Abstract
Human nuclear type IB topoisomerase (Top1) inhibitors are widely used and powerful anti-cancer agents. In this study, we introduce and validate a single-molecule supercoil relaxation assay as a molecular pharmacology tool for characterizing therapeutically relevant Top1 inhibitors. Using this assay, we determined the effects on Top1 supercoil relaxation activity of four Top1 inhibitors; three clinically relevant: camptothecin, LMP-400, LMP-776 (both indenoisoquinoline derivatives), and one natural product in preclinical development, lamellarin-D. Our results demonstrate that Top1 inhibitors have two distinct effects on Top1 activity: a decrease in supercoil relaxation rate and an increase in religation inhibition. The type and magnitude of the inhibition mode depend both on the specific inhibitor and on the topology of the DNA substrate. In general, the efficacy of inhibition is significantly higher with supercoiled than with relaxed DNA substrates. Comparing single-molecule inhibition with cell growth inhibition (IC50) measurements showed a correlation between the binding time of the Top1 inhibitors and their cytotoxic efficacy, independent of the mode of inhibition. This study demonstrates that the single-molecule supercoil relaxation assay is a sensitive method to elucidate the detailed mechanisms of Top1 inhibitors and is relevant for the cellular efficacy of Top1 inhibitors.
Keywords: Single-molecule, camptothecin, indenoisoquinoline, magnetic tweezers, DNA topology
Introduction
DNA topoisomerases are highly conserved enzymes that play critical roles in maintaining genomic stability (1–3). Genomic DNA is organized in a highly compact state. Consequently, accumulations of inter- and intra- DNA molecule entanglements such as DNA supercoils, catenanes, and knots inevitably arise in DNA metabolism. DNA topoisomerases (topos) are uniquely able to relieve these DNA topological problems through a well-conserved catalytic activity – DNA cleavage and religation (1, 2, 4).
Due to the potentially highly toxic single- or double-strand DNA breaks generated during their normal catalytic cycle, topoisomerases are widely used as targets for anti-cancer agents that primarily act as catalytic poisons by trapping the topoisomerase in a cleavage complex with DNA (4–8). Camptothecin (CPT) (Fig. 1A) was the first human topoisomerase IB (Top1) inhibitor discovered, and currently there are two U.S. Food and Drug administration approved camptothecin derivatives (Topotecan and Irinotecan) in clinical use (4). The general paradigm for cytotoxicity caused by Top1-targeted anti-cancer agents is based on extensive studies demonstrating that CPT poisons Top1 by trapping a DNA-Top1 cleavage complex (Top1cc) potentially leading to a double strand break (7, 9, 10). In addition to this well-established mode of action, an alternative or complementary mode of CPT action has been observed in which uncoiling of positive supercoils by Top1 is hindered, resulting in increased cellular levels of positive supercoiling that may contribute to CPT cytotoxicity (11).
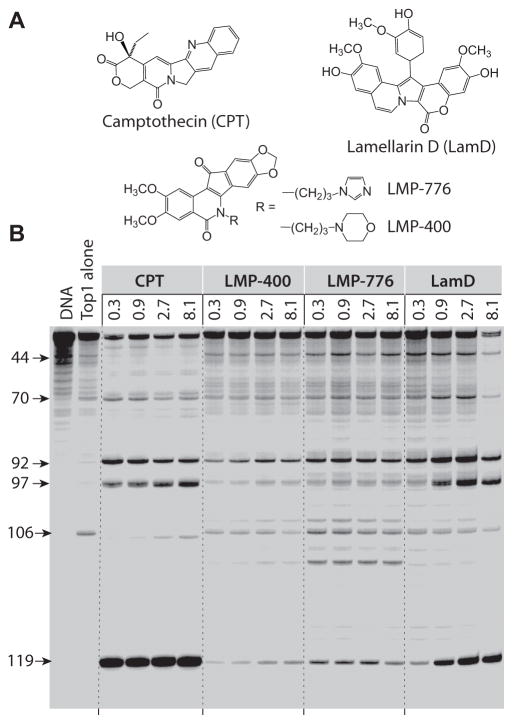
Figure 1.
Comparison of Top1 mediated DNA cleavage induced by camptothecin (CPT), LMP-400, LMP-776 and lamellarin D (LamD). (a) Compound structures. (b) (lane 1) DNA alone; (lane 2) Top1 alone; (lane 3–16) Top1 + indicated drugs at 0.3, 0.9, 2.7, and 8.1 μM, respectively. The numbers on the left and arrows indicate cleavage site positions. A 117-bp DNA substrate was used in this assay, but the cleavage sites are numbered to be consistent with the 161-bp DNA substrate traditionally used in this assay in order to facilitate comparison (25).
Although CPT derivatives are among the most effective anti-cancer agents, their chemical instability, high cellular efflux as well as the development of CPT resistance, spurred an extensive search for and development of alternative and improved Top1 inhibitors (7, 12). To date, a number of natural and synthetic compounds have shown promising Top1 inhibition and cytotoxicity, and two non-camptothecin derivatives of indenoisoquinolines (LMP-400 and LMP-776) are in Phase 1 clinical trials (12).
Top1 inhibitors are typically characterized by their in vitro and in vivo inhibition efficacy and by their anti- proliferative activities in human cancer cell lines. A common methodology employed to test the degree of Top1 inhibition in vitro is a DNA cleavage assay in which formation of stable Top1ccs induced by a drug is quantified by measuring cleaved or nicked DNA visualized on a gel (13).
DNA cleavage assays are typically performed with linear DNA substrates that provide a simple quantitative analysis of Top1 cleavage and, if performed with 3′-end labeled DNA, mapping of sequence-dependent cleavage information (shown in Fig. 1B) (13). Similar assays employing supercoiled DNA as a substrate can provide additional insight related to not only Top1 catalytic inhibition but also to Top1-poisoning by inhibitors and potential roles of DNA topology in the processes of relaxation and inhibition (9, 14, 15). Moreover, since DNA is supercoiled in vivo, in vitro measurements with supercoiled DNA substrates may more faithfully reflect the in vivo situation, at least with respect to the influence of DNA topology. Despite the success of the ensemble DNA cleavage assays in providing a measure of Top1 poisoning by Top1 inhibitors, they reveal little mechanistic detail concerning how the drug affects and inhibits the catalytic activity of Top1. More recently, single-molecule techniques have been employed to investigate Top1 supercoil relaxation activity and the effects of CPT and CPT derivatives in real time (11, 16, 17). Single-molecule techniques in which individual molecules are controlled and measured in real time can provide exquisitely detailed information on the physical properties of biopolymers and biophysical activities of enzymes (18). Magnetic tweezers has been a particularly useful tool to study DNA topology and elucidate mechanisms of DNA topoisomerases as it affords the ability to precisely control and measure DNA twist and writhe (19, 20).
