Abstract
We previously reported that patients with lung adenocarcinomas with KRAS gene mutations and strong proliferating activity had poorer outcomes, even in the early stage of the disease. The aim of the present study was to elucidate the potential molecular basis of these highly malignant lung tumors by focusing on S100 proteins (S100A2, S100A7, and S100A11), which are downstream targets of oncogenic KRAS and promoters of tumor progression. The immunohistochemical expression of S100 proteins was examined in 179 primary lung adenocarcinomas, and the potential relationships between their levels and clinicopathologic factors were analyzed. Among the three subtypes, S100A11 levels were significantly higher in adenocarcinomas with KRAS mutations and strong proliferating activity. They were also higher in adenocarcinomas with poorly differentiated tumors. Furthermore, higher levels of S100A11 were associated with shorter disease-free survival. These results suggest that the up-regulation of S100A11 plays a role in tumor progression, particularly in KRAS-mutated lung adenocarcinomas.
Introduction
Lung cancer is one of the most common causes of cancer-related death in the developed world [1–2]. A large proportion of patients, even those with early stage non-small-cell lung cancer, die due to recurrent disease [3–4]. Although some lung tumors are sensitive to conventional chemotherapeutic agents or certain molecular targeting agents, many are not [5–6]. Thus, a deeper understanding of the molecular basis of lung carcinogenesis is needed in order to develop novel therapeutic strategies.
We previously reported that patients with lung adenocarcinomas with KRAS gene mutations and strong proliferating activity had poorer outcomes, even in the early stage of the disease [6]. Therefore, the aim of the present study was to elucidate the potential molecular basis of these highly malignant lung tumors by focusing on S100 proteins. The S100 protein family binds to calcium and modulates the transmission of various cellular signals. We previously demonstrated that S100A2, S100A7, and S100A11 were downstream targets of oncogenic KRAS (Tables 1 and 2) [7–8]. The expression of S100 proteins was also shown to be altered in various cancers [9–20] (laryngeal [10], breast [11], lung [12–13], gastric [14], colorectal [15], hepatocellular [16], pancreatic [17], prostate [18], kidney [19], and bladder cancers [20]) including lung cancers [12–13], and these proteins have been suggested to promote the progression of carcinogenesis by modulating cell migration and proliferative activity [21].
Table 1. Downstream targets up-regulated by oncogenic KRAS with gene chip microarray analysis.
| Mock | KRAS/G12 | KRAS/V12 | Ratio | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Accession | Map | Signal Value | Flags | Signal Value | Flags | Signal Value | Flags | V12/G12 | V12/MOCK |
| PPBP | R64130 | 4q12-q13 | 0.060 | A | 0.106 | A | 24.380 | P | 230.647 | 406.330 |
| NAV3 | NM_014903 | 12q14.3 | 0.180 | A | 0.307 | A | 32.910 | P | 107.024 | 182.831 |
| IL1RL1 | NM_003856 | 2q12 | 0.123 | A | 1.000 | A | 17.379 | P | 17.379 | 141.224 |
| ESM1 | NM_007036 | 5q11.2 | 0.170 | A | 0.404 | A | 22.068 | P | 54.680 | 129.812 |
| SERPINB2 | NM_002575 | 18q21.3 | 0.353 | P | 0.100 | P | 36.969 | P | 36.969 | 104.676 |
| CCL3 | NM_002983 | 17q11-q21 | 0.420 | A | 0.932 | A | 43.922 | P | 47.122 | 104.577 |
| MMP1 | NM_002421 | 11q22.3 | 0.823 | P | 1.000 | P | 82.574 | P | 82.574 | 100.284 |
| NICE-1 | NM_019060 | 1q21 | 0.952 | A | 1.000 | A | 82.501 | P | 82.501 | 86.700 |
| IL24 | NM_006850 | 1q32 | 0.205 | P | 1.000 | P | 14.916 | P | 14.916 | 72.744 |
| IL1B | NM_000576 | 2q14 | 0.589 | P | 1.000 | P | 32.344 | P | 32.344 | 54.881 |
| PI3 | NM_002638 | 20q12-q13 | 0.270 | A | 0.327 | A | 14.277 | P | 43.698 | 52.877 |
| S100A7 | NM_002963 | 1q21 | 0.300 | A | 0.625 | A | 9.611 | P | 15.387 | 32.035 |
| HSD17B2 | NM_002153 | 16q24.1-q24.2 | 0.160 | A | 0.231 | A | 5.041 | P | 21.857 | 31.504 |
| FOS | BC004490 | 14q24.3 | 0.480 | A | 0.951 | A | 11.526 | P | 12.116 | 24.013 |
Accession, gene bank accession number; Map, chromosome locus; MOCK, mock-transduced NHBE-T; G12, wild-type KRAS-transduced NHEB-T; V12, oncogenic mutant KRAS-transduced NHBE-T.
