Abstract
Gorlin syndrome (GS) is an autosomal dominant disorder that predisposes affected individuals to developmental defects and tumorigenesis, and caused mainly by heterozygous germline PTCH1 mutations. Despite exhaustive analysis, PTCH1 mutations are often unidentifiable in some patients; the failure to detect mutations is presumably because of mutations occurred in other causative genes or outside of analyzed regions of PTCH1, or copy number alterations (CNAs). In this study, we subjected a cohort of GS-affected individuals from six unrelated families to next-generation sequencing (NGS) analysis for the combined screening of causative alterations in Hedgehog signaling pathway-related genes. Specific single nucleotide variations (SNVs) of PTCH1 causing inferred amino acid changes were identified in four families (seven affected individuals), whereas CNAs within or around PTCH1 were found in two families in whom possible causative SNVs were not detected. Through a targeted resequencing of all coding exons, as well as simultaneous evaluation of copy number status using the alignment map files obtained via NGS, we found that GS phenotypes could be explained by PTCH1 mutations or deletions in all affected patients. Because it is advisable to evaluate CNAs of candidate causative genes in point mutation-negative cases, NGS methodology appears to be useful for improving molecular diagnosis through the simultaneous detection of both SNVs and CNAs in the targeted genes/regions.
Introduction
Nevoid basal cell carcinoma syndrome (NBCCS; OMIM 109400), also known as Gorlin syndrome or basal cell nevus syndrome, is an autosomal dominant disorder that predisposes affected individuals to developmental defects, including bifid ribs and palmar or plantar pits, as well as tumorigenesis, including the development of basal cell carcinoma, medulloblastoma, or keratocystic odontogenic tumors (formerly known as odontogenic keratocysts) [1–3]. Heterozygous germline PTCH1 mutations have been found in patients with Gorlin syndrome, and the spectrum of associated features is thought to arise from PTCH1 malfunction [4, 5].
Diverse PTCH1 mutations have been identified in Japanese Gorlin syndrome patients [6]. Although numerous germline PTCH1 mutations have been reported, no clusters of mutations have been identified [6, 7]. Despite exhaustive analysis, PTCH1 mutations are often unidentifiable in some patients [6]; for example, Sanger sequencing of PTCH1 exons 2–23, encompassing the entire coding region, detected mutations in approximately 40%–70% of individuals with a typical clinical presentation of Gorlin syndrome [8, 9]. This failure to detect mutations is presumably because of copy number alterations (CNAs), including gross deletions, within or around the exons of PTCH1, or mutations of PTCH1 outside the analyzed regions such as those within introns or regulatory elements. In addition, mutations in rare causative genes other than PTCH1, such as PTCH2 [10, 11] and SUFU [12], have also been detected in individuals suffering from Gorlin syndrome.
Because mutations, or single nucleotide variations (SNVs), as well as CNAs of PTCH1 and other genes related to the Hedgehog signaling pathway-related genes with numerous exons can be responsible for Gorlin syndrome, we conducted hybridization and polymerase chain reaction (PCR)-based next generation sequencing (NGS) analyses in a cohort of Gorlin syndrome-affected individuals from six unrelated families [13, 14] for a combined screening of causative alterations in candidate genes.
Materials and Methods
Patients and DNA samples
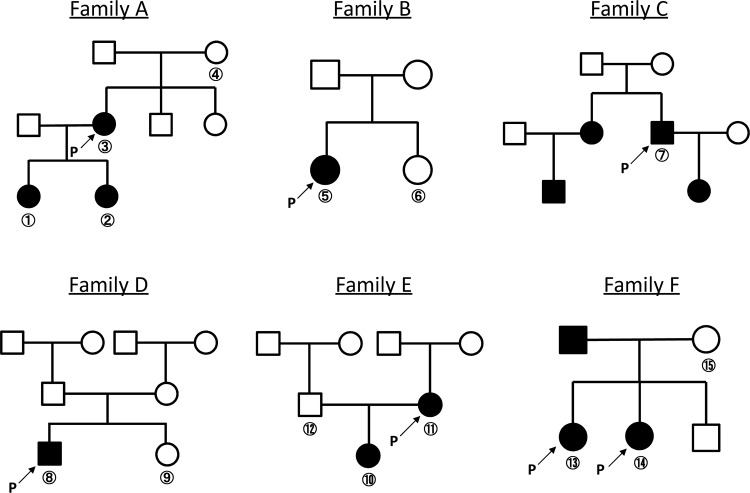
We investigated 15 clinically evaluated Japanese individuals from six pedigrees, including 10 patients with Gorlin syndrome and five unaffected family members (Fig 1). Our group previously reported the clinicopathologic manifestations of all patients [13, 14], and this information is included in Table 1 with the corresponding patient identification codes.
