Abstract
Cecropins are the most potent induced peptides to resist invading microorganisms. In the present study, two full length cDNA encoding cecropin2 (Px-cec2) and cecropin3 (Px-cec3) were obtained from P. xylostella by integrated analysis of genome and transcriptome data. qRT-PCR analysis revealed the high levels of transcripts of Px-cecs (Px-cec1, Px-cec2 and Px-cec3) in epidermis, fat body and hemocytes after 24, 30 and 36 h induction of Metarhizium anisopliae, respectively. Silencing of Spätzle and Dorsal separately caused the low expression of cecropins in the fat body, epidermis and hemocytes, and made the P.xylostella larvae more susceptible to M. anisopliae. Antimicrobial assays demonstrated that the purified recombinant cecropins, i.e., Px-cec1, Px-cec2 and Px-cec3, exerted a broad spectrum of antimicrobial activity against fungi, as well as Gram-positive and Gram-negative bacteria. Especially, Px-cecs showed higher activity against M. anisopliae than another selected fungi isolates. Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) revealed that cecropins exerted the vital morphological alterations to the spores of M. anisopliae. Based on our results, cecropins played an imperative role in resisting infection of M. anisopliae, which will provide the foundation of biological control of insect pests by using cecorpins as a target in the future.
Introduction
All organisms living in the natural world might have the risk of being pathogen invasion; insects are not of the exception. Insects living in complex environment combat infections, which relies mostly on the innate immunity comprising of cellular and humoral immune responses [1]. The immune deficiency (IMD) triggered by Gram-negative bacteria, including the upper pattern recognition receptors i.e., PGRP-LC and PGRP-LE, and the downstream signaling molecules i.e., dFADD, Dredd, dTAK1, dIKK complex and Relish [2]. Toll pathways are activated by Gram-positive bacteria and fungi [3,4] in which certain gene expression products are released into the hemolymph, which resist the invasion of pathogens.
The typical effectors in the intracellular signal transduction pathways are antimicrobial peptides (AMPs) [5]. Most of these are small cationic molecules consisting of 12–50 amino acids [6,7], which are able to disrupt bacterial membranes via non-specific electrostatic interactions with the membrane lipids. AMPs have broad-spectrum antibacterial property [8], active against bacteria and/or fungi, as well as some parasites and viruses [9]. According to the molecular structure, insect AMPs can be classified into four families: cysteine-loss residues peptides (cecropin and moricin), cysteine-rich peptides (defensin and drosomycin), proline-rich peptides (apidaecin, drosocin, and lebocin), and glycine-rich peptides/proteins (attacin and gloverin) [10–12]
Cecropins are one of basic antimicrobial peptides produced by insects [13]. Cecropin has been initially isolated from the hemolymph of Hyalophora cecropia during 1980 [14], subsequently, different forms of cecropins have been isolated from other insect species, such as Bombyx mori [15], Ceratitis capitata [16], Glossina morsitans morsitans [17], Manduca sexta [18], Trichoplusia ni [19] and also from one mammal, the pig [20]. In Drosophila melanogaster, the cecropin multigenes family consists of both four functional genes (Cecropin A1, A2, B and C), two pseudo-genes (Cecropin 1 and 2) and the functional genes coding for cecropins [21,22]. On bacterial infection, all functional genes are expressed mainly in the fat bodies at different developmental stages [23]. Cecropin A1 and A2 are mainly expressed in larvae and adults, while, B and C are mostly expressed during the pupal stage [22]. At low concentrations (0.1 to 5 μM), cecropins exhibit lytic antibacterial activities against several Gram-negative and Gram-positive bacteria, but not against eukaryotic cells [24–26].
The diamondback moth (DBM), P. xylostella (Lepidoptera: Plutellidae) is a worldwide pest, causing serious damage about 4–5 billion USD annually to cruciferous crops [27], such as cabbage, broccoli, and cauliflower [28]. Due to its high fecundity, overlapping generations, genetic plasticity and selection pressure to various insecticides [29–31], entomopathogenic fungi such as M. anisopliae has been used as a biological control agent for a long time to reduce pesticide residues and ensure food safety [32]. Cecropins are the terminal effectors which can be activated by the infection of entomopathogenic fungus. However, the mechanism of the interaction of M. anisopliae with the innate immunity of P. xylostella is unclear.
By decoding the genomic sequence of P. xylostella [33], more immune-related genes will be found and further research will be needed to confirm the modulation of cecropins expression in P. xylostella. The objective of the current study was to investigate, whether cecropins have any role against the successful invasion of M. anisopliae in P. xylostella, and if so what is their impact and mechanism in P. xylostella?
Materials and Methods
Insects and microorganisms
P. xylostella were reared on an artificial diet at 25 ± 2°C in 14 h: 10 h light: dark photoperiod and 60–70% relative humidity. D. melanogaster Schneider S2 cells were kindly provided by Prof. Wenqing Zhang (Sun-yat Sen University, Guangzhou, China) and were maintained in an incubator at 27°C with Schneider's Drosophila medium (Gibco, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco, USA).
Salmonella choleraesuis (Smith), Escherichia coli DH5α, Pseudomonas fluorescens (Flugge), Bacillus cereus (Flugge), Staphylococcus aureus Rosenbach and fungi, Botrytis cinerea (Klotzsch), Penicillium crustosum Thom, Peronophythora litchi, Colletotrichum orbicular Berk, Fusarium oxysporum Schlecht, Colletotrichum gloeosporioiees Penz. were obtained from the Research Institute of Microbiology, Guangzhou, China, while the insect pathogenic fungus, M. anisopliae (MaQ 10) was provided by Dr. Qiongbo Hu (South China Agricultural University, Guangzhou, China), which was kept in China Center for Type Culture Collection (No. CCTCCM 208173).
The bacteria S. aureus, B. cereus, E.coli DH5α, P. fluorescent, S. choleraesuis were grown on LB broth at 37°C to mid-log bacteria(2–7×105CFU/ml). The fungi, M. anisopliae, B. cinerea, P. crustosum, P. litchi, C. orbicular, F. oxysporum, and C. gloeosporioiees were grown on potato dextrose agar (PDA) plates and incubated at 26 ± 2°C for 10 days. The conidia were then harvested in deionized water containing 0.05% Tween-80 to a final concentration of 1×109 conidia/ml. Spore viability was determined before preparation of final concentration by spreading 0.2 ml suspension on PDA and estimating the number of germinated propagules after 24 h of incubation at room temperature.
Cloning of cecropin genes
Total RNA was extracted from the fat body of each instar P. xylostella after 24h treatment with M. anisopliae using Trizol reagent according to the manufacturer’s protocol (Invitrogen, USA). First-strand cDNA was synthesized with 2μg of total RNA in combination with oligo-dT18 primer and Super-script III reverse transcriptase (TaKaRa, Japan) was used to remove the genomic DNA. Px-cec2 and Px-cec3 Unigenes sequences were obtained from P. xylostella transcriptome and the PCR reactions were performed according to the following conditions: 5 min at 94°C, 30 cycles at 94°C for 30 sec, 55°C (Px-cec2) or 56°C (Px-cec3) for 30 sec, 72°C for 30 sec and 8 min at 72°C. 2 μg of mRNA was used to prepare the 5′- and 3′-RACE cDNAs using the SMART RACE cDNA Amplification Kit (TaKaRa, Japan). 3′—UTR region was amplified by 3′-RACE using the following touchdown PCR: 5 cycles of 94°C 30 sec, 72°C 3 min; 5 cycles of 94°C 30 sec, 70°C 30 sec, 72°C 1 min; 25 cycles of 94°C for 30 sec, 62°C (Px-cec2) or 65°C (Px-cec3) for 30 sec, and 72°C for 1 min, while for 5′—UTR, annealing temperature was changed to 65°C (Px-cec2) or 63°C (Px-cec3). All primers Px-cec2-F1/Px-cec-R and Px-cec3-F1/ Px-cec-R for 3′ -UTR and Px-cec-F/Px-cec-R2, Px-cec-F/Px-cec-R2 for 5′ -UTR are shown in Table 1.