Flags indicate whether gene expression was present (P) or absent (A). Genes whose signal values were more than 20-fold higher in KRAS/V12 cells than in mock- and/or KRAS/G12 cells are extracted from our date of a gene chip microarray analysis previously described [7]. The genes sorted are listed in this table.
Table 2. Downstream targets up-regulated by oncogenic KRAS with proteomic analysis.
| Signal Intensity | Signal Ratio | |||||||
|---|---|---|---|---|---|---|---|---|
| Protein | Accession | Map | P value | MOCK | G12 | V12 | V12/G12 | V12/MOCK |
| S100A2 | NM_005978 | 1q21.33 | 0.0001214 | 0.400 | 1.005 | 1.665 | 1.657 | 4.163 |
| VIME | NM_003380 | 10p13 | 0.00000091 | 0.368 | 0.754 | 1.424 | 1.889 | 3.870 |
| EEF2 | NM_001961 | 19p13.3 | 0.00000313 | 0.454 | 0.922 | 1.515 | 1.643 | 3.337 |
| GC | NM_000583 | 4q12-q13 | 0.001 | 0.480 | 0.934 | 1.582 | 1.694 | 3.296 |
| CTSL1 | NM_001912 | 9q21.33 | 0.0003414 | 0.635 | 0.708 | 1.708 | 2.412 | 2.690 |
| VCP | NM_007126 | 9p13.3 | 0.002 | 0.599 | 1.010 | 1.551 | 1.536 | 2.589 |
| ENOA | NM_001428 | 1p36.2 | 0.0001339 | 0.641 | 0.936 | 1.560 | 1.667 | 2.434 |
| S100A11 | NM_005620 | 1q21 | 0.002 | 0.569 | 0.761 | 1.291 | 1.696 | 2.269 |
| CTSL1 | NM_001912 | 9q21.33 | 0.001 | 0.730 | 0.802 | 1.551 | 1.934 | 2.125 |
Accession, gene bank accession number; Map, chromosome locus; MOCK, mock-transduced NHBE-T; G12, wild-type KRAS-transduced NHEB-T; V12, oncogenic mutant KRAS-transduced NHBE-T.
Proteins whose signal intensities were more than 2-fold higher in KRAS/V12 cells than in mock- and/or KRAS/G12 cells are extracted from our date of a comprehensive proteomic analysis previously described [8]. The proteins sorted are listed in this table.
We here examined 179 surgically resected primary lung adenocarcinomas for the immunohistochemical expression of S100 proteins (S100A2, S100A7, and S100A11), and analyzed the potential relationships between their levels and clinicopathologic factors.
Materials and Methods
Primary Lung Cancer
All 179 lung cancers examined were cases of stage I adenocarcinoma that underwent radical surgical resection at the Kanagawa Cardiovascular and Respiratory Center (Yokohama, Japan). The research plan was approved by the Ethics Committees of Yokohama City University and Kanagawa Cardiovascular and Respiratory Center. Written informed consent for the research use of resected materials was obtained from all subjects providing materials.
Histologic subtypes were classified according to the 2011 IASLC/ATS/ERS classification of lung adenocarcinoma [22], and tumor stages were determined according to the international TNM classification system (seventh edition of UICC)[23].
Western blotting
Tissue lysates were subjected to SDS-polyacrylamide gel electrophoresis, and transferred onto PVDF membranes (Amersham, Piscataway, NJ). The membranes were then incubated with nonfat dry milk in Tris-buffered saline containing Tween-20 (TBS-T) in order to block non-immunospecific protein binding, and then with a primary antibody against S100 A11 (Santa Cruz, Santa Cruz, CA), and beta-actin (Sigma, St. Louis, MO). After washing with TBS-T, the membranes were incubated with animal-matched HRP-conjugated secondary antibodies (Amersham). Immunoreactivity was visualized with an enhanced chemiluminescence system (Amersham).