Fig 1. Family trees of the analyzed families affected by Gorlin syndrome.
The shaded circles and squares represent affected individuals. Arrows with “P” represent the probands in each family.
Table 1. Patients and unaffected family members.
| Family ID | Individual No. | Patient / Unaffected member | Age | Gender | Symptoms agree with Kimonis criteria | Other symptoms | JOPM* No. | JOPM* Family ID | PLOS** No. | PLOS** Family ID |
|---|---|---|---|---|---|---|---|---|---|---|
| A | 1 | Patient | 13 | F | KCOT, Rib anomaly, Skin pits, Thoracic deformity | - | 9 | 8 | 26 | 3 |
| 2 | Patient | 10 | F | KCOT, Rib anomaly, Skin pits, Ovarian fibroma | Ovarian cyst | 10 | 8 | 25 | 3 | |
| 3 | Patient | 35 | F | BCC, KCOT, Rib anomaly, Skin pits, Calcification of the falx cerebri | Multiple nevi | 11 | 8 | 27 | 3 | |
| 4 | Unaffected | - | F | 8 | 3 | |||||
| B | 5 | Patient | 10 | F | KCOT, Rib anomaly, Skin pits, Vertebrate anomaly, | Dermoid cyst (skin), Glioma, Mandibular protrusion, Hydrocephalus, Psychomotor retardation, Macrocephaly | 2 | 2 | ||
| 6 | Unaffected | - | F | 2 | ||||||
| C | 7 | Patient | 65 | M | BCC, KCOT, Skin pits | Multiple nevi, Epilepsy | 15 | 10 | 36 | - |
| D | 8 | Patient | 15 | M | KCOT, Rib anomaly, Skin pits, Calcification of the falx cerebri | Multiple nevi, Mandibular protrusion | 6 | 6 | ||
| 9 | Unaffected | - | F | 6 | ||||||
| E | 10 | Patient | 10 | F | KCOT, Rib anomaly, Skin pits, Calcification of the falx cerebri | Language development disorder, Ventricular distention | 18 | 12 | 23 | 2 |
| 11 | Patient | 13 | F | KCOT, Skin pits, Calcification of the falx cerebri | Unknown | 19 | 12 | 24 | 2 | |
| 12 | Unaffected | - | M | 12 | 2 | |||||
| F | 13 | Patient | 13 | F | KCOT, Rib anomaly, Ocular hypertelorism | Multiple nevi, Mental retardation, Agenesis of corpus callosum, Prolonged retention of deciduous tooth | 12 | 9 | ||
| 14 | Patient | 12 | F | KCOT, Thoracic deformity | Multiple nevi, Prolonged retention of deciduous tooth | 13 | 9 | |||
| 15 | Unaffected | - | F | 9 |
KCOT, keratocystic odontogenic tumors.
* Reference No.13
** Reference No. 14.
Peripheral blood samples for study were collected after obtaining informed consent from all participants or their legal guardians, and genomic DNA was extracted using Wizard® Genomic DNA Purification Kits (Promega, Madison, WI). The study was approved by the ethical committees of Tokyo Medical and Dental University and Tokushima University. For those agreeing to participate in this study, written consent was obtained from patients 20 years of age or older, and from the parent of minor children.
Design of custom targeted exome for NGS
A custom HaloPlex panel (Agilent Technologies, Santa Clara, CA) was designed using Agilent’s SureDesign tool (www.agilent.com/genomics/suredesign) to capture all exons with 25 bp of the flanking intronic sequences for eight genes in the Hedgehog signaling pathway: PTCH1, PTCH2, SHH, SUFU, SMO, GLI, GLI2, and GLI3. The entire custom design comprised 119 targets with a total size of 42.893 Kb, 3 283 total amplicons, and a final probe design size of 80.612 Kb, with a total theoretical coverage of 99.98% for the targeted regions.
Sequence capture and NGS
Sequence capture was performed using the HaloPlex Target Enrichment System for Ion Torrent sequencing according to the manufacturer’s instructions (Protocol Version D, Agilent Technologies). Pooled samples for multiplex sequencing were sequenced using the Ion Personal Genome Machine System (Thermo Fisher Scientific, Waltham, MA) with 318 chips (Thermo Fisher Scientific).