Table 1. List of Primers and their sequences used in experiment.
| Gene name | Sequence(5’-------3’) | function |
|---|---|---|
| Px-cec-R | CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | UPM |
| Px-cec-F | CTAATACGAC TCACTATAGGGC | UPM |
| Px-cec2-F1 | CAGGTGGAATCCGTTCAA | Px-cec2 3 ′ RACE PCR |
| Px-cec2-R2 | TGGAGTTGGCTTGTCCTATC | Px-cec2 5 ′RACE PCR |
| Px-cec3-F1 | GCTCCCAGGTGGAAAGGC | Px-cec3 3 ′ RACE PCR |
| Px-cec3-R2 | CTCCATCCCGGATGTGTCGTCC | Px-cec3 5 ′RACE PCR |
| Actin-qF | TGGCACCACACCTTCTAC | qRT-PCR |
| Actin-qR | CATGATCTGGGTCATCTTCT | qRT-PCR |
| Px-cec1-qF | GCCAAGGTGGAAGCCGTTTA | qRT-PCR |
| Px-cec1- qR | TATAGAAGTGGCTTGTCCGATGA | qRT-PCR |
| Px-cec2-qF | TTCGTGTTGGTGGCGGTATTC | qRT-PCR |
| Px-cec2-qR | GCAGGTCTAGCGATGGAGTTG | qRT-PCR |
| Px-cec3-qF | TACTTCTTCTTCACGGTTGTCG | qRT-PCR |
| Px-cec3-qR | CCCAATATGCTGGATGCTTGTC | qRT-PCR |
| GFP-F | GGATCCTAATACGACTCACTATAGGAAGGGCGAGGAGCTGTTCACCG | dsGFP |
| GFP-R | GGATCCTAATACGACTCACTATAGGCAGCAGGACCATGTGATCGCGC | dsGFP |
| Spa-RNAiF | GGATCCTAATACGACTCACTATAGGACGCTGCCTCCAACGCTTCT | RNAi |
| Spa-RNAiR | GGATCCTAATACGACTCACTATAGGTCCTCGCATACCAGTCCCT | RNAi |
| Dor-RNAiF | GGATCCTAATACGACTCACTATAGGCTGAAGCGAAAGCGACAGAAACC | RNAi |
| Dor-RNAiR | GGATCCTAATACGACTCACTATAGGCTGCTGCGCCATAGGAGCCATA | RNAi |
| Px-cec1-ExpF | GCGAGATCTAAGCCGTTTAAAAAATTGGA | Recombinant expression |
| Px-cec1-ExpR | GCCTCGAGTTTGCCAGTAGGTCTGGCTATAG | Recombinant expression |
| Px-cec2-ExpF | GCGAGATCTAAATTGGAGCGAGTGGGACAGC | Recombinant expression |
| Px-cec2-ExpR | GCCTCGAGCGGCAGGTCTAGCGATGGAGTT | Recombinant expression |
| Px-cec3-ExpF | GCGAGATCTTACTTCTTCTTCACGGTTGTCG | Recombinant expression |
| Px-cec3-ExpR | GCCTCGAGTCCCATTATGCTGGATGCTTGT | Recombinant expression |
Spa, Spätzle; Dor: Dorsal; AGATCT, restriction enzyme Bgl II; CTCGAG, restriction enzyme Xho I
Sequence analysis
The cDNA sequences of cecropins were analyzed with bioinformatics analysis tools. Homology searches of cDNA were performed by using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The translation of Px-cec2, Px-cec3 and the deduced amino acid sequence was performed with ExPASY (http://www.expasy.ch/) while, the sequence alignment was performed by Clustal X. 2.0 (http://www.ebi.ac.uk/tools/clustalw2). The signal peptide was analyzed by SignalP (http://www.expasy.ch/SingalP) and the theoretical isoelectric point (pI) was calculated by the compute pI software (http://www.expasy.org/tools/pi-tool.html). A neighbor-joining phylogenetic tree was constructed based on the amino acids sequence by using MEGA 6.0.
qRT-PCR of cecropins
To assess the mRNA expression level of Pxcecs in the 4th instar P. xylostella larvae after the induction of M. anisopliae fat body, epidermis and hemocytes were isolated from M. anisopliae induced P. xylostella larvae after 6, 12, 18, 24, 30, 36, 42, 48 h and washed 3 times in 1×PBS buffer. Tween-80 was used to inject control. The cDNA was synthesized on the basis of the manufacturer’s protocol using the PrimeScript™ RT Master Mix (Perfect Real Time) (TaKaRa, Japan). The primers for qRT-PCR amplification i.e., Px-cec1-qF/qR, Px-cec2-qF/qR and Px-cec3-qF/qR (Table 1) were designed according to ORF of cecropin genes. The final reaction mixture contained 1μl of each primer, 12.5 μl of SYBR® Green PCR Master Mix and 2 μl cDNA. All quantitative reactions were subjected to: 95°C for 3 min followed by 39 cycles at 95°C for 10 sec, 58°C for 30 sec, and ending with 95°C for 10 sec. While as an internal control, the partial fragment of muscle-actin gene (GenBank accession No. AB282645) was also amplified from the same cDNAs with the Actin qF/qR primers (Table 1). Melting curve analysis was applied to all reactions to ensure homogeneity of each reaction product. Three biological replications (n = 3) were performed for each reaction and the 2-ÄÄCt method was used to measure the relative transcription levels.
Recombinant expression and purification of Pxcecs
The cDNA sequence encoding mature Pxcecs was amplified by using specific primers i.e., Px-cecs-ExpF/ExpR (Table 1). The PCR fragments, purified by agarose gel electrophoresis were digested with Bgl II and Xho I enzymes, ligated into Bgl II /Xho I-digested expression vector pMT/BiP/V5-His A (Invitrogen, USA) and later on transformed into the competent DH5α cells. Whereas, the recombinant expression vectors (pMT-Cec1, pMT-Cec2, pMT-Cec3) were confirmed by DNA sequencing.
D. melanogaster Schneider S2 cells was maintained in an incubator at 27°C in Schneider cell Culture Media (Invitrogen, USA), supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen, USA) containing 1% penicillin-streptomycin solution (Sigma, USA). For transfection assay, S2 cells were placed on cover slips in six-well plates overnight at a density of 1×10 6 cells/mL in serum-free medium (Hyclone, USA) and were then incubated overnight, 1 μg plasmid and 6 μl Lipofectamine LTX (Gibco, USA) transfection reagent were used for transient transfection based on the manufacturer’s instructions. Construction stable S2 cell lines was performed essentially in the same manner as described previously [34].