In this study, we employed a single-molecule supercoil relaxation assay using magnetic tweezers to test four Top1 inhibitors, camptothecin (CPT), LMP-776 (indimitecan) and -400 (indotecan), and lamellarin D (LamD) (Fig. 1A). The two indenoisoquinolines, LMP-400 (NSC 724998) and -776 (NSC 725776) are particularly relevant because they are currently in Phase I clinical trials at the National Institutes of Health (NCT01051635) (12). Lamellarin D is a naturally occurring compound extracted from a sea mollusk. It exhibits potent Top1 inhibition activity (21), and recently was shown to preferentially poison mitochondrial Top1 (Top1mt) in cancer cells (22). Single-molecule supercoil relaxation measurements provided detailed information on how individual Top1 inhibitors affect the catalytic activity of Top1. We identified two robust characteristics of Top1 activity that changed in the presence of drug – (i) religation inhibition and (ii) relaxation rate reduction. From these measures of inhibitor interactions, we quantified inhibitor binding efficiency and the lifetime of the inhibitor stabilizing Top1-DNA cleavage complex as metrics to quantify Top1 inhibition. To compare the results from the single-molecule assay with conventional assays, e.g., DNA cleavage assays (13, 15), we performed DNA cleavage assays with the four inhibitors using linear DNA and three different DNA topological substrates: negatively supercoiled [(−) supercoils], positively supercoiled [(+) supercoils], and relaxed circular DNA. Overall, we found that the inhibitor binding efficiency, DNA-cleavage complex induction efficiency, and lifetime of the inhibitor-stabilized cleavage complex, for each of the four inhibitors were linearly correlated with their respective cytotoxicity (IC50). Our study demonstrates that single-molecule supercoil relaxation assays provide molecular level details of the changes in catalytic activity of Top1due to the binding of inhibitors and could be used as a sensitive tool to screen Top1 inhibitors alone or in conjunction with DNA cleavage assay prior to in vivo screening.
Material and methods
Drug and enzymes
CPT, LMP400, and LMP776 were from Developmental Therapeutics Program (DTP), DCTD, NCI, NIH. Lamellarin D (LamD) was a generous gift from Dr. Carmen Cuevas Marchante and José M. Fernandez Sousa-Faro, PharmaMar. Human nuclear Top1 was purified from baculovirus infected SF9 cells as previously described (23).
Top1 cleavage assay with linear pSK DNA
Briefly, a 117-bp DNA oligonucleotide (Integrated DNA Technologies) encompassing the previously identified Top1 cleavage sites in the 161-bp fragment from pBluescript SK(−) phagemid DNA was employed. This 117-bp oligonucleotide contains a single 5′-cytosine overhang, which was 3′-end-labeled by fill-in reaction with [32P] dGTP in React 2 buffer (50 mM Tris-HCl, pH 8.0, 100 mM MgCl2, 50 mM NaCl) with 0.5 units of DNA polymerase I (Klenow fragment, New England BioLabs). Unincorporated [32P] dGTP was removed using mini Quick Spin DNA columns (Roche, Indianapolis, IN), and the eluate containing the 3′-end-labeled DNA substrate was collected. Approximately 2 nM of radiolabeled DNA substrate was incubated with recombinant Top1 in 10 μL of reaction buffer [10 mM Tris-HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, and 15 μg/mL BSA] at 25°C for 20 min in the presence of the indicated drug concentrations. Reactions were terminated by adding SDS (0.5% final concentration) followed by the addition of two volumes of loading dye (80% formamide, 10 mM sodium hydroxide, 1 mM sodium EDTA, 0.1% xylene cyanol, and 0.1% bromphenol blue). Aliquots of each reaction mixture were subjected to 20% denaturing PAGE. Gels were dried and visualized by using a phosphoimager and ImageQuant software (Molecular Dynamics). Cleavage sites were numbered as previously described in the 161-bp fragment (24, 25).
DNA supercoil relaxation assay
We prepared “coilable” DNA molecules and sample for performing DNA supercoil relaxation assay as previously described (17, 23, 26). Measurements of supercoil relaxation by Top1 (50–500 pM) were performed in topoisomerase buffer (10 mM Tris pH 8, 50 mM KCl, 10 mM MgCl2, 0.3% w/v BSA, 0.04% Tween-20, 0.1 mM EDTA, and 5 mM DTT) with varying inhibitor concentrations (0 – 10 μM) and at two different DNA twist densities: −0.008 (negative supercoils) and 0.008 (positive supercoils), corresponding to a tension applied on the DNA of 0.2 pN (17). Top1 relaxation activity was measured by tracking the height of the bead at 200 Hz using video tracking routines as described previously (17). Religation inhibition induced by an inhibitor was quantified from the ratio of futile to productive supercoiling attempts, i.e, the fraction of events for which there was no change in DNA linking number (ΔLk) when the magnets were rotated (22). For comparison to the frequency in the absence of inhibitor, the relative change in religation inhibition values were calculated compared to the values obtained in the absence of inhibitor (Relative change = [Value (drug presence)−Value(drug absence)]/Value(drug absence). The rates of supercoil relaxation by Top1 were obtained by analyzing the DNA extension change using custom written software in Igor Pro Version 6.13 (Wavemetrics) (17). The slow relaxation-rate event probability was determined by calculating the fraction of events with rates less than 30% of the mean rate obtained in the absence of inhibitor (22) and then compared to those in the absence of drug to obtain the relative changes in low rate probability. The results of this analysis were relatively insensitive to the precise choice of the threshold for low rate determination.