SNV analysis and validation
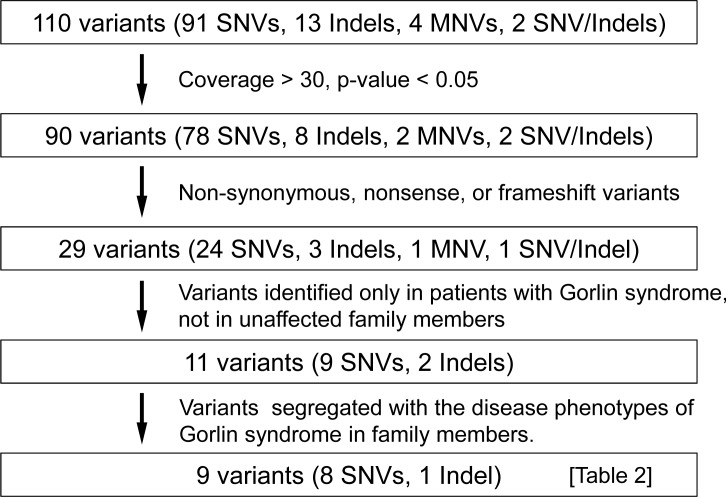
Data were analyzed using Torrent Suite Software v4.2.1 (Thermo Fisher Scientific) and Ion Reporter Software v4.6 (Thermo Fisher Scientific). The flow chart used to select candidate genes is shown in Fig 2. Annotated variants were selected according to the criterion that the causal variants should be segregated in patients with Gorlin syndrome-affected family members. Candidates for pathogenic variants within the PTCH1 were confirmed by Sanger sequencing. The pathogenicity of missense variants was assessed using tools for prediction of possible impact of an amino acid substitution on the structure and function of a human protein, such as Sorting Intolerant From Tolerant (SIFT) v5.1.1 (http://sift.bii.a-star.edu.sg/), PolyPhen-2 v2.2.2r398 (http://genetics.bwh.harvard.edu/pph2/index.shtml), MutationTaster (http://www.mutationtaster.org/index.html). SIFT examines the degree of conservation for amino acid residues across species, and calculated probabilities are used to partition ‘tolerated’ (normalized probability >0.05) from ‘damaging’ (normalized probability ≤0.05) substitutions for prediction of a phenotypic effect. PolyPhen-2 finds change in protein structure and function, and provides both a qualitative prediction (one of 'probably damaging', 'possibly damaging', 'benign' or 'unknown') and a score. MutationTaster checks evolutionary conservation, change in protein structure and function, and additionally effects on splicing or mRNA expression, and predicts the disease potential: ‘disease causing’, which is probably deleterious, ‘disease causing automatic’, which is known to be deleterious, ‘polymorphism’, which is probably harmless, and ‘polymorphism automatic’, which is known to be harmless. Identified SNVs was also evaluated by comparing with known alterations reported in mutation databases, such as the Human Gene Mutation Database (HGMD professional 2015.2, http://www.hgmd.cf.ac.uk/ac/index.php), and ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/).
Fig 2. Flow chart indicating the validation process for variants.
After four validation steps, nine genetic variants were selected, as shown in Table 2.
CNA analysis and validation
The coverage of every target region in each sample was normalized and compared to the average normalized data from all samples from unaffected family members analyzed in the same run to determine the relative coverage ratio [15]. Deletions and duplications were suspected to be present within a genomic interval if this ratio fell below 0.7 or rose above 1.3, respectively. CNA validation and detailed mapping of altered regions were performed using an Affymetrix CytoScan HD chromosome microarray platform (Affymetrix, Santa Clara, CA), which provides 906 600 polymorphic (SNP) and 946 000 non-polymorphic (CNA) markers, according to the manufacturer’s recommendations. The raw data were processed using Chromosome Analysis Suite software (ChAS, Affymetrix), and the output data were interpreted with the UCSC Genome Browser (http://genome.ucsc.edu; GRCh37/hg19 assembly) [16].
Results
In the NGS analysis, which applied our customized Hedgehog pathway-related gene panel to our cohort of Gorlin syndrome patients and their unaffected family members, a mean coverage of 282X was obtained per sample and target region. We observed an average of 92.48% of bases at >1X coverage, 81.68% of bases at >20X coverage, and 64.62% of bases at >100X coverage. Specific SNVs causing inferred amino acid changes in PTCH1, PTCH2, and GLI2 were found in four, one, and one families (seven, one, and one affected individuals), respectively, whereas CNAs within or around PTCH1 were found in two families in which no possible causative SNVs were detected (Table 2).
