Abstract
Ezrin is a member of the ERM (ezrin, radixin, moesin) family of proteins and functions as a linker between the plasma membrane and the actin cytoskeleton. Ezrin is a key driver of tumor progression and metastatic spread of osteosarcoma. We discovered a quinoline-based small molecule, NSC305787 that directly binds to ezrin and inhibits its functions in promoting invasive phenotype. NSC305787 possesses a very close structural similarity to commonly used quinoline-containing antimalarial drugs. On the basis of this similarity and of recent findings that ezrin has a likely role in the pathogenesis of malaria infection, we screened antimalarial compounds in an attempt to identify novel ezrin inhibitors with better efficacy and drug properties. Screening of Medicines for Malaria Venture (MMV) Malaria Box compounds for their ability to bind to recombinant ezrin protein yielded 12 primary hits with high selective binding activity. The specificity of the hits on ezrin function was confirmed by inhibition of the ezrin-mediated cell motility of osteosarcoma cells. Compounds were further tested for phenocopying the morphological defects associated with ezrin suppression in zebrafish embryos as well as for inhibiting the lung metastasis of high ezrin-expressing osteosarcoma cells. The compound MMV667492 exhibited potent anti-ezrin activity in all biological assays and had better physicochemical properties for druglikeness than NSC305787. The drug-like compounds MMV020549 and MMV666069 also showed promising activities in functional assays. Thus, our study suggests further evaluation of antimalarial compounds as a novel class of anti-metastatic agents for the treatment of metastatic osteosarcoma.
Introduction
The vast majority of deaths associated with cancer are due to the metastatic spread of cancer cells from a primary tumor to distant sites (1). Osteosarcoma is a highly metastatic cancer of bone that afflicts children, adolescents and young adults, with the majority of patients having micrometastasis at the time of initial diagnosis (2). Although, the survival of patients with metastatic disease at diagnosis remains to be poor with 5-year survival rates being less than 20%, significant improvements have been achieved in the management of localized tumors through development of multimodality approaches that resulted in 5-year overall survival of 60%-78% (3-7). However, recurrent osteosarcoma occurs in 30-40% of those patients initially diagnosed with localized disease (8). Development of metastasis to the lungs remains the most common cause of death in patients with osteosarcoma. Hence, a mechanistic understanding of the metastatic process and development of molecularly targeted therapeutics aimed at preventing such disseminated disease may provide additional improvements in disease outcomes for patients with metastatic disease (9).
Ezrin is a member of the ERM (ezrin, radixin, moesin) protein family that functions as a linker protein between F-actin in the cortical layer and membrane-associated proteins on the cell surface (10). Ezrin serves as a key regulator of diverse cellular processes such as formation and organization of cell-surface structures, maintenance and determination of cell shape and modulation of cell adhesion, migration and signaling pathways (10, 11). ERM proteins are characterized by the presence of three distinct regions including N-terminal membrane-associated domain (called N-ERMAD or N-terminal ERM-association domain), followed by a long central α-helical region and a C-terminal (called C-ERMAD or C-terminal ERM-association domain) domain, which is able to bind both actin filaments in the cytoskeleton and N-ERMAD. In the current understanding of ezrin regulation, the self-association of the protein by head-to-tail joining of the molecule is believed to mask the respective ligand binding sites, which leads to a dormant closed conformation. The activation of ezrin from its closed conformation to the open form has been proposed to occur in a two-step process that involves phosphorylation of a conserved threonine residue (Thr-567) in the C-ERMAD of ezrin and binding to membrane phosphatidylinositol 4,5-biphosphate via its N-terminal domain (12). This conformational change is regarded as critical for binding of ezrin to membrane proteins including transmembrane receptors, small GTPase regulators and adaptors (10-12). Thus, ezrin acts as a scaffolding protein and can assemble highly specific and regulated clusters of membrane proteins at the cell cortex. Independent from its canonical role as a cytoskeletal scaffolding protein at the plasma membrane, our recent findings suggest that ezrin has a likely role in early phases of protein translation initiation as part of a ribonucleoprotein complex (13). We also recently demonstrated that ezrin interacts with proteins involved in protein translation initiation and stress granule dynamics. Among those proteins, we identified DDX3, a DEAD-box RNA helicase involved in multiple aspects of RNA metabolism from transcription to translation, as a direct binding partner of ezrin (14).
The elevated levels of ezrin have been shown to confer advantageous metastatic behavior in a mouse model of osteosarcoma (15) and high ezrin expression has been linked to poor outcome in patients that suffer from osteosarcoma (16). Many clinical studies have also implicated the role of ezrin in tumor progression of a variety of cancers including carcinomas of the lung, breast, colon, pancreas, endometrium and ovary, cutaneous and uveal melanomas, oral squamous cell carcinomas, rhabdomyosarcoma, brain tumors and soft tissue sarcomas (17, 18). We have recently discovered a small molecule inhibitor of ezrin NSC305787, which directly binds to ezrin and inhibits its function associated with the metastatic phenotype in both in vitro and in vivo experimental models (19).
NSC305787 contains an aromatic quinoline core structure with a bulky aliphatic adamantyl group in its northeast quadrant and a piperidine unit in the southeast region (Supplemental Figure 1). Examination of the structure of NSC305787 revealed that it shares significant structural features with commonly used quinoline-based antimalarial agents such as quinine, quinidine, mefloquine, choloroquine and amodiaquine (Supplemental Figure 1), which are mostly active against the inthraerythrocytic stages of the malaria parasite (20). The close structural resemblance between NSC305787 and antimalarial agents, therefore, has led us to screen the quinoline-family of antimalarial drugs including quinine, quinidine, mefloquine, chloroquine and amodiaquine as well as Medicines for Malaria Venture (MMV) Malaria Box content as ezrin inhibitors. The MMV Open Access Malaria Box constitutes 200-drug like and 200-probe like compounds selected from 19,000 structurally unique molecules with the confirmed activity against the blood-stage of Plasmodium falciparum (21-24).
Consequently, we screened the antimalarial compounds for their direct binding ability to ezrin. We followed this primary screen with secondary functional assays using in vitro, in vivo and ex vivo experimental models. Here, we identify compounds from MMV400 library as novel small molecule inhibitors of ezrin blocking cell motility and invasion.
Materials and Methods
Open Access Malaria Box
The Malaria Box was provided in 100% DMSO at 10 mM concentration in a 96 well format, with five plates each containing 80 compounds. The Plate mapping corresponds to the May 2012 shipment. Detailed information regarding the Malaria Box is described in Spangenberg et al. (24). Antimalarial compounds that showed potent binding activity to recombinant ezrin protein were purchased from commercial sources (MMV666069 from ChemBridge Corp., #67378588; MMV665977 from Vitas-M Laboratory, Ltd., #STK701059; MMV020243 from ChemBridge Corp., #9290708; MMV020549 from ChemDiv Inc., #E950-0299; MMV396680 from ChemDiv Inc., #G420-0124; MMV667492 from Vitas-M Laboratory, Ltd., #STK529340; MMV000448 from Vitas-M Laboratory, Ltd., #STK589099; MMV006172 from Vitas-M Laboratory, Ltd., #STK746950; MMV666103 from Asinex Ltd., #BAS02538915).
