Abstract
The γ-tubulin complex constitutes a key component of the microtubule-organizing center and nucleates microtubule assembly. This complex differs in complexity in different organisms: the budding yeast contains the γ-tubulin small complex (γTuSC) composed of γ-tubulin, GCP2 and GCP3, whereas animals contain the γ-tubulin ring complex (γTuRC) composed of γTuSC and three additional proteins, GCP4, GCP5 and GCP6. In Trypanosoma brucei, the composition of the γ-tubulin complex remains elusive, and it is not known whether it also regulates assembly of the subpellicular microtubules and the spindle microtubules. Here we report that the γ-tubulin complex in T. brucei is composed of γ-tubulin and three GCP proteins, GCP2-GCP4, and is primarily localized in the basal body throughout the cell cycle. Depletion of GCP2 and GCP3, but not GCP4, disrupted the axonemal central pair microtubules, but not the subpellicular microtubules and the spindle microtubules. Furthermore, we showed that the γTuSC is required for assembly of two central pair proteins and that γTuSC subunits are mutually required for stability. Together, these results identified an unusual γ-tubulin complex in T. brucei, uncovered an essential role of γTuSC in central pair protein assembly, and demonstrated the interdependence of individual γTuSC components for maintaining a stable complex.
Introduction
Microtubules are critically important components of the cytoskeleton in eukaryotic organisms and play essential roles in regulating intracellular transport, organelle positioning, and spatial and temporal organization of the cell (Nogales, 2001). The cytoskeleton of Trypanosoma brucei, an early branching parasitic protozoan, is characterized by an array of subpellicular microtubules located underneath the plasma membrane and is crucial for maintaining cell shape and cell polarity and for positioning single-copied organelles (Gull, 1999). Microtubules in T. brucei also constitute a vital part of the flagellum axoneme that contains a canonical 9+2 array of microtubules with nine outer microtubule doublets surrounding two central pair microtubules (Gull, 1999). The single flagellum in a trypanosome cell is nucleated by the basal body that adopts a canonical nine-triplet microtubule array, reminiscent of the centriole in animals, and acts as the cell’s microtubule-organizing center (MTOC) (Gull, 1999). The basal body in trypanosomes is composed of a mature basal body, which nucleates the flagellum, and an adjacent pro-basal body, which becomes matured during the S-phase of the cell cycle and starts to nucleate new flagellum assembly (Sherwin & Gull, 1989). Upon maturation of the existing pro-basal body, two additional pro-basal bodies adjacent to the two mature basal bodies emerge, and following cell cycle progression, the two pairs of mature basal body–pro-basal body start to be separated (Sherwin & Gull, 1989), which is driven by the elongation of the new flagellum and its attachment zone filament (Absalon et al., 2007). Given that γ-tubulin is found in the spindle poles (He et al., 2005, Scott et al., 1997), T. brucei may also possess intra-nuclear MTOCs that nucleate spindle microtubule assembly, but our knowledge about these MTOCs is very limited.
Microtubule nucleation is known to be mediated by the γ-tubulin ring complex (γTuRC), which is a multi-subunit protein complex that varies in size and composition in different organisms, with organisms of higher complexity usually composing of more subunits (Kollman et al., 2011). γTuRC in humans is composed of γ-tubulin, five additional subunits named Gamma-tubulin Complex Proteins 2 to 6 (GCP2-GCP6), and two non-GCP family proteins, MOZART1 and MOZART2 (aka GCP8) (Teixido-Travesa et al., 2010, Hutchins et al., 2010), all of which are essential for microtubule nucleation. In invertebrate animals such as Drosophila, γTuRC is composed of γ-tubulin and five GCP proteins, GCP2-GCP6, but only γ-tubulin and the orthologs of GCP2 and GCP3 are essential (Verollet et al., 2006). Unlike metazoa, however, Saccharomyces cerevisiae and its closely related yeasts only retain Spc97 and Spc98 (Geissler et al., 1996, Knop et al., 1997, Vinh et al., 2002), which are GCP2 and GCP3 orthologs, respectively, thereby forming the so-called γ-tubulin small complex (γTuSC) that is believed to constitute the core of the microtubule nucleation machinery in eukaryotes. The five GCP proteins contain two motifs that are unique to the GCP proteins, and all bind to γ-tubulin directly (Kollman et al., 2011).
γ-tubulin from T. brucei exhibits high sequence homology to its yeast and human homologs (Scott et al., 1997) and is required for nucleation of the central pair microtubules in the flagellum axoneme (McKean et al., 2003). However, little is known about the molecular composition of the γ-tubulin complex in trypanosomes and the interplay among its components. It is also not known whether the γ-tubulin complex is required for nucleating subpellicular microtubules and spindle microtubules. A recent report identified the putative GCP2 homolog in T. brucei and showed that it is essential for cell viability and cell motility (Sheriff et al., 2014), but no evidence was presented to support it is a bona fide component of the γ-tubulin complex, and its potential role in microtubule assembly was not investigated. In this study, we carried out tandem affinity purification and identified the trypanosome γ-tubulin complex, which is composed of γ-tubulin and three GCP proteins, GCP2, GCP3 and GCP4. Our results suggest that only the γTuSC plays essential roles in T. brucei and is required for nucleation of central pair microtubules and for assembly of central pair proteins in the flagellar axoneme.
Results
Identification of the γ-tubulin complex in T. brucei by tandem affinity purification
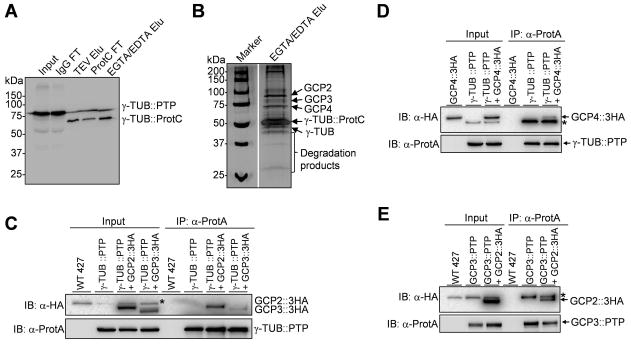
To identify the trypanosome γ-tubulin complex, we carried out tandem affinity purification. To this end, γ-tubulin was tagged with a C-terminal PTP (Protein A-TEV-Protein C) epitope (Schimanski et al., 2005) at one of its endogenous loci in the procyclic form. Through a two-step affinity purification (Fig. 1A), several distinct protein bands were co-precipitated with PTP-tagged γ-tubulin (Fig. 1B). Mass spectrometry analysis of individual protein bands identified GCP2 (Tb927.10.9770), GCP3 (Tb927.11.11340) and GCP4 (Tb927.6.2400), in addition to the γ-tubulin bait (Fig. 1B). In a reciprocal tandem affinity purification experiment with GCP3 as the bait, γ-tubulin, GCP2 and GCP4 were all identified by subsequent mass spectrometry (data not shown), which further confirmed the formation of a complex by the four proteins. Moreover, we also carried out co-immunoprecipitation to confirm the association of GCP proteins with γ-tubulin, and the results showed that γ-tubulin was indeed capable of pulling down each of the three GCP proteins from trypanosome lysate (Fig. 1C, D). Additionally, GCP3 was also able to pull down GCP2 from trypanosome cell lysate (Fig. 1E). Altogether, these results suggest that the γ-tubulin complex in T. brucei consists of γ-tubulin, GCP2, GCP3 and GCP4.
Figure 1. Identification of the trypanosome γ-tubulin complex.