Immunohistochemistry
Tumor sections were cut from formalin-fixed, paraffin-embedded tissue blocks. Sections were deparaffinized, rehydrated, and incubated with 3% hydrogen peroxide, followed by 5% goat serum to block endogenous peroxidase activities and non-immunospecific protein binding. Sections were boiled in citrate buffer (0.01M, pH6.0) for 15 minutes to retrieve masked epitopes and then incubated with primary antibodies against S100A2, S100A7, and S100A11 (Santa Cruz) as well as Ki-67 (DAKO, Ely, UK). Immunoreactivity was visualized using an Envision detection system (DAKO), and nuclei were counterstained with hematoxylin. The immunohistochemical expression levels of S100 proteins were evaluated by a scoring system as described in the Results section. The labeling index of Ki-67 was calculated as the proportion of cells with positive nuclei among 500–1000 cancer cells. Ki-67 labeling indices of <10% and > = 10% were classified as low and high levels based on the findings of our previous study [6].
Statistical Analysis
Differences in scores among the groups classified based on clinicopathologic subjects were analyzed by the Mann-Whitney or Kruskal-Wallis test. The relationships between scores and clinicopathologic subjects were analyzed using a multiple linear regression model. The post-operative disease-free span was defined as the period ranging from the date of surgery to the date when recurrence was diagnosed. An observation was censored at the last follow-up if the patient was alive or had died of a cause other than lung cancer. Recurrence curves were plotted using the Kaplan-Meier method, and the absolute risk of recurrence at five years was estimated from these curves. Differences in the disease-free survival spans were analyzed using the Log-rank test. P values less than 0.05 were considered significant. All statistical analyses were performed using SPSS software (SPSS for Windows Version 10.0; SPSS; Chicago, IL, USA).
Results
Immunohistochemical expression of S100 proteins
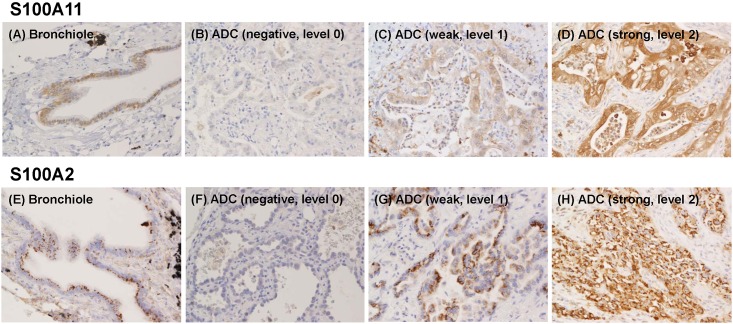
The S100A11 protein was expressed in the nuclei and cytoplasm of the normal epithelial cells of bronchioles (Fig 1A), while the S100A2 protein was expressed predominantly on the apical side of the cytoplasm in a granular form (Fig 1E).
Fig 1. Immunohistochemical examination of S100A11 and S100A2 protein expression levels in tumors and non-tumorous epithelia from lung adenocarcinoma (ADC) patients undergoing surgical resection.
Representative photographs from normal bronchioles (A, E) and tumors, in which the expression of S100A11 and S100A2 was negative (B, F), weak (C, G) and strong (D, H), are shown.
S100 protein levels varied in neoplastic cells, and even in individual tumors. The expression of these proteins was higher in some neoplastic cells than in bronchiolar epithelial cells (Fig 1D and 1H), whereas other cells expressed them weakly (Fig 1C and 1G) or at an undetectable level (Fig 1B and 1E). On the other hand, the S100A7 protein was only expressed in a few adenocarcinoma cells. It was strongly expressed in the keratinizing cells of squamous cell carcinomas (S1 Fig). Thus, the S100A7 protein was excluded from the following analysis.