Cell lines and culturing
Mouse K7M2 and K12 cell lines were kindly provided by Dr. Chand Khanna from National Cancer Institute (NCI/NIH, MD, USA) in 2007; no authentication was done by the authors. Cells were maintained in DMEM supplemented with 10% fetal bovine serum in a humidified atmosphere of 5% CO2 at 37°C.
Cytotoxicity assay
For cytotoxicity experiments, cells were plated on T-75 flasks and grown until they reached at 70-80% confluency. Cell proliferation was monitored in xCELLigence Real-Time Cell Analysis (RTCA) (ACEA Biosciences Inc., San Diego, CA, USA) system using E-Plate 96 or E-Plate 16 (ACEA Biosciences Inc., San Diego, CA, USA) that are integrated with gold microelectrode arrays in the bottom of each well. 100 μl of culture media was added into the plates, and the plates were inserted into the station for background measurement. Cells were then seeded into the wells (5,000 cells/well in 100 μl) and left to attach overnight. The medium was then removed and replaced with 100 μl of fresh medium containing indicated concentrations of compounds. Impedance was measured every 10 minutes during 48 hr and is represented as cell index.
Preparation of recombinant ezrin protein
Recombinant human ezrin was purified as described previously (25). Escherichia coli strain M15 [pREP4] (Qiagen, Valencia, CA, USA) was used for the expression of untagged, full-length ezrin from pQE16 expression vector. Ezrin was purified by using a hydroxyapatite (Bio-Rad, Hercules, CA, USA) column followed by cation-exchange on a SP-Sepharose fast flow (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA) column in AKTA Explorer Chromatography System (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA). Protein purity was determined by SDS-PAGE followed by Coomassie staining.
Surface plasmon resonance (SPR)
The primary screen of the MMV library was performed on a Biacore 3000 instrument using a CM5 chip coupled with ezrin on one flow cell and glideosome associated protein GAP50 (26) as a negative control on another flow cell. Compounds from the MMV Malaria Box were dissolved to a final concentration of 10 μM in HBS-P (10 mM HEPES, pH 7.4, 150 mM NaCl, %0.05 (v/v) surfactant P20) and passed over all four flow cells. The running buffer contained 1% DMSO with HBS-P to match the DMSO level of the samples.
After initial screening of antimalarial compounds, the small molecules that showed at least three-fold difference in relative binding to ezrin compared to GAP50 were further evaluated to identify their binding affinities using Biacore T200 instrument at room temperature. First flow cell was left empty for background signal subtraction and second flow cell of a CM5 sensor chip was used for immobilization of the purified recombinant ezrin protein (~10,000 RU) by amine coupling method in sodium acetate buffer pH 4.5. NSC305787 was used as a positive control in these SPR experiments. HBS-P (10 mM HEPES, pH 7.4, 150 mM NaCl, %0.05 (v/v) surfactant P20) containing DMSO between 2% and 10% final concentration was used as the running buffer. Kinetic analysis was done by injecting seven different concentrations of compounds (0.78, 1.56, 3.13, 6.25, 12.5, 25 and 50 μM) over ezrin-captured and control surfaces in triplicates. Each injection was done with 60 seconds association time and 180 seconds dissociation time. KD values for compound-ezrin interactions were obtained using BiaEvaluation software (version 1.0).
Immunoprecipitation (IP) and immunoblotting (IB)
IP experiments were performed on lysates of K7M2 mouse osteosarcoma cells as described previously (14). Total cell lysates were prepared in phospho-lysis buffer (50 mM HEPES pH 7.9, 100 mM NaCl, 4.0 mM sodium pyrophosphate, 10 mM EDTA, 10 mM sodium fluoride and 1% Triton X-100) containing 2.0 mM sodium vanadate, 1.0 mM PMSF, 4.0 μg/ml aprotinin, 4.0 μg/ml leupeptin and 1.0 μg/ml calyculin A. Lysates were incubated on ice for 30 min, and then cleared by centrifugation at 16,000xg for 10 min at 4°C. An equal amount of protein lysate (approximately 1000-1500 μg/1.0 ml phospho-lysis buffer) was incubated with 2.0 μl of anti-ezrin antibody (Sigma Aldrich, St. Louis, MO, USA; #E8897) or the equal amount of control mouse IgG (Santa Cruz Biotechnology, Santa Cruz, CA, USA; #sc-2025) overnight at 4°C on a rotating axis, followed by incubation with BSA-blocked Protein A/G Plus-Agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 1 hr. Subsequent to washing of the pelleted beads three times with 0.3 ml (each) IP-wash buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1.0 mM EGTA, 1.0 mM EDTA, 1% Triton X-100, 0.5% Nonidet P-40, 0.2 mM sodium vanadate and 0.2 mM PMSF), the bound proteins were eluted by boiling samples in SDS-PAGE sample buffer for 5 minutes. The immune complexes were then analyzed by western blot analyses. IB experiments were performed on both K7M2 and K12 cells as previously described (14, 27). Equal amount of protein samples were separated by SDS-PAGE and then transferred to an Immobilon-P membrane (Millipore, Billerica, MA, USA). In order to block non-specific binding sites, the membrane was incubated in 5% nonfat dry milk diluted in TTBS (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.05% Tween 20) for 1 hr at room temperature followed by the addition of primary antibody for 2 hr at room temperature. Dilutions for primary antibodies were as follows: anti-ezrin antibody (Sigma Aldrich, St. Louis, MO, USA; #E8897) at 1:2500, anti-phospho ezrin(Thr567)/Radixin(Thr564)/Moesin(Thr558) antibody (Cell Signaling Technology, Danvers, MA, USA; #3141) at 1:1000 and anti-actin-HRP (Santa Cruz Biotechnology, Santa Cruz, CA, USA; #sc-1615) at 1:5000. The membrane was then rinsed three times with TTBS and incubated for 1 hr at room temperature in HRP conjugated anti-mouse (GE Healthcare Bio Sciences, Pittsburgh, PA, USA; #NA931V) or HRP-conjugated anti-rabbit (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA; #NA934V) secondary antibody diluted at 1:5000 in TTBS. The blot was washed three times in TTBS and immunoreactive bands were revealed by using Millipore Immobilon Western Chemiluminescent HRP substrate according to the manufacturer’s instructions (Millipore Corporation, Billerica, MA, USA). Chemiluminescence was detected using a Fujifilm LAS-3000 imaging system.