(A). Tandem affinity purification of the γ-tubulin complex from procyclic trypanosomes. PTP-tagged γ-tubulin (γ-TUB::PTP) and protein C-tagged γ-tubulin (γ-TUB::ProtC) after TEV protease digestion were detected with anti-Protein C antibody in input, IgG Sepharose column flow-through (IgG FT), eluate after TEV protease digestion (TEV Elu), anti-Protein C column flow-through (ProtC FT), and final EGTA/EDTA eluate (EGTA/EDTA Elu). (B). Silver staining of purified proteins. The final EGTA/EDTA eluate was separated on a 10% SAS-PAGE and stained with silver solution. Individual protein bands were excised and analyzed by LC-MS/MS. Proteins identified by LC-MS/MS are indicated on the right side of the gel. (C). Co-immunoprecipitation of GCP2::3HA and GCP3::3HA with γ-tubulin::PTP. (D) Co-immunoprecipitation of GCP4::3HA with γ-tubulin::PTP. (E). Co-immunoprecipitation of GCP2::3HA with GCP3::PTP.
Intriguingly, by homology-based search of T. brucei genome database, we identified a fourth GCP-like protein (Tb927.10.6790), which is homologous to GCP3. We named it GCP3L for GCP3-Like protein. GCP3L encodes a protein of 1727 amino acids with a calculated molecular mass of 188.7 kDa, which is much larger than the other GCP proteins. GCP3L was not co-immunoprecipitated with γ-tubulin (Fig. 1B) and GCP3 (data not shown) in our tandem affinity purifications. Whether GCP3L is a component of the γ-tubulin complex in trypanosomes remains to be determined. If GCP3L is indeed a component of the γ-tubulin complex, it may associate with the complex through weak interactions or transiently at certain cell cycle stages. Moreover, T. brucei apparently lacks the homolog of GCP6, which is believed to be added recently to the animal and fungal lineages (Kollman et al., 2011). It appears that the molecular composition of the trypanosome γ-tubulin complex is different from that in fungi, such as Saccharomyces cerevisiae and its closed related yeasts (Geissler et al., 1996, Knop et al., 1997), and that in animals, such as Drosophila, Xenopus, and humans (Kollman et al., 2011).
The γ-tubulin complex is concentrated near the basal bodies throughout the cell cycle
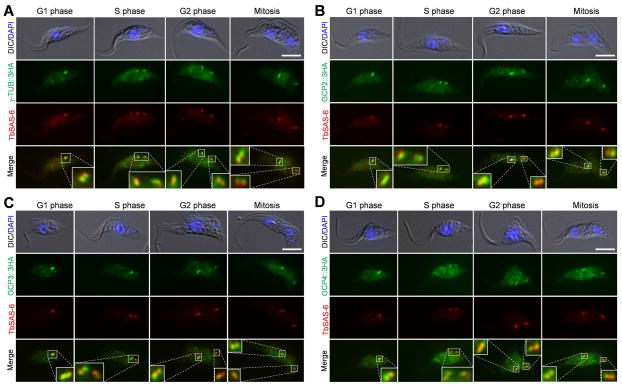
The localization of γ-tubulin in trypanosomes has been investigated previously (Scott et al., 1997, He et al., 2005), but conclusive results were not obtained. Using a polyclonal anti-γ-tubulin antibody, γ-tubulin was localized to multiple subcellular locations, including the basal body, the spindle poles and a small subset of subpellicular microtubules (Scott et al., 1997). When γ-tubulin was tagged with YFP or a Ty epitope and stably overexpressed in trypanosome, however, both tagged forms were only localized to the basal body in G1 cells, and Ty-tagged form was localized to the basal body and the spindle poles in a mitotic cell (He et al., 2005). To clarify the localization of γ-tubulin as well as to determine the subcellular localization of GCP2, GCP3 and GCP4, we tagged each of these proteins with a C-terminal triple HA epitope at one of their respective endogenous loci and carried out immunofluorescence microscopy with anti-HA antibody and anti-TbSAS-6 antibody, which detects the basal body cartwheel in both the mature basal body and the pro-basal body (Hu et al., 2015). The results showed that all four proteins were detected in the vicinity of the mature basal body and the pro-basal body throughout the cell cycle, as shown by their partial co-localization with TbSAS-6 (Fig. 2). Additionally, all four proteins appeared to be also detected in some punctate structures in the cell body at all of the cell cycle stages, but they were not enriched in the spindle poles during the mitotic phase (Fig. 2). On the detergent-extracted cytoskeletons, the punctate structures detected by all four proteins were significantly decreased, but the four proteins were still localized to the vicinity of mature basal bodies and pro-basal bodies (Supplementary Fig. 1). These results suggest that γ-tubulin and its associated proteins are concentrated in the basal body region throughout the cell cycle.
Figure 2. Subcellular localization of γ-tubulin and its associated proteins in procyclic trypanosomes.
γ-tubulin (A) and its associating GCP proteins, GCP2 (B), GCP3 (C), and GCP4 (D), were each tagged with a C-terminal triple HA epitope from one of its endogenous loci in procyclic trypanosomes. Cells were immunostained with FITC-conjugated anti-HA mAb and anti-TbSAS-6 pAb. Scale bars: 5 μm.
GCP2 and GCP3, but not GCP4, are essential for cell viability
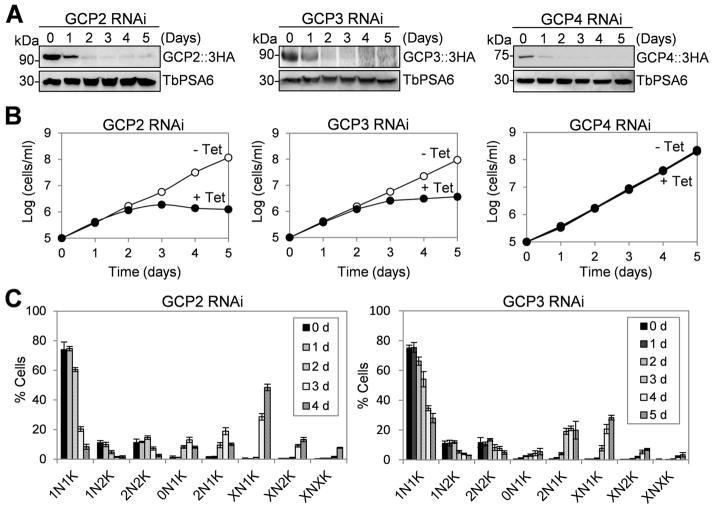
To investigate whether the three γ-tubulin–associated proteins are essential in T. brucei, RNAi was carried out in the procyclic form of T. brucei. To monitor the efficiency of RNAi, the three GCP proteins were each endogenously tagged with a triple HA epitope in their respective RNAi cell lines for western blotting with anti-HA antibody. Upon RNAi induction for 2 days, the level of GCP2 was significantly decreased, whereas GCP3 and GCP4 levels were each decreased to undetectable levels (Fig. 3A). Depletion of GCP2 and GCP3 caused severe growth inhibition, but knockdown of GCP4 exerted little effect on cell proliferation (Fig. 3B), suggesting that GCP2 and GCP3, but not GCP4, are essential for cell viability in the procyclic form of T. brucei. Therefore, only GCP2 and GCP3 were further characterized in this study. The numbers of different cell types upon RNAi induction were counted, which showed that initially there was an emergence of cells with two nuclei and one kinetoplast (2N1K) and subsequently a prominent accumulation of cells with multiple (>2) nuclei and one kinetoplast (XN1K) (Fig. 3C), suggesting inhibited kinetoplast segregation and defective cytokinesis. Given that γ-tubulin is essential for viability of procyclic trypanosomes (McKean et al., 2003), these results suggest that only the γTuSC plays essential roles in trypanosomes.