Table 2. SNVs identified in affected patients but not in unaffected family members.
| Family ID | Indivisual No. | Gene | Mutation | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (Affected vs. unaffected) | Nucleic acid change | Type | AA cange | Exon | SIFT prediction (score) | PolyPhen-2 prediction (score) | MutationTaster prediction | Frequency in HGVD | dbSNP | Reported as pathogenic alteration | |||
| A | 1–3 vs. 4 | PTCH1 | NM_000264.3(PTCH1_v001) | c.1502A>G | Missense | p.Q501R | 10 | Damaging (0.00) | Probably damaging (0.967) | Disease_causing | - | - | - |
| PTCH1 | NM_000264.3(PTCH1_v001) | c.2222C>T | Missense | p.A741V | 14 | Damaging (0.01) | Possibly damaging (0.787) | Disease_causing | 0.004 | rs2227971 | - | ||
| PTCH1 | NM_000264.3(PTCH1_v001) | c.3953C>T | Missense | p.P1318L | 23 | Tolerated (0.13) | Benign (0.014) | Disease_causing | 0.0001 | - | - | ||
| B | 5 vs. 6 | PTCH1 | NM_000264.3(PTCH1_v001) | c.2619C>A | Nonsense | p.Y873* | 16 | - | - | - | - | - | Boutet et al., 2003 |
| C | 7 | PTCH1 | NM_000264.3(PTCH1_v001) | c.3394T>C | Missense | p.S1132P | 20 | Damaging (0.01) | Probably damaging (1.000) | Disease_causing | - | - | Reifenberger et al., 2001 |
| PTCH2 | NM_001166292.1(PTCH2_v001) | c.221G>A | Missense | p.R74H | 2 | Damaging (0.01) | Possibly damaging (0.933) | Disease_causing | 0.006 | - | - | ||
| PTCH2 | NM_001166292.1(PTCH2_v001) | c.524G>T | Missense | p.R175L | 4 | Tolerated (0.11) | Probably damaging (0.990) | Disease_causing | 0.006 | - | - | ||
| D | 8 vs. 9 | PTCH1 | NM_000264.3(PTCH1_v001) | c.1591_1601del | Frameshift | p.I531Gfs*92 | 9 | - | - | - | - | - | - |
| GLI2 | NM_005270.4(GLI2_v001) | c.1906G>C | Missense | p.A636P | 11 | Tolerated (0.26) | Possibly damaging (0.877) | Polymorphism | - | - | - | ||
| E | 10,11 vs. 12 | No | |||||||||||
| F | 13,14 vs. 15 | No | |||||||||||
Causative genetic alterations identified in each family
Family A
In family A, we identified three mutations in PTCH1 that had been inferred to cause amino acid changes (Table 2). Among these, the p.Q501R and p.A741V changes in PTCH1 were predicted to be functionally pathogenic in silico, but have not been reported in either HGMD or ClinVar. Because p.A741V is listed in the Human Genetic Variation Database (HGVD, http://www.genome.med.kyoto-u.ac.jp/SnpDB/index.html) at a low frequency (allele frequency = 0.004) and in dbSNP (http://www.ncbi.nlm.nih.gov/SNP/, rs2227971), we considered it a non-causative mutation with respect to Gorlin syndrome. In addition, p.Q501R mutation occurred right next to the 4th transmembrane domain (p.502–522). In HGMD, 26 and 2 of 53 missense mutations reported as disease-causing mutations locate within and right next to any of twelve transmembrane domains of PTCH1, respectively. Since only 251 and 24 of 1447 amino acids locate within or right next to transmembrane domains, transmembrane domains of the PTCH1 protein seem to be hotspots for missense mutations contributing to functional defects. Therefore, we concluded that p.Q501R was a novel missense mutation causative for Gorlin syndrome in family A.
Family B
In family B, we identified a nonsense mutation in PTCH1 (p.Y873*). As this mutation was previously reported to be causative for NBCCS by Boutet et al. [17], we concluded that this alteration was responsible for Gorlin syndrome in family B.