Chemotaxis/cell migration assay
Cell migration experiments were performed using xCELLigence Real-Time Cell Analysis (RTCA) (ACEA Biosciences Inc., San Diego, CA, USA) system as described previously (28). This method allows label-free monitoring of cell migration continuously in real-time via electrical impedance readout. Cell migration experiments were performed using 16-well plates (CIM-16, ACEA Biosciences Inc., San Diego, CA, USA), which consists of an upper and a lower chamber separated by a microporous membrane containing randomly distributed 8-μm pores. Briefly, 160 μl of DMEM media containing 10% fetal bovine serum was loaded in the lower wells as chemoattractant and the upper chamber was attached. The serum-free media in the lower well was used as a negative control for cell motility. Upper wells were filled with 50 μl serum-free media and the plate was left in the RTCA DP device at 37°C for 1 hr to pre-equilibrate. Following measurement of background signal generated by cell-free media, 50 μl of K7M2 or K12 cell suspension (1.5 × 105 cells) in serum free media was placed into the top wells. Next, 50 μl of serum-free media containing appropriate concentrations of NSC305787 and the antimalarial compounds were added into the wells. The final concentration of DMSO in cell suspension was 1%. The assembled plate was then transferred to the RTCA-DP machine and data was collected every 10 min over 21 hr. The electrical impedance measured by the device as a dimensionless parameter termed cell index was used to calculate % motility of compound-treated osteosarcoma cells compared to vehicle-treated control.
Zebrafish embryo development assay
Zebrafish (Danio rerio) were raised and maintained in Georgetown University zebrafish facility using standard husbandry techniques as described previously (29). A translation-blocking anti-ezrin morpholino (MO), which has been previously described and validated by Link et al. 2006 (30), was synthesized by Gene-Tools, LCC (Philomath, OR, USA). Embryos were collected and staged according to both hours post-fertilization (hpf) at 28.5°C and by morphological criteria (31). For chemical screening, zebrafish embryos were arrayed in 96-well plates and compounds were added at 10-50 μM concentrations at 3 hpf. Embryos were scored for epiboly defects and/or characterized for other morphological defects at time points corresponding to between 7.0-8.5 hpf and 1, 3 and 5 days post fertilization (dpf), respectively. All animal procedures were approved by the Institutional Animal Care and Use Committee of Georgetown University.
Pulmonary metastasis assay (PuMA)
The PuMA assay was performed as previously described (32). Green fluorescent protein (GFP)-expressing K7M2 cells (5×105) were injected to three Balb/c mice through tail vein. The mice were euthanized by CO2 inhalation after 15 min of tumor cell injection. The trachea was exposed and infused with a culture medium/agarose solution using a gravity perfusion apparatus. The inflated lungs were removed and placed in cold PBS containing 100 U/ml penicillin and 100 μg/ml streptomycin at 4°C for 20 minutes to solidify the agarose. Axial sections of 1–2 mm in thickness were cut from each lobe. Two lung sections from each of three animal (in total six lung sections per treatment) placed on a piece of sterile Gelfoam (Pfizer-Pharmacia & Upjohn Co., Kalamazoo, MI, USA) that had been preincubated in culture medium. Compounds were added immediately to the culture media at day 0 of the culture period. Lung sections were incubated at 37°C in a humidified atmosphere with 5% CO2. The medium was replaced with fresh culture medium with or without compounds every other day. Animal care and use was in accordance with the guidelines of the NIH Animal Care and Use Committee, and approved by the Institutional Animal Care and Use Committee of Georgetown University.
A Nikon ECLIPSE Ti fluorescence inverted microscope (Nikon Instruments, Melville, NY, USA) equipped with a DS-Fi1c high-definition cooled color camera head was used to capture images of GFP-positive tumor cells at 10x magnification. Images were processed and analyzed by NIS-Elements BR 3.0 imaging software (Nikon Instruments, Melville, NY, USA). Metastatic burden was quantified in the lung sections at day 8 by measuring the total fluorescent area of each lung section as μm2 occupied by metastatic cells. The fluorescent area for each lung section was normalized to 100 μm2 for day 0 to allow quantitative evaluation of metastatic progression over time.
Histological analysis
Lung tissues were fixed in 10% buffered formalin overnight at 4°C and embedded in paraffin using standard techniques. Serial sections of 5 μm thickness were cut from paraffin-embedded tissues and mounted on glass slides. Tissue sections were stained with hematoxylin and eosin (H&E). H&E-stained slides were then examined for the viability and structural integrity of pulmonary architecture. Images of H&E stained lung sections were taken at 10x objective using Olympus BX40 microscope equipped with ProgRes C5 digital camera.
Statistical analysis
All data are reported as mean ± standard deviation of n observations. Statistical evaluation was performed by an unpaired Student's t test using the GraphPad Prism version 6.0c (GraphPad software, La Jolla, CA, USA), taking a probability P<0.05 as significant.
Results
Initial screening of MMV Malaria Box compounds for direct binding to ezrin
We used surface plasmon resonance technology to screen compounds that directly bind to purified recombinant ezrin protein that was immobilized on a sensor chip. We first screened antimalarial drugs, quinine, quinidine, mefloquine, amodiaquine and chloroquine for their direct binding ability to ezrin due to their structural similarity to our lead compound, NSC305787 (Supplemental Figure 1). However, none of the compounds produced any significant binding to ezrin compared to NSC305787.
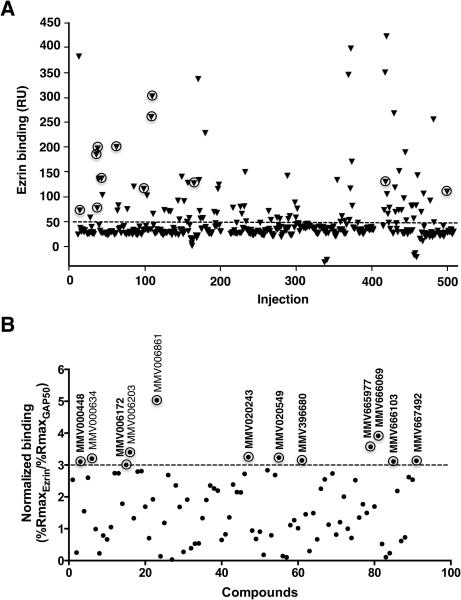
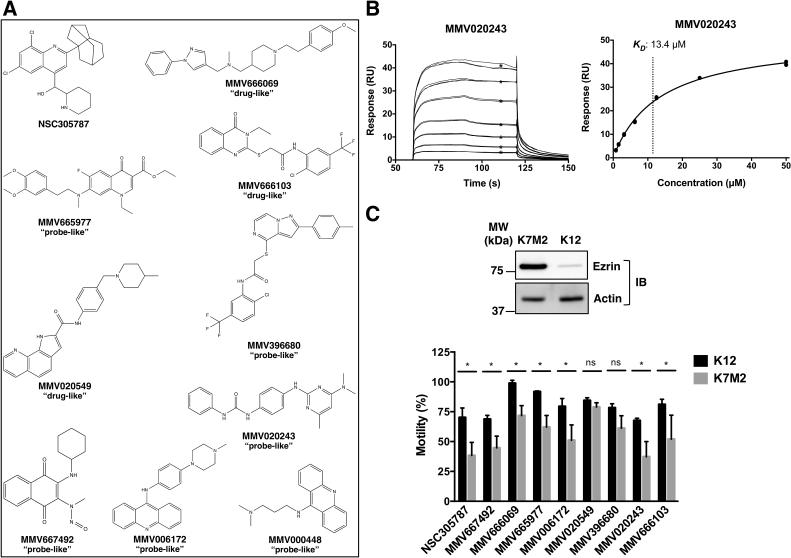
We then extended our screen to the MMV400 library, of which 91 compounds demonstrated varying degrees of binding to ezrin above the background level (Figure 1A). We used a negative control protein, GAP50, in order to eliminate non-specific binders. The hit selection of Malaria Box screening was based on a threshold of three-fold difference in relative binding of a compound to ezrin with respect to GAP50. Based on this selection criterion, 12 hits were identified out of 91 compounds, of which four were “drug-like” and the remaining eight were “probe-like” (Figure 1B). Among 12 primary hits, we subsequently determined the binding affinities of 9 commercially available compounds (Figure 2A) for ezrin by detailed SPR studies in a Biacore T200 instrument. For this multiple concentration secondary analysis, we excluded MMV396680 and MMV000448 due to their poor solubility. This analysis yielded KD values ranging from 2.1±1.0 to 29.4±4.4 μM for seven hit compounds (Table 1, Figure 2B and Supplemental Figure 2).