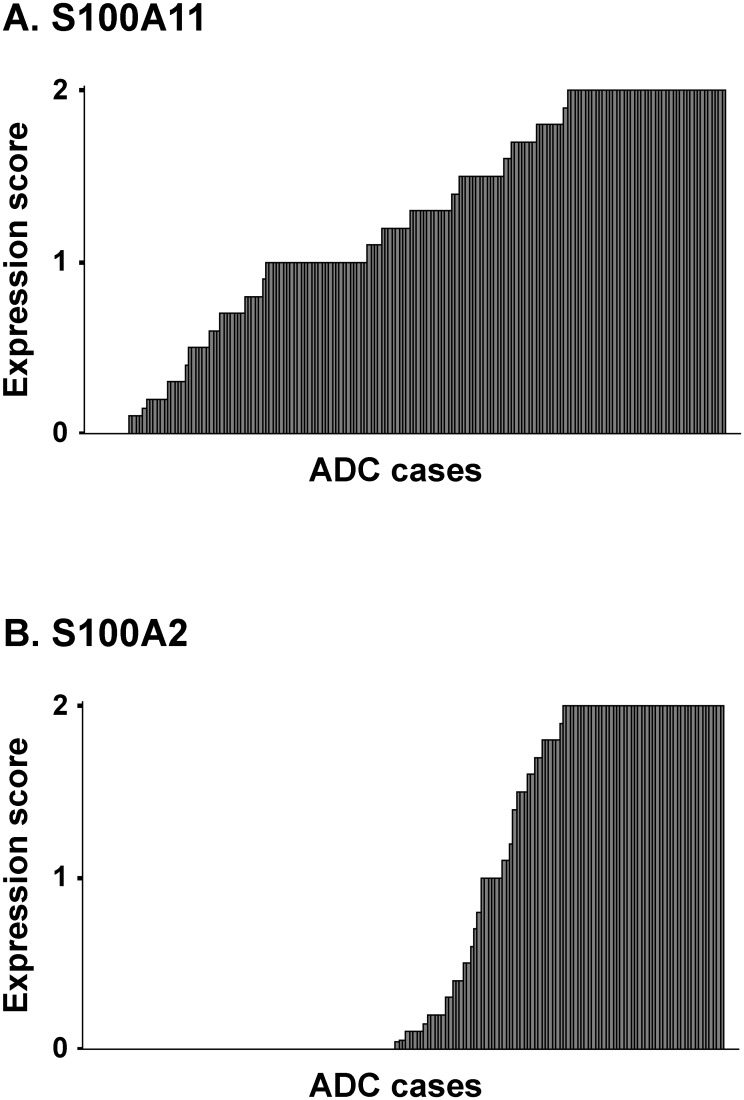
The expression levels of S100A2 and S100A11 were classified into negative (level 0), weak (level 1), and strong (level 2). The weak level was defined as the level almost equivalent to that in bronchiolar epithelial cells. The strong level was defined as an unequivocally stronger level than that in bronchiolar epithelial cells. The immunohistochemical expression score was determined as the average level in a maximal tumor section (if 30%, 10%, and 60% of neoplastic cells in the maximal tumor section were negative, weak, and strong levels, respectively, the average level was calculated as “0.3x0+0.1x1+0.6x2 = 1.3”). The expression levels of S100A2 and S100A11 in all the tumors examined are shown in Fig 2A and 2B.
Fig 2. The S100A11 and S100A2 expression levels in all tumors.
Median expression scores were 1.30 in S100A11 (A) and 0.10 in S100A2 (B). S100A11 was not expressed in 8 cases (A), while S100A2 was not expressed in nearly half of the cases examined (83 cases) (B). ADC, Adenocarcinoma.
The validity of the immunohistochemical evaluation was supported by the Western blot analysis because the S100 protein levels evaluated by immunohistochemistry and those by Western blotting were roughly parallel (The result of a Western blot for S100A11 was shown in S2 Fig).
S100 protein levels and clinicopathologic factors
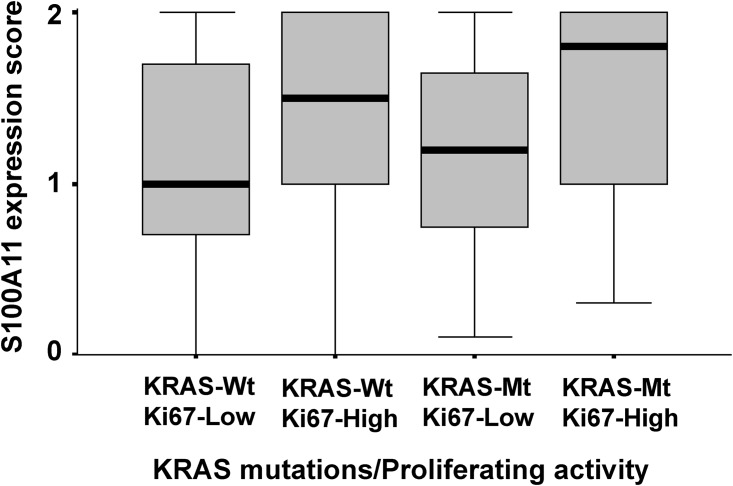
S100A11 levels were significantly higher in adenocarcinomas with KRAS mutations and strong proliferating activity (P = 0.038 in the Kruskal-Wallis test, Fig 3). They were also significantly higher in adenocarcinomas with poorly differentiated tumors (P = 0.021 in the Kruskal-Wallis test, Table 3) and lymphatic or vascular involvement (P = 0.026 in the Mann-Whitney test, Table 3). A multiple linear regression analysis showed that poorly differentiated tumors correlated with higher S100A11 levels (P = 0.039, Table 4). On the other hand, S100A2 levels were not associated with KRAS mutations, proliferating activity, or any of the other clinicopathologic factors (data not shown).