Family C
In family C, we identified missense mutations in PTCH1 and PTCH2 (Table 2). The p.R175L mutation in PTCH2 was predicted to be “Tolerated” by SIFT, and both p.R175L and p.R74H mutations in PTCH2 were observed in the HGVD at low frequencies (allele frequency = 0.006 for both variations), suggesting those alterations to be rare benign variants. As the p.S1132P mutation of PTCH1 was reported to be causative for NBCCS [18], we concluded that this alteration was responsible for Gorlin syndrome in family C.
Family D
In family D, we identified a possibly deleterious frameshift mutation in PTCH1 (p.I531Gfs*92) resulting in a truncated protein lacking 8 of 12 transmembrane domains that had not been reported in HGMD or ClinVar. On the other hand, the p.A636P mutation in GLI2 was predicted as “Tolerated” and a “Polymorphism” by SIFT and MutationTaster, respectively. Accordingly, we concluded that p.I531Gfs*92 was a novel PTCH1 mutation causative of Gorlin syndrome in family D.
Families E and F
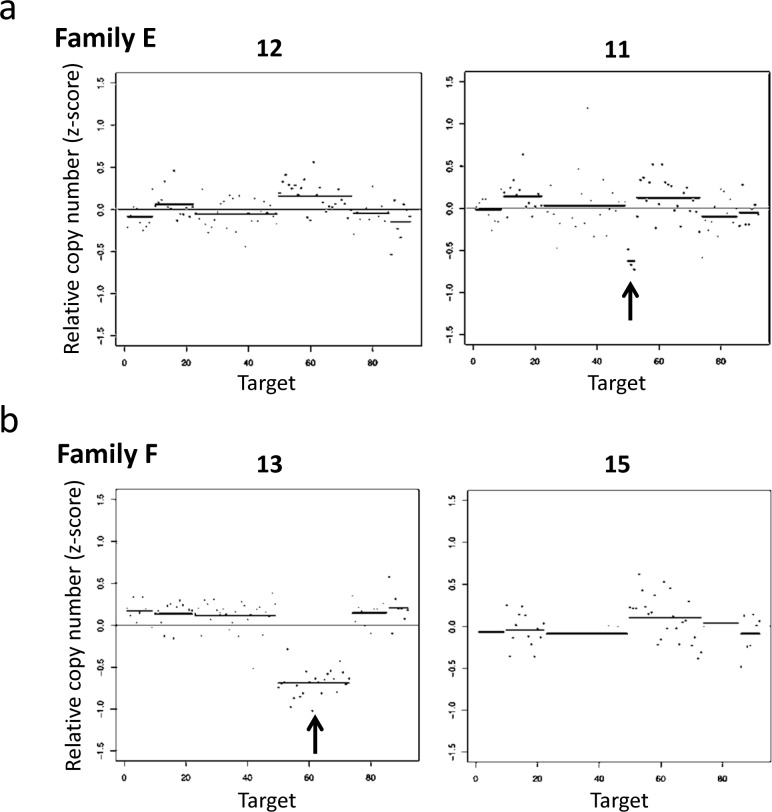
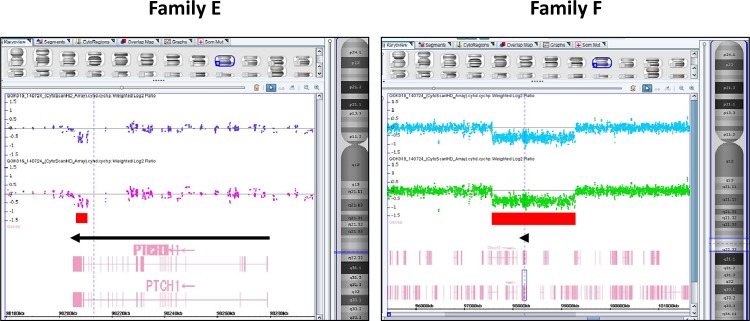
In families E and F, we were unable to identify any causative SNVs within the target gene coding regions, but rather found deletions within the PTCH1 gene (Fig 3). A deletion from exon 22 to the 3'-UTR of PTCH1 was observed in family E (Fig 3A), whereas all exons of PTCH1 were deleted in family F (Fig 3B). These deletions were confirmed using an array-based copy number analysis (Fig 4). The size of the deletion in family E, as determined by an array-based method, was approximately 3.8 kb (chr9:98,207,091–98,210,940) within PTCH1 (from exon 23 to the part of 3’-UTR). As informative probes were not arranged around exons 21 and 22 on the Affymetrix CytoScan HD array platform, it is possible that the deleted region was underestimated by array-based methods relative to the NGS-based method. In family F, a relatively large deletion (approximately 1.7 Mb) (chr9:97,579,146–99,280,739) containing several genes from C9orf3 to CDC14B, including full-length PTCH1, was detected. These alterations have not been previously reported in HGMD or ClinVar, indicating them to be novel deletions causative of Gorlin syndrome. The array-based karyotype results of patients with Gorlin syndrome in families E and F were arr[hg19] 9q22.32(98,207,091–98,210,940)×1 and arr[hg19] 9q22.32(97,579,146–99,280,739)×1, respectively.