Figure 1.
Screening of MMV400 library by surface plasmon resonance technology for direct binding to ezrin. (A) Purified recombinant ezrin protein was immobilized on a sensor chip and small molecules were individually injected over the surface at a single concentration in a Biacore 3000 instrument. A negative control protein GAP50 was also immobilized on a neighboring flow cell in the same sensor chip to eliminate false positives resulting from non-specific protein binding. The data points represented with triangles encircled with rings represent the primary hits with relatively high specific binding for ezrin identified through filtering of the data as explained in panel B. (B) Compound filtering was implemented as a mean to prioritize the hits based on their specific binding to ezrin over GAP50, which yielded a total of 12 primary hits. The analyte binding capacity or theoretical maximum response (Rmax for maximum response) of each compound were calculated for both ezrin and GAP50. The relative binding of each compound (response units, RU) to ezrin and GAP50 were normalized to Rmax, which was taken as 100%. Any molecule demonstrating a threshold of 3-fold difference in relative binding to ezrin in terms of normalized response with respect to the negative control protein GAP50 was then selected as a primary hit. 9 compounds out of 12 primary hits used in this study are shown in bold.
Figure 2.
Surface plasmon resonance binding kinetics of selected hits from MMV400 library binding to ezrin. (A) Chemical structures are given for NSC305787 and nine antimalarial compounds identified through SPR screen from MMV400 library. (B) A representative sensorgram showing binding of MMV020243 to immobilized ezrin at seven concentrations in triplicates ranging from 0.78 to 50.0 μM is presented with the steady-state affinity curve given in the right panel. Black ∗ represent binding levels for each concentration that were used to calculate the steady state affinity from the curve in the right panel. The final KD value calculated from n=6 separate experiments is given in Table 1. C. Inhibition of cell migration in osteosarcoma cells by the known anti-ezrin molecule NSC305787 and selected hits from MMV400 library. Upper Panel: Immunoblotting showed that highly metastatic K7M2 cells express high levels of ezrin relative to the low metastatic K12 cells. Lower Panel: Cell migration experiments were performed using xCELLigence electric cell impedance system in CIM-Plates 16 with 10% serum serving as the chemoattractant in the lower chamber. Cell migration was monitored in real-time for a period of 21 h. Data was normalized to control treatment (DMSO), which was taken as 100%. NSC305787 and the selected hits from SPR screening except MMV020549 and MMV396680 showed greater inhibition of K7M2 motility compared to its less aggressive counterpart K12 cells. K7M2 and K12 cells were treated with 5.0 μM of MMV667492, MMV666069, MMV665977, MMV020549, MMV020243, MMV666103 and MMV396680 and 3.0 μm of NSC305787 and MMV006172 for 21 hour. Values are presented as the means ± standard deviations of n separate experiments as given in Supplemental Figure 4. (*; p<0.05, ns; not significant, compared to control; using a Student’s unpaired t test).
Table 1.
Summary of the biological activities of antimalarial agents from MMV400 library for inhibition of ezrin function
| Compound ID | KD (μM)a | Migration inhibitionb |
Ezrin MO-like phenotypec |
Prevention of lung metastasisd |
MMV400 library set |
Lipinski&Veber drug criteria violation counte |
|---|---|---|---|---|---|---|
| NSC305787 | 6.6 ± 3.6 (n=6) | yes | potent | yes | not applicable | 1 |
| MMV667492 | 29.4 ± 4.4 (n=2) | yes | potent | yes | probe-like | 0 |
| MMV020549 | 10.7 ± 0.4 (n=3) | no* | potent | yes | drug-like | 0 |
| MMV666069 | 2.1 ± 1.0 (n=5) | yes | moderate/subtle | yes | drug-like | 0 |
| MMV020243 | 14.8 ± 3.9 (n=6) | yes | moderate/subtle | no | probe-like | 0 |
| MMV665977 | 3.1 ± 1.6 (n=5) | yes | no | yes | probe-like | 0 |
| MMV666103 | 2.6 ± 1.0 (n=4) | yes | no | nd** | drug-like | 0 |
| MMV006172 | 6.1 ± 2.8 (n=3) | yes | no | nd | probe-like | 1 |
| MMV396680 | nd | no | moderate/subtle | nd | probe-like | 1 |
| MMV000448 | nd | nd | no | nd | probe-like | 0 |
Binding affinities of compounds for ezrin were determined by SPR. The results are expressed as mean ± standard deviations. n indicates number of separate experiments for calculation of the final KD values.
Compounds demonstrated greater inhibition of high-ezrin expressing K7M2 OS cell migration than less aggressive K12 cells.
Compounds produced ezrin MO-like phenotypes in zebrafish embryonic developmental assays.
Compounds inhibited metastatic progression of GFP-expressing osteosarcoma cells in ex vivo lung organ culture assay.
Values represent number of conditions violated over total six conditions.
According to the Lipinski’s and Veber’s druglikeness criteria, a compound should have a low molecular weight (≤500 daltons), low partition coefficient (cLogP ≤5), low number of H-bond donor (≤5) and acceptors (≤10), low number of rotatable bonds (≤10) and low polar surface area (≤140).
no: not observed
nd: not determined
We evaluated the 9 primary hits based on their physicochemical properties to determine whether they conform to Lipinski’s ‘Rule of Five’ for druggability (33). Molecular weight (≤500 daltons), partition coefficient (cLogP ≤5) and the number of hydrogen bond donor (≤5) and accepting groups (≤10) present in the compound are used to predict whether or not a compound is likely to be orally bioactive based on its absorption or permeation. Veber et al. (34) have also proposed additional rules including the number of rotatable bonds (≤10) and the polar surface area (≤140) to predict the compounds with good oral bioavailability. Computational analysis of the compounds using MolInspiration software (http://www.molinspiration.com) revealed that all the primary hits except MMV006172 and MMV396680 had structural features that satisfy all six conditions of the above methodology (Table 1). Thus, although 6 antimalarial compounds were initially categorized as “probe-like” in MMV Malaria Box (Figure 2A), 4 of them hold therapeutic promise for further functional assays and lead optimization studies. The lead compound NSC305787 did not meet all of the Lipinski and Veber druglikeness criteria either due to its high computed partition coefficient (cLogP) of 6.30 (Table 1).
