1.65 ± 0.11 for the upper band (p < 0.01), range 1.45- to 1.95-fold; 2.33 ± 0.54 for the lower band (p = 0.09), range 1.27- to 3.46-fold; and 1.98 ± 0.30 for the averaged bands (p 0.05), range 1.43- to 2.53-fold (Fig. 1B).
Fig 1.
Northern blotting confirms differential expression of ITM2B. (A) Autoradiograph of a Northern blot prepared from total RNA of BJAB cells transiently transfected with an expression (study) construct (S) containing the BCL6 zinc fingers or the vector into which the S construct was cloned (control, C). Top panel: Hybridization with a cDNA fragment of ITM2B obtained from subtractive hybridization; its ~1.7 and ~1.2 kb transcripts are noted. Bottom panel: The blot was stripped and rehybridized with a human β-actin probe. Differential expression of ITM2B normalized to β-actin for this blot is 2-fold for the upper band and 3-fold for the lower band. (B) For the four Northern blots performed, densitometry indicates that differential expression of ITM2B (S lane) as compared with the control (C) is, by paired t test, mean ± SEM, 1.65 ± 0.11 for the upper band (p < 0.01), range 1.45- to 1.95-fold; 2.33 ± 0.54 for the lower band (p = 0.09), range 1.27-to 3.46-fold; and 1.98 ± 0.30 for the averaged bands (p 0.05), range 1.43- to 2.53-fold.
3.3. The BCL6 protein binds to the high-affinity BCL6 binding site in the first intron of ITM2B
We identified an exact match to the consensus binding site for BCL6 ([A/T]TC[C/T][A/T][A/C]GA) [25] within the first intron of the ITM2B gene (TTCCTAGA, nt 48,811,973–48,811,980). This sequence was present in both of the protein-encoding transcripts of the gene. Tiling ChIP-on-chip experiments have shown that BCL6 preferentially localizes to this element [25].
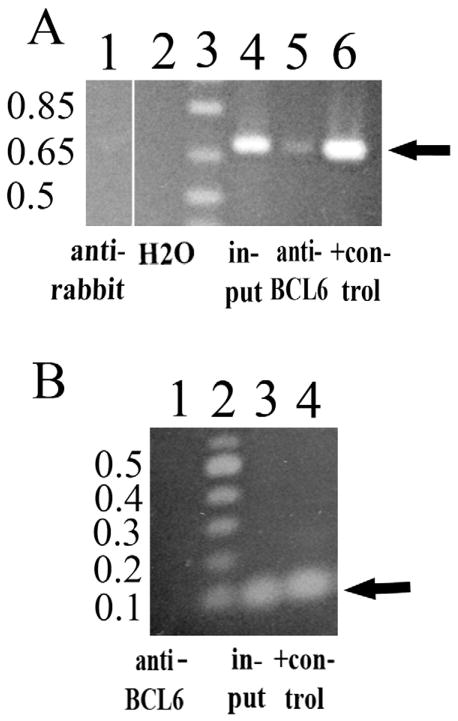
Two ChIP assays were performed and showed that in a B cell lymphoma line, endogenous BCL6 was bound to the region in the first intron of the ITM2B gene containing the BCL6 consensus binding site because DNA from this region was enriched in chromatin immunoprecipitated with anti-BCL6 (Fig. 2A, lane 5). This genomic region was not enriched in material immunoprecipitated with anti-rabbit IgG (Fig. 2A, lane 1), while a band of the correct size was observed with input (lane 4) and the positive control (genomic DNA, lane 6). No binding of BCL6 was detected to the coding region of ITM2B, which does not contain a putative binding site (Fig. 2B, lane 1), while a band in the correct location was observed with input (lane 3) and the positive control (genomic DNA, lane 4).
Fig 2.
The BCL6 protein binds to ITM2B in BJAB cells (ChIP assay). (A) The first intronic region of ITM2B containing the BCL6 consensus binding site (0.697 kb). BCL6 is bound to the region of the first intron of ITM2B containing the putative BCL6 binding site (arrow) because DNA from this area is enriched in chromatin immunoprecipitated with anti-BCL6 (lane 5). Rabbit antibody, lane 1 (control) does not enrich this genomic region. The positive (+) control (lane 6) is genomic DNA amplified with the same primers. Lane 3 is 1 Kb Plus DNA Ladder (Invitrogen™, Life Technologies, Grand Island, NY), and lane 4 is input. All of the lanes are from the same gel. (B) ITM2B coding region (0.1 kb, arrow). Binding of BCL6 to the coding region of ITM2B (lane 1) is not detected, as there is no putative binding site in this region. Input (lane 3) and the positive (+) control (genomic DNA, lane 4) amplified with the same primers (within the second exon of ITM2B) show the expected 100 bp product (arrow). Lane 2 is 100 bp DNA Ladder (New Engl. Biolabs, Ipswich, MA).