In order to obtain recombinant cecropins, the cell culture medium containing stable S2 cells expressing Pxcecs were collected every 24 h after induction of 250 μM copper sulfate in 150-cm2 flasks to induce the protein. Cell culture medium was combined after 10 days collection, cell debris was removed by centrifugation at 1,000g for 10 min and cell-free medium was incubated overnight at 4°C and later on the protein was detected by 30% SDS–PAGE. Purification of recombinant cecropins was performed according to the method reported previously [35].
RNAi of Spätzle and Dorsal
In order to minimize any off-target effect during RNAi, the gene fragments used for dsRNA synthesis comprised the sequence where a 19 bp consecutive identical cDNA sequences with Spätzle and Dorsal were not found. Double-stranded RNA (dsRNA) for Spätzle and Dorsal RNAi was prepared by using T7 RiboMAX™ Express RNAi System (Promega, USA) according to the manufacturer’s protocol. A dsRNA segment corresponding to the green fluorescent protein (GFP) was also synthesized as a negative control as described previously. dsRNAs were confirmed by gel electrophoresis in 1.0% agarose and the concentrations of dsRNAs were determined by spectrophotometer (Nanodrop 1000, Thermo Scientific) and stored at -20°C for further use.
The 4th instar P. xylostella larvae were injected with 1μl dsRNA Spätzle (100ng/1μl, thirty larvae), dsRNA Dorsal (100ng/1μl, thirty larvae) and dsRNA GFP (100ng/1μl, thirty larvae) respectively. After 24 h of RNAi treatment, the repeated injection of 1μl M. anisopliae spore suspension (1×109 conidia/ml) was again injected to stimulate the immune response. After 36 h of induction, qRT-PCR was performed to analyze the expression of cecropins in three tissues of P. xylostella.
Bioassay on pathogenicity of M. anisopliae
Fourth instar larvae of P. xylostella were treated with dsRNA specific to PxSpa and PxDor as described above. After 24 h since the second dsRNA injection, the larvae were dipped into M. anisopliae spores (1×109 conidia/ml) suspension for 30 sec, and then reared normally under similar conditions as described earlier. The control larvae were treated with 0.05% tween-80 solution only. Each treatment was replicated three times.
Antimicrobial activity assays
The antimicrobial activity of the purified recombinant Pxcecs was tested against several Gram-positive and Gram-negative bacteria and fungi. The minimal growth inhibition concentration (MIC) was determined using a liquid growth inhibition assay and expressed as the lowest final concentration of the peptide at which no growth was observed [35], while the cell density was measured by monitoring the absorbance at 494 nm.
Scanning electron microscopy
In order to clarify recombinant cecropins interacting with M. anisopliae spores. 30 μM recombinant proteins i.e., Px-cec1, Px-cec2 and Px-cec3 were incubated with mid-log phase M. anisopliae spores (1×109 conidia/ml) at 37°C for 3 h and collected by centrifugation at 3,000g for 5 min. The supernatants were later on inoculated by immersion for 5 sec in 2 ml conidial suspension. The precipitates were fixed in 2.5% glutaraldehyde in 0.1 M phosphate buffer for 3 h, and washed three times with 0.1 M phosphate buffer and then dehydrated in ascending series of ethanol (50, 70, 80, 90 and 100%, 15 min each). The samples were dried at room temperature for few seconds and mounted on SEM stubs with double-sided carbon tape. Dried samples were sputtered with gold and observed with the SEM under Quanta 200 FEG at high-vacuum mode.
Transmission electron microscopy of M. anisopliae
The M. anisopliae spores (1×106 conidia/ml) were treated with 30 μM Px-cec1, Px-cec2 and Px-cec3 at 37°C for 30 min. The supernatant was collected by centrifugation (5,000g for 5 min) and cold glutaraldehyde (0.5%, v/v, in 0.1 M sodium cacodylate buffer, pH 7.4) was added for 2 h at 4°C as described previously [5]. The fixed M. anisopliae spores were observed by using a Jeol JEM 1200 transmission electron microscope operating at 80 kV.
Statistical Analysis
All the data were presented as relative mRNA expression. The relative expression level of Cecropins was calculated by using the CFX96 Real-Time system (Bio-Rad, USA). Statistical analysis were performed using SAS 12.0 statistical software. Significant differences among different treatments were determined by using Duncan’s multiple range test (DMRT) at the 95% confidence interval (p<0.001).
Results
Cloning of cecropin genes from P. xylostella
Two full-length cDNA sequences of Px-cec2 and Px-cec3 were isolated from transcriptome and 5′ and 3′ RACE from of fungi infected P. xylostella in Fig 1. The nucleotide sequence of Px-cec2 (GenBank accession No. GU391356) consisted of 198 bp open reading frame (ORF) encoding a predicted polypeptide of 65 amino acids, while the deduced mature protein (Px-cec2) comprised of 39 amino acid residues having a predicted molecular weight of 4.1 kDa and a theoretical isoelectric point (pI) of 10.44. Whereas, the nucleotide sequence of Px-cec3 (GenBank accession No. KF960048) contained 186 bp ORF with 35 amino acids and a predicted molecular mass of 3.59 kDa and 10.57 pI (Fig 1).
Fig 1. Nucleotide sequence of Px-cec2 and Px-cec3 cDNA from P. xylostella and their deduced amino acids.

The putative signal peptide is boxed, while the mature peptide is indicated in bold type, while the predicted transmembrane domain is shaded. The stop codon is marked as an asterisk.
Sequence analysis of P. xylostella cecropins
The multiple sequence alignment of cecropins from P. xylostella showed high conservation of the mature cecropins (S1 Fig). Identical amino acids of four peptides at the same position are shown with asterisk, indicating these positions to have a single, fully conserved residue, however, these four genes also showed dissimilar sites. The phylogenetic tree indicated homology of Px-cecs with several Lepidopteran’s cecropins such as cecropin D from Agrius convolvuli (Linnaeus) and Spodoptera litura (Fabricius), moreover, both Px-cec2 and Px013797 (P. xylostella Genome Database) showed maximum similarity with Px-cec1, suggesting the similar function (S2 Fig).
qRT-PCR of P. xylostella cecropins
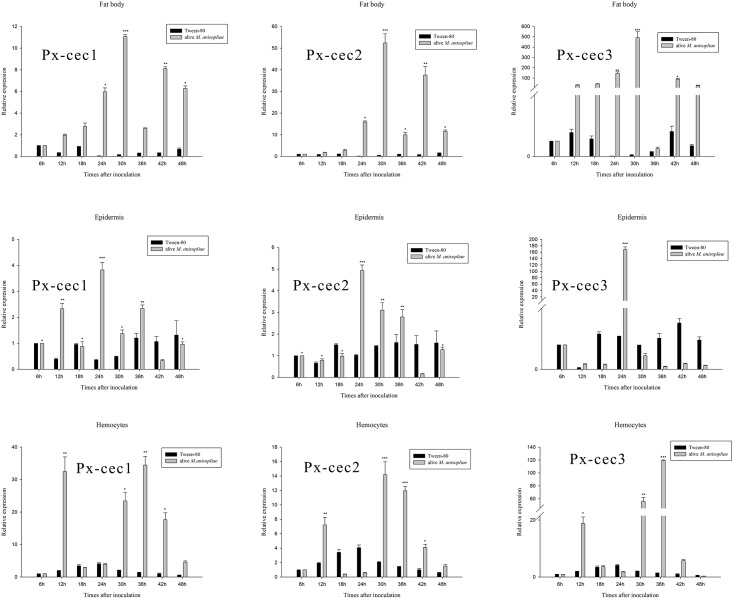
The qRT-PCR results indicated Px-cecs to be mainly expressed in fat body and epidermis after 30 and 24 h induction of M. anisopliae, respectively. Px-cec1 and Px-cec3 were highly expressed after 36 h, while Px-cec2 was highly expressed 30h after the inoculation of M. anisopliae in hemocytes (Fig 2). Moreover, the results from Fig 2 depicted the sensitivity of Px-cec3 to M. anisopliae with maximum expression in all tested tissues.