Inhibition of ezrin-mediated cell motility in osteosarcoma cells
We first employed an osteosarcoma cell culture method to confirm the hits from the primary screen on the basis of their ability to inhibit ezrin-mediated cell motility. K7M2 murine osteosarcoma cells are highly aggressive and metastatic to the lungs and were derived from less aggressive K12 cells originating from spontaneously occurring osteosarcoma in BALB/c mouse (2). K7M2 cells express higher levels of ezrin protein than K12 (Figure 2C), which leads to enhanced metastatic phenotype of K7M2 cells (15, 32). Initially, we determined the cytotoxicity profiles of each compound for both cell lines to identify the non-toxic dose limits prior to investigating their ability to inhibit ezrin-mediated cell motility (Supplemental Figure 3). Since cell viability is not affected by the alterations in ezrin level, concentrations that reduce the cell viability were considered as non-specific response and irrelevant to the ezrin inhibition. Therefore, all experiments were performed at appropriate concentrations of individual drugs and time intervals that do not affect cell viability. Compound MMV000448 was discarded at this stage because of its relatively high cytotoxicity towards both K12 and K7M2 cell lines (Supplemental Figure 3).
We used a real-time cell monitoring technology based on electrical cell impedance in a label-free environment to explore the potential inhibitory effect of compounds on migration of K12 and K7M2 cells. We used a Boyden-like dual chamber system consisting of an upper chamber and a lower chamber separated by a 8-μm pore-sized membrane. As cells migrate from the upper chamber to lower chamber in response to serum, they contact and adhere to the gold microelectrode sensors on the underside of the membrane, which results in an increase in electrical impedance. Figure 2C shows the % motility of K7M2 and K12 cells in the presence of primary hits. Data was normalized to control treatment, which was taken as 100%. The addition of anti-ezrin compound NSC305787 decreased the migration of K12 and K7M2 cells to 70.2±8.0% and 38.5±10.8% of the control, respectively. All the primary hits except MMV020549 and MMV396680 exhibited the same pattern as NSC305787 with a more marked reduction in the motility of K7M2 cells compared to that of K12 cells at varying degrees (Figure 2C and Supplemental Figure 4).
Inhibition of cell motility during zebrafish embryonic development
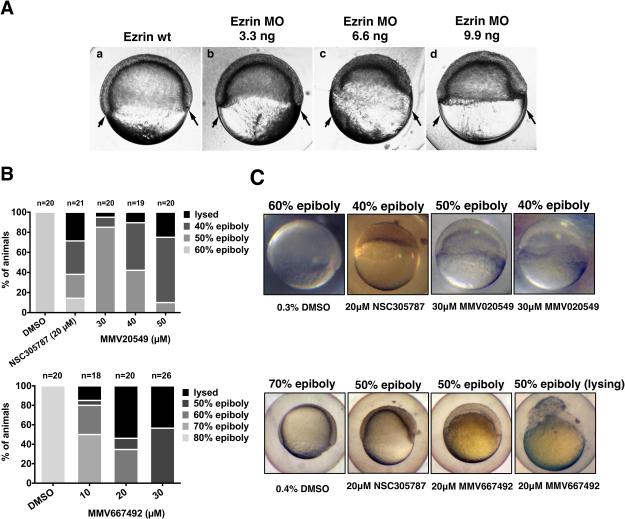
The functional analysis of ezrin2 (the zebrafish homolog) protein in zebrafish developmental model revealed that ezrin2 is required for germ layer morphogenesis during gastrulation (30). Injection of antisense MO oligonucleotides designed to knockdown ezrin2 expression shows specific phenotypes in zebrafish characterized by the reduced epiboly movements and in the most severe cases, the complete detachment of the blastoderm from the yolk cell at early gastrulation (30). Figure 3A shows the ezrin MO-induced inhibition of epiboly. Compared to uninjected wild-type siblings at 60% epiboly, MO injected embryos show reduced epiboly and a thickening of the blastoderm. The failure of gastrulation is seen at the initiation of involution as an exaggerated thickening at the blastoderm margins. Severe ezrin morphant phenotypes are characterized by strong epiboly defects resulting in cells piling up at the animal pole (19). This phenotype was used as a guide to screen small molecules by observing embryonal development in 96-well plate format. Thus, in addition to the chemotaxis assays, we tested the potential biological effects of 9 primary hits on a zebrafish embryonic phenotype assay. We initially screened the compounds at 10 and 30 μM concentrations, evaluating phenotypes at germ rising stage, 1 dpf, 3 dpf and 5 dpf. A brief summary of the findings from this initial screening is presented in Supplemental Table 1 and Supplemental Figure 5. Treatment with 10 μM NSC305787 mimicked the ezrin-knock down phenotype, which resulted in 100% epiboly failure with ultimate lysis of embryos by 1 dpf. Among the hit compounds tested, embryos treated with MMV020549 and MMV667492 at 10 and 30 μM concentrations developed the best ezrin MO-like phenotypes, which were characterized by morphological defects including delayed development, convergence extension defects, apc-mutant like phenotypes, deformed embryos and eventually the lysis of the embryos. Following treatment with MMV666069, MMV020243 and MMV396680, the embryos exhibited subtle defects with no swim bladder, curved embryo phenotypes and slight increase in mortality, respectively, whereas MMV665977, MMV006172, MMV666103 and MMV000448-treated embryos developed normally.
Figure 3.
Phenotypic screening of selected hits from MMV400 library for reduced cell motility phenotypes in zebrafish embryonic developmental assays. (A) Ezrin morphant embryos are shown at the time that wild-type siblings have reached the 60% epiboly stage (a). Arrows mark the location of marginal regions in wild-type embryos at 60% epiboly. Embryos were injected at the single cell stage with 3.3 ng ezrin MO (b), 6.6 ng ezrin MO (c), or 9.9 ng ezrin MO (d). Ezrin MO injections result in retarded epiboly, a thickened blastoderm, and defective involution of the blastoderm. (B) Small molecule treated embryos were scored for epiboly defects at early stages of development between 7.0 and 8.5 hpf. Embryos treated with small molecule inhibitor of ezrin, NSC305787, completely mimicked ezrin-MO-injected embryos. Treatment of the embryos with MMV Malaria Box compounds MMV020549 and MMV667492 resulted in delay of the epiboly progression around the yolk with increasing concentration of drugs compared to vehicle treated group. (C) Representative images of embryos treated with NSC305787 and MMV Malaria Box compounds MMV020549 and MMV667492 from Panel B are presented with different degrees of epiboly defects, compared to vehicle-treated controls, observed between 7.0 and 8.5 hpf.