DNA cleavage assay supercoiled substrates
DNA cleavage assays were adapted from the method of McClendon and Osheroff (15). The DNA cleavage assay was repeated at least three times for individual drugs and DNA topological states. Reactions were carried out at 37° C for 10 min in a thermocycler (Eppendorf) and stopped by adding 2 μl of 5 % SDS and 1 μl of 375 mM EDTA (pH 8.0). To remove Top1, 2 μl of Proteinase K (0.8 unit/μl, NEB) was added and the reaction was incubated at 45° C for 1 hr to digest Top1. pBR322 (NEB) was used as (−) supercoils substrates. (+) supercoils substrates were generated by incubating negative supercoiled pBR322 with Archaeo-globus fulgidus reverse gyrase following the protocol of McClendon et al (27) while relaxed pBR322 was generated by relaxing negatively supercoiled pBR322 with Top1. To quantify the DNA cleavage activity 0.4 nM of DNA from individual samples were mixed with 5 μl of 6x DNA loading dye (Crytalgen) that were heated at 60°C for 5 min and loaded on to 1% DNA agarose gel (1X TBE containing 0.4 μg/ml ethidium bromide). 0.4 μg/ml Ethidium bromide was included in the gel to achieve separation between nicked and the other topological states of the DNA. The gel was run for 2 hours and 40 min in 1X TBE running buffer at 70 V and then soaked for 3 hours in TE buffer containing 0.2 ppm Sybr-green dye (Sigma) to enhance the detection sensitivity of DNA. The gel image was obtained by using BioSpectrum (UVP) and analyzed by custom written software in LabVIEW (National Instruments) and Igor Pro (Wavemetrics) to estimate the amount of DNA in individual bands in each lane.
Results
Differential trapping of Top1-DNA cleavage complexes (Top1cc) by CPT, LMP-400, LMP-776 and LamD
We compared the drug-induced Top1cc in a routine biochemical assay (13, 28) using a 118 base pair 3′-end-labeled linear DNA fragment derived from the pSK plasmid. The four drugs trapped Top1cc at submicromolar concentrations, but differed in the distribution and intensity of their selective cleavage sites across the DNA sequence, which is related to the selective interaction of each drug with the DNA bases flanking the Top1cc (Fig. 1) (29).
Two different modes of human Top1 inhibition
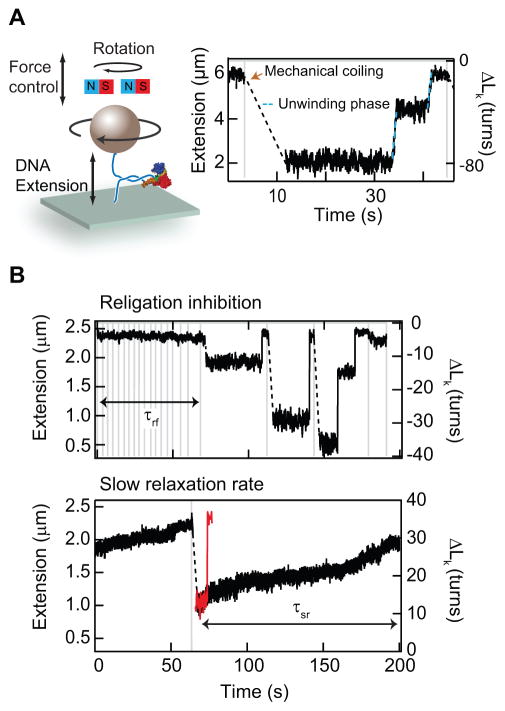
Next we characterized the effects of the four Top1 inhibitors by comparing Top1 catalytic activity in the absence of and presence of increasing concentrations of inhibitor using a magnetic-tweezers-based single-molecule DNA supercoil relaxation assay. In this assay, DNA supercoils are generated by rotating a 1 μm magnetic particle tethered to the surface by a long dsDNA (either 11 or 23 kb), using a magnet assembly above the sample chamber (Fig. 2A) (17, 23). The chirality of DNA supercoils is determined by the direction of magnet rotation, i.e., clockwise (counter-clockwise) rotation generates negative (positive) supercoils. DNA twist density (related to the torque on the DNA) can be varied by changing the applied tension on the DNA. DNA extension is measured by tracking the bead height relative to the surface (23). Supercoiling of relaxed DNA initially increases the twist with little change in DNA extension. At a force-dependent critical supercoiling value the DNA buckles forming a plectoneme. Further rotation increases the extent of the plectoneme, i.e., the DNA writhe, without changing the twist or torque in the DNA. Consequently, the torque remains constant as long as plectonemes remain. The DNA extension increases as Top1 relaxes supercoiled DNA (Fig. 2A) (17). In the presence of inhibitors, two distinctive changes were observed in Top1 catalytic activity (Fig. 2B). First, there was a significant increase in the frequency and duration of religation inhibition, i.e., periods when the DNA molecule could not be supercoiled by rotating the magnets, which we attribute to drug intercalation at the cleavage site and the formation of a stable cleavage complex (29–31). Second, the frequency of slow relaxation-rate events increased, consistent with the slow relaxation mode first observed in single-molecule measurements of topotecan inhibition (11). To quantify and compare the effects of the four Top1 inhibitors, we measured the frequency of, i.e., the fraction of events, that displayed religation inhibition and slow relaxation-rate events and the time durations over which religation inhibition (τrf) and slow relaxation-rate (τsl) events persisted (Fig. 2B).
Figure 2.
Overview of single-molecule Top1 DNA supercoil relaxation and inhibition assay. (a) Cartoon of magnetic tweezers instrument (not to scale). A magnetic bead is attached to the surface via a rotationally constrained DNA molecule. DNA tension and supercoiling are controlled by the height and rotation of magnets above the sample chamber. During Top1 supercoil relaxation assay, the DNA is supercoiled by rotating the magnets, which results in a linear decrease in the DNA extension once the DNA forms a plectoneme. As Top1 relaxes DNA supercoils, the DNA extension increases, indicated as the unwinding phase (blue dashed line). (b) Two distinctive effects of inhibitors on Top1 catalytic activity: First, the inhibitor can prevent DNA religation, resulting in relatively long but finite duration (life time = τrf) events during which the DNA cannot be supercoiled (Top panel). Second, the inhibitor can decrease the relaxation rate. The red trace shows DNA relaxation without CPT for comparison with the black trace that shows dramatically reduced relaxation rate in the presence of CPT. The duration of these slow relaxation-rate events is τsl (Bottom panel).