Fig 3. Graphical representations of DNA copy number alterations across target regions (1p33-p34, 2q14, 7p13, 7q32.3, 7q36, 9q22.3, 10q24.32, and 12q13.2-q13.3), studied via targeted NGS-based exome analysis.
(a) The copy number loss shows a region from exon 22 to the 3'-UTR of PTCH1 in a Gorlin syndrome patient (right panel; individual No. 11, Family E), but not in an unaffected family member (left panel; individual No. 12, Family E). (b) The copy number loss shows a sequencing region of 9q22.3 in a Gorlin syndrome patient (left panel; individual No. 13, Family F), but not in an unaffected family member (right panel; individual No. 15, Family F).
Fig 4. Image of deleted region detected by an array-based method using an Affymetrix CytoScan HD chromosome microarray platform.
Left panel: The deletion size was approximately 3.8 kb within the PTCH1 (from exon 23 to part of the 3’-UTR) in family E (upper and lower lanes are individual No. 10 and No. 12, respectively). Right panel: A relatively large deletion (approximately 1.7-Mb) containing genes from C9orf3 to CDC14B, including full-length PTCH1, was detected in family F (upper and lower lanes are individual No. 13 and No. 14, respectively).
Collectively, the Gorlin syndrome phenotypes were consistent with heterozygous PTCH1 defects caused by various types of mutations or deletions in all investigated families with this disease.
Discussion
Numerous germline PTCH1 mutations, including nonsense, missense, frameshift, and splicing mutations as well as gross insertions and deletions, have been reported in Gorlin syndrome patients. Miyashita et al. (http://www.med.kitasato-u.ac.jp/∼molgen/sub10.html) genotyped a panel of Gorlin syndrome patients, finding PTCH1 mutations within various percentages of the patient group: 52.4% were frameshift, 14.3% were nonsense, 11.1% were splicing, and 9.5% were missense mutations. Despite exhaustive analysis, PTCH1 mutations could not be identified in some patients. This failure to detect mutations was presumably due to large deletions or the existence of mutations outside of the regions analyzed or in genes other than PTCH1 [6]. Although mutations in PTCH2 and SUFU had been previously implicated in Gorlin syndrome, none of the prior studies that comprehensively analyzed the Hedgehog signaling pathway-related genes identified causative genes other than those three genes.
In this study, we used NGS to simultaneously screen all exons of 8 Hedgehog signaling pathway genes in 15 individuals from 6 families, including Gorlin syndrome patients and their unaffected family members. Through targeted resequencing of all coding exons with flanking exon-intron junctions, as well as simultaneous evaluation of the copy number status using alignment map files obtained via NGS, we found that Gorlin syndrome phenotypes could be explained by PTCH1 mutations or deletions in all affected patients. The p.Y873* and p.S1132P mutations of PTCH1 were previously reported to be causative for NBCCS [17, 18]. Another missense mutation, a small deletion, and two gross deletions were newly identified alterations possibly responsible for Gorlin syndrome in the affected families. Although the p.P1318L PTCH1 mutation was previously reported by our group [14], we concluded this mutation not to be causative because it was predicted to be functionally “tolerated” by SIFT and have been reported in HGVD as a rare variation. Those results suggest that an NGS-based approach to the simultaneous detection of candidate for causative alterations in putative target genes with in silico annotation could effectively identify genetic alterations responsible for Gorlin syndrome. Possibly disease-causing alteration was detected in all cases tested in this study. However, if mutations existed outside of regions targeted for NGS analysis, they would be missed through this approach. Further examinations using larger cohorts containing both familial and sporadic cases will be needed to verify the validity of this approach.