In light of the initial findings, we tested the effect of MMV020549 and MMV667492 in detail at different time points. MMV020549 exhibited a dose dependent inhibition of epiboly and gastrulation between 7.0 and 8.5 hpf (Figure 3B and C). MMV667492 showed a strong anti-ezrin-like phenotype in a dose range of 10 to 30 μM (Figure 3B and C). We observed no evidence of gastrulation occurring in MMV667492-treated embryos.
Prevention of metastatic growth in a lung organ culture assay
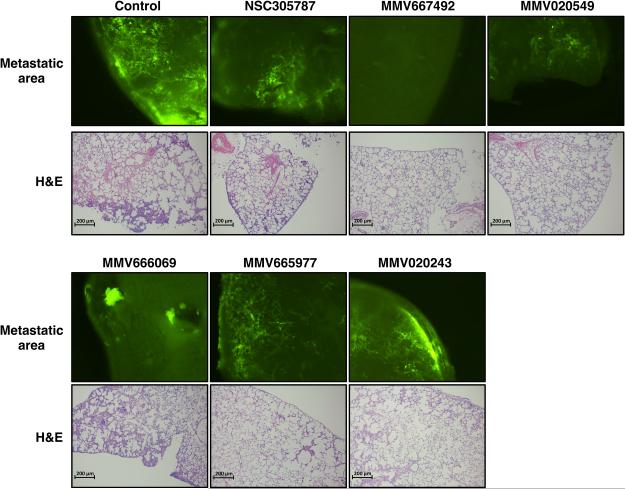
Osteosarcoma has a high propensity for early metastasis, with tumor cells being more prone to metastasize to the lungs than the other tissues (16, 35). We tested the potential anti-ezrin activity of our hits in a pulmonary metastasis assay (PuMA) utilizing ex vivo cultures of lung tissue. This method allows the assessment of metastatic progression of cancer cells reaching to the lung (32). Green florescent protein (GFP)-expressing K7M2 mouse osteosarcoma cells were injected into tail-vein of Balb/c mice. Following injection of tumor cells, the mice were euthanized and the lungs were infused with a mixture of culture medium and agarose to maintain the lung structure. The lungs were then allowed to cool to solidify the agarose medium solution, and axial sections were made from each lung lobe. Lung sections were placed on sterile Gelfoam sections bathing in culture media and immediately treated with either vehicle or compounds. Metastatic foci growing in lung slices were then visualized and quantitated based on GFP intensity using a fluorescence microscope (Figure 4 and 5).
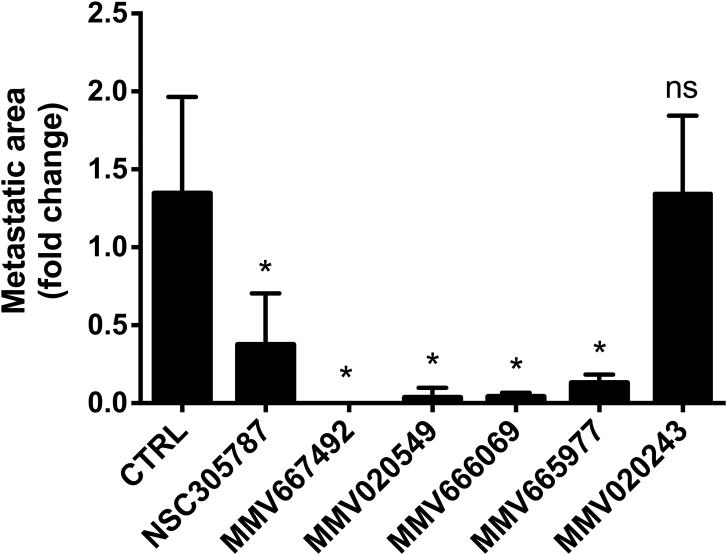
Figure 4.
Inhibition of osteosarcoma metastatic growth in lung organ culture by selected hits from MMV400 library. Green fluorescent protein-positive K7M2 osteosarcoma cells were delivered to three BALB/C mice by tail-vein injection, and the lungs were dissected. Two lung sections from each of three animals (in total six lung sections per treatment) were grown in culture media, and metastatic progression was monitored using a fluorescence microscope. 10 μM of the anti-ezrin compound NSC305787 and 30 μM of antimalarial compounds were added to the culture media at day 0 of the culture period. Images were acquired at day 8 and the area of fluorescence signal in each section was quantified. Data represents fold change in fluorescent area at day 8 of drug-treated lung sections as compared to vehicle-treated control and is shown as mean ± standard deviation of total six lung sections from three mice (*; p<0.05 compared to control; using a Student’s unpaired t test).
Figure 5.
Representative images for lung metastatic growth of GFP-expressing K7M2 cells and the corresponding H&E staining of lung sections treated with either vehicle or anti-ezrin compounds in PuMA are presented on the upper and bottom panels, respectively.
Exposure of lung tissue slices to the anti-ezrin compound NSC305787 significantly inhibited the metastatic growth of high-ezrin expressing K7M2 cells compared to the vehicle-treated group. When the lung tissues were treated with MMV667492 and MMV020549, the most active hits found in the zebrafish embryonic phenotype assays, we observed a significant decrease in the number of GFP-expressing metastatic foci compared to vehicle-treated control group (Figure 4 and 5). We also tested the potential anti-metastatic activities of MMV666069 and MMV020243, which demonstrated subtle to moderate effects in zebrafish embryonic developmental assays. MMV666069 significantly inhibited the metastatic growth of K7M2 osteosarcoma cells and produced a distinguishing phenotype in the live fluorescent imaging of lung sections (Figure 5). On the other hand, a large number of metastatic colonies containing high-ezrin expressing K7M2 osteosarcoma cells were evident in lung tissues treated with MV020243 (Figure 4 and 5). Interestingly, MMV665977, an ineffective compound found in zebrafish embryonic development assays, significantly inhibited the metastatic growth of K7M2 cells in lung tissues as well (Figure 4 and 5).
We performed histological examinations of lung tissue slices treated with compounds to confirm the maintenance of lung architecture over a 8-day period of drug treatment in ex vivo pulmonary metastasis assay. We observed structurally intact pulmonary architecture (Figure 5), which confirms that the observed inhibition of metastatic growth by compounds was not due to the deterioration of lung tissue.