Fig 2. qRT-PCR analysis of relative expression of cecropin genes from fat body, epidermis, hemocytes after inoculation of M. anisopliae.
Actin was used as an internal control. The mRNA levels of cecropins were highly expressed in 30 h after induction of M. anisopliae in fat body and 24 h in epidermis, Px-cec1, Px-cec2 and Px-cec3 showed maximum expression after 36 h, 30 h, 36 h in hemocytes, respectively. In addition, Px-cec3 depicted more sensitivity to M. anisopliae than Px-cec1 and Px-cec2. Relative expression levels of 6 h was arbitrarily set at 1. Three biological replications (n = 3) were conducted, and the 2-ΔΔCt method was used to measure the relative transcription levels. Means with different number of asterisk are significantly different (P<0.05) (Duncan’s Multiple Range Test) among different time after treated with alive M. anisopliae. *: different with the lowest expression after treated with M. anisopliae; **: significant different with the lowest expression after treated with M. anisopliae; ***: highly significant different with the lowest expression after treated with M. anisopliae.
RNAi effect on expressional level of Px-cecs
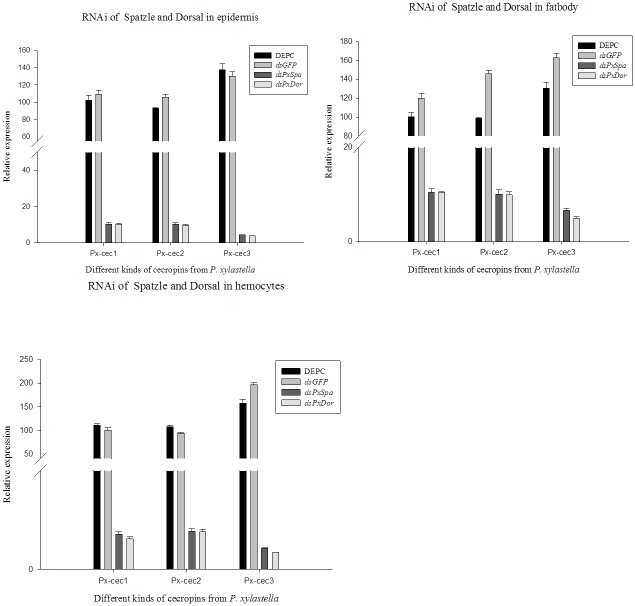
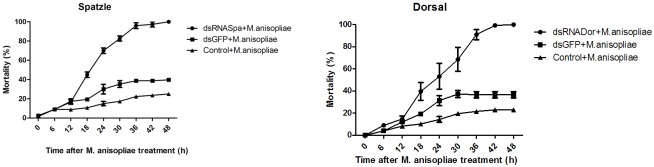
The qRT-PCR results revealed that the expression level of both Spätzle and Dorsal decreased significantly after 24 till 60 h (S3 and S4 Figs). On the other hand, Px-cecs were weakly detected in 36 h group. The mRNA level of Px-cec1 was remarkably decreased in fat body, Px-cec2 in hemocytes, while, Px-cec3 dropped rapidly in all three tissues (Fig 3). The results demonstrated cecropins to be presumably involved in the regulation by Toll pathways in P. xylostella. Meanwhile, after treated by dsRNA to Pxspa and Pxdor separately, which made the 4th instar larvae more susceptible to M. anisopliae infection. Control larvae showed less than 60% mortality to M. anisopliae (1×109 conidia/ml), while the larvae treated by dsRNA (Pxspa and Pxdor) showed significantly increased susceptibility and resulted in almost 100% mortality within 2 days post infection (Fig 4). The results suggested that Pxcecs are efficient factors regulated by Toll signal pathway against the fungal infection in P. xylostella.
Fig 3. The knock-down of Spätzle and Dorsal independently by RNAi.
qRT-PCR analysis of cecropins on fat body, epidermis and hemocytes in P. xylostella after 36 h of RNAi. The relative expression levels of cecropins mRNA was different after treatments. The mRNA level of Px-cec1 was remarkably decreased in fat body, Px-cec2 in hemocytes, while Px-cec3 was dropped rapidly as compared to other tissues. Actin was used as an internal control. Each bar represents the mean ± S.E. (n = 3).
Fig 4. The analysis on susceptibility of P. xylostella larvae to alive M. anisopliae after knockdown of PxSpa and PxDor.
After dsRNA treatment, the larvae were treated with M. anisopliae spores (1×109 conidia/ml). Mortality was recorded every 6h. Each treatment was replicated three times and each treatment consisted of 30 larvae.
Expression and purification of cecropins
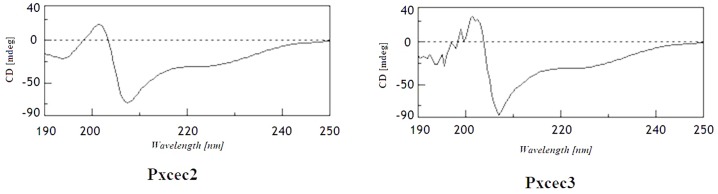
The cDNA fragment encoding mature Pxcecs were amplified by PCR and inserted into the Drosophila expression vector pMT/BiP/V5-HisA. Following transient transfection of Drosophila S2 cells with pMT-Pxcecs and induction expression with final concentration of 250μM copper sulfate, a recombinant protein was detected by SDS-PAGE in both medium and cell lysate. In order to purify the protein, stable transformant strains were selected by pcoBlast and purified recombinant cecropins were analyzed by Tricine-SDS-PAGE (Fig 5), which revealed that Pxcecs were purified to homogeneity with molecular mass of 3.8 kDa. The homogeneities were confirmed by MALDI-TOF-MS analysis (data not shown). The purified Pxcecs were also analyzed using far-UV CD at room temperature. The relative secondary structure contents of recombinant Pxcec2 and Pxcec3 mainly comprised of alpha-helices with some random coil in 10mM phosphate buffer (pH 7.2)(Fig 6), which were almost identical to the secondary structure of Px-cec1, reported previously by our lab [5].
Fig 5. SDS-PAGE analysis of recombinant protein from Drosophila S2 cells.
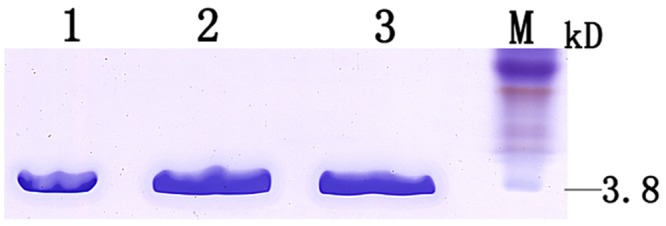
M: Kaleidoscope polypeptide weight protein marker; 1: recombinant protein of Px-cec1; 2: recombinant protein of Px-cec2; 3: recombinant protein of Px-cec3.The result showed that there was a single band corresponding to the expected size of 3.8 kDa.