Fig 3. S100A11 expression levels in adenocarcinomas with KRAS mutations and proliferating activity.
The thickened lines indicate the median score of S100 A11 expression, which was 1.00 in KRAS Wild-type/Ki-67 Low cases (n = 94), 1.50 in KRAS Wild-type/Ki-67 High cases (n = 62), 1.20 in KRAS Mutated-type/Ki-67 Low cases (n = 11) and 1.80 in KRAS Mutated-type/Ki-67 High cases (n = 11). S100A11 expression levels were significantly higher in adenocarcinomas with KRAS mutations and strong proliferating activity (P = 0.038 in the Kruskal-Wallis test). Wt, Wild-type; Mt, Mutated-type.
Table 3. Relationship between S100A11 expression and clinicopathologic characteristics of stage I lung adenocarcinomas.
| Subjects | No. | Expression score (median) | P-value |
|---|---|---|---|
| Age | |||
| <65 | 63 | 1.20 | P = 0.617 |
| > = 65 | 116 | 1.30 | |
| Gender | |||
| Male | 89 | 1.20 | P = 0.845 |
| Female | 90 | 1.30 | |
| Histologic Grade | |||
| WEL | 42 | 1.00 | P = 0.021 |
| MOD | 90 | 1.30 | |
| POR | 47 | 1.50 | |
| Histologic Subtype | |||
| AIS/MIA | 42 | 1.00 | P = 0.064 |
| LEP | 43 | 1.20 | |
| CAN | 59 | 1.50 | |
| PAP/MPAP | 17 | 1.40 | |
| SOL | 12 | 1.25 | |
| MUC | 6 | 1.35 | |
| Lymphatic or vascular involvement | |||
| Absent | 129 | 1.20 | P = 0.026 |
| Present | 50 | 1.50 |
No., number of cases; WEL, well differentiated; MOD, moderately differentiated; POR, poorly differentiated carcinomas; AIS, adenocarcinoma in situ; MIA, minimally invasive adenocarcinoma; LEP, lepidic predominant; ACN, acinar predominant; PAP, papillary predominant, SOL, solid predominant; MUC, invasive mucinous adenocarcinoma.
Histologic subtypes were classified according to the 2011 IASLC/ ATS/ETS classification of lung adenocarcinoma.
P, significant level for Mann-Whitney or Kruskal-Wallis test.
Table 4. Multiple linear regression analysis of relationships between S100A11 expression and clinicopathologic characteristics of stage I lung adenocarcinomas.
| Subjects | Coefficient B | Standard Error | Standardized Coefficient β | P-value |
|---|---|---|---|---|
| Age | ||||
| <65 | 1 | |||
| > = 65 | 2.069 | 9.944 | 0.016 | 0.835 |
| Gender | ||||
| Male | 1 | |||
| Female | 2.878 | 9.612 | 0.023 | 0.765 |
| Histologic Grade | ||||
| WEL | 1 | |||
| MOD | 19.767 | 12.240 | 0.156 | 0.108 |
| POR | 31.061 | 14.925 | 0.216 | 0.039 |
| Lymphatic or vascular involvement | ||||
| Absent | 1 | |||
| Present | 13.261 | 11.532 | 0.094 | 0.252 |
WEL, well differentiated; MOD, moderately differentiated; POR, poorly differentiated carcinomas.
Histologic subtypes were excluded from factors due to the strong correlation that existed with histologic grade (Spearman’s coefficient 0.567, P<0.001).