Structural and shape comparison analysis of identified ezrin inhibitors
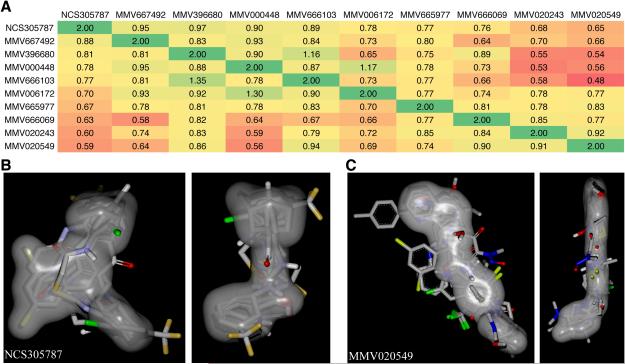
We performed structural and shape comparison analysis of ezrin inhibitors using the OpenEye software ROCS (Rapid Overlay of Compound Structures) (36). A heat map was generated based on the combined shape complementarity and chemical Tanimoto score as calculated by ROCS. This analysis revealed that the identified ezrin inhibitors from MMV400 library fall into two shape classes (Figure 6). It is likely that two binding sites exist on ezrin, which are occupied by these compounds. However, extended compounds such as MMV020549 can adopt a very compact form and fit into the shape constraints of NCS305787 as well. These computational predictions will have to be experimentally validated in crystal structure or NMR studies.
Figure 6.
Structural and shape comparison of identified ezrin inhibitors from MMV400 library. A) A heat map was generated based on the combined shape complementarity and chemical Tanimoto score as calculated by the OpenEye software ROCS (36). A score of 2 for a molecule represents 100% identity in shape and chemical properties. B) Overlay of all inhibitors based on the shape of NCS305787 in grey in two views rotated by 90 degrees to each other. C) Overlay of all inhibitors based on the shape of MMV020549 in grey in two views rotated by 90 degrees to each other.
Discussion
Metastasis remains the major cause of cancer mortality. Even though our understanding of molecular events that drive metastasis has improved for the past two decades, there is still lack of efficient therapeutics targeting metastatic progression. Extensive research on metastasis biology has led to the identification of a number of molecular pathways and cellular mechanisms with druggable targets in several cancers (37-39). Ezrin is one such protein with a cytoskeleton-membrane linker function that has been identified as a major driver of osteosarcoma metastasis (15, 16). Earlier work in our laboratory has led to the discovery of a quinoline-based small molecule inhibitor of ezrin, NSC305787, which possesses a very close structural similarity to the quinoline family of antimalarial agents. Here, we present the findings of our screening of antimalarial compounds in an attempt to find novel scaffolds with better activity and improved physicochemical properties for targeting ezrin-dependent metastatic phenotype. The overall comparative results for screening assays are summarized in Table 1.
Our findings revealed a naphtoquinone-based antimalarial compound, MMV667492 (Figure 2A), as the most potent inhibitor of ezrin-mediated cell motility and invasive phenotype in all functional assays. The remaining compounds were found to be inactive in at least one of the functional assays. Among those compounds, MMV020549, which is composed of piperidine and a pyrroloquinoline carboxamide (Figure 2A), was active in all but the cell motility assay of osteosarcoma cells, whereas MMV666069 gave significant responses in all biological activity assays with a moderate/subtle morphogenic effect in zebrafish embryonic developmental assays (Table 1). Both compounds have been assigned to drug-like category in MMV400 library. MMV020243, which is composed of a pyrimidine ring, a phenyl group and a second phenylurea ring (Figure 2A), exhibited more potent inhibition of cell motility toward high-ezrin expressing cells and produced curved embryo phenotypes in zebrafish developmental assays, whereas it was not active in preventing ex vivo lung metastasis (Table 1). MMV665977 (an oxoquinoline derivative), MMV666103 (an oxoquinazoline-derivative) and MMV006172 (an acridine-derivative) (Figure 2A) were all inactive in mimicking the ezrin MO phenotype in zebrafish developmental assays but showed a more preferential-inhibition of the motility of high-ezrin expressing K7M2 cells. MMV665977 also significantly prevented ex vivo lung metastasis of high-ezrin expressing K7M2 osteosarcoma cells. MMV396680, a pyrazolo-pyrazine derivative, had a slight toxic effect on embryonic development of zebrafish and exhibited only a subtle increase in preferential inhibition of the motility of K7M2 cells compared to that of K12 cells. MMV000448, an acridine derivative, was excluded from cell motility assays due to its high cytotoxicity, and was inactive in zebrafish embryonic developmental assays (Table 1).
Although a clear structure-activity relationship was not discernible among compounds, MMV667492, as the most potent anti-ezrin compound found in this study, displayed a high level of molecular similarity to NSC305787 with a two fused-ring structure in the center and with a bulky aliphatic group in its north quadrant and hydrophilic groups capable of hydrogen bonding in the south region (Figure 2A). Our primary screening based on SPR analysis produced two acridine-containing antimalarial compounds with very close structures, MMV000448 and MMV006172 (Figure 2A), and none of which was confirmed as a potent ezrin inhibitor through subsequent functional assays (Table 1).
KD values of antimalarial agents for binding to ezrin were found comparable in the low μM ranges, however, no correlation was found between binding affinity and ability to inhibit ezrin function in different assays (Table 1). It should be kept in mind that binding of these small molecules might occur at multiple sites on ezrin, therefore, a high binding affinity to a particular site might not reflect a strong inhibition of ezrin function. Since ezrin exerts its diverse biological functions through protein-protein interactions, binding of compounds to different sites on ezrin might lead to distinct local conformational changes, which in turn may result in higher or lower preference of ezrin in binding to different set of proteins. This might also account for the differences in biological activities of compounds in different functional assays, as there might be potential differences in binding partners of ezrin in mouse and zebrafish. Since we tested the binding affinities of our compounds using recombinant human ezrin expressed in E. coli, they may also exhibit differences in binding to the mouse ezrin or the zebrafish homolog of ezrin, ezrin2 (30). In addition, these compounds might differentially affect the phosphorylation status of ezrin particularly on Thr567, Tyr146, Tyr353 and Tyr477 residues, each of which might be associated with specific signaling pathways leading to different cellular responses. Due to the high level of homology existing between ERM family proteins, the observed biological effects might also partially result from the inhibition of radixin and moesin function. Importantly, for the experiments involving culturing of the cells or tissues, high serum protein binding may also have an impact on efficacy of the compounds, since it can affect the free fraction that is responsible for the biological action. Finally, the differential response to compounds may reflect their differential permeability and/or uptake across the cell membranes and their intracellular metabolism.
The most notable compounds identified through our screen were MMV667492, MMV020549 and MMV666069, all of which exhibited superior activity over the original lead compound NSC305787 in inhibiting the pulmonary metastatic growth (Figure 4 and 5). These compounds successfully satisfied all of the Lipinski’s and Veber’s druglikeness criteria, whereas NSC305787 failed to meet the criteria of low cLogP (Table 1). Of those compounds, MMV666069 produced subtle but not a strong ezrin MO like phenotype in zebrafish embryonic developmental assays. Apart from the above-mentioned reasons accounting for the differences in biological activities of compounds between different assays, this might also be related to the limitations of the assay. Zebrafish embryos develop at least up to 48 hpf within a protective chorion, which is a 1.5–2.5 μm thick acellular envelope consisting of three layers; outer, middle and innermost layers. Thus, chorion may provide a potential barrier for the uptake of some particular chemicals and particulates (40).