Fig 6. Analysis of Pxcec2 and Pxcec3 using far-UV CD spectra under 10 mM phosphate buffer (pH 7.2) at room temperature.
Antimicrobial activity assays of recombinant Pxcecs
Antimicrobial activity of recombinant Pxcecs were tested against several microbial strains, including Gram-negative, Gram-positive and fungi. The findings of minimal inhibitory concentration (MIC) showed that Pxcecs exhibited high antimicrobial activity against the tested strains (Table 2). Pxcecs illustrated a higher efficacy against Gram-negative than Gram-positive bacteria and fungi. Among the tested microorganisms, E. coli DH5α proved to be the most sensitive strain to Pxcecs, with MIC values of 0.1(Px-cec1), 0.5(Px-cec2) and 0.2(Px-cec3) μM, respectively. While for the fungi isolates, Pxcecs showed higher activity against M. anisopliae, with MIC values of 4.5(Px-cec1), 5.1(Px-cec2) and 6.5 (Px-cec3) μM, than another selected isolates of fungi.
Table 2. Antimicrobial activity of cecropins from P. xylostella.
| Microorganisms | MIC (μM) | ||
|---|---|---|---|
| Px-cec1 | Px-cec2 | Px-cec3 | |
| Gram-positive | |||
| Staphylococcus aureus | 2.1 | 5.0 | 2.5 |
| Bacillus cereus | 2.6 | 3.5 | 2.7 |
| Gram-negative | |||
| Escherichia coli DH5α | 0.1 | 0.5 | 0.2 |
| Pseudomonas fluorescent | 1.1 | 1.9 | 1.3 |
| Salmonella choleraesuis | 2.1 | 2.5 | 2.2 |
| Fungi | |||
| Metarhizium anisopliae | 4.5 | 5.1 | 6.5 |
| Botrytis cinerea | 15.0 | 10.8 | 11.0 |
| Penicillium crustosum | 13.0 | 11.9 | 12.0 |
| Peronophythora litchi | 14.0 | 15.0 | 12.0 |
| Colletotrichum gloeosporioiees Penz. | 17.3 | 18.2 | 19.0 |
| Colletotrichum orbicucar | 12.5 | 25.0 | 20.0 |
| Fusarium oxysporum | 8.0 | 12.0 | 9.0 |
MIC, minimal growth inhibition concentrations; Px-cec1, the recombinant protein cecropin 1; Px-cec2, the recombinant protein cecropin 2 Px-cec3, the recombinant protein cecropin3
Effect of cecropins on M. anisopliae spore morphology
The treatment of M. anisopliae spores with cecropins at 30 μM for 3 h, showed a few differences in the morphology. Cecropins induced spores remained short, while, the cell wall of the spores showed wrinkles and experienced blebbing as compared to the control (Fig 7). The morphological changes of M. anisopliae spore after treatment with 30μM Px-cecs were also examined using TEM, which showed that the cell membrane appeared to be wafery and the cellular cytoplasmic contents were dissolved and unclear. In addition, the entire cell membrane was disrupted, causing the cellular cytoplasmic contents to leak after interacted with Px-cec1, Px-cec2 and Px-cec3 (Fig 8). Conversely the untreated spores had a bright and normal smooth surface with clear cellular cytoplasmic contents.
Fig 7. SEM analysis of the spore of M. anisopliae interacted with cecropins.
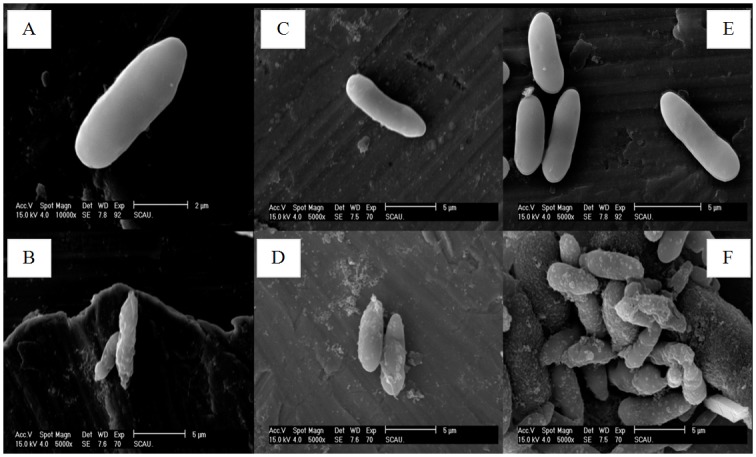
A, C and E: CK (naive M. anisopliae spore); B, D and F: the spore of M. anisopliae interacted with Px-cec1, Px-cec2 and Px-cec3, respectively. The SEM analysis showed that M.anisopliae spore became short and wrinkled (B, D and F) after interacted with cecropins from P. xylostella as compared to the untreated spore (A, C and E), which had a bright and normal smooth surface.
Fig 8. TEM showing the effect of cecropins from P. xylostella on the spores of M. anisopliae.
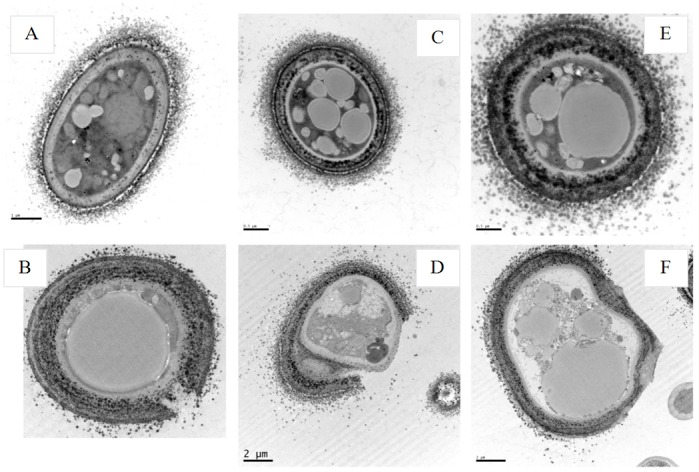
A, C and E: CK (naive M. anisopliae spore); B, D and F: the spore of M. anisopliae interacted with Px-cec1, Px-cec2 and Px-cec3, respectively. The TEM images revealed that the cell membrane of the spore became wafery and the cellular cytoplasmic contents were dissolved and became vague (B, D and F). The entire cell membrane was disrupted, causing the cellular cytoplasmic contents to leak out after interaction with Px-cec1 and Px-cec2 (B and D), the untreated spore (A, C and E) had a bright and normal smooth surface and the cellular cytoplasmic content was clear.
Discussion
Cecropins, the first antibacterial peptides discovered in animals containing α-helix[36,37]. In this study, two full length cDNA encoding Px-cec2 and Px-cec3 from P. xylostella were cloned and the recombinant proteins i.e., Px-cec1, Px-cec2 and Px-cec3 were purified from Drosophila S2 cells. The antibacterial activity assay showed recombinant proteins to have strong activity against M. anisopliae.