The chirality of DNA supercoils modulates the effects of Top1 inhibitors
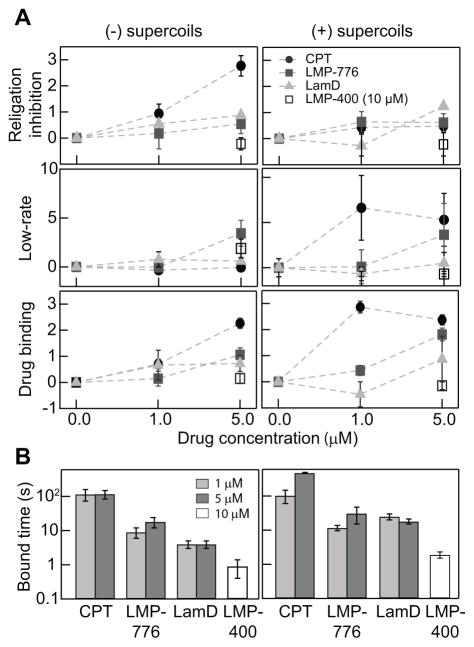
We used the religation-inhibition and slow relaxation-rate metrics to quantify the relative efficiency of the four inhibitors at two different inhibitor concentrations (1 and 5 μM) and two DNA supercoil chiralities (positively and negatively supercoiled) (Fig. 3). Individual inhibitors exhibited inhibition metric profiles that depended to different degrees on DNA supercoil chirality (Fig. 3). For CPT, religation inhibition was the dominant effect on (−) supercoils as the probability increased up to three-fold higher than in the absence of inhibitor (Fig. 3A). On the other hand, an increase in low relaxation-rate events was the dominant effect on (+) supercoils, consistent with observations of topotecan inhibition (11). Religation inhibition by LMP-776, although lower than that by CPT, showed a similar trend. Despite the overall similarities, the increase in the frequency of low relaxation-rate events for (−) as well as (+) supercoils indicates that the detailed interactions of LMP-776 at the Top1 cleavage site and the related effects on the Top1-DNA interactions are likely different from those of CPT. Despite sharing closely related chemical composition and structure (Fig. 1A), LMP-400 inhibition efficiency was significantly lower than that of LMP-776, requiring a concentration of 10 μM to achieve minimal inhibition (Fig. 3). LamD showed a comparable religation inhibition activity to LMP-776 but a negligible increase in the frequency of low relaxation-rate events. As religation inhibition and low relaxation-rate events both reflect the formation of Top1cc, we combined these two effects to estimate the overall binding probability of individual inhibitors. For CPT, the binding probability for (+) supercoils was superior to that of (−) supercoils as it appeared to saturate at 1 μM. The binding of LMP-776 was almost independent of supercoil chirality while that of LamD slightly favored (−) supercoils. The overall binding of LMP-776 and LamD were lower than for CPT but higher than for LMP-400, which shows very little inhibitor binding even at 10 μM.
Figure 3.
Single-molecule measurements of DNA topology-dependent Top1 inhibition. (a) Two modes of Top1 inhibition and effective binding probability as a function of inhibitor concentration and chirality of DNA supercoiling. Religation inhibition (Top panel). Note that religation inhibition values are relative to values in the absence of inhibitor. Relative change in the probability of low relaxation-rate events (Middle panel). Inhibitor binding determined from combining the probabilities of religation inhibition and low relaxation-rate events (Bottom panel). (b) Inhibitor binding time for different concentrations of each inhibitor for both positively and negatively supercoiled DNA.
Following a similar line of logic, we estimated the overall inhibitor binding lifetime by combining the durations of persistent religation inhibition and low relaxation-rate events (Fig. 3B). CPT lifetime was independent of DNA supercoil chirality and significantly longer than those of the other inhibitors, indicating a 10–100-fold higher binding stability. The lifetime of LMP-776 was on the order of 10 s for both (+) and (−) supercoils. A similar lack of chiral effects on binding was observed for LMP-400, though the lifetime was on the order of 1 s. The lifetime of LamD on (−) supercoils was slightly less than that of LMP-776, but they were comparable for (+) supercoils indicating that binding probability is different from binding stability since LamD shows lower binding probability for (+) supercoils than does LMP-776.
Supercoiled DNA cleavage assay reveals inhibitor efficacy is modulated by DNA topology
To understand how Top1 inhibition activity determined from single-molecule measurements relates to ensemble DNA cleavage measurements, we performed ensemble DNA cleavage assays using three different topological states of circular DNA as substrates: positive, negative and relaxed DNA with a range of drug concentrations (0–50 μM) (Fig. 4). In a conventional DNA cleavage assay, Top1 inhibition efficacy is estimated from the amount of cleaved DNA substrate as cleavage is correlated with both the stability and efficiency of Top1-DNA cleavage complex formation. Thus, the estimated efficacy represents a combination of drug binding affinity and cleavage complex stability. DNA cleavage assays are typically performed with linear DNA (Fig. 1B) (13). Although topotecan- and LamD-stabilized cleavage have been tested with supercoiled DNA (15, 21), a detailed comparison of cleavage efficiency of Top1 inhibitors for different DNA topological states has not been undertaken.
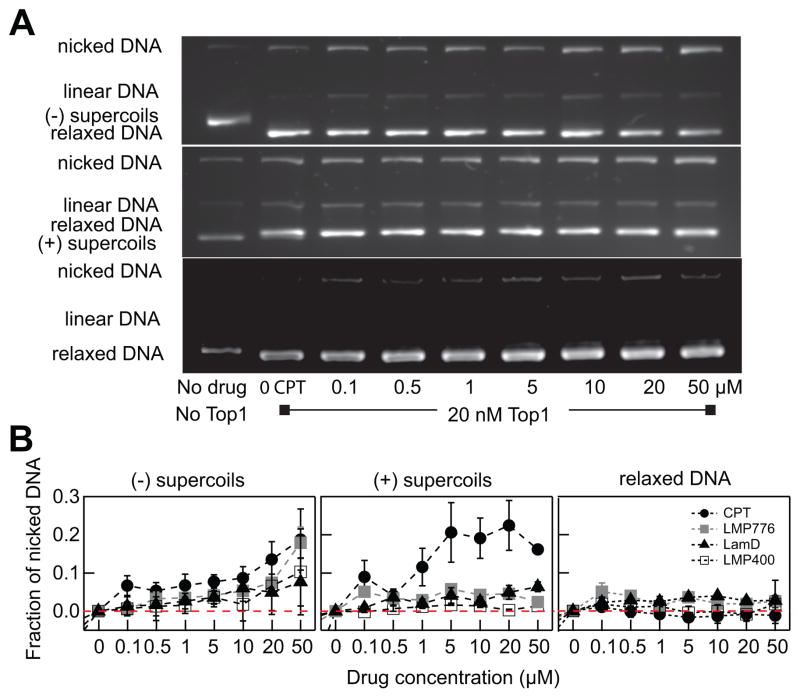
Figure 4.