Despite reports of numerous germline PTCH1 mutations in patients with Gorlin syndrome, no clustering of mutations has been identified [7]. Previous studies revealed a lack of genotype–phenotype correlations between mutations in PTCH1 and the major clinical features of Gorlin syndrome [8, 19]. On the other hand, large deletions containing the PTCH1 might cause distinguishing symptoms such as mental retardation [20, 21] because of the deletion of other genes included in the regions around PTCH1. In our study, the 1.7-Mb gene deletion detected in patient 13 from family F, who exhibited mental retardation, included PTCH1 and seven other Refseq genes: C9orf3 (NM_032823), FANCC (NM_001243743), ERCC6L2 (NM_001010895), HSD17B3 (NM_000197), SLC35D2 (NM_007001), HABP4 (NM_014282), and CDC14B (NM_003671). Notably, among these, hyaluronan binding protein 4 (HABP4), encoded by HABP4, is known to bind to FXR1 (fragile X mental retardation-related protein 1),[22] suggesting that HABP4 haploinsufficiency due to heterozygous deletion was involved in the observed pathogenesis (i.e., mental retardation) observed in the Gorlin syndrome patient from family F. This haploinsufficiency might have disrupted the interaction of HABP4 with FXR1, although only one of the two affected family members clearly exhibited this phenotype.
In summary, we systematically screened individuals with familial Gorlin syndrome for causative genetic alterations using hybridization and PCR-based NGS, and found that various PTCH1 mutations, including large deletions, could explain the disease phenotypes. Because it is advisable to examine the CNAs of candidate causative genes in point mutation-negative cases, NGS methodology appears to be useful for improving the molecular diagnosis of Gorlin syndrome through the simultaneous detection of both SNVs and CNAs in targeted genes/regions.
Acknowledgments
This work was supported by JSPS KAKENHI Grant Number 26293304 (I.I.) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
The authors thank Professor Ken-ichi Kozaki for useful discussion, and the Bioresource Laboratory, Tokyo Medical and Dental University and the Support Center for Advanced Medical Sciences, Institute of Biomedical Sciences, Tokushima University for technical assistances.
Data Availability
Data are contained at ClinVar, accessions SCV000255957 - SCV000255962.
Funding Statement
This work was supported by JSPS KAKENHI Grant Number 26293304 (I.I.) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
References
- 1. Gorlin RJ. Nevoid basal cell carcinoma syndrome. Dermatol Clin. 1995;13(1):113–25. . [PubMed] [Google Scholar]
- 2. Gorlin RJ, Goltz RW. Multiple nevoid basal-cell epithelioma, jaw cysts and bifid rib. A syndrome. N Engl J Med. 1960;262:908–12. 10.1056/NEJM196005052621803 . [DOI] [PubMed] [Google Scholar]
- 3. Rayner CR, Towers JF, Wilson JS. What is Gorlin's syndrome? The diagnosis and management of the basal cell naevus syndrome, based on a study of thirty-seven patients. Br J Plast Surg. 1977;30(1):62–7. . [DOI] [PubMed] [Google Scholar]
- 4. Hahn H, Wicking C, Zaphiropoulous PG, Gailani MR, Shanley S, Chidambaram A, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85(6):841–51. . [DOI] [PubMed] [Google Scholar]
- 5. Johnson RL, Rothman AL, Xie J, Goodrich LV, Bare JW, Bonifas JM, et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272(5268):1668–71. . [DOI] [PubMed] [Google Scholar]
- 6. Fujii K, Miyashita T. Gorlin syndrome (nevoid basal cell carcinoma syndrome): update and literature review. Pediatr Int. 2014;56(5):667–74. 10.1111/ped.12461 . [DOI] [PubMed] [Google Scholar]
- 7. Lindström E, Shimokawa T, Toftgård R, Zaphiropoulos PG. PTCH mutations: distribution and analyses. Hum Mutat. 2006;27(3):215–9. 10.1002/humu.20296 . [DOI] [PubMed] [Google Scholar]
- 8. Wicking C, Shanley S, Smyth I, Gillies S, Negus K, Graham S, et al. Most germ-line mutations in the nevoid basal cell carcinoma syndrome lead to a premature termination of the PATCHED protein, and no genotype-phenotype correlations are evident. Am J Hum Genet. 1997;60(1):21–6. [PMC free article] [PubMed] [Google Scholar]