The cecropin genes of D. melanogaster and Anopheles gambiae have been categorized into four types i.e., A, B, C and D, while, B. mori cecropin genes have been divided into five types (A, B, C, D and E) [38]. However, according to the present study, all sequences from P. xylostella have been categorized into three types (Px-cec1, Px-cec2 and Px-cec3), moreover Px-cec2 and Px-cec3 are the first to be reported and characterized.
In B. mori, the cecropin A and B genes showed highest expression in the fat bodies and hemocytes, while, the cecropin D was expressed only in the fat bodies and hemocytes [39]. On the other hand, all functional genes were expressed, mainly in fat bodies after bacterial infection, although at different stages of development in D. melanogaster [23], but the expression of cecropins after fungal induction has not been reported in P. xylostella. To understand the role of cecropins in mRNA level against fungal induction in P. xylostella, three tissues i.e., fat body, epidermis and hemocytes were isolated from M. anisopliae infected larvae. qRT-PCR investigation showed that Px-cec1 and Px-cec3 were mainly expressed in fat body, while Px-cec2 in hemocytes (Fig 2). Interestingly, the transcript level of Px-cec3 was higher than those of Px-cec1 and Px-cec2 in the three selected tissues. Based on above results, P. xylostella cecropins displayed specific expressional profiles at different time in the three selected tissues, which demonstrated Px-cec3 to take a predominant position to resist the fungal infection. Moreover, the time required to achieve the highest expression in epidermis was earlier than fat body and hemocytes revealing M. anisopliae adherence to epidermis at first, then to fat body and hemocytes.
Insects and microbes partially share the same environment, including D. melanogaster and B. mori. D. melanogaster has evolved sensitive mechanisms for pathogen recognition and various strategies to defend against bacteria, fungi, parasites and viruses [40]. Expression of AMP genes in D. melanogaster is regulated by the two signal transduction pathways [41]. Imd pathways may be taking the roles to regulate the expression of diptericins, while, Toll pathways transduct the signals to trigger the expression of cecropins, drosomycins etc. At present, the Toll signaling pathways has been well studied in D. melanogaster, but less characterized in other insect species [41], meanwhile, Spätzle and Dorsal have been reported to activate Toll pathways in D. melanogaster [42,43]. To verify insect pests Toll pathways in vivo, RNAi in P. xylostella was performed. Our results confirmed that cecropins were mainly regulated by Toll pathways and participate in innate immunity after M. anisopliae infection in P. xylostella. Importantly, qRT-PCR indicated that Px-cec3 owned more sensitivity towards M. anisopliae with maximum expression in fat body, epidermis and hemocytes. In addition, the mRNA level of Px-cec1 was strongly decreased in hemocytes, while Px-cec2 and Px-cec3 were notably decreased in the fat body which depicted cecropins to exhibit tissue-specific expression in P. xylostella.
Purified Px-cec1 has already been obtained by constructed vector pET-32a (+) (Novagen, USA) in the previous study [5]. In the present study, the recombinant proteins Px-cec1, Px-cec2 and Px-cec3 were expressed and purified from Drosophila S2 cells, which enhanced the quality of protein, while the activity of Px-cec1 against pathogenic bacteria has already been tested in our lab. In this research, seven fungi were added for testing. Recombinant protein Pxcecs showed strong antibacterial activity against Gram-positive, Gram-negative bacteria and fungi, which affirmed cecropins to have broad-spectrum antibacterial property. Recombinant cecropins showed lower MIC values against Gram-negative bacteria than other tested strains (Table 2). In comparison to the cecropins tested in our lab, Pxcecs illustrated higher antibacterial activity against P. crustosum, in contrast to already reported cecropins against P. crustosum from Musca domestica [44].
In the previous study, the detail mechanism of microbial cell lysis by the antimicrobial peptides is not well understood. It is believed that antimicrobial peptides may form an ion channel or pore on the cell membrane prior to cell death[45,46]. Cecropins interacted with bacterial membranes resulting in the formation of ion channels, so as to kill the microorganisms[47]. SEM and TEM of Px-cec1 on S. aureus have been already reported from our laboratory [5].In the current study, to elucidate the mechanisms of Px-cecs against fungi, we used SEM and TEM to assay the morphological changes of the spores of M. anisopliae induced by cecropins treatment. We found that cell surface of spores treated with cecropins was damaged and disrupted and gross leakage of cytoplasmic contents was also observed. Our results suggested that the fungi spores membrane is an important target of antimicrobial peptide and also illustrated cecropins playing crucial role in innate immunity of P. xylostella.
In short, this study is the first to our knowledge to examine the biological activity of Px-cec2 and Px-cec3 and widen the horizon of Px-cec1. The results of qRT-PCR suggested that Px-cec1, Px-cec2 and Px-cec3 were differentially expressed after induction of M. anisopliae. Recombinant proteins Pxcecs displayed high resistance towards all tested microorganisms. Based on our results, cecropins resisted the infection of fungi and for the effective fungal applications as pesticides or the blockage of signal transduction pathways in P. xylostella, it is necessary to reduce its ability to resist against entomopathogenic fungi, which will in turn improve the effectiveness of fungi.
Supporting Information
Conserved amino acid residues among four genes (black background), identical residues among three genes (pink background), and same residues among two genes are shown on blue background.
(TIF)
Drosophila melanogaster cecropin C (AAB82507); Drosophila melanogaster cecropin B (AAF57027.1); Aedes aegypti cecropin A (AAF59831); Manduca sexta cecropin 6 (AAO74638); Agrius convolvuli cecropin D precursor (ACX37671); Hyalophora cecropia cecropin partial(AAP93872); P. xylostella cecropin 1 (ADA13281); P. xylostella cecropin 2 (ADC54851); Px013797(derived from P. xylostella Genome Database); Pseudoplusia includens cecropin A (AAR99379); Papilio xuthus cecropin (ACR82292); Bombyx mori cecropin precursor(NP-001037392); P. xylostella cecropin 3 (KF960048); Hyphantria cunea cecropin A3 (AAB39003); Hyphantria cunea cecropin A (AID51414); Bombyx mori cecropinB precursor (NP-001096031); Spodoptera litura cecropin D partial(ABQ51092); Hyalophora cecropia cecropin B(AAA29184); Trichoplusia ni cecropin B (ABV68872); Helicoverpa armigera cecropin (AAX51304); Antheraea pernyi cecropin B(P01509); Tribolium castaneum cecropin 2 precursor (NP-001164146).
(TIF)
qRT-PCR analysis of Spätzle after RNAi on fat body, epidermis and hemocytes in P. xylostella from 12 h to 60 h. The relative expression levels of Spätzle mRNA was different after treatments, Means with two asterisks are statistically different (p<0.001) (Duncan’s Multiple Range Test) among treatments; Actin was used as an internal control. Each bar represents the mean ± S.E. (n = 3).
(TIFF)
qRT-PCR analysis of Dorsal after RNAi on fat body, epidermis and hemocytes in P. xylostella from 12 to 60 h. The relative expression levels of Dorsal mRNA was different after treatments, Means with two asterisks are statistically different (p<0.001) (Duncan’s Multiple Range Test) among treatments; Actin was used as an internal control. Each bar represents the mean ± S.E. (n = 3).
(TIF)
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31371989, 31071734) and the National High Technology Research and Development Program (‘863’ Program) of China (2012AA101505).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (31371989, 31071734) and the National High Technology Research and Development Program (‘863’ Program) of China (2012AA101505).