DNA topology modulates the level of Top1cc formation induced by Top1 inhibitors. (a) Visualization of Top1cc formation with increasing CPT concentration (0– 50 μM) with three different DNA substrates: negative supercoiled pBR322 (top), positive supercoiled pBR322 (middle), and relaxed pBR322 DNA (bottom). (b) Fraction of nicked DNA as a function of drug concentration for three different DNA topological substrates. The amount of nicked DNA indicates the amount of Top1cc trapped by an inhibitor. Red dotted line indicates no difference in the fraction of nicked DNA relative to measurements without inhibitor.
To probe the effect of DNA topology on the formation of cleavage complexes, the assay had to be performed under conditions that minimized the relaxation of substrate by Top1. To probe this pre-steady state phase, we employed a 1:2 ratio of DNA to Top1 and ran the reaction for only 10 min at 37°C. Top1cc formation was estimated from the fraction of nicked DNA obtained by agarose gel electrophoresis analysis of each reaction (Fig. 4A). The nicked fraction was then normalized by the nicked fraction obtained in the absence of inhibitor to obtain relative changes. Consistent with single-molecule measurements, the overall efficiency of CPT was higher than those of the other drugs tested. For (−) supercoils, the fraction of nicked DNA gradually increased for increasing CPT concentrations, whereas for (+) supercoils the nicked fraction increased sharply up to a CPT concentration of 5 μM, after which it remained relatively constant (Fig. 4B). This chirality-dependent CPT efficiency is similar to the chirality-dependent CPT binding efficiency observed in the single-molecule assay indicating that CPT is a more effective Top1 inhibitor with positively rather than negatively supercoiled DNA (Fig. 3 and 4). The nicked fraction measured for the other three inhibitors was higher for (−) supercoils than for (+) supercoils, particularly at high drug concentrations (>10 μM). At lower inhibitor concentrations (<10 μM), changes in the fraction of nicked DNA were minimal for all drugs except for CPT. Presumably the ensemble DNA cleavage assay may not be sensitive enough to detect small changes observed in the single-molecule assay. Interestingly, all drugs were ineffective in Top1 inhibition with relaxed DNA as the nicked DNA fractions are negligible independent of drug concentration and type.
Strong correlation between inhibitor binding lifetimes, cleavage complex formation, and IC50 values
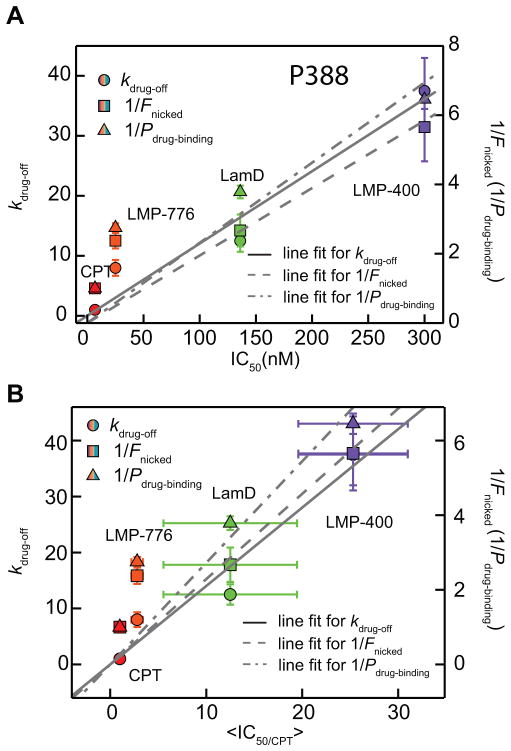
The in vitro measures of Top1 inhibition are a convenient screening tool, but may not reflect the in vivo efficacy due to complex cellular and biochemical processes that may influence the stability and availability of Top1 inhibitors in a drug- and cell type-dependent manner. Thus, it is useful to investigate if there are correlations between single-molecule, in vitro, and cellular measurements. In general, the in vivo efficacy can be evaluated based on the level of cytotoxicity characterized by the inhibitor IC50 value. We compared the single-molecule inhibition results to published IC50 values from 4 different cell lines: P388 (murine leukemia cells), HCT116 (Human colon cells), MCF-7 (human breast cancer cells) and CEM (human leukemic lymphoid cells) (21, 24). Importantly, growth inhibition for all four cell lines was shown to specifically depend on cellular Top1 inhibition (21, 24). The single-molecule inhibition measurements provide multiple inhibition metrics including the frequency of slow relaxation-rate events, the frequency of religation inhibition, and the inhibitor binding lifetime. Cellular Top1 inhibition and subsequent cytotoxicity could in principle be dominated by one or more of these observed effects and the frequency of slow relaxation-rate event has been suggested for CPT derivatives (11). We calculated individual drug unbinding rates (koff), inverse binding probability (1/Pbinding), and inverse DNA nicking probability (1/Fnicked) by combining results from (+) and (−) supercoils measured at 1 μM drug concentration and compared these metrics to IC50 values from four cell lines listed above (Fig. 5A and Supplementary Figure S1) (21, 24).
Figure 5.
Correlation between single-molecule measures of inhibition and cellular measures of cytotoxicity. (a) The drug off-rate (kdrug-off, solid circles), the inverse drug binding probability (1/Pdrug-binding, solid triangles), and the inverse nicked DNA fraction (1/Fnicked, solid squares) for each inhibitor plotted versus IC50 (P388 cell line). Individual linear fits provide the Pearson correlation coefficient ranging from 0 to 1, Pr, as the level of linear correlation between in-vitro quantities and cytotoxicity. Pr measured for kdrug-off was 0.97, for 1/Pdrug-binding was 0.97, and for 1/Fnicked was 0.96. (b) kdrug-off (solid circles), 1/Pdrug-binding (solid triangles), and 1/Fnicked (solid squares) for each inhibitor plotted versus <IC50/CPT>. <IC50/CPT> indicates the average IC50 relative to that of CPT for each cell line. The x-axis error bars correspond to the standard error from IC50/CPT values of different cell lines for individual inhibitors. Pr measured for kdrug-off was 0.97, for 1/Pdrug-binding was 0.95, and for 1/Fnicked was 0.97. Individual IC50 values and their sources are listed in Table 1.