3.4. The BCL6 protein represses transcription from the BCL6 consensus binding site in the first intron of ITM2B
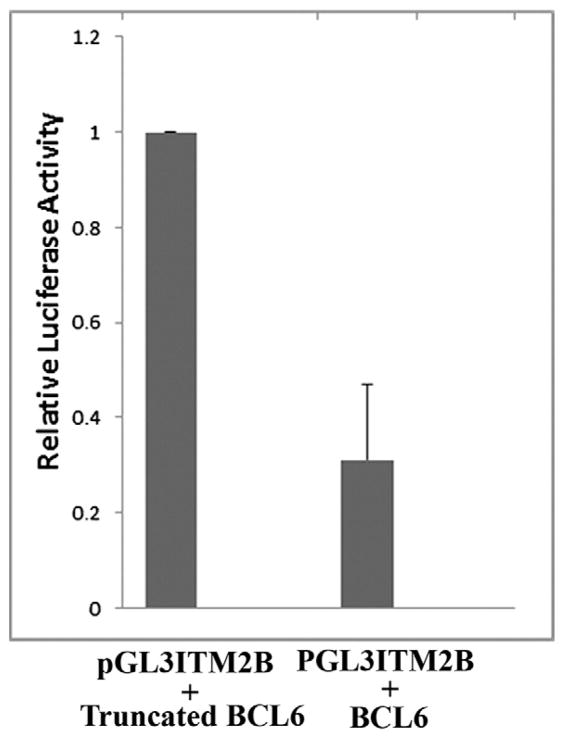
Transient transfections performed in NIH3T3 cells revealed that relative luciferase levels, depicted in Fig. 3 from three independent experiments, indicated significant repression (p 0.0001) from the BCL6 consensus binding site in the first intron of ITM2B by the full-length BCL6 protein (mean ± SD = 0.31 ± 0.16) as compared with a truncated control that cannot bind DNA (for comparison, the absolute value of the control was set at 1). These studies show that BCL6 directly represses ITM2B and provide evidence for transcriptional regulation of ITM2B by BCL6.
Fig 3.
The BCL6 protein represses transcription from sequences containing the BCL6 consensus binding site in the first intron of ITM2B. Relative luciferase levels in NIH3T3 cells show significant repression (p 0.0001) of transcription (mean ± SD = 0.31 ± 0.16) by the full-length BCL6 protein from sequences encompassing the BCL6 consensus binding site in the first intron of the ITM2B gene as compared with a truncated control lacking the BCL6 zinc fingers, which are needed to bind DNA. The data were normalized with results for the control designated as 1.
3.5. Knockdown of the BCL6 protein increases endogenous ITM2B expression
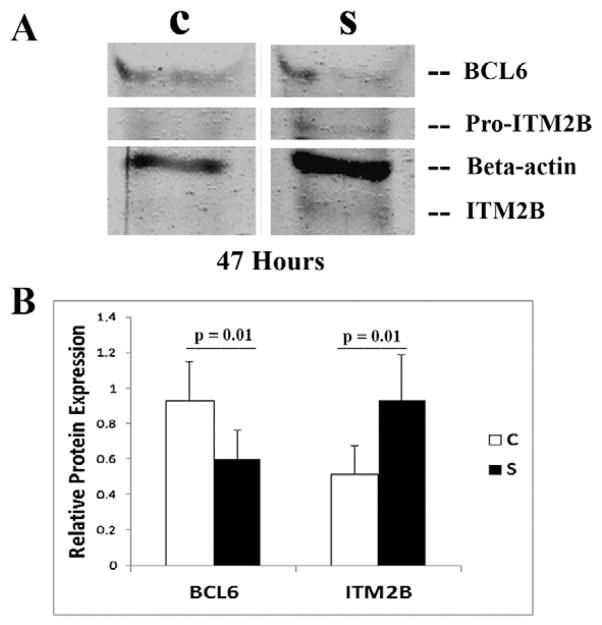
Transient transfection of siRNA duplexes targeting endogenous BCL6 in BJAB cells led to knockdown of BCL6 protein levels that could be detected by 24 h and persisted until at least 47 h (Fig. 4A, confirming sufficient knockdown of BCL6 with the siRNA, as demonstrated previously [23]). Data were analyzed from four independent experiments. The pro-peptide of ITM2B was detected at a molecular mass of ~50 kDa in all four experiments, and mature ITM2B (lacking the pro-peptide) was detected at a lower molecular weight [26]. Relative band intensities on Western blots were normalized to β-actin, which was used to determine the amount of protein loaded. Between 42 and 47 h, overall knockdown of the BCL6 protein was 35.2% (range, 29% to 56%; mean ± SEM for control siRNA was 0.93 ± 0.22 as compared with 0.60 ± 0.17 for cells transfected with BCL6 siRNA, p = 0.01). Higher levels of ITM2B protein were noted at the same time points (overall, a 1.8-fold increase in endogenous ITM2B in the siRNA-BCL6-transfected cells as compared with control cells, range 1.64 to 4.9-fold, mean ± SEM for control siRNA 0.51 ± 0.17 as compared with 0.93 ± 0.26 for cells transfected with BCL6 siRNA, p = 0.01, Fig. 4B). These observations confirm that an increase in endogenous ITM2B expression is a direct effect of BCL6 knockdown.