References
- 1. Hultmark D.Drosophila immunity: paths and patterns. Current Opinion in Immunology. 2003; 15(1): 12–19. [DOI] [PubMed] [Google Scholar]
- 2. Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, et al. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proceedings of the National Academy of Sciences.1995; 92(21): 9465–9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Engel E, Viargues P, Mortier M, Taillebourg E, Couté Y. Identifying USPs regulating immune signals in Drosophila: USP2 deubiquitinates Imd and promotes its degradation by interacting with the proteasome. Cell Communication and Signaling.2014; 12: 41–43. 10.1186/s12964-014-0041-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Charroux B, Royet J. Drosophila immune response: From systemic antimicrobial peptide production in fat body cells to local defense in the intestinal tract. Fly.2014; 4(1):40–47. [DOI] [PubMed] [Google Scholar]
- 5. Jin F, Sun Q, Xu X, Li L, Gao G. cDNA cloning and characterization of the antibacterial peptide cecropin 1 from the diamondback moth, Plutella xylostella L. Protein Expression and Purification. 2012;85(2):230–238. 10.1016/j.pep.2012.08.006 [DOI] [PubMed] [Google Scholar]
- 6. Maltseva AL, Kotenko ON, Kokryakov VN, Starunov VV, Krasnodembskaya AD. Expression pattern of arenicins—the antimicrobial peptides of polychaete Arenicola marina. Frontiers in Physiology.2014; 5: 497–503. 10.3389/fphys.2014.00497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Forde E, Devocelle M. Pro-Moieties of Antimicrobial Peptide Prodrugs. Molecules. 2015; 20(1): 1210–1227. 10.3390/molecules20011210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Park SC, Park Y, Hahm KS. The role of antimicrobial peptides in preventing multidrug-resistant bacterial infections and biofilm formation. International Journal of Molecular Sciences. 2011; 12(9): 5971–5992. 10.3390/ijms12095971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nicolas E, Nappi AJ, Lemaitre B. Expression of antimicrobial peptide genes after infection by parasitoid wasps in Drosophila . Developmental and Comparative Immunology. 1996; 20(3): 175–181. [DOI] [PubMed] [Google Scholar]
- 10. Imler JL, Hoffmann JA. Signaling mechanisms in the antimicrobial host defense of Drosophila . Current Opinion in Microbiology. 2000; 3(1): 16–22. [DOI] [PubMed] [Google Scholar]
- 11. Reddy KV, Yedery RD, Aranha C. Antimicrobial peptides: premises and promises. International Journal of Antimicrobial Agents. 2004; 24(6): 536–547. [DOI] [PubMed] [Google Scholar]
- 12. Xu XX, Jin FL, Wang YS, Freed S, Hu QB. Molecular cloning and characterization of gloverin from the diamondback moth, Plutella xylostella L. and its interaction with bacterial membrane. World Journal of Microbiology & Biotechnology. 2015; 31(10): 1529–1541. [DOI] [PubMed] [Google Scholar]
- 13. Steiner H, Andreu D, Merrifield RB. Binding and action of cecropin and cecropin analogues: Antibacterial peptides from insects. Biochimica et Biophysica Acta (BBA)—Biomembranes. 1988; 939(2): 260–266. [DOI] [PubMed] [Google Scholar]
- 14. Steiner H, Hultmark D, Engstrom A, Bennich H, Boman HG. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature. 1981;182(11): 246–248. [PubMed] [Google Scholar]
- 15. Yamano Y, Matsumoto M, Inoue K, Kawabata T, Morishima I. Cloning of cDNAs for cecropins A and B, and expression of the genes in the silkworm, Bombyx mori .Bioscience Biotechnology and Biochemistry. 1994;58(8):1476–8. [DOI] [PubMed] [Google Scholar]
- 16. Rosetto M, Manetti AG,Marchini D, Dallai R, Telford JL, Baldari CT. Sequences of two cDNA clones from the medfly Ceratitis capitata encoding antibacterial peptides of the cecropin family. Gene. 1993; 134(2): 241–243. [DOI] [PubMed] [Google Scholar]
- 17. Kaaya GP Flyg C, Boman HG. Insect immunity: Induction of cecropin and attacin-like antibacterial factors in the haemolymph of Glossina morsitans morsitans . Insect Biochemistry and Molecular Biology. 1987; 17(2): 309–315. [Google Scholar]
- 18. Kang D L G, Gunne H, Steiner H. A Family of Bacteria-regulated, Cecropin D-like Peptides from Manduca sexta. Insect Biochemistry and Molecular Biology. 1996; 263(36): 19424–19429. [PubMed] [Google Scholar]
- 19. Kang D, Liu G, Gunne H, Steiner H. PCR differential display of immune gene expression in Trichoplusia ni . Insect Biochemistry and Molecular Biology. 1996; 26(2): 177–184. [DOI] [PubMed] [Google Scholar]
- 20. Lee JY, Boman A, Sun CX, Andersson M, Jörnvall H, Jörnvall H, et al. Boman HGAntibacterial peptides from pig intestine: isolation of a mammalian cecropin. Proceedings of the National Academy of Sciences. 1989; 86(23): 9159–9162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kylsten P, Samakovlis C, Hultmark D. The cecropin locus in Drosophila; a compact gene cluster involved in the response to infection. The EMBO Journal. 1990; 9(1): 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tryselius Y, Samakovlis C, Kimbrell DA, Hultmark D. CecC, a cecropin gene expressed during metamorphosis in Drosophila pupae. European Journal of Biochemistry. 1992; 204(1): 395–399. [DOI] [PubMed] [Google Scholar]
- 23. Ponnuvel KM, Subhasri N, Sirigineedi S, Murthy GN, Vijayaprakash NB. Molecular evolution of the cecropin multigene family in silkworm Bombyx mori. Bioinformation. 2010; 5(3): 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen HM, Wang W, Smith D, Chan SC. Effects of the anti-bacterial peptide cecropin B and its analogs, cecropins B-1 and B-2, on liposomes, bacteria, and cancer cells. Biochimica et biophysica Acta. 1997; 1336(2): 171–179. [DOI] [PubMed] [Google Scholar]
- 25. Jaynes JM JG, Jeffers GW, White KL, Enright FM. In vitro cytocidal effect of lytic peptides on several transformed mammalian cell lines. Peptide Research. 1989; 2(2): 157–160. [PubMed] [Google Scholar]
- 26. Jan PS, Huang HY, Chen HM. Expression of a synthesized gene encoding cationic peptide cecropin B in transgenic tomato plants protects against bacterial diseases. Applied and Environmental Microbiology. 2010; 76(3): 769–775. 10.1128/AEM.00698-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Basukriadi A, Wilkins RM. Oviposition deterrent activities of Pachyrhizus erosusseed extract and other natural products on Plutella xylostella (Lepidoptera: Plutellidae).Journal of Insect Science 2014; 10.1093/jisesa/ieu106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tang W, Yu L, He W, Yang G, Ke F. DBM-DB: the diamondback moth genome database. Database. 2014; 10.1093/database/bat087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. BrancoI MC, GatehouseII AG. Insecticide Resistance in Plutella xylostella (L.) (Lepidoptera:Yponomeutidae) in the Federal District, Brazil. Anais da Sociedade Entomológica do Brasil. 1997; 26(1): 75–79. [Google Scholar]