Single-molecule and in vitro measurements of inhibition are generally correlated with IC50 values obtained with different cell lines (Fig. 5A and S1). However, cell-line specific IC50 values for each inhibitor make it difficult to globally compare their cytotoxicity with in vitro measures of inhibition. Therefore, we first considered P388, the only cell line for which the IC50 values of all four drugs were determined (21, 24). For each inhibitor we plotted the unbinding rate (kdrug-off), inverse binding probability (1/Pbinding), and inverse nicking probability (1/Fnicked) against its corresponding P388 IC50 value (Fig. 5A). There is a strong correlation between the three in vitro metrics and the corresponding IC50 values. For kdrug-off and 1/Pbinding obtained from the single-molecule assay, linear fits resulted in a slope of 0.12 ± 0.01 and Pearson correlation coefficient, Pr = 0.97; and a slope of 0.023 ± 0.004 and Pr = 0.97 respectively. For 1/Fnicked obtained from the DNA cleavage assay, a linear fit returned a slope of 0.019 ± 0.003 and Pr = 0.96. To determine if this correlation holds among different cell lines, the IC50 values of all tested drugs were normalized by the CPT IC50 value for each cell line since CPT is the reference Top1 inhibitor without other known cellular targets driving its cytotoxicity (32). We calculated the average normalized IC50, <IC50/CPT> for each inhibitor. <IC50/CPT> values for LMP-400 and -776 were calculated based on P388, HCT-116, and MCF-7 cell lines and <IC50/CPT> values for LamD were based on P388, and CEM (21, 24). We plotted unbinding rates (kdrug-off), inverse binding probability (1/Pbinding), and inverse nicking probability (1/Fnicked) as a function of calculated <IC50/CPT> values as shown in Fig. 5B. Consistent with the correlations observed for the P388 cell line, all three in vitro metrics are correlated with the <IC50/CPT> values. For kdrug-off and 1/Pbinding obtained from the single-molecule assay, the linear fits resulted in a slope of 1.4 ± 0.1 with a Pearson correlation coefficient, Pr = 0.97; and a slope of 0.27 ± 0.04 and Pr = 0.97 respectively. For 1/Fnicked obtained from the DNA cleavage assay, a linear fit returned a slope of 0.23 ± 0.04 and Pr = 0.95.
Discussion
In this study, we demonstrate that a single-molecule DNA supercoil relaxation assay can be employed as a tool for probing Top1 inhibition and its molecular effects in detail. We find that inhibitor binding efficiency and binding lifetime estimated from the single-molecule measurements are strongly correlated with cytotoxicity (IC50) of Top1 inhibitors measured in cell culture, indicating that single-molecule approaches alone or in parallel with conventional biochemical inhibition assays can provide an initial rapid screening step to determine Top1 inhibition efficiency of test compounds.
Underlying mechanism of religation inhibition and decreased relaxation-rate by Top1 inhibitors
Since CPT was identified as the first Top1 inhibitor, numerous experimental and theoretical studies have been undertaken to elucidate the molecular mechanism and cytotoxicity of CPT and its derivatives (7, 9–11, 14, 17, 31, 33–38).
In the established model, CPT binds at the interface between Top1 and the DNA nick at which it is intercalated, thereby preventing DNA religation while inducing a stable Top1cc until it is released. The efficiency of generating nicked DNA is well explained within this model. However, recent single-molecule experiments revealed an intriguing change in Top1 relaxation activity in the presence of topotecan and CPT (11, 17). Instead of simply inhibiting religation, the overall relaxation rate of positive, but not negative, supercoiled DNA decreased significantly. The basis for this new phenomenon was explained through molecular dynamics simulations demonstrating that the presence of CPT enhances the rotational energy barrier height for unwinding (+) supercoils whereas it lowers the rotational energy barrier height for unwinding (−) supercoils (38). This finding provides additional implications for the effects of CPT in vivo as the significant decrease in relaxation leads to accumulation of (+) supercoils which could potentially adversely affect numerous cellular activities and contribute to cytotoxicity (11, 39). These results raised the possibility that accumulation of (+) supercoils is an important component of Top1 inhibitor induced cytotoxicity.
Our results indicate that inhibition by LMP-776 and LamD is dominated by religation inhibition and the probability of slow relaxation rate events remains low except for very high concentrations of LMP-776 (5 μM). This finding confirms that religation inhibition without a substantial decrease in relaxation rate is sufficient to induce cytotoxicity in vivo (33, 35). More generally, whereas there was a strong correlation between the inhibitor unbinding rate (kdrug-off) and the IC50 value of the four inhibitors we measured, the correlation between the decrease in relaxation rate and the IC50 value was exceedingly weak (Supplementary Figure S2). Our interpretation of these findings is that the decrease in relaxation rate and the inhibition of religation are two manifestations of the formation of a covalent cleavage complex, the probability of formation and lifetime of which are the physiologically important parameters in determining cytotoxicity. As a result, there is a strong correlation between the IC50 values and inhibitor bound time, which includes the contributions of both decreased relaxation rate and religation inhibition, but a comparatively weak correlation between the IC50 values and either individual manifestation of inhibition.