Figure 3. GCP2 and GCP3, but not GCP4, are essential in procyclic trypanosomes.
(A). Western blotting to monitor the GCP protein level in GCP RNAi cells. GCP2, GCP3, and GCP4 were each tagged with a triple HA epitope from their respective endogenous locus in cells harboring the respective RNAi construct. Western blotting was carried out with anti-HA mAb to detect 3HA-tagged GCP proteins. The same membrane was re-probed with anti-TbPSA6, which detects the α6 subunit of the proteasome, for loading control. (B). Clonal cell lines harboring individual GCP RNAi constructs were incubated with (+ Tet) or without (− Tet) tetracycline for 5 days, and cell growth was monitored by daily counting the cells. (C) Tabulation of cells with different numbers of nucleus (N) and kinetoplast (K) in non-induced control and GCP RNAi-induced cells. About 200 cells from each cell line were counted. The error bars represent S.D. calculated from three independent experiments.
Depletion of GCP2 and GCP3 impairs new flagellum biogenesis
To investigate the effect of GCP2 and GCP3 RNAi on flagellum biogenesis, we immunostained the control and RNAi cells with L8C4 to label the flagellum. Since the new flagellum is nucleated from the mature basal body (Sherwin & Gull, 1989), cells were co-immunostained with YL 1/2 to label the mature basal body (Sherwin et al., 1987). We counted the numbers of flagellum and mature basal body in 2N1K cells, which emerged first upon RNAi induction and may be the primary defect caused by GCP2 and GCP3 RNAi. The results showed that ~24% of the 2N1K cells from GCP2 RNAi and GCP3 RNAi contained only one flagellum and one mature basal body, whereas ~16% of the 2N1K cells from GCP2 RNAi and ~30% of the 2N1K cells from GCP3 RNAi contained one flagellum and two mature basal bodies (Fig. 4A–B). The rest of the 2N1K cells, ~60% in GCP2 RNAi and 56% in GCP3 RNAi, contained two flagella and multiple (≥2) mature basal bodies, but the new flagellum appeared to be shorter than the new flagellum in the control 2N2K cells (Fig. 4A–C).
Figure 4. Depletion of GCP2 and GCP3 impairs flagellum assembly.
(A). Biogenesis of flagellum in GCP2 RNAi cells and GCP3 RNAi cells. Non-induced control and RNAi-induced cells were immunostainined with L8C4 and YL 1/2 to label flagella and mature basal bodies, respectively. Arrows indicate the old flagellum, solid arrowheads indicate the new flagellum, and open arrowheads show the mature basal body. Scale bars: 5 μm. (B). Quantification of flagellum and mature basal body in GCP2-deficient 2N1K cells and GCP3-deficient 2N1K cells. Data were presented as the mean percentage ± S.D. of about 200 of 2N1K cells counted from each of the three independent experiments. (C). Flagellum length of the control 2N2K cells and the 2N1K cells from GCP2 and GCP3 RNAi cells. Flagellum was immunostainined with the L8C4 antibody. About 200 of 2N2K cells from the non-induced control cells and 200 each of the 2N1K cells from GCP2 and GCP3 RNAi cells were randomly selected for measuring the length of new and old flagella with ImageJ. Shown is the average flagellum length, and error bars represent S.D. calculated from three independent RNAi experiments for both GCP2 and GCP3 RNAi. (D). Basal body and flagellum in GCP2- and GCP3-deficient multinucleated (>2) cells. Mature basal body was immunostained with YL 1/2 antibody, and flagellum was immunolabeled with the L8C4 antibody. Arrows indicate the old flagellum, solid arrowheads indicate the small new flagellum, and open arrowheads show the basal body. Scale bars: 5 μm. (E). Quantification of basal body and flagellum in GCP2- and GCP3-deficient polyploid cells. Data were presented as the mean percentage ± S.D. of about 100 polyploid cells counted from each of the three independent experiments.
The lack of a new flagellum or formation of a shorter new flagellum in the 2N1K cells may be attributed to either a delay or defects in new flagellum biogenesis. If there was a delay, then a full-length new flagellum should be detected in cells after longer RNAi induction. To investigate which hypothesis is correct, we counted the numbers of flagella in XN1K cells, which became prominent after longer RNAi induction and were derived from the 2N1K cells. About 50% of the XN1K cells in GCP2 RNAi and ~43% of the XN1K cells in GCP3 RNAi contained only one flagellum, whereas the rest of the XN1K cells possessed two or three flagella (Fig. 4D, E). In the XN1K cells with multiple flagella, the old flagellum was of full-length (Fig. 4D, arrows), but the new flagellum was always much shorter (Fig. 4D, solid arrowheads). All of the XN1K cells, no matter how many flagella they possess, contained multiple (>2) mature basal bodies that were clustered around the single kinetoplast (Fig. 4D, open arrowheads). These results confirmed that biogenesis of the new flagellum was impaired upon GCP2 or GCP3 depletion. Additionally, the production of 2N1K cells with two mature basal body and one flagellum (~16% in GCP2 RNAi cells and ~30% in GCP3 RNAi cells) (Fig. 4A, B) and XN1K cells with multiple mature basal bodies and one flagellum (Fig. 4D, E) suggest that although the pro-basal bodies in these cells have been matured, they were not capable of nucleating new flagellum assembly. Moreover, in both GCP2 RNAi and GCP3 RNAi cells, ~20% of the cells contained a detached new flagellum (data not shown).
GCP2 and GCP3 are required for nucleation of central pair microtubules
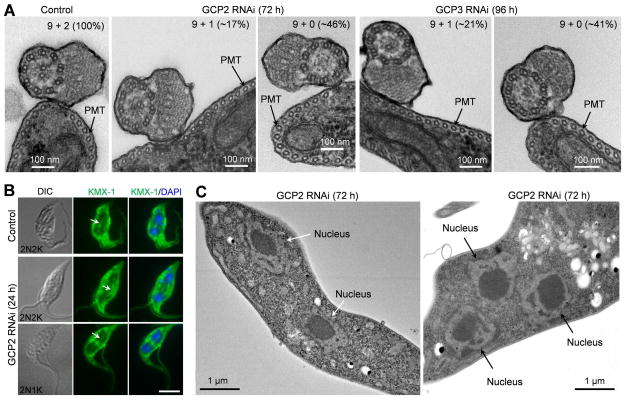
γ-tubulin is required for nucleation of central pair microtubules in the new flagellum (McKean et al., 2003). To investigate whether GCP2 and GCP3 RNAi also affected central pair microtubule assembly, we examined the flagellum axoneme by electron microscopy, which showed that ~17% of the axonemes in GCP2 RNAi cells and ~21% of the axonemes in GCP3 RNAi cells missed one central pair microtubule (9+1 array), and ~45% in GCP2 RNAi cells and ~41% in GCP3 RNAi cells missed both central pair microtubules (9+0 array) (Fig. 5A). However, the axonemal outer doublet microtubules and the subpellicular microtubules apparently were not affected by GCP2 and GCP3 RNAi (Fig. 5A). These results, together with the previous work (McKean et al., 2003), suggest that γTuSC is required for nucleation of central pair microtubules, but not the outer doublet microtubules in the axoneme.
Figure 5. Effect of GCP2 and GCP3 RNAi on the assembly of axonemal microtubules, subpellicular microtubules and spindle microtubules.