- 30. Santos VC, de Siqueira HA, da Silva JE, de Farias MJ. Insecticide resistance in populations of the diamondback moth, Plutella xylostella (L.) (Lepidoptera: Plutellidae), from the state of Pernambuco, Brazil. Neotropical Entomology. 2011; 40(2): 264–270. [DOI] [PubMed] [Google Scholar]
- 31. Raymond B, Wright DJ, Bonsall MB. Effects of host plant and genetic background on the fitness costs of resistance to Bacillus thuringiensis . Heredity. 2011; 106(2): 281–288. 10.1038/hdy.2010.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Han PF, Jin FL, Dong XL, Fan JQ, Qiu BL, Ren SX. Transcript and protein profiling analysis of the destruxin A-induced response in larvae of Plutella xylostella . PLoS One. 2013; 8(4):e60771 10.1371/journal.pone.0060771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xia X, Zheng D, Zhong H, Qin B, Gurr GM. DNA sequencing reveals the midgut microbiota of diamondback moth, Plutella xylostella (L.) and a possible relationship with insecticide resistance. PLoS One. 2013. 8(7):e68852 10.1371/journal.pone.0068852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhong X, Xu XX, Yi HY, Lin C, Yu XQ. A Toll-Spätzle pathway in the tobacco hornworm, Manduca sexta . Insect Biochemistry and Molecular Biology. 2012; 42(7): 514–524. 10.1016/j.ibmb.2012.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jin F, Xu X, Zhang W, Gu D. Expression and characterization of a housefly cecropin gene in the methylotrophic yeast, Pichia pastoris . Protein Expression and Purification. 2006; 49(1): 39–46. [DOI] [PubMed] [Google Scholar]
- 36. Liu ZY, Xu T, Zheng ST, Zhang LT, Zhang FC. Study on the interaction mechanism of antimicrobial peptide Cecropin-XJ in Xinjiang silkworm and Staphylococcus aureus DNA by spectra. Guang Pu Xue Yu Guang Pu Fen Xi. 2008; 28(3): 612–616. [PubMed] [Google Scholar]
- 37. Xia L, Liu Z, Ma J, Sun S, Yang J. Expression, purification and characterization of cecropin antibacterial peptide from Bombyx mori in Saccharomyces cerevisiae. Protein Expression and Purification. 2013; 90(1): 47–54. 10.1016/j.pep.2013.02.013 [DOI] [PubMed] [Google Scholar]
- 38. Hong SM, Kusakabe T, Lee JM, Tatsuke T, Kawaguchi Y. Structure and expression analysis of the cecropin-E gene from the silkworm, Bombyx mori . Bioscience, Biotechnology, & Biochemistry. 2008; 72(8): 1992–1998. [DOI] [PubMed] [Google Scholar]
- 39. Yang J, Furukawa S, Sagisaka A, Ishibashi J, Taniai K, Shono T, et al. cDNA cloning and gene expression of cecropin D, an antibacterial protein in the silkworm, Bombyx mori. Biochemistry &Molecular Biology. 1999; 122(4): 409–414. [DOI] [PubMed] [Google Scholar]
- 40. Lemaitre B, Hoffmann JA. The Host Defense of Drosophila melanogaster . Annual Review of Immunology. 2007; 25: 697–743. [DOI] [PubMed] [Google Scholar]
- 41. Zhong X, Xu XX, Yi HY, Lin C, Yu XQ. A Toll-Spatzle pathway in the tobacco hornworm, Manduca sexta . Insect Biochemistry and Molecular Biology. 2012; 42(7): 514–524. 10.1016/j.ibmb.2012.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Morisato D, Anderson KV. The spätzle gene encodes a component of the extracellular signaling pathway establishing the dorsal-ventral pattern of the Drosophila embryo. Cell. 1994; 76(4): 677–688. [DOI] [PubMed] [Google Scholar]
- 43. Ferreira AG, Naylor H, Esteves SS, Pais IS, Martins NE. The Toll-dorsal pathway is required for resistance to viral oral infection in Drosophila . PLoS Pathog.2014;10(12):e1004507 10.1371/journal.ppat.1004507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xu X, Jin F, Yu X, Ji S, Wang J. Expression and purification of a recombinant antibacterial peptide, cecropin, from Escherichia coli . Protein expression and purification. 2007; 53(2): 293–301. [DOI] [PubMed] [Google Scholar]
- 45. Epand RF, Mowery BP, Lee SE, Stahl SS, Lehrer RI. Dual mechanism of bacterial lethality for a cationic sequence-random copolymer that mimics host-defense antimicrobial peptides. Journal of Molecular Biology. 2008; 379(1): 38–50. 10.1016/j.jmb.2008.03.047 [DOI] [PubMed] [Google Scholar]
- 46. Sung WS, Lee J, Lee DG. Fungicidal effect and the mode of action of piscidin 2 derived from hybrid striped bass. Biochemical and Biophysical Research Communications. 2008; 371(3): 551–555. 10.1016/j.bbrc.2008.04.107 [DOI] [PubMed] [Google Scholar]
- 47. Christensen B, Fink J, Merrifield RB, Mauzerall D. Channel-forming properties of cecropins and related model compounds incorporated into planar lipid membranes. Proceedings of the National Academy of Sciences. 1988; 85(14): 5072–5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Conserved amino acid residues among four genes (black background), identical residues among three genes (pink background), and same residues among two genes are shown on blue background.
(TIF)
Drosophila melanogaster cecropin C (AAB82507); Drosophila melanogaster cecropin B (AAF57027.1); Aedes aegypti cecropin A (AAF59831); Manduca sexta cecropin 6 (AAO74638); Agrius convolvuli cecropin D precursor (ACX37671); Hyalophora cecropia cecropin partial(AAP93872); P. xylostella cecropin 1 (ADA13281); P. xylostella cecropin 2 (ADC54851); Px013797(derived from P. xylostella Genome Database); Pseudoplusia includens cecropin A (AAR99379); Papilio xuthus cecropin (ACR82292); Bombyx mori cecropin precursor(NP-001037392); P. xylostella cecropin 3 (KF960048); Hyphantria cunea cecropin A3 (AAB39003); Hyphantria cunea cecropin A (AID51414); Bombyx mori cecropinB precursor (NP-001096031); Spodoptera litura cecropin D partial(ABQ51092); Hyalophora cecropia cecropin B(AAA29184); Trichoplusia ni cecropin B (ABV68872); Helicoverpa armigera cecropin (AAX51304); Antheraea pernyi cecropin B(P01509); Tribolium castaneum cecropin 2 precursor (NP-001164146).
(TIF)
qRT-PCR analysis of Spätzle after RNAi on fat body, epidermis and hemocytes in P. xylostella from 12 h to 60 h. The relative expression levels of Spätzle mRNA was different after treatments, Means with two asterisks are statistically different (p<0.001) (Duncan’s Multiple Range Test) among treatments; Actin was used as an internal control. Each bar represents the mean ± S.E. (n = 3).
(TIFF)
qRT-PCR analysis of Dorsal after RNAi on fat body, epidermis and hemocytes in P. xylostella from 12 to 60 h. The relative expression levels of Dorsal mRNA was different after treatments, Means with two asterisks are statistically different (p<0.001) (Duncan’s Multiple Range Test) among treatments; Actin was used as an internal control. Each bar represents the mean ± S.E. (n = 3).
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.