In the DNA cleavage assay, the overall efficiency of forming the cleavage complex is a convolution of the protein-DNA Kd and the inhibitor Kd, which are difficult to directly determine from the single-molecule measurements. The ensemble measurements thus provide a related but alternative measure of inhibition efficiency. For example, neither the dramatically reduced relaxation rate with (+) supercoils nor the increased religation inhibition with (−) supercoils caused by CPT observed in the single-molecule assay (Fig. 3) is apparent in the cleavage assay. However, the fraction of nicked DNA for (+) supercoils (Fig. 4B; middle panel) is similar to the low relaxation-rate event probability for (+) supercoils (Fig. 3B, middle; right panel) and the fraction of nicked DNA for (−) supercoils (Fig. 4B; left panel) is similar to the religation inhibition probability (Fig. 3B; top; left panel). Thus, the nicked DNA fraction is related to the drug binding efficiency, consistent with the single-molecule results. The one exception to this general trend is LMP-776, which exhibits relatively high binding efficiency for (+) supercoils (Fig. 3B; bottom) that is not reflected in the fraction of nicked DNA (Fig. 4). Interestingly, the fraction of relaxed DNA increases with increasing LMP-776 concentration suggesting that the drug binding may enhance the effective binding affinity of Top1 and in turn increase the overall relaxation of DNA despite the slow relaxation rate as observed with CPT. LamD, which is an effective inhibitor of mitochondrial topoisomerase IB (Top1mt) in addition to nuclear Top1 (22), appeared to hinder the relaxation of (+) supercoils at high concentrations (Supplementary Figure S3). This finding was seemingly at odds with the observation that moderate LamD concentrations (~1 μM) slightly enhanced the relaxation rate (Fig. 3B; middle; left). The ensemble DNA cleavage measurements indicate that the efficiency of inhibition is influenced by the topology of the DNA (Fig. 4). It was surprising to find essentially no DNA cleavage-complex formation with relaxed DNA for all four inhibitors. This may be due to low relative binding affinity of Top1 for linear DNA (40), though recent measurements performed at low Top1 concentrations suggest there is very little difference in affinity between relaxed and supercoiled DNA (41). Alternatively, the lack of inhibitor stimulated cleavage on relaxed DNA may reflect the influence of torque on the occupancy of the cleaved but not rotating kinetic state postulated in the recently proposed “kinetic clutch” model for Top1 relaxation and religation (17). In this scenario, in the absence of a driving torque, i.e., DNA supercoiling, Top1 bound to linear DNA will equilibrate among three states: (i) ligated, (ii) cleaved but rotationally constrained and able to bind inhibitor, and (iii) cleaved but rotationally unconstrained and unable to bind inhibitor. Previous estimates of the kinetics linking these states suggest that state ii, in which an inhibitor can bind, has a low probability in comparison to the other two states in the absence of a driving torque. During relaxation of supercoiled DNA, there is a second kinetic pathway from state iii back to state ii that shifts the equilibrium towards state ii, which would favor inhibitor binding. Whereas the current data do not permit an unambiguous dissection of which, if either, of these possibilities make the dominate contributions to the observed topological differences in inhibitor efficiency, the fact that the relative effects of DNA topology differ among the four inhibitors, and that some of these differences depend on the chirality of supercoiling, suggest that the underlying mechanisms reflect something other than topology dependent Top1 binding affinity. Independent of the underlying mechanistic basis for the topological effect, on a practical level these results suggest that DNA topology should be considered when evaluating the efficiency of Top1 inhibitors in vitro.
Single-molecule assay limitations and future directions
Our study shows that the single-molecule assay provides detailed measures of Top1 activity changes induced by inhibitors and combining the single-molecule assay with the DNA cleavage assays provides comprehensive information on the effects and mechanisms of Top1 inhibitors. The high correlation between inhibitor binding efficiency or bound lifetime and IC50 for the four inhibitors we tested suggests that the single-molecule DNA relaxation assay may provide an efficient tool to screen Top1 inhibitor efficiency. Nonetheless, the deviations between in vitro and in vivo Top1 inhibition efficiency should be noted, particularly for LMP-776. These differences reflect the more complex chemical and physiological nature in cellular conditions, which are difficult to recapitulate with in vitro measurements. For example LMP-776, due to its robust chemical stability and low susceptibility to cellular efflux compared to CPT or CPT derivatives enables it to outperform the in vitro estimation (12).
Supplementary Material
Table 1.
IC50 values for indicated cell lines treated with corresponding Top1 inhibitors.
Acknowledgments
Financial support
This research was supported by the Intramural Research Programs of the National Heart, Lung, and Blood Institute (HL001056 to K.C. Neuman), and the National Cancer Institute, Center for Cancer Research, National Institutes of Health (Z01-BC 006161 to Y. Pommier).
We thank Lauren Kim for help with DNA gel analysis.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
References
- 1.Champoux JJ. DNA topoisomerases: Structure, Function, and Mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–40. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 3.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q Rev Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 4.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010;17:421–33. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu LF. DNA topoisomerase poisons as antitumor drugs. Annu Rev Biochem. 1989;58:351–75. doi: 10.1146/annurev.bi.58.070189.002031. [DOI] [PubMed] [Google Scholar]
- 6.Fortune JM, Osheroff N. Topoisomerase II as a target for anticancer drugs: when enzymes stop being nice. Prog Nucleic Acid Res Mol Biol. 2000;64:221–53. doi: 10.1016/s0079-6603(00)64006-0. [DOI] [PubMed] [Google Scholar]
- 7.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 8.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338–50. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsiang YH, Hertzberg R, Hecht S, Liu LF. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem. 1985;260:14873–8. [PubMed] [Google Scholar]