Fig 4.
siRNA duplexes specifically targeting BCL6 lead to an increase in endogenous ITM2B expression. (A) Representative Western blot showing BCL6, β-actin (as previously reported [23]), and ITM2B protein expression at 47 h in BJAB cells transiently transfected with siRNA duplexes [C, control siRNA; S, study (BCL6) siRNA]. The ITM2B propeptide (pro-ITM2B) is detected at ~50 kDa, and mature ITM2B (lacking the propeptide) is noted at a somewhat lower molecular weight [26]. (B) The graph depicts the relative protein expression of BCL6 and ITM2B in C and S cells after normalization for β-actin expression. (Left) BCL6 protein expression (mean ± SEM: control siRNA, 0.93 ± 0.22; BCL6 siRNA, 0.60 ± 0.17; p = 0.01). (Right) ITM2B protein expression (mean ± SEM: control siRNA, 0.51 ± 0.17; BCL6 siRNA, 0.93 ± 0.26; p = 0.01).
3.6. Reciprocal relationship between BCL6 and ITM2B protein expression in human lymphomas
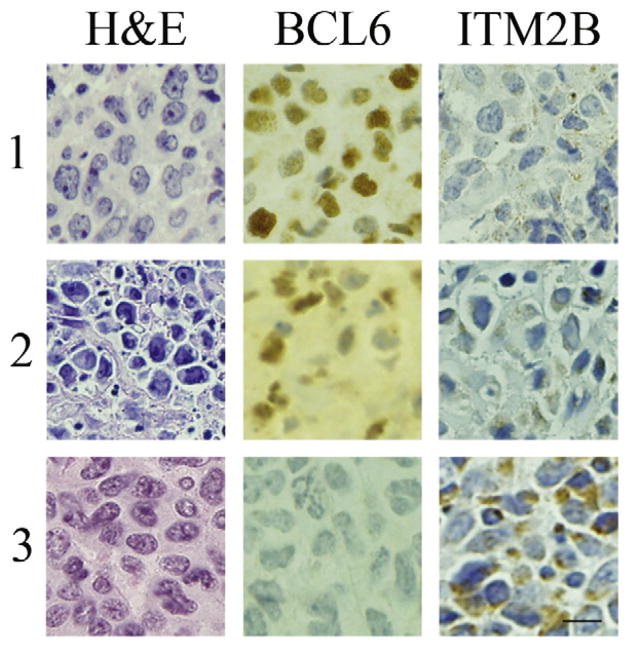
The relationship between BCL6 and ITM2B protein expression was studied in 16 human lymphomas (10 B cells, 6 T cells) by immunohistochemical staining (Fig. 5). B cell lymphomas: nine of the B cell lymphomas were positive for BCL6 and negative for ITM2B (representative example, Fig. 5, panel 1). The tenth B cell tumor was variably positive or negative for BCL6; the BCL6 positive areas were negative for ITM2B and vice-versa. T-cell lymphomas: four T-cell lymphomas were positive for BCL6 and negative for ITM2B (representative example Fig. 5, panel 2). Two T-cell lymphomas were negative for BCL6 and positive for ITM2B (Fig. 5, panel 3).
Fig 5.
Immunohistochemistry of representative human lymphomas depicting an inverse relationship between BCL6 and ITM2B expression. In each case, the H&E stain reveals effacement of normal tissue architecture with replacement by malignant cells. Panels 1 and 2 depict B- and T-cell lymphomas, respectively, that are BCL6 positive (brown nuclear staining) and ITM2B negative. Panel 3 shows a BCL6-negative T-cell lymphoma that is ITM2B positive (brown, granular cytoplasmic staining). The bar (lowest right panel) indicates 20 μm.
4. Discussion
The human ITM2B gene (formerly called BRI2) has been mapped to chromosome 13q14.3. Wild-type ITM2B is a type II transmembrane protein containing 266 amino acids. Northern blotting identifies two transcripts, 1200 nt and 1700 nt long, respectively, originating from poly(A) signals at positions 1114 and 1593 [12]. The apparent molecular mass of the ITM2B protein (42–44 kDa) is higher than predicted from the number and composition of amino acids, suggesting that it is glycosylated [27]. It is a substrate for regulated intramural proteolysis and is processed by furin and ADAM10 (a disintegrin and metalloproteinase domain 10) as well as other proteases in a fashion similar to Notch to release secreted fragments [26]. The pro-peptide of ITM2B can be detected at a molecular mass of ~50 kDa on Western blots, and the mature variant lacking the pro-peptide is noted at a slightly lower molecular weight [26]. Alternative splicing generates a long and short form of ITM2B. Interestingly, the short form induces apoptosis independently of p53 in hematopoietic cell lines [13], a function that is similar to that of some other targets of BCL6, e.g., PDCD2 [28].