(A). Transmission electron microscopy of the flagellar axoneme of non-induced control, GCP2 RNAi, and GCP3 RNAi cells. Percentage of axonemal microtubule array (9+2, 9+1, and 9+0) in non-induced control, GCP2 RNAi, and GCP3 RNAi cells were counted from ~120 sections for each of the two cell lines. PMT: subpellicular microtubules. (B). Mitotic spindle structure in non-induced control and GCP2 RNAi cells. Cells were immunostained with KMX-1 to label the mitotic spindle (arrows) in 2N2K and 2N1K cells. Scale bar: 5 μm. (C). Morphology of the nucleus in GCP2 RNAi cells visualized under electron microscope. Shown are a bi-nucleate cell and a tri-nucleate cell. Note that the nuclei were of normal shape and were well segregated, and nuclear envelope was intact.
To investigate whether γTuSC deficiency disrupted mitotic spindle formation, we immunostained the cells with the KMX-1 antibody, and the results showed that in the 2N2K and 2N1K cells from GCP2 RNAi, the mitotic spindle was clearly detected, which leads to nuclear segregation (Fig. 5B, arrows). The nuclei appeared to be normal in shape, contained intact nuclear membrane, and were well separated, as visualized under light microscope (Fig. 4A, 5B) and electron microscope (Fig. 5C). Similar observations were also made in GCP3 RNAi cells (data not shown). These results suggest that GCP2 and GCP3 are not required for nucleating spindle microtubule assembly.
γTuSC is required for assembly of central pair proteins to the flagellum
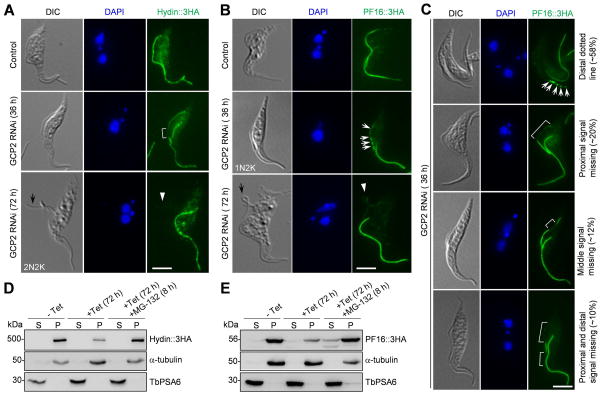
The effect of γTuSC depletion on the assembly of the central pair microtubules in the new flagellum prompted us to investigate the potential impact on the localization of central pair proteins to the new flagellum. Three central pair proteins, Hydin, PF16, and PF20, have been identified in trypanosomes (Ralston et al., 2006, Branche et al., 2006, Dawe et al., 2007). We attempted to tag the three proteins with a triple HA epitope from their respective endogenous locus in γTuSC RNAi cells. Only Hydin and PF16 were successfully tagged, so our effort was focused on these two proteins. RNAi of each of the three γTuSC subunits (γ-tubulin, GCP2 and GCP3) exerted almost identical effect on Hydin and PF16. Therefore, only the result from GCP2 RNAi was presented. Cells were induced with tetracycline for 36 and 72 hours and immunostained with anti-HA antibody. The results showed that upon GCP2 RNAi for 36 h, Hydin::3HA level was reduced from the middle portion of the new flagellum (Fig. 6A, white bracket), and PF16::3HA level was decreased at multiple locations in the new flagellum (Fig. 6B, white arrows). After RNAi induction for 72 h, levels of Hydin::3HA and PF16::3HA in the new flagellum were reduced to very low levels (Fig. 6A, B, white arrowheads). We further examined PF16::3HA in the new flagellum after RNAi for 36 h, and found that PF16::3HA level appeared to be randomly reduced in the new flagellum from either the proximal region, both the proximal and distal regions, the middle portion, or multiple locations (Fig. 6C, white arrows and white brackets). These results suggest that GCP2 RNAi disrupted the assembly of Hydin and PF16 to the new flagellum. Given that levels of Hydin and PF16 appeared to be randomly reduced from the new flagellum, GCP2 RNAi may also compromise the stability of Hydin and PF16 in the new flagellum.
Figure 6. GCP2 RNAi disrupts Hydin and PF16 assembly to the new flagellum.
(A, B). Effect of GCP2 RNAi on the localization of Hydin (A) and PF16 (B). Hydin and PF16 were each endogenously tagged with a triple HA epitope in cells harboring the GCP2 RNAi construct. Non-induced control and GCP2 RNAi-induced cells (36 h and 72 h) were immunostained with anti-HA antibody to detect Hydin::3HA and PF16::3HA. The white bracket shows the reduced Hydin::3HA signal in the middle portion of the new flagellum, the white arrows show the portions of the new flagellum with reduced PF16::3HA signal, and the white arrowheads indicate the signal of Hydin::3HA and PF16::3HA in the detached flagellum (black arrows). Scale bars: 5 μm. (C). GCP2 RNAi caused random depletion of PF16::3HA from the new flagellum at earlier time points of RNAi induction. White arrows show the PF16::3HA signal (as multiple dots) at the distal portion of the new flagellum, and white brackets outline the portions of the new flagellum with much reduced PF16::3HA signal. Scale bar: 5 μm. (D, E). Effect of GCP2 RNAi on the stability of Hydin (D) and PF16 (E) in cytosolic and the cytoskeletal fractions. Non-induced control and GCP2 RNAi cells treated without and with MG-132 were lysed in PEME buffer containing 1% NP-40 for cytoskeleton preparation. The soluble cytosolic fraction (S) and cytoskeletal pellet fraction (P) were separated by centrifugation, loaded onto a SDS–PAGE gel and transferred onto a PVDF membrane for western blotting with anti-HA antibody to detect Hydin::3HA and PF16::3HA. The same blot was probed with anti-TbPSA6 as the cytosolic marker and with anti-α-tubulin as the cytoskeletal marker. Similar results were obtained in three independent experiments.
The decrease of Hydin and PF16 levels in the new flagellum upon GCP2 RNAi led us to hypothesize that the two proteins were either degraded or mis-localized to the cytosol. To test which hypothesis is correct, cellular fractionation was carried out to separate the cytosolic fraction and the cytoskeletal fraction, and western blotting with anti-HA antibody showed that Hydin and PF16 associated tightly with the flagellum upon detergent treatment and were significantly decreased after GCP2 RNAi for 72 h (Fig. 6D, E). When MG-132, a potent inhibitor of the proteasome, was added to GCP2 RNAi cells for 8 h, levels of Hydin and PF16 were significantly increased in the pellet fraction, although PF16 level in the cytosolic fraction was also increased (Fig. 6D, E). Similar results were also obtained in γ-tubulin RNAi and GCP3 RNAi cells (data not shown). These results suggest that when Hydin and PF16 were not assembled to the new flagellum, they were degraded.
GCP2 and GCP3 are required for cell motility
Although the previous work showed that RNAi of γ-tubulin produced immotile cells (McKean et al., 2003), detailed analysis of motility defects was not carried out. The defective assembly of central pair microtubules caused by GCP2 and GCP3 depletion suggests that cell motility probably was also impaired. To test this possibility and to quantitatively characterize the motility defect, sedimentation assay and motility tracing were carried out. Sedimentation assays showed that both the GCP2 RNAi cells and the GCP3 RNAi cells lost cell motility (Fig. 7A). Tracing the motility of cells by time-lapse video microscopy showed that there were two distinct types of motility-defective cells: immotile cells and tumbler cells (Fig. 7B and Supplementary Movies 2, 3, 5, and 6), in contrast to the non-induced control cells that were highly motile (Fig. 7B and Supplementary Movies 1 and 4). The immotile cells constituted about 56% and 51% of the total population in GCP2 RNAi cells and GCP3 RNAi cells, respectively, and the tumbler cells constituted about 35% and 34% of the total population in GCP2 RNAi and GCP3 RNAi cells, respectively (Fig. 7C). These results suggest that GCP2 and GCP3 are required for cell motility, which likely is attributed to the defective assembly of the central pair microtubules.