S100A11 protein levels and disease-free survival
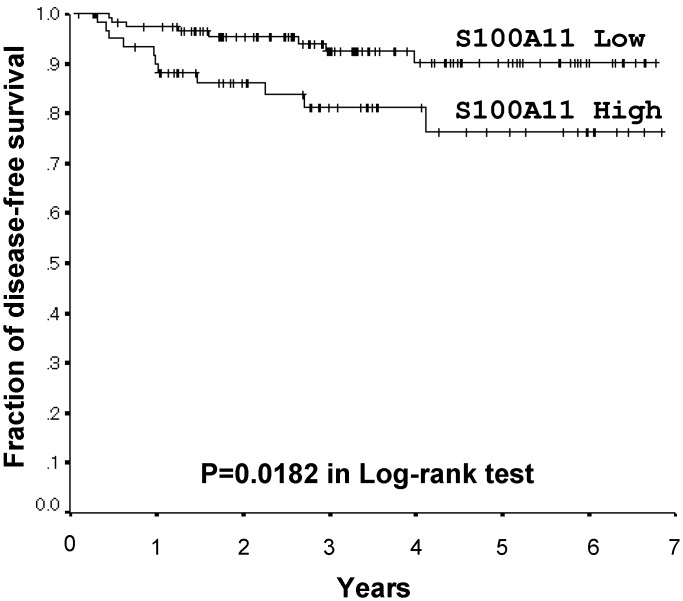
Since the results obtained suggested that the up-regulation of S100A11 was involved in tumor progression, particularly in KRAS-mutated lung adenocarcinomas, its prognostic value was subsequently verified. One hundred and seventy-nine patients with lung adenocarcinomas at stage I were examined. S100A11 expression scores of <1.65 and > = 1.65 were classified as low and high based on a receiver operating characteristic curve (area under the curve 0.629, 95% confidential interval 0,501–0.757). One hundred and eighteen patients were low-expressers, while 61 were high-expressers. Five-year disease-free survival rates were 90.1% and 76.3% in low- and high-S100A11 expressers, respectively. The strong expression of S100A11 correlated with shorter disease-free survival in post-operative lung adenocarcinomas (p = 0.0182 in the Log-rank test, Fig 4). On the other hand, a correlation was not found between S100A11 expression levels and the site of recurrent disease (data not shown).
Fig 4. Kaplan-Meier Disease-free survival curves by S100A11 expression levels for stage I lung adenocarcinomas.
Five-year disease-free survival rates were 76.3% in high-S100A11 expressers (n = 61) and 90.1% in low-S100A11 expressers (n = 118). The strong expression of S100A11 correlated with shorter disease-free survival in post-operative lung adenocarcinomas (p = 0.0182 in the Log-rank test).
Discussion
Among the S100 proteins (S100A2, S100A7, and S100A11) examined in the present study, S100A11 levels were significantly higher in adenocarcinomas with KRAS mutations and strong proliferating activity (Fig 3). They were also higher in adenocarcinomas with poorly differentiated tumors (Tables 3 and 4). Furthermore, the strong expression of S100A11 correlated with shorter disease-free survival (Fig 4). These results suggested that alterations in the expression of S100A11 played a role in tumor progression, particularly in KRAS-mutated lung adenocarcinomas.
S100A11, also called S100C or calgizzarin, belongs to the S100 family of multi-gene calcium-binding proteins. It is located on 1q21 with 15 other S100 family members and has a molecular weight of 11.7kDa [24]. S100 family members regulate important intracellular events such as enzyme activities, the dynamics of cytoskeletal constituents, and Ca2+ concentrations, in order to modulate cell growth and motility as well as differentiation [9]. The up-regulation of S100A11 has been reported in various cancers [9], such as laryngeal [10], breast [11], lung [12–13], gastric [14], colorectal [15], pancreatic [16], and prostate cancers [17], and has frequently been associated with cancer progression, implicating its oncogenic role [10–17]. Tian T et al. previously demonstrated that S100A11 was up-regulated in a highly metastatic lung cancer cell line through a proteomic analysis [13]. Wang et al. also reported that S100A11 levels increased with progression of the disease stage in colorectal cancer [15]. Our results are consistent with these findings.
However, previous studies demonstrated that S100A11 expression levels were reduced in tumors with higher malignant activity in kidney [19] and bladder cancers [20]. Memon AA et al. showed that S100A11 was down-regulated in a higher-graded cell line of bladder cancer by a proteome analysis, and the weak expression of S100A11 was associated with poor survival [20]. This discrepancy implies the complexity of cancer progression. For example, several cancer-related proteins, such as transforming growth factor-beta, have opposite functions as inhibitors or promoters in different stages of cancer progression [25]. Therefore, the potential role of the aberrant expression of S100A11 in carcinogenesis may differ among various types of malignancies.
In summary, the results of the present study suggested that the up-regulation of S100A11 played a role in tumor progression, particularly in KRAS-mutated lung adenocarcinomas. The biological function of S100A11 still remains unclear. Further investigations are warranted in order to elucidate the mechanisms by which S100A11 promotes the progression of lung cancer.