BCL6 encodes a POZ/zinc finger transcriptional repressor, which regulates gene expression by interactions with co-repressors that recruit histone deacetylases and induce epigenetic remodeling and heterochromatin formation [29,30]. BCL6 expression is not limited to B cells; BCL6 also contributes to the normal function of T-cell subsets [5,6], macrophages [31], progenitor cells during normal hematopoiesis, and to the function of cancer stem cells in myeloid leukemia [30]. Further, BCL6 has been found to be involved in interactions with other genes that affect neural functions such as G protein signaling in neuron-like cells [32] and differentiation of the cerebral cortex in a mouse model [33].
The BCL6 and ITM2B proteins are both expressed in many tissues, not only in lymphoid and hematopoietic cells [13,29] but also in the brain [12,34,35]. In human brain and mouse brain, ITM2B has been found to be expressed in olfactory neurons, the cerebral cortex, hippocampus, and cerebellum. In the mouse, BCL6 expression is found in these regions as well [36–40]. In humans, although BCL6 is expressed in the brain [34], to our knowledge, its regional distribution has not been characterized. Since ITM2B and BCL6 have a similar regional expression pattern in the mouse, at least, experiments to study the possible colocalization and regulation of ITM2B by BCL6 in the human brain and in neuron-derived cells would be of great interest. Additionally, testing the effects of BCL6 in a three-dimensional human neural cell culture model of Alzheimer’s disease such as that recently published by Choi et al. [41] would be very exciting.
Working with a B lymphoma cell line, we have presented the following observations in support of the concept that the ITM2B gene is a target of BCL6 repression: (a) subtractive hybridization in this cell line, in which BCL6 repressive effects were inhibited, followed by Northern blotting, revealed differential expression of ITM2B; (b) ChIP assays showed that BCL6 binds to the high-affinity consensus binding site identified in the first intron of the ITM2B gene; (c) in transient transfection assays the full-length BCL6 protein caused significant repression (3-fold) from the BCL6 consensus binding site in the first intron of ITM2B as compared with a truncated control lacking the BCL6 zinc fingers that cannot bind DNA; (d) knockdown of BCL6 by siRNA duplexes led to a significant increase in endogenous ITM2B protein; and (e) immunohistochemical staining revealed a reciprocal relationship between BCL6 and ITM2B protein expression in B- and T-cell human lymphomas.
The short form of the ITM2B protein appears to play a role in induction of apoptosis [13]. Through repression of specific target genes, it is very likely that BCL6 inhibits apoptosis [11,14,23,28]. Thus, the deregulation of BCL6, as is known to be associated with lymphomagenesis, likely leads to perturbation of apoptosis events, perhaps resulting in abnormal growth of cells. Only about 50% of B cell lymphomas can be cured with current therapies; T-cell lymphomas are even more resistant to treatment than B cell tumors, and most patients die from these neoplasms. The inhibition or upregulation of specific targets could permit the selection of patients with B- or T-cell lymphomas who might benefit most (personalized therapy) and reduce the unwanted side effects of more global immunosuppressive and cytotoxic treatments.
No approved specific molecular targeted treatment is available for FBD, FDD or AD, which may share common molecular pathways leading to dementia [35]. Familial dementias are caused by mutations in the gene that encodes the amyloid precursor protein (APP) [42] and in genes that regulate the processing of APP [43], which include ITM2B; the ITM2B protein binds to APP and inhibits its processing [16,18,44]. Fotinopoulou et al. [44] reported an interaction between the amyloid precursor protein (APP) and ITM2B, which led to a reduction in APP secretion as well as amyloid β (Aβ) peptides. Their data suggested that ITM2B inhibits the activity of α-secretase. Matsuda et al. demonstrated that maturation and transport of ITM2B along the secretory pathway is essential for generating an inhibitor of the processing of the APP. These data establish ITM2B as a potentially important molecular target for an anti-amyloid therapy in AD [16–18,44].
Peng et al. [45] suggested a model for how the processing of ITM2B and APP are related—these authors found that recombinant human ITM2B, residues 90–236, binds to ABri23 (the main component of amyloid deposits in FBD) as well as to Aβ1–40 (which is cleaved from the N-terminal extracellular part of APP), inhibiting its aggregation and fibril formation. They found that the binding of ABRi23 to ITM2B(90–236) is stronger than that of Aβ1–40, which blocks interactions between ITM2B and APP. Proteolytic release of the ABRi23 peptide results in making the ITM2B pocket available for binding to the Aβ region of APP. Tamayev et al. indicated that reducing β-cleavage of the APP is an appropriate approach for treating human dementias [19]. In a mouse model, Tamayev et al. showed that memory deficits due to the FBD ITM2B mutation are due to loss of ITM2B function rather than amyloidosis [20]. Similarly, in a mouse knock-in model of FDD, loss of ITM2B function, not the amyloid cascade, resulted in synaptic and memory deficits [46]. Consistent with this finding, the levels of several amyloid-β protein precursor (AβPP) metabolites, including Aβ, were found to be increased in the brain from a patient with FDD [47]. Further, Tamayev et al. reported that APP heterozygosity prevents memory and synaptic dysfunctions in FDD knock-in mice, suggesting that FDD pathogenesis may be due to APP and/or toxic APP metabolites [48]. APP haploinsufficiency also prevented memory deficits in a murine knock-in model of FBD [49].