Figure 7. GCP2 and GCP3 RNAi compromises cell motility.
(A). Sedimentation assays to monitor cell motility of GCP2 RNAi and GCP3 RNAi cells. The parental 29-13 strain, the non-induced control and GCP2 RNAi and GCP3 RNAi cells after tetracycline induction for 24, 48, and 72 hours were each incubated in cuvettes and cell density (optical density) was determined and plotted against the time of incubation (h). Data represent the average OD600 value ± S.D. from three independent experiments. (B). Monitoring the motility of non-induced control cells, GCP2 RNAi cells (48 h), and GCP3 RNAi cells (72 h) by time-lapse video microscopy. White dashed lines indicate the posterior tip of the cells at the start of time-lapse video microscopy. Scale bar: 10 μm. (C). Tabulation of cells scored as runner (white), tumbler (grey) or immotile (black) in non-induced control cells, GCP2 RNAi cells, and GCP3 RNAi cells. n, number of cells counted.
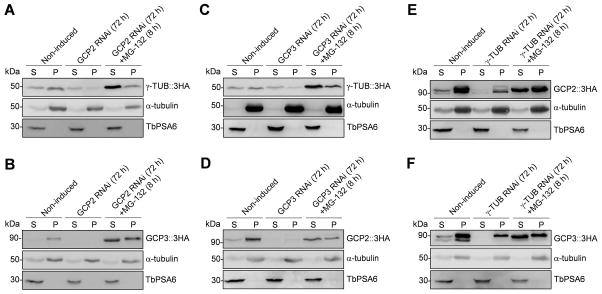
γTuSC components are interdependent for complex formation
We next investigated the effect of RNAi of individual γTuSC subunits on the stability of their partners. We separated the cytosolic and cytoskeletal fractions by treating the cells with detergent, and carried out western blotting to monitor the levels of γTuSC subunit proteins in the RNAi cells. We found that in GCP2 RNAi cells, the level of γ-tubulin in the pellet fractions was significantly decreased (Fig. 8A). To investigate whether γ-tubulin was degraded in GCP2 RNAi cells, we treated the RNAi cells with MG-132. Western blotting showed that the level of γ-tubulin in the cytosolic fraction and pellet fraction were increased as compared to the RNAi cell alone (Fig. 8A), suggesting that γ-tubulin was indeed degraded in GCP2 RNAi cells. We further examined the effect of GCP2 RNAi on GCP3, and found that GCP3 in GCP2 RNAi cells were reduced to undetectable levels, but was stabilized in the presence of MG-132 (Fig. 8B), suggesting that, like γ-tubulin, GCP3 was also degraded upon GCP2 depletion. Similarly, in GCP3 RNAi cells, both γ-tubulin and GCP2 were degraded (Fig. 8C, D), and in γ-tubulin RNAi cells, both GCP2 and GCP3 were degraded (Fig. 8E, F). Together, these results suggest that each of the three γTuSC subunits is required to maintain the stability of its partners in the complex.
Figure 8. γTuSC components are interdependent for stability.
(A) Effect of GCP2 RNAi on γ-tubulin stability. γ-tubulin was endogenously tagged with a triple HA epitope in GCP2 RNAi cells. Preparation of cytosolic (S) and cytoskeletal (P) fractions for western blotting was described in the Materials and Methods section. α-tubulin and TbPSA6 served as the loading controls for the cytoskeletal and cytosolic fractions, respectively. (B). Effect of GCP2 RNAi on GCP3 stability. GCP3 was endogenously tagged with a triple HA epitope in GCP2 RNAi cells. (C) Effect of GCP3 RNAi on γ-tubulin stability. γ-tubulin was endogenously tagged with a triple HA epitope in GCP3 RNAi cells (D) Effect of GCP3 RNAi on GCP2 stability. GCP2 was endogenously tagged with a triple HA epitope in GCP3 RNAi cells (E). Effect of γ-tubulin RNAi on GCP2 stability. GCP2 was endogenously tagged with a triple HA epitope in γ-tubulin RNAi cells. (F). Effect of γ-tubulin RNAi on GCP3 stability. GCP2 was endogenously tagged with a triple HA epitope in γ-tubulin RNAi cells. For all of the western blots, at least three independent experiments were carried out, and the results were very similar.
Discussion
In this paper we reported the purification and functional characterization of the γ-tubulin complex in T. brucei. Through tandem affinity purification, we identified three GCP-like proteins, GCP2, GCP3 and GCP4, as γ-tubulin partners (Fig. 1). Functional studies demonstrated that GCP2 and GCP3, but not GCP4, are required for cell viability in procyclic trypanosomes (Fig. 3), suggesting that trypanosomes employ an essential γTuSC for nucleating microtubule assembly. Given that S. cerevisiae and its closely related yeasts have lost GCP4, GCP5 and GPC6, but are still capable of nucleating microtubule assembly (Knop et al., 1997, Geissler et al., 1996, Vinh et al., 2002) and that Drosophila GCP4, GCP5 and GCP6 are dispensable for microtubule nucleation (Verollet et al., 2006), it is believed that γTuSC represents the core of the microtubule nucleation machinery in eukaryotes and is sufficient for proper microtubule organization, albeit with a much lower activity than γTuRC (Kollman et al., 2011).
Through epitope tagging, we showed that γ-tubulin and its associated GCP proteins are concentrated in the vicinity of basal bodies throughout the cell cycle (Fig. 2 and Supplementary Fig. 1). They are also distributed in the cell body, but are not enriched in the spindle poles during mitosis (Fig. 2). Due to the limited resolution of fluorescence microscopy, the precise location of the γ-tubulin complex in the basal body region cannot be unambiguously determined, but the complex appears to be distal to the basal body cartwheel (Fig. 2 and Supplementary Fig. 1). Contrary to the previous reports (Scott et al., 1997; He et al., 2005), however, epitope-tagged γTuSC subunits were not detected in the spindle poles in any mitotic cells examined by us (Fig. 2 and Supplemental Fig. 1). Similarly, in Leishmania, another kinetoplastid parasite closely related to T. brucei, and Giardia, γ-tubulin was also not found in the spindle poles (Libusova et al., 2004, Nohynkova et al., 2000). Moreover, in T. brucei cells depleted of γTuSC subunits, the mitotic spindle was still formed, and chromosome segregation occurred normally (Fig. 5), thus arguing against a role of the γTuSC in nucleating spindle microtubules in T. brucei. Spindle assembly in eukaryotes involves two distinct pathways, the centrosome-mediated pathway and the chromatin-mediated pathway. T. brucei is known to lack centrosomes, and the centriole cartwheel protein SAS-6 is exclusively localized in the basal body (Hu et al., 2015). Although direct evidence is lacking to support the presence of the chromatin-mediated pathway in T. brucei, the fact that the proteins required for the chromatin-mediated spindle assembly pathway but not the centrosome-mediated pathway are expressed in T. brucei (Li, 2012) hints that it is likely present. Additionally, although γTuSC components are also detected as punctate structures on the microtubule cytoskeleton (Fig. 2 and Supplementary Fig. 1), depletion of GCP2 and GCP3 exerted little effect on the subpellicular microtubules (Fig. 5A). Therefore, γTuSC in T. brucei may be primarily required for nucleation of the central pair microtubules.