- 10.Stewart L, Redinbo MR, Qiu X, Hol WGJ, Champoux JJ. A Model for the Mechanism of Human Topoisomerase I. Science. 1998;279:1534–41. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 11.Koster DA, Palle K, Bot ESM, Bjornsti M-A, Dekker NH. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–7. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- 12.Pommier Y, Cushman M. The indenoisoquinoline noncamptothecin topoisomerase I inhibitors: update and perspectives. Mol Cancer Ther. 2009;8:1008–14. doi: 10.1158/1535-7163.MCT-08-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dexheimer TS, Pommier Y. DNA cleavage assay for the identification of topoisomerase I inhibitors. Nat Protocols. 2008;3:1736–50. doi: 10.1038/nprot.2008.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaxel C, Kohn KW, Yves P. Topoisomerase I Interaction with SV4O DNA in the presence and absence of camptothecin. Nucleic Acids Res. 1988;16:11157–70. doi: 10.1093/nar/16.23.11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McClendon AK, Osheroff N. The Geometry of DNA Supercoils Modulates Topoisomerase-Mediated DNA Cleavage and Enzyme Response to Anticancer Drugs†. Biochem. 2006;45:3040–50. doi: 10.1021/bi051987q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434:671–4. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 17.Seol Y, Zhang H, Pommier Y, Neuman KC. A kinetic clutch governs religation by type IB topoisomerases and determines camptothecin sensitivity. Proc Natl Acad Sci USA. 2012;109:16125–30. doi: 10.1073/pnas.1206480109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Meth. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neuman KC. Single-molecule Measurements of DNA Topology and Topoisomerases. J Biol Chem. 2010;285:18967–71. doi: 10.1074/jbc.R109.092437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charvin G, Strick TR, Bensimon D, Croquette V. TRACKING TOPOISOMERASE ACTIVITY AT THE SINGLE-MOLECULE LEVEL. Annu Rev Biophys Biomol Struct. 2005;34:201–19. doi: 10.1146/annurev.biophys.34.040204.144433. [DOI] [PubMed] [Google Scholar]
- 21.Facompré M, Tardy C, Bal-Mahieu C, Colson P, Perez C, Manzanares I, et al. Lamellarin D: A Novel Potent Inhibitor of Topoisomerase I. Cancer Res. 2003;63:7392–9. [PubMed] [Google Scholar]
- 22.Khiati S, Seol Y, Agama K, Rosa ID, Agrawal S, Fesen K, et al. Poisoning of Mitochondrial Topoisomerase I by Lamellarin D. Mol Pharmacol. 2014;86:193–9. doi: 10.1124/mol.114.092833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seol Y, Neuman KC. Single-Molecule Measurements of Topoisomerase Activity with Magnetic Tweezers. In: Mashanov GI, Batters C, editors. Single Molecule Enzymology. Humana press; 2011. pp. 229–41. [DOI] [PubMed] [Google Scholar]
- 24.Antony S, Agama KK, Miao Z-H, Takagi K, Wright MH, Robles AI, et al. Novel Indenoisoquinolines NSC 725776 and NSC 724998 Produce Persistent Topoisomerase I Cleavage Complexes and Overcome Multidrug Resistance. Cancer Res. 2007;67:10397–405. doi: 10.1158/0008-5472.CAN-07-0938. [DOI] [PubMed] [Google Scholar]
- 25.Pourquier P, Ueng L-M, Fertala J, Wang D, Park H-J, Essigman JM, et al. Induction of reversible complexes between eukaryotic DNA topoisomerase I and DNA-containing oxidative base damages. J Biol Chem. 1999;274:8516–23. doi: 10.1074/jbc.274.13.8516. [DOI] [PubMed] [Google Scholar]
- 26.Seol Y, Neuman KC. Magnetic Tweezers for Single-Molecule Manipulation. In: Peterman EJ, Wuite G, editors. Single Molecule Enzymology. Humana press; 2011. pp. 265–93. [DOI] [PubMed] [Google Scholar]
- 27.McClendon AK, Rodriguez AC, Osheroff N. Human topoisomerase II alpha rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J Biol Chem. 2005;280:39337–45. doi: 10.1074/jbc.M503320200. [DOI] [PubMed] [Google Scholar]
- 28.Beck DE, Agama K, Marchand C, Chergui A, Pommier Y, Cushman M. Synthesis and Biological Evaluation of New Carbohydrate-Substituted Indenoisoquinoline Topoisomerase I Inhibitors and Improved Syntheses of the Experimental Anticancer Agents Indotecan (LMP400) and Indimitecan (LMP776) J med chem. 2014;57:1495–512. doi: 10.1021/jm401814y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pommier Y, Marchand C. Interfacial inhibitors: targeting macromolecular complexes. Nat Rev Drug Discov. 2012;11:25–36. doi: 10.1038/nrd3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaxel C, Capranico G, Kerrigan D, Kohn KW, Pommier Y. Effect of local DNA sequence on topoisomerase I cleavage in the presence or absence of camptothecin. J Biol Chem. 1991;266:20418–23. [PubMed] [Google Scholar]
- 31.Staker BL, Hjerrild K, Feese MD, Behnke CA, Burgin AB, Stewart L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc Natl Acad Sci USA. 2002;99:15387–92. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pommier Y. Camptothecins and Topoisomerase I: A Foot in the Door. Targeting the Genome Beyond Topoisomerase I with Camptothecins and Novel Anticancer Drugs: Importance of DNA Replication, Repair and Cell Cycle Checkpoints. Current Medicinal Chemistry -Anti-Cancer Agents. 2004;4(5):429–34. doi: 10.2174/1568011043352777. [DOI] [PubMed] [Google Scholar]
- 33.Bjornsti M-A, Benedetti P, Viglianti GA, Wang JC. Expression of human DNA topoisomerase I in Yeast cells lacking yeast DNA topoisomerase I: restoration of sensitivity of the cells to the antitumor drug camptothecin. Cancer Res. 1989;49:6318–23. [PubMed] [Google Scholar]
- 34.Hertzberg RP, Caranfa MJ, Hecht SM. On the mechanism of topoisomerase I inhibition by camptothecin: Evidence for binding to an enzyme-DNA complex. Biochem. 1989;28:4629–38. doi: 10.1021/bi00437a018. [DOI] [PubMed] [Google Scholar]
- 35.Nitiss JL, Wang JC. Mechanisms of cell killing by drugs that trap covalent complexes between DNA topoisomerases and DNA. Mol Pharmacol. 1996;50:1095–102. [PubMed] [Google Scholar]
- 36.Laco GS. Evaluation of Two Models for Human Topoisomerase I Interaction with dsDNA and Camptothecin Derivatives. PLoS ONE. 2011;6:e24314. doi: 10.1371/journal.pone.0024314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pommier Y. DNA Topoisomerase I Inhibitors: Chemistry, Biology, and Interfacial Inhibition. Chem Rev. 2009;109:2894–902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wereszczynski J, Andricioaei I. Free energy calculations reveal rotating-ratchet mechanism for DNA supercoil relaxation by topoisomerase IB and its inhibition. Biophys J. 2010;99:869–78. doi: 10.1016/j.bpj.2010.04.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ray Chaudhuri A, Hashimoto Y, Herrador R, Neelsen KJ, Fachinetti D, Bermejo R, et al. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat Struct Mol Biol. 2012;19:417–23. doi: 10.1038/nsmb.2258. [DOI] [PubMed] [Google Scholar]
- 40.Madden KR, Stewart L, Champoux JJ. Preferential binding of human topoisomerase I to superhelical DNA. EMBO J. 1995;14:5399–409. doi: 10.1002/j.1460-2075.1995.tb00224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Litwin TR, Sola M, Holt IJ, Neuman KC. A robust assay to measure DNA topology-dependent protein binding affinity. Nucleic Acids Res. 2015;43:e43. doi: 10.1093/nar/gku1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.