In addition, Tamayev and D’Adamio [50] reported that as ITM2B binds to APP and inhibits APP cleavage by secretases, processing of APP is increased in FDD due to the loss of ITM2B. Processing of APP by β-secretase results in the products amino-terminal-soluble APPβ (sAPPβ) and β-carboxyl-terminal (β-CTF); γ-secretase processing of the latter product yields Aβ, which is considered the main cause of AD, but inhibition of γ-secretase did not ameliorate the synaptic/memory deficits of FDD knock-in mice. Thus, the authors [50] suggested that APP and ITM2B interact, that APP mediates FDD neuropathology, and that sAPPβ and/or β-CTF (not Aβ) are the cause of dementia. Lombino et al. [43] showed that mutation of a single residue of APP, the intracellular amino acid Thr668, which prevents phosphorylation at this residue, can prevent memory and synaptic plasticity deficits in the knock-in mouse model of FDD. APPPThr668 is enriched in patients with AD [51], suggesting a pathogenic role for phosphorylation at this site. Because this mutation does not appear to alter essential functions of APP during development [52], it has been suggested [43] that targeting the role of Thr668, and perhaps its phosphorylation, may offer a safe therapeutic approach to dementias.
Of additional interest are the recent findings by Del Campo et al. [15], indicating that the ITM2B-containing BRICHOS-domain is increased up to 3-fold in the hippocampus of patients with early stages of AD as compared with controls, and loss of ITM2B function in AD is supported by the decreased presence of ITM2B-amyloid precursor protein complexes in the hippocampus of these patients. Thus, a further understanding of the genes, such as BCL6, that function as regulators of the ITM2B protein may provide insights for the development of new, specific molecular therapies that could benefit patients who will die from these diseases.
Acknowledgments
We are very grateful to Dr. J. Turner for the use of his laboratory’s cell culture facilities, luminometer, and sonicator. We thank Dr. L. Shen (Turner laboratory) for helpful discussions and D. Lane (Hematopathology Laboratory) for BCL6 immunohistochemistry.
This work was supported by the Department of Pathology at The University of Chicago (to B.W.B.), University of Chicago Cancer Center, support grant P30 CA014599 (to B.W.B.), and Hematology Research Funds at The University of Chicago donated by S. Samsky and E. Lanzl (to J.M.B.).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Baron BW, Nucifora G, McCabe N, Espinosa R, III, Le Beau MM, McKeithan TW. Identification of the gene associated with the recurring chromosomal translocations t(3;14)(q27;q32) and t(3;22)(q27;q11) in B-cell lymphomas. Proc Natl Acad Sci U S A. 1993;90:5262–5266. doi: 10.1073/pnas.90.11.5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang CC, Ye BH, Chaganti RS, Dalla-Favera R. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc Natl Acad Sci U S A. 1996;93:6947–6952. doi: 10.1073/pnas.93.14.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seyfert VL, Allman D, He Y, Staudt LM. Transcriptional repression by the proto-oncogene BCL-6. Oncogene. 1996;12:2331–2342. [PubMed] [Google Scholar]
- 4.Baron BW, Desai M, Baber LJ, Paras L, Zhang Q, Sadhu A, Duguay S, Nucifora G, McKeithan TW, Zeleznik-Le N. BCL6 can repress transcription from the human immunodeficiency virus type 1 promoter/enhancer region. Genes Chromosom Cancer. 1997;19:14–21. [PubMed] [Google Scholar]
- 5.Chang CC, Vlad G, D’Agati VD, Liu Z, Zhang QY. BCL6 is required for differentiation of Ig-like transcript 3-Fc-induced CD8+ T suppressor cells. Immunology. 2010;185:5714–5722. doi: 10.4049/jimmunol.1001732. [DOI] [PubMed] [Google Scholar]
- 6.Liu X, Yan X, Zhong B, Nurieva RI, Wang A, Wang X, Martin-Orozco N, Wang Y, Chang SH, Esplugues E, Flavell RA, Tian Q, Dong C. Bcl6 expression specifies the T follicular helper cell program in vivo. J Exp Med. 2012;209:1841–1852. doi: 10.1084/jem.20120219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basso K, Schneider C, Shen Q, Holmes AB, Setty M, Leslie C, Dalla-Favera R. BCL6 positively regulates AID and germinal center gene expression via repression of miR-155. J Exp Med. 2012;209:2455–2465. doi: 10.1084/jem.20121387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sena P, Mariani F, Benincasa M, De Leon MP, Di Gregorio C, Mancini S, Cavani F, Smargiassi A, Palumbo C, Roncucci L. Morphological and quantitative analysis of BCL6 expression in human colorectal carcinogenesis. Oncol Rep. 2014;31:103–110. doi: 10.3892/or.2013.2846. [DOI] [PubMed] [Google Scholar]