Our RNAi results suggest that GCP2 and GCP3 are both required for flagellum biogenesis. This conclusion was supported by the fact that in GCP2 and GCP3 RNAi cells, the new flagellum was either not assembled at all (in ~41% of the 2N1K cells and ~43% of the XN1K cells) or was partially assembled, thus being shorter than the new flagellum of the control cells (Fig. 4). The failure to synthesize the new flagellum in some of the 2N1K cells could be due to the fact that assembly of the new flagellum had not initiated when RNAi was induced. Therefore, new flagellum formation was completely inhibited in those cells. The major defect in the flagellum of GCP2 and GCP3 RNAi cells appeared to be the missing of one or two central pair microtubules in ~60% of the axonemes examined (Fig. 5), which is in good agreement with that of γ-tubulin RNAi (McKean et al., 2003). This provides another line of evidence to support that GCP2 and GCP3 function together with γ-tubulin to nucleate the central pair microtubules in T. brucei. Cells with a shorter flagellum were also observed after PF16 RNAi in T. brucei (Absalon et al., 2007) and in central pair-deficient Chlamydomonas cells (Lechtreck et al., 2013), but were not observed after Hydin RNAi in T. brucei (Dawe et al., 2007).
An interesting finding made in this report is the requirement of γTuSC for assembly of two central pair proteins, PF16 and Hydin, in the new flagellum, in which the central pair microtubules were disrupted (Fig. 6). It suggests that assembly of central pair microtubules is necessary to maintain the two proteins in the flagellum axoneme. It is unclear whether other central pair proteins are similarly affected. However, since the trypanosome central pair proteome is not determined, this cannot be investigated. To date, three central pair proteins, Hydin, PF16 and PF20, have been identified in T. brucei, all of which appear to be required to maintain the orientation of the central pair microtubules and are involved in the assembly of the central pair microtubules (Ralston et al., 2006, Branche et al., 2006, Dawe et al., 2007). In Chlamydomonas reinhardtii, a number of central pair proteins have been identified and characterized. Intriguingly, in the deletion mutant of PF19, in which the central pair microtubules were not assembled, levels of Hydin and several other central pair proteins were decreased (Lechtreck et al., 2013). It appears that assembly of the central pair microtubules in Chlamydomonas is also required for assembly of central pair proteins, similar to that in trypanosomes.
Our results also suggest that individual γTuSC subunits are mutually required for maintaining a stable complex (Fig. 8). It is likely that formation of γTuSC is necessary to maintain the stability of each of its three subunits. However, it remains unclear why the stability of γTuSC subunits was affected in the absence of their partner, but it appears that in the absence of one subunit, the other γTuSC subunits accumulated in the cytosol, where they were degraded by the proteasome (Fig. 8). Similar findings have been reported previously in other protein complexes in T. brucei (Wei et al., 2013, Wei et al., 2014). In the TbKIN-C–TbKIN-D complex and the TbCentrin3–TbIAD5-1 complex, depletion of one component of the protein complex invariably destabilized its partner (Wei et al., 2013, Wei et al., 2014). It is unclear whether this is a general control mechanism employed by trypanosome cells to maintain the integrity of protein complexes, but it would be interesting to investigate whether other protein complexes in trypanosomes are similarly regulated.
Experimental Procedures
Trypanosome cell lines and RNAi
The procyclic 29-13 cell line (Wirtz et al., 1999) was grown in the SDM-79 medium supplemented with 10% fetal bovine serum (Atlanta Biologicals, Inc), 15 μg/ml G418, and 50 μg/ml hygromycin. The procyclic 427 cell line was cultured at 27°C in SDM-79 medium supplemented with 10% fetal bovine serum.
For RNAi of individual γ-tubulin complex components, a 500–600bp DNA fragment from the coding region of each of the four γ-tubulin complex genes was cloned into the pZJM vector (Wang et al., 2000). The resulting plasmids were electroporated into the 29-13 cell line according to our published procedures (Wei et al., 2014). Transfectants were selected under 2.5 μg/ml phleomycin and cloned by limiting dilution. RNAi was induced by incubating the cells with 1.0 μg/ml tetracycline.
In situ epitope tagging of proteins
For endogenous epitope tagging of γ-tubulin complex proteins, PF16 (Branche et al., 2006, Ralston et al., 2006), and Hydin (Dawe et al., 2007), a 400~500-bp DNA fragment corresponding to the C-terminal coding region of each of these genes was cloned into the pC-3HA-PAC vector. The resulting construct was linearized by restriction digestion within the gene fragment with appropriate restriction enzymes and electroporated into the 29-13 cell line harboring the RNAi construct. Transfectants were selected with 1.0 μg/ml puromycin and cloned by limiting dilution. Successful gene tagging was confirmed by PCR and subsequent sequencing of the PCR fragment as well as by western blotting with anti-HA antibody (Sigma-Aldrich).
Tandem affinity purification and LC-MS/MS
For tandem affinity purification of the γ-tubulin complex from T. brucei, a 500-bp DNA fragment corresponding to the C-terminal coding region of γ-tubulin was PCR amplified from the genomic DNA and cloned into the pC-PTP-NEO vector (Schimanski et al., 2005). The resulting construct, pC-γ-TUB::PTP-NEO, was linearized and transfected into the 427 cell line. Transfectants were selected under 40 μg/ml G418 and cloned by limiting dilution. Tandem affinity purification was carried out according to our published procedures (Hu et al., 2012b, Wei et al., 2014). Briefly, about 2 L cell culture (5 × 106/ml) were harvested, centrifugated at 5,000 rpm for 30 min, and lysed by thorough sonication in IP buffer (25 mM Tris-HCl pH 7.6, 500 mM NaCl, 1 mM DTT, 1% NP-40, and protease inhibitor cocktail). The cell lysate was cleared by centrifugation at 10,000 rpm for 30 min, and the supernatant was loaded onto the IgG sepharose beads (Invitrogen). After overnight cleavage with the TEV protease, the eluate was incubated with the Protein C agarose beads (Invitrogen). The bound proteins were eluated with EGTA/EDTA, precipitated by incubating with 20 μl StrataClean resin, separated on SDS-PAGE, and stained with silver solution (Pierce).
Protein bands were excised from the SDS-PAGE gel, treated with trypsin, and analyzed on an LTQ Orbitrap XL (Thermo-Fisher Scientific) interfaced with an Eksigent nan0-LC 2D plus ChipLC system (Eksigent Technologies, Dublin, CA) at the Proteomics Core Facility of the University of Texas Health Science Center at Houston. Data analysis was performed according to our previous publication (Wei et al., 2014). Raw data files were searched against Trypanosoma brucei database (version 4) using Mascot search engine. The search conditions used peptide tolerance of 10 p.p.m. and MS/MS tolerance of 0.8 Da with the enzyme trypsin and two missed cleavages.
Co-immunoprecipitation
Cells harboring the pC-γ-TUB::PTP-NEO construct was transfected with pC-GCP2::3HA-PAC, pC-GCP3::3HA-PAC, and pC-GCP4::3HA-PAC, respectively. Successful transfectants were selected under 1.0 μg/ml puromycin in addition to 40 μg/ml G418, and cloned by limiting dilution. For co-immunoprecipitation of GCP2 and GCP3, the wild-type 427 cells were first transfected with pC-GCP3::PTP-NEO, and transfectants were cloned by limiting dilution. Subsequently, the cells containing the pC-GCP3::PTP-NEO construct were transfected with pC-GCP2::3HA-PAC, and transfectants were also cloned by limiting dilution.
Co-immunoprecipitation was carried out as described previously (Wei et al., 2014). Briefly, cells were lysed by thorough sonication in IP buffer, and the cleared cell lysate was incubated with IgG sepharose beads at 4°C for 1 hr. Beads were then washed six times with IP buffer, and bound proteins were eluted with 10% SDS. Eluted proteins were then separated by SDS-PAGE, transferred onto a PVDF membrane, and blotted with anti-HA antibody (Sigma-Aldrich) to detect 3HA-tagged GCP proteins. The same membrane was then re-probed with anti-Protein A antibody (Sigma-Aldrich) to detect PTP-tagged γ-tubulin or GCP3.