- 9. Soufir N, Gerard B, Portela M, Brice A, Liboutet M, Saiag P, et al. PTCH mutations and deletions in patients with typical nevoid basal cell carcinoma syndrome and in patients with a suspected genetic predisposition to basal cell carcinoma: a French study. Br J Cancer. 2006;95(4):548–53. 10.1038/sj.bjc.6603303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fan Z, Li J, Du J, Zhang H, Shen Y, Wang CY, et al. A missense mutation in PTCH2 underlies dominantly inherited NBCCS in a Chinese family. J Med Genet. 2008;45(5):303–8. 10.1136/jmg.2007.055343 . [DOI] [PubMed] [Google Scholar]
- 11. Fujii K, Ohashi H, Suzuki M, Hatsuse H, Shiohama T, Uchikawa H, et al. Frameshift mutation in the PTCH2 gene can cause nevoid basal cell carcinoma syndrome. Fam Cancer. 2013;12(4):611–4. 10.1007/s10689-013-9623-1 . [DOI] [PubMed] [Google Scholar]
- 12. Smith MJ, Beetz C, Williams SG, Bhaskar SS, O'Sullivan J, Anderson B, et al. Germline mutations in SUFU cause Gorlin syndrome-associated childhood medulloblastoma and redefine the risk associated with PTCH1 mutations. J Clin Oncol. 2014;32(36):4155–61. 10.1200/JCO.2014.58.2569 . [DOI] [PubMed] [Google Scholar]
- 13. Shimada Y, Morita K, Kabasawa Y, Taguchi T, Omura K. Clinical manifestations and treatment for keratocystic odontogenic tumors associated with nevoid basal cell carcinoma syndrome: a study in 25 Japanese patients. J Oral Pathol Med. 2013;42(3):275–80. 10.1111/j.1600-0714.2012.01202.x . [DOI] [PubMed] [Google Scholar]
- 14. Shimada Y, Katsube K, Kabasawa Y, Morita K, Omura K, Yamaguchi A, et al. Integrated genotypic analysis of hedgehog-related genes identifies subgroups of keratocystic odontogenic tumor with distinct clinicopathological features. PLoS One. 2013;8(8):e70995 10.1371/journal.pone.0070995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Okamoto N, Naruto T, Kohmoto T, Komori T, Imoto I. A novel PTCH1 mutation in a patient with Gorlin syndrome. Human Genome Variation. 2014;1:14022 10.1038/hgv.2014.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao W, Higaki T, Eguchi-Ishimae M, Iwabuki H, Wu Z, Yamamoto E, et al. DGCR6 at the proximal part of the DiGeorge critical region is involved in conotruncal heart defects. Human Genome Variation. 2015;2:15004 10.1038/hgv.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boutet N, Bignon YJ, Drouin-Garraud V, Sarda P, Longy M, Lacombe D, et al. Spectrum of PTCH1 mutations in French patients with Gorlin syndrome. J Invest Dermatol. 2003;121(3):478–81. 10.1046/j.1523-1747.2003.12423.x . [DOI] [PubMed] [Google Scholar]
- 18. Reifenberger J, Arnold N, Kiechle M, Reifenberger G, Hauschild A. Coincident PTCH and BRCA1 germline mutations in a patient with nevoid basal cell carcinoma syndrome and familial breast cancer. J Invest Dermatol. 2001;116(3):472–4. 10.1046/j.1523-1747.2001.01279-2.x . [DOI] [PubMed] [Google Scholar]
- 19. Bale AE, Gailani MR, Leffell DJ. The Gorlin syndrome gene: a tumor suppressor active in basal cell carcinogenesis and embryonic development. Proc Assoc Am Physicians. 1995;107(2):253–7. . [PubMed] [Google Scholar]
- 20. Nagao K, Fujii K, Saito K, Sugita K, Endo M, Motojima T, et al. Entire PTCH1 deletion is a common event in point mutation-negative cases with nevoid basal cell carcinoma syndrome in Japan. Clin Genet. 2011;79(2):196–8. 10.1111/j.1399-0004.2010.01527.x . [DOI] [PubMed] [Google Scholar]
- 21. Fujii K, Ishikawa S, Uchikawa H, Komura D, Shapero MH, Shen F, et al. High-density oligonucleotide array with sub-kilobase resolution reveals breakpoint information of submicroscopic deletions in nevoid basal cell carcinoma syndrome. Hum Genet. 2007;122(5):459–66. 10.1007/s00439-007-0419-y . [DOI] [PubMed] [Google Scholar]
- 22. Gonçalves KeA, Bressan GC, Saito A, Morello LG, Zanchin NI, Kobarg J. Evidence for the association of the human regulatory protein Ki-1/57 with the translational machinery. FEBS Lett. 2011;585(16):2556–60. 10.1016/j.febslet.2011.07.010 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are contained at ClinVar, accessions SCV000255957 - SCV000255962.