- 9.Sato T, Tran TH, Peck AR, Girondo MA, Liu C, Goodman CR, Neilson LM, Freydin B, Chervoneva I, Hyslop T, Kovatich AJ, Hooke JA, Shriver CD, Fuchs SY, Rui H. Prolactin suppresses a progestin-induced CK5-positive cell population in luminal breast cancer through inhibition of progestin-driven BCL6 expression. Oncogene. 2014;33:2215–2224. doi: 10.1038/onc.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xin N, Fu L, Shao Z, Guo M, Zhang X, Zhang Y, Dou C, Zheng S, Shen X, Yao Y, Wang J, Wang J, Cui G, Liu Y, Geng D, Xiao C, Zhang Z, Dong R. RNA interference targeting Bcl-6 ameliorates experimental autoimmune myasthenia gravis in mice. Mol Cell Neurosci. 2014;58:85–94. doi: 10.1016/j.mcn.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 11.Baron BW, Anastasi J, Thirman MJ, Furukawa Y, Fears S, Kim DC, Simone F, Birkenbach M, Montag A, Sadhu A, Zeleznik-Le N, McKeithan TW. The human programmed cell death-2 (PDCD2) gene is a target of BCL6 repression: Implications for a role of BCL6 in the down-regulation of apoptosis. Proc Natl Acad Sci U S A. 2002;99:2860–2865. doi: 10.1073/pnas.042702599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pittois K, Deleersnijder W, Merregaert J. cDNA sequence analysis, chromosomal assignment and expression pattern of the gene coding for integral membrane protein 2B. Gene. 1998;217:141–149. doi: 10.1016/s0378-1119(98)00354-0. [DOI] [PubMed] [Google Scholar]
- 13.Fleischer A, Rebollo A. Induction of p53-independent apoptosis by the BH3-only protein ITM2Bs. FEBS Lett. 2004;557:283–287. doi: 10.1016/s0014-5793(03)01519-9. [DOI] [PubMed] [Google Scholar]
- 14.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 15.Del Campo M, Hoozemans JJ, Dekkers LL, Rozemuller AJ, Korth C, Muller-Schiffmann A, Scheltens P, Blankenstein MA, Jimenez CR, Veerhuis R, Teunissen CE. BRI2-BRICHOS is increased in human amyloid plaques in early stages of Alzheimer’s disease. Neurobiol Aging. 2014;35:1596–1604. doi: 10.1016/j.neurobiolaging.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Matsuda S, Giliberto L, Matsuda Y, Davies P, McGowan E, Pickford F, Ghiso J, Frangione B, D’Adamio L. The familial dementia BRI2 gene binds the Alzheimer gene amyloid-β precursor protein and inhibits amyloid-β production. J Biol Chem. 2005;280:28912–28916. doi: 10.1074/jbc.C500217200. [DOI] [PubMed] [Google Scholar]
- 17.Matsuda S, Giliberto L, Matsuda Y, McGowan EM, D’Adamio L. BRI2 inhibits amyloid β-peptide precursor protein processing by interfering with the docking of secretases to the substrate. J Neurosci. 2008;28:8668–8676. doi: 10.1523/JNEUROSCI.2094-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuda S, Matsuda Y, Snapp EL, D’Adamio L. Maturation of BRI2 generates a specific inhibitor that reduces APP processing at the plasma membrane and in endocytic vesicles. Neurobiol Aging. 2011;32:1400–1408. doi: 10.1016/j.neurobiolaging.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamayev R, Matsuda S, Arancio O, D’Adamio L. β- but not γ-secretase proteolysis of APP causes synaptic and memory deficits in a mouse model of dementia. EMBO Mol Med. 2012;4:171–179. doi: 10.1002/emmm.201100195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamayev R, Giliberto L, Li W, d’Abramo C, Arancio O, Vidal R, D’Adamio L. Memory deficits due to familial British dementia BRI2 mutation are caused by loss of BRI2 function rather than amyloidosis. J Neurosci. 2010;30:14915–14924. doi: 10.1523/JNEUROSCI.3917-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura T, Yamazaki Y, Saiki Y, Moriyama M, Largaespada DA, Jenkins NA, Copeland NG. Evi9 encodes a novel zinc finger protein that physically interacts with BCL6, a known human B-cell proto-oncogene product. Mol Cell Biol. 2000;20:3178–3186. doi: 10.1128/mcb.20.9.3178-3186.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi SI, Vidal R, Frangione B, Levy E. Axonal transport of British and Danish amy-loid peptides via secretory vesicles. FASEB J. 2004;18:373–375. doi: 10.1096/fj.03-0730fje. [DOI] [PubMed] [Google Scholar]