Immunofluorescence microscopy
Cells were washed once with PBS, settled onto glass coverslips for 30 min, fixed with cold methanol (−20°C) for 30 min, and finally rehydrated with PBS for 10 min. After blocking with 3% BSA in PBS for 1 h, coverslips were incubated with primary antibodies for 1 h. The following primary antibodies were used: FITC-conjugated anti-HA mAb (1:400 dilution, Sigma-Aldrich), anti-HA antibody (1:400 dilution, Sigma-Aldrich), L8C4 (anti-PFR2 mAb, 1:50 dilution) (Kohl et al., 1999), YL 1/2 (1:1,000 dilution, Millipore), KMX-1 (1:100 dilution, Millipore), and anti-TbSAS-6 pAb (1:400 dilution) (Hu et al., 2015). After washing three times, coverslips were incubated with FITC- or Alexa Fluor 594-conjugated secondary antibody for 1 h. After washing three times with PBS, cells were mounted with DAPI-containing VectaShield mounting medium (Vector Labs). Slides were examined under an inverted fluorescence microscope (Olympus IX71) equipped with a cooled CCD camera (model Orca-ER, Hamamatsu) and a PlanApo N 60× 1.42-NA DIC objective. Images were acquired using the Slidebook5 software (Intelligent Imaging Innovations).
Transmission electron microscopy
Procedures for preparation of thin sections of trypanosome cells for electron microscopy have been published previously (Hu et al., 2012a, Hu et al., 2012b, Wei et al., 2014). Briefly, non-induced control and GCP2 and GCP3 RNAi cells were fixed in glutaraldehyde, treated with Millonig’s buffer, incubated with 2% OsO4 at 4°C for 60 min, dehydrated with ethanol, and finally embedded in resin. The 120 nm thin sections were cut using a Leica Ultracut-R microtome and a diamond knife (Daitome-U.S.), placed on 150 mesh copper grids (EMS), and stained with uranyl acetate. Thin sections were then rinsed with distilled water and incubated with Renold’s lead citrate. Grids were imaged using a JEOL 1400 TEM at 60kv and captured with a Gatan CCD camera.
Protein stability assay
Cells induced for RNAi for 64 h were split to two flasks. One flask was incubated with 1.0 μg/ml tetracycline, and the other was incubated with 1.0 μg/ml tetracycline and 50 μg/ml MG-132 (Selleckchem) for an additional 8 h. Cells (1×107) were then lysed in PEME buffer (100 mM PIPES, pH 6.9, 2 mM EGTA, 0.1 mM EDTA, 1 mM MgSO4) containing 1% NP-40, centrifuged to separate cytosolic (S) and cytoskeletal (P) fractions. An equal volume of the PEME buffer containing 1% NP-40 was then added to the cytoskeletal fraction. Both fractions were mixed with SAS-PAGE sampling buffer, boiled for 5 min, and an equal volume of both samples was loaded onto a SDS-PAGE gel for western blotting with anti-HA antibody to detect 3HA-tagged proteins. The same blots were then re-probed with anti-α-tubulin mAb (Sigma-Aldrich) and anti-TbPSA6 pAb, which recognizes the α6 subunit of the 26S proteasome (Li et al., 2002), as the cytoskeleton and cytosol markers, respectively. Protein band intensity in cytosolic and cytoskeletal fractions was measured by ImageJ and normalized with that of the cytosolic and cytoskeletal loading controls, respectively.
Supplementary Material
Supplementary Figure 1. Localization of γ-tubulin and its associated GCP proteins on the cytoskeletons of procyclic trypanosomes. γ-tubulin (A) and its associating GCP proteins, GCP2 (B), GCP3 (C), and GCP4 (D), were each tagged with a C-terminal triple HA epitope from one of its endogenous loci in procyclic trypanosomes. Cells were treated with PEME buffer containing 1% NP-40, and then immunostained with FITC-conjugated anti-HA mAb and anti-TbSAS-6 pAb. Scale bars: 5 μm.
Supplementary Movie 1. Motility of non-induced control of the GCP2 RNAi cell line. Live cells were spread onto a chamber on a glass slide, and time-lapse video was recorded under an automated confocal microscope.
Supplementary Movie 2. Tumbler cells found in GCP2 RNAi cells induced for 48 hours.
Supplementary Movie 3. Immotile cells found in GCP2 RNAi cells induced for 48 hours.
Supplementary Movie 4. Motility of non-induced control of the GCP3 RNAi cell line.
Supplementary Movie 5. Tumbler cells found in GCP3 RNAi cells induced for 72 hours.
Supplementary Movie 6. Immotile cells found in GCP3 RNAi cells induced for 72 hours.
Abbreviated summary.
The γ-tubulin complex nucleates microtubule assembly in eukaryotes. In different organisms, this complex differs in complexity: the lower eukaryotes contain a γ-tubulin small complex, whereas the higher eukaryotes contain a γ-tubulin ring complex. Trypanosoma brucei, an early branching protozoan, possesses an unusual γ-tubulin complex, but only the γ-tubulin small complex is essential for cell viability and is required for nucleation of central pair microtubules and for assembly of central pair-associated proteins in the flagellar axoneme.
Acknowledgments
We thank Dr. George Cross of Rockefeller University for providing the 29-13 cell line, Dr. Paul Englund of Johns Hopkins University for the pZJM vector, Dr. Arthur Günzl of University of Connecticut Health Center for the pC-PTP-NEO vector, and Dr. Keith Gull of the University of Oxford for the L8C4 antibody. This work was supported by NIH grants AI101437 and AI108657.
Footnotes
Conflict of interest: we declare no competing financial interest.