The infection of erythrocytes by the malaria parasite Plasmodium falciparum results in extensive remodeling of cytoskeleton and plasma membrane of the host cell by the parasite-coded proteins. Although the function of the majority of the exported proteins of parasite are not known, recent studies demonstrate that some of which interact with the components of host cytoskeleton (41). Parish et al. (42) have demonstrated that a member of the P. falciparum PHIST family proteins binds to the erythrocyte cytoskeleton associated protein Band 4.1, also known as 4.1R. The protein Band 4.1 presents significant structural homology with ezrin, both of which share a N-terminal FERM domain followed by a long α-helical region (43). Recently, it has been demonstrated through phosphoproteome analysis that the malaria parasite P. vivax inhibits erythroid cell growth through down-regulating the phosphorylation of ezrin, α-actinin-1 and Rho kinase, which indicates the involvement of ezrin-dependent cellular mechanisms in suppression of erythropoiesis by P. vivax (44). These findings suggest that our MMV400 library hits might exert their antimalarial effects via inhibition of erythrocytic ezrin function required for parasite-cell association but not through a direct inhibitory effect on the parasite growth.
We examined the effects of hits from MMV400 library including MMV667492, MMV020549 and MMV666069 and NSC305787 on phosphorylation status of ezrin in K7M2 osteosarcoma cells. Both MMV20549 and MMV666069 significantly increased ezrin T567 phosphorylation, whereas treatment of cells with MMV667492 produced only a moderate increase in phospho-ezrin levels (Supplemental Figure 6). None of the small molecules altered total ezrin protein levels in the cell. We did not observe any change in phospho-ezrin levels with NSC305787 treatment. Although the current understanding of ezrin function is based on its activation by phosphorylation on T567 residue followed by subsequent membrane localization, our recent findings highlights a novel function of ezrin in regulation of gene translation that is distinct from its known functions at the plasma membrane as a cytoskeletal scaffolding protein (13, 14). On the basis of the aforementioned study demonstrating that the malaria parasite P. vivax proteins inhibit erythroid cell growth by preventing the phosphorylation of ezrin T567 (44), our present findings suggest that the MMV400 library hits might exert their antimalarial effects through increasing phospho-ezrin levels in the cell.
In summary, we have identified novel small molecule inhibitors of ezrin from MMV Malaria Box that selectively target ezrin function as anti-invasive agents. Although more studies need to be undertaken to determine the mechanism of action of these antimalarial compounds, they hold promise as a novel class of anti-metastatic agents for the treatment of metastatic osteosarcoma and raise the possibility that some of the antimalarial compounds might function through regulating ezrin activity in host cells.
Supplementary Material
Acknowledgements
We would like to thank Dr. Chand Khanna from National Cancer Institute (NCI/NIH, MD, USA) for osteosarcoma cell lines. We would like to thank Dr. Abraham T. Kallarakal for helping with the Biacore experiments in determination of the binding affinities of compounds.
Funding Sources: This work was supported by the US Department of Defense grant W81XWH-10-1-0137 (to A. Uren). Additional support was provided by the Johns Hopkins Malaria Research Institute and the Bloomberg Family Foundation (to J. Bosch) and the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1232825 (to D.D. Colón-López). We wish to thank the Biacore Molecular Interaction Shared Resource and the Animal Model Shared Resource at the Lombardi Comprehensive Cancer Center (Georgetown University), which are supported by a grant P30 CA51008 from the National Cancer Institute.
Footnotes
Conflict of Interest: Georgetown University has filed a patent application for using NSC305787 and related compounds in cancer therapy, where AU and JAT are listed as inventors.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nature reviews Cancer. 2014;14:722–35. doi: 10.1038/nrc3838. [DOI] [PubMed] [Google Scholar]
- 3.Link MP, Goorin AM, Miser AW, Green AA, Pratt CB, Belasco JB, et al. The effect of adjuvant chemotherapy on relapse-free survival in patients with osteosarcoma of the extremity. The New England journal of medicine. 1986;314:1600–6. doi: 10.1056/NEJM198606193142502. [DOI] [PubMed] [Google Scholar]
- 4.Meyers PA, Heller G, Healey JH, Huvos A, Applewhite A, Sun M, et al. Osteogenic sarcoma with clinically detectable metastasis at initial presentation. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1993;11:449–53. doi: 10.1200/JCO.1993.11.3.449. [DOI] [PubMed] [Google Scholar]
- 5.Harris MB, Gieser P, Goorin AM, Ayala A, Shochat SJ, Ferguson WS, et al. Treatment of metastatic osteosarcoma at diagnosis: a Pediatric Oncology Group Study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1998;16:3641–8. doi: 10.1200/JCO.1998.16.11.3641. [DOI] [PubMed] [Google Scholar]
- 6.Kim HJ, Chalmers PN, Morris CD. Pediatric osteogenic sarcoma. Current opinion in pediatrics. 2010;22:61–6. doi: 10.1097/MOP.0b013e328334581f. [DOI] [PubMed] [Google Scholar]
- 7.Meyers PA, Healey JH, Chou AJ, Wexler LH, Merola PR, Morris CD, et al. Addition of pamidronate to chemotherapy for the treatment of osteosarcoma. Cancer. 2011;117:1736–44. doi: 10.1002/cncr.25744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daw NC, Chou AJ, Jaffe N, Rao BN, Billups CA, Rodriguez-Galindo C, et al. Recurrent osteosarcoma with a single pulmonary metastasis: a multi-institutional review. British journal of cancer. 2015;112:278–82. doi: 10.1038/bjc.2014.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eccles SA, Welch DR. Metastasis: recent discoveries and novel treatment strategies. Lancet. 2007;369:1742–57. doi: 10.1016/S0140-6736(07)60781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nature reviews Molecular cell biology. 2002;3:586–99. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- 11.Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nature reviews Molecular cell biology. 2010;11:276–87. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McClatchey AI. ERM proteins at a glance. Journal of cell science. 2014;127:3199–204. doi: 10.1242/jcs.098343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Briggs JW, Ren L, Nguyen R, Chakrabarti K, Cassavaugh J, Rahim S, et al. The ezrin metastatic phenotype is associated with the initiation of protein translation. Neoplasia. 2012;14:297–310. doi: 10.1593/neo.11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Celik H, Sajwan KP, Selvanathan SP, Marsh BJ, Pai AV, Saygideger Kont Y, et al. Ezrin binds to DEAD-box RNA helicase DDX3 and regulates its function and protein level. Molecular and cellular biology. 2015;35:3145–3162. doi: 10.1128/MCB.00332-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khanna C, Wan X, Bose S, Cassaday R, Olomu O, Mendoza A, et al. The membrane-cytoskeleton linker ezrin is necessary for osteosarcoma metastasis. Nature medicine. 2004;10:182–6. doi: 10.1038/nm982. [DOI] [PubMed] [Google Scholar]
- 16.Ren L, Khanna C. Role of ezrin in osteosarcoma metastasis. Advances in experimental medicine and biology. 2014;804:181–201. doi: 10.