- 23.Baron BW, Zeleznik-Le N, Baron MJ, Theisler C, Huo D, Krasowski MD, Thirman MJ, Baron RM, Baron JM. Repression of the PDCD2 gene by BCL6 and the implications for the pathogenesis of human B and T cell lymphomas. Proc Natl Acad Sci U S A. 2007;104:7449–7454. doi: 10.1073/pnas.0701770104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baron BW, Anastasi J, Hyjek EM, Baron JM. PIM1 gene cooperates with human BCL6 gene to promote the development of lymphomas. Proc Natl Acad Sci U S A. 2012;109:5735–5739. doi: 10.1073/pnas.1201168109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ci W, Polo JM, Cherchietti L, Shaknovich R, Wang L, Yang SN, Ye K, Farinha P, Horsman DE, Gascoyne RD, Elemento O, Melnick A. The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood. 2009;113:5536–5548. doi: 10.1182/blood-2008-12-193037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin L, Fluhrer R, Reiss K, Kremmer E, Saftig P, Haass C. Regulated intramembrane proteolysis of Bri2 (Itm2b) by ADAM 10 and SPPL2a/Sppl2b. J Biol Chem. 2008;283:1644–1652. doi: 10.1074/jbc.M706661200. [DOI] [PubMed] [Google Scholar]
- 27.Tsachaki M, Serlidaki D, Fetani A, Zarkou V, Rozani I, Ghiso J, Efthimiapoulos S. Glycosylation of BRI2 on asparagine 170 is involved in its trafficking to the cell surface but not in its processing by furin or ADAM10. Glycobiology. 2011;21:1382–1388. doi: 10.1093/glycob/cwr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baron BW, Hyjek E, Gladstone B, Thirman MJ, Baron JM. PDCD2, a protein whose expression is repressed by BCL6, induces apoptosis in human cells by activation of the caspase cascade. Blood Cells Mol Dis. 2010;45:169–175. doi: 10.1016/j.bcmd.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 29.Dent AL, Vasanwala FH, Toney LM. Regulation of gene expression by the proto-oncogene BCL-6. Crit Rev Oncol Hematol. 2002;41:1–9. doi: 10.1016/s1040-8428(01)00164-0. [DOI] [PubMed] [Google Scholar]
- 30.Green MR, Vicente-Duenas C, Romero-Camarero I, Long Liu C, Dai B, Gonzalez-Herrero I, Garcia-Ramirez I, Alonso-Escudero E, Iqbal J, Chan WC, Campos-Sanchez E, Orfao A, Pintado B, Flores T, Blanco O, Jimenez R, Martinez-Climent JA, Criado FJ, Cenador MB, Zhao S, Natkunam Y, Lossos IS, Majeti R, Melnick A, Cobaleda C, Alizadeh AA, Sanchez-Garcia I. Transient expression of Bcl6 is sufficient for oncogenic function and induction of mature B-cell lymphoma. Nat Commun. 2014;5:3904–3916. doi: 10.1038/ncomms4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toney LM, Cattoretti G, Graf JA, Merghoub T, Pandolfi PP, Dalla-Favera R, Ye BH, Dent AL. BCL-6 regulates chemokine gene transcription in macrophages. Nat Immunol. 2000;1:214–220. doi: 10.1038/79749. [DOI] [PubMed] [Google Scholar]
- 32.Yang J, Huang J, Chatterjee TK, Twait E, Fisher RA. A novel mechanism involving coordinated regulation of nuclear levels and acetylation of NF-YA and Bcl6 activates RGS4 transcription. J Biol Chem. 2010;285:29760–29769. doi: 10.1074/jbc.M110.121459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bedogni F, Hodge RD, Elsen GE, Nelson BR, Daza RAM, Beyer RP, Bammler TK, Rubenstein JLR, Hevner RF. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc Natl Acad Sci U S A. 2010;107:13129–13134. doi: 10.1073/pnas.1002285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allman D, Jain A, Dent A, Maile RR, Selvaggi T, Kehry MR, Staudt LM. BCL-6 expression during B-cell activation. Blood. 1996;87:5257–5268. [PubMed] [Google Scholar]
- 35.Del Campo M, Teunissen CE. Role of BRI2 in dementia. J Alzheimers Dis. 2014;40:481–494. doi: 10.3233/JAD-131364. [DOI] [PubMed] [Google Scholar]
- 36.Bajalica-Lagercrantz S, Piehl F, Farnebo F, Larsson C, Lagercrantz J. Expression of the BCL6 gene in the pre- and postnatal mouse. Biochem Biophys Res Commun. 1998;247:357–360. doi: 10.1006/bbrc.1998.8551. [DOI] [PubMed] [Google Scholar]
- 37.Pickford F, Onstead L, Camacho-Prihar C, Hardy J, McGowan E. Expression of mBRI2 in mice. Neurosci Lett. 2003;338:95–98. doi: 10.1016/s0304-3940(02)01356-3. [DOI] [PubMed] [Google Scholar]
- 38.Akiyama H, Kondo H, Arai T, Ikeda K, Kato M, Iseki E, Schwab C, McGeer PL. Expression of BRI, the normal precursor of the amyloid protein of familial British dementia, in human brain. Acta Neuropathol. 2004;107:53–58. doi: 10.1007/s00401-003-0783-1. [DOI] [PubMed] [Google Scholar]
- 39.Otaki JM, Fearon DT, Yamamoto H. The proto-oncogene BCL-6 is expressed in olfactory sensory neurons. Neurosci Res. 2005;53:189–200. doi: 10.1016/j.neures.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 40.Korutla L, Wang P, Jackson TG, Mackler SA. NAC1, a POZ/BTB protein that functions as a corepressor. Neurochem Int. 2009;54:245–252. doi: 10.1016/j.neuint.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 41.Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, D’Avanzo C, Chen H, Hooli B, Asselin C, Muffat J, Klee JB, Zhang C, Wainger BJ, Peitz M, Kovacs DM, Woolf CJ, Wagner SL, Tanzi RE, Kim DY. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature. 2014;515:274–278. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bertram L, Lill CM, Tanzi RE. The genetics of Alzheimer disease: back to the future. Neuron. 2010;68:270–281. doi: 10.1016/j.neuron.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 43.Lombino F, Biundo F, Tamayev R, Arancio O, D’Adamio L. An intracellular threonine of amyloid-β precursor protein mediates synaptic plasticity deficits and memory loss. PLoS One. 2013;8:e57120. doi: 10.1371/journal.pone.0057120. Epub Feb. 22, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fotinopoulou A, Tsachaki M, Vlavaki M, Poulopoulos A, Rostagno A, Frangione B, Ghiso J, Efthimiopoulos S. BRI2 interacts with amyloid precursor protein (APP) and regulates amyloid β (Aβ) production. J Biol Chem. 2005;280:30768–30772. doi: 10.1074/jbc.C500231200. [DOI] [PubMed] [Google Scholar]
- 45.Peng S, Fitzen M, Jornvall H, Johansson J. The extracellular domain of Bri2 (ITM2B) binds the ABri peptide (1–23) and amyloid β-peptide (Aβ1-40): implications for Bri2 effects on processing of amyloid precursor protein and Aβ aggregation. Biochem Biophys Res Commun. 2010;393:356–361. doi: 10.1016/j.bbrc.2009.12.122. [DOI] [PubMed] [Google Scholar]
- 46.Tamayev R, Matsuda S, Fa M, Arancio O, D’Adamio L. Danish dementia mice suggest that loss of function and not the amyloid cascade causes synaptic plasticity and memory deficits. Proc Natl Acad Sci U S A. 2010;107:20822–20827. doi: 10.1073/pnas.1011689107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsuda S, Tamayev R, D’Adamio L. Increased AβPP processing in familial Danish dementia patients. J Alzheimers Dis. 2011;27:385–391. doi: 10.3233/JAD-2011-110785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamayev R, Matsuda S, Giliberto L, Arancio O, D’Adamio L. APP heterozygosity averts memory deficit in knockin mice expressing the Danish dementia BRI2 mutant. EMBO J. 2011;30:2501–2509. doi: 10.1038/emboj.2011.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamayev R, D’Adamio L. Memory deficits of British dementia knock-in mice are prevented by APP haploinsufficiency. J Neurosci. 2012;32:5481–5485. doi: 10.1523/JNEUROSCI.5193-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamayev R, D’Adamio L. Inhibition of γ-secretase worsens memory deficits in a genetically congruous mouse model of Danish dementia. Mol Neurodegener. 2012;7:19–25. doi: 10.1186/1750-1326-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shin RW, Ogino K, Shimabuku A, Taki T, Nakashima H, Ishihara T, Kitamoto T. Amyloid precursor protein cytoplasmic domain with phospho-Thr668 accumulates in Alzheimer’s disease and its transgenic models: a role to mediate interaction of Aβ and tau. Acta Neuropathol. 2007;113:627–636. doi: 10.1007/s00401-007-0211-z. [DOI] [PubMed] [Google Scholar]
- 52.Barbagallo APM, Wang Z, Zheng H, D’Adamio L. The intracellular threonine of amyloid precursor protein that is essential for docking of Pin1 is dispensable for developmental function. PLoS One. 2011;6:e18006. doi: 10.1371/journal.pone.0018006. [DOI] [PMC free article] [PubMed] [Google Scholar]