Supporting Information
Representative photographs from normal bronchioles (A), adenocarcinoma (B) and squamous cell carcinoma (C) are shown. S100A7 was not expressed in the normal epithelial cells of bronchioles (A), and was only expressed in a few adenocarcinoma (ADC) cells (B). On the other hand, it was strongly expressed in the keratinizing cells of squamous cell carcinomas (SQC) (C).
(TIFF)
Protein lysates from tumors (T) and non-tumorous tissues (N) of lung adenocarcinoma tissues of patients undergoing surgical resection were subjected to a Western blot analysis for S100A11 and β-actin (ACTB) in the representative cases (A). The signal intensities of the bands were evaluated by NIH imaging. S100A11 levels were normalized to those of ACTB. Normalized levels are shown (B). The immunohistochemical expression of S100A1 in the same tumors was shown (C). S100 protein levels evaluated by immunohistochemistry and those by Western blot were roughly parallel. Cs, Case.
(TIFF)
Acknowledgments
We especially thank Emi Honda and Misa Otara (Department of Pathology, Kanagawa Cardiovascular and Respiratory Center) for their technical assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by the Ministry of Education, Culture, Sports, and Science of Japan (Tokyo Japan), and Yokohama Medical Facility (Yokohama, Japan). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hoffman PC, Mauer AM, Vokes EE. Lung cancer. Lancet 2000;355:479–485. [DOI] [PubMed] [Google Scholar]
- 2. Spira A, Ettinger DS. Multidisciplinary management of lung cancer. N Engl J Med 2004;350:379–392. [DOI] [PubMed] [Google Scholar]
- 3. Yang P, Allen MS, Aubry MC, Wampfler JA, Marks RS, Edell ES, et al. Clinical features of 5,628 primary lung cancer patients: experience at mayo clinic from 1997 to 2003. Chest 2005;128:452–462. [DOI] [PubMed] [Google Scholar]
- 4. Asamura H, Goya T, Koshiishi Y, Sohara Y, Eguchi K, Mori M, et al. ; Japanese Joint Committee of Lung Cancer Registry. A Japanese lung cancer registry study: prognosis of 13,010 resected lung cancers. J Thorac Oncol 2008;3:46–52. 10.1097/JTO.0b013e31815e8577 [DOI] [PubMed] [Google Scholar]
- 5. Okudela K, Woo T, Kitamura H. KRAS gene mutations in lung cancer: particulars established and issues unresolved. Pathol Int 2010;60:651–660. 10.1111/j.1440-1827.2010.02580.x [DOI] [PubMed] [Google Scholar]
- 6. Woo T, Okudela K, Yazawa T, Wada N, Ogawa N, Ishiwa N, et al. Prognostic value of KRAS mutations and Ki-67 expression in stage I lung adenocarcinomas. Lung Cancer 2009;65:355–362. 10.1016/j.lungcan.2008.11.020 [DOI] [PubMed] [Google Scholar]
- 7. Okudela K, Yazawa T, Woo T, Sakaeda M, Ishii J, Mitsui H, et al. Down-regulation of DUSP6 expression in lung cancer: its mechanism and potential role in carcinogenesis. Am J Pathol 2009;175(2):867–81. 10.2353/ajpath.2009.080489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Okudela K, Katayama A, Woo T, Mitsui H, Suzuki T, Tateishi Y, et al. Proteome analysis for downstream targets of oncogenic KRAS-the potential participation of CLIC4 in carcinogenesis in the lung. PLoS One 2014;9(2):e87193 10.1371/journal.pone.0087193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Salama I, Malone PS, Mihaimeed F, Jones JL. A review of the S100 proteins in cancer. Eur J Surg Oncol. 2008;34(4):357–64. [DOI] [PubMed] [Google Scholar]
- 10. Wang C, Zhang Z, Li L, Zhang J, Wang J, Fan J, et al. S100A11 is a migration-related protein in laryngeal squamous cell carcinoma. Int J Med Sci. 2013;10(11):1552–9. 10.7150/ijms.5986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cross SS, Hamdy FC, Deloulme JC, Rehman I. Expression of S100 proteins in normal human tissues and common cancers using tissue microarrays: S100A6, S100A8, S100A9 and S100A11 are all overexpressed in common cancers. Histopathology. 2005;46(3):256–69. [DOI] [PubMed] [Google Scholar]
- 12. Hao J, Wang K, Yue Y, Tian T, Xu A, Hao J, et al. Selective expression of S100A11 in lung cancer and its role in regulating proliferation of adenocarcinomas cells. Mol Cell Biochem. 2012;359(1–2):323–32. 10.1007/s11010-011-1026-8 [DOI] [PubMed] [Google Scholar]