References
- Absalon S, Kohl L, Branche C, Blisnick T, Toutirais G, Rusconi F, Cosson J, Bonhivers M, Robinson D, Bastin P. Basal body positioning is controlled by flagellum formation in Trypanosoma brucei. PLoS ONE. 2007;2:e437. doi: 10.1371/journal.pone.0000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branche C, Kohl L, Toutirais G, Buisson J, Cosson J, Bastin P. Conserved and specific functions of axoneme components in trypanosome motility. J Cell Sci. 2006;119:3443–3455. doi: 10.1242/jcs.03078. [DOI] [PubMed] [Google Scholar]
- Dawe HR, Shaw MK, Farr H, Gull K. The hydrocephalus inducing gene product, Hydin, positions axonemal central pair microtubules. BMC Biol. 2007;5:33. doi: 10.1186/1741-7007-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S, Pereira G, Spang A, Knop M, Soues S, Kilmartin J, Schiebel E. The spindle pole body component Spc98p interacts with the gamma-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. Embo J. 1996;15:3899–3911. [PMC free article] [PubMed] [Google Scholar]
- Gull K. The cytoskeleton of trypanosomatid parasites. Annu Rev Microbiol. 1999;53:629–655. doi: 10.1146/annurev.micro.53.1.629. [DOI] [PubMed] [Google Scholar]
- He CY, Pypaert M, Warren G. Golgi duplication in Trypanosoma brucei requires Centrin2. Science. 2005;310:1196–1198. doi: 10.1126/science.1119969. [DOI] [PubMed] [Google Scholar]
- Hu H, Hu L, Yu Z, Chasse AE, Chu F, Li Z. An orphan kinesin in trypanosomes cooperates with a kinetoplastid-specific kinesin to maintain cell morphology by regulating subpellicular microtubules. J Cell Sci. 2012a;125:4126–4136. doi: 10.1242/jcs.106534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Liu Y, Zhou Q, Siegel S, Li Z. The centriole cartwheel protein SAS-6 in Trypanosoma brucei is required for pro-basal body biogenesis and flagellum assembly. Eukaryot Cell. 2015 doi: 10.1128/EC.00083-15. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Hu H, Li Z. A kinetoplastid-specific kinesin is required for cytokinesis and for maintenance of cell morphology in Trypanosoma brucei. Mol Microbiol. 2012b;83:565–578. doi: 10.1111/j.1365-2958.2011.07951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins JR, Toyoda Y, Hegemann B, Poser I, Heriche JK, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J, Conrad C, Comartin D, Schleiffer A, Sarov M, Pozniakovsky A, Slabicki MM, Schloissnig S, Steinmacher I, Leuschner M, Ssykor A, Lawo S, Pelletier L, Stark H, Nasmyth K, Ellenberg J, Durbin R, Buchholz F, Mechtler K, Hyman AA, Peters JM. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, Schiebel E. The spindle pole body component Spc97p interacts with the gamma-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. Embo J. 1997;16:1550–1564. doi: 10.1093/emboj/16.7.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl L, Sherwin T, Gull K. Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J Eukaryot Microbiol. 1999;46:105–109. doi: 10.1111/j.1550-7408.1999.tb04592.x. [DOI] [PubMed] [Google Scholar]
- Kollman JM, Merdes A, Mourey L, Agard DA. Microtubule nucleation by gamma-tubulin complexes. Nat Rev Mol Cell Biol. 2011;12:709–721. doi: 10.1038/nrm3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, Gould TJ, Witman GB. Flagellar central pair assembly in Chlamydomonas reinhardtii. Cilia. 2013;2:15. doi: 10.1186/2046-2530-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. Regulation of the cell division cycle in Trypanosoma brucei. Eukaryot Cell. 2012;11:1180–1190. doi: 10.1128/EC.00145-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zou CB, Yao Y, Hoyt MA, McDonough S, Mackey ZB, Coffino P, Wang CC. An easily dissociated 26 S proteasome catalyzes an essential ubiquitin-mediated protein degradation pathway in Trypanosoma brucei. J Biol Chem. 2002;277:15486–15498. doi: 10.1074/jbc.M109029200. [DOI] [PubMed] [Google Scholar]
- Libusova L, Sulimenko T, Sulimenko V, Hozak P, Draber P. gamma-Tubulin in Leishmania: cell cycle-dependent changes in subcellular localization and heterogeneity of its isoforms. Exp Cell Res. 2004;295:375–386. doi: 10.1016/j.yexcr.2004.01.009. [DOI] [PubMed] [Google Scholar]
- McKean PG, Baines A, Vaughan S, Gull K. Gamma-tubulin functions in the nucleation of a discrete subset of microtubules in the eukaryotic flagellum. Curr Biol. 2003;13:598–602. doi: 10.1016/s0960-9822(03)00174-x. [DOI] [PubMed] [Google Scholar]
- Nogales E. Structural insight into microtubule function. Annu Rev Biophys Biomol Struct. 2001;30:397–420. doi: 10.1146/annurev.biophys.30.1.397. [DOI] [PubMed] [Google Scholar]
- Nohynkova E, Draber P, Reischig J, Kulda J. Localization of gamma-tubulin in interphase and mitotic cells of a unicellular eukaryote, Giardia intestinalis. Eur J Cell Biol. 2000;79:438–445. doi: 10.1078/0171-9335-00066. [DOI] [PubMed] [Google Scholar]
- Ralston KS, Lerner AG, Diener DR, Hill KL. Flagellar motility contributes to cytokinesis in Trypanosoma brucei and is modulated by an evolutionarily conserved dynein regulatory system. Eukaryot Cell. 2006;5:696–711. doi: 10.1128/EC.5.4.696-711.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimanski B, Nguyen TN, Gunzl A. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot Cell. 2005;4:1942–1950. doi: 10.1128/EC.4.11.1942-1950.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott V, Sherwin T, Gull K. gamma-tubulin in trypanosomes: molecular characterisation and localisation to multiple and diverse microtubule organising centres. J Cell Sci. 1997;110(Pt 2):157–168. doi: 10.1242/jcs.110.2.157. [DOI] [PubMed] [Google Scholar]
- Sheriff O, Lim LF, He CY. Tracking the biogenesis and inheritance of subpellicular microtubule in Trypanosoma brucei with inducible YFP-alpha-tubulin. Biomed Res Int. 2014;2014:893272. doi: 10.1155/2014/893272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwin T, Gull K. The cell division cycle of Trypanosoma brucei brucei: timing of event markers and cytoskeletal modulations. Philos Trans R Soc Lond B Biol Sci. 1989;323:573–588. doi: 10.1098/rstb.1989.0037. [DOI] [PubMed] [Google Scholar]
- Sherwin T, Schneider A, Sasse R, Seebeck T, Gull K. Distinct localization and cell cycle dependence of COOH terminally tyrosinolated alpha-tubulin in the microtubules of Trypanosoma brucei brucei. J Cell Biol. 1987;104:439–446. doi: 10.1083/jcb.104.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixido-Travesa N, Villen J, Lacasa C, Bertran MT, Archinti M, Gygi SP, Caelles C, Roig J, Luders J. The gammaTuRC revisited: a comparative analysis of interphase and mitotic human gammaTuRC redefines the set of core components and identifies the novel subunit GCP8. Mol Biol Cell. 2010;21:3963–3972. doi: 10.1091/mbc.E10-05-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verollet C, Colombie N, Daubon T, Bourbon HM, Wright M, Raynaud-Messina B. Drosophila melanogaster gamma-TuRC is dispensable for targeting gamma-tubulin to the centrosome and microtubule nucleation. J Cell Biol. 2006;172:517–528. doi: 10.1083/jcb.200511071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinh DB, Kern JW, Hancock WO, Howard J, Davis TN. Reconstitution and characterization of budding yeast gamma-tubulin complex. Mol Biol Cell. 2002;13:1144–1157. doi: 10.1091/mbc.02-01-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Morris JC, Drew ME, Englund PT. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J Biol Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- Wei Y, Hu H, Lun ZR, Li Z. The cooperative roles of two kinetoplastid-specific kinesins in cytokinesis and in maintaining cell morphology in bloodstream trypanosomes. PLoS ONE. 2013;8:e73869. doi: 10.1371/journal.pone.0073869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Hu H, Lun ZR, Li Z. Centrin3 in trypanosomes maintains the stability of a flagellar inner-arm dynein for cell motility. Nat Commun. 2014;5:4060. doi: 10.1038/ncomms5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Localization of γ-tubulin and its associated GCP proteins on the cytoskeletons of procyclic trypanosomes. γ-tubulin (A) and its associating GCP proteins, GCP2 (B), GCP3 (C), and GCP4 (D), were each tagged with a C-terminal triple HA epitope from one of its endogenous loci in procyclic trypanosomes. Cells were treated with PEME buffer containing 1% NP-40, and then immunostained with FITC-conjugated anti-HA mAb and anti-TbSAS-6 pAb. Scale bars: 5 μm.
Supplementary Movie 1. Motility of non-induced control of the GCP2 RNAi cell line. Live cells were spread onto a chamber on a glass slide, and time-lapse video was recorded under an automated confocal microscope.
Supplementary Movie 2. Tumbler cells found in GCP2 RNAi cells induced for 48 hours.
Supplementary Movie 3. Immotile cells found in GCP2 RNAi cells induced for 48 hours.
Supplementary Movie 4. Motility of non-induced control of the GCP3 RNAi cell line.
Supplementary Movie 5. Tumbler cells found in GCP3 RNAi cells induced for 72 hours.
Supplementary Movie 6. Immotile cells found in GCP3 RNAi cells induced for 72 hours.