- 13. Tian T, Hao J, Xu A, Hao J, Luo C, Liu C, et al. Determination of metastasis-associated proteins in non-small cell lung cancer by comparative proteomic analysis. Cancer Sci. 2007;98(8):1265–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mori M, Shimada H, Gunji Y, Matsubara H, Hayashi H, Nimura Y, et al. S100A11 gene identified by in-house cDNA microarray as an accurate predictor of lymph node metastases of gastric cancer. Oncol Rep. 2004;11(6):1287–93. [PubMed] [Google Scholar]
- 15. Wang G, Wang X, Wang S, Song H, Sun H, Yuan W, et al. Colorectal cancer progression correlates with upregulation of S100A11 expression in tumor tissues. Int J Colorectal Dis. 2008;23(7):675–82. 10.1007/s00384-008-0464-6 [DOI] [PubMed] [Google Scholar]
- 16. Song HY, Liu YK, Feng JT, Cui JF, Dai Z, Zhang LJ, et al. Proteomic analysis on metastasis-associated proteins of human hepatocellular carcinoma tissues. J Cancer Res Clin Oncol. 2006;132(2):92–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xiao MB, Jiang F, Ni WK, Chen BY, Lu CH, Li XY, et al. High expression of S100A11 in pancreatic adenocarcinoma is an unfavorable prognostic marker. Med Oncol. 2012;29(3):1886–91. 10.1007/s12032-011-0058-y [DOI] [PubMed] [Google Scholar]
- 18. Rehman I, Azzouzi AR, Cross SS, Deloulme JC, Catto JW, Wylde N, et al. Dysregulated expression of S100A11 (calgizzarin) in prostate cancer and precursor lesions. Hum Pathol. 2004;35(11):1385–91. [DOI] [PubMed] [Google Scholar]
- 19. Kondo A, Sakaguchi M, Makino E, Namba M, Okada S, Huh NH. Localization of S100C immunoreactivity in various human tissues. Acta Med Okayama. 2002;56(1):31–4. [DOI] [PubMed] [Google Scholar]
- 20. Memon AA, Sorensen BS, Meldgaard P, Fokdal L, Thykjaer T, Nexo E. Down-regulation of S100C is associated with bladder cancer progression and poor survival. Clin Cancer Res. 2005;11:606–11. [PubMed] [Google Scholar]
- 21. Matsubara D, Niki T, Ishikawa S, Goto A, Ohara E, Yokomizo T, et al. Differential expression of S100A2 and S100A4 in lung adenocarcinomas: clinicopathological significance, relationship to p53 and identification of their target genes. Cancer Sci. 2005;96:844–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 2011;6(2):244–85. 10.1097/JTO.0b013e318206a221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goldstraw P, Crowley J, Chansky K, Giroux DJ, Groome PA, Rami-Porta R, et al. ; International Association for the Study of Lung Cancer International Staging Committee; Participating Institutions. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol 2007;2(8):706–14. [DOI] [PubMed] [Google Scholar]
- 24. Todoroki H, Kobayashi R, Watanabe M, Minami H, Hidaka H. Purification, characterization, and partial sequence analysis of a newly identified EF-hand type 13-kDa Ca(2+)-binding protein from smooth muscle and non-muscle tissues. J Biol Chem. 1991;266(28):18668–73. [PubMed] [Google Scholar]
- 25. Roberts AB, Wakefield LM. The two faces of transforming growth factor beta in carcinogenesis. Proc Natl Acad Sci U S A. 2003;100(15):8621–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative photographs from normal bronchioles (A), adenocarcinoma (B) and squamous cell carcinoma (C) are shown. S100A7 was not expressed in the normal epithelial cells of bronchioles (A), and was only expressed in a few adenocarcinoma (ADC) cells (B). On the other hand, it was strongly expressed in the keratinizing cells of squamous cell carcinomas (SQC) (C).
(TIFF)
Protein lysates from tumors (T) and non-tumorous tissues (N) of lung adenocarcinoma tissues of patients undergoing surgical resection were subjected to a Western blot analysis for S100A11 and β-actin (ACTB) in the representative cases (A). The signal intensities of the bands were evaluated by NIH imaging. S100A11 levels were normalized to those of ACTB. Normalized levels are shown (B). The immunohistochemical expression of S100A1 in the same tumors was shown (C). S100 protein levels evaluated by immunohistochemistry and those by Western blot were roughly parallel. Cs, Case.
(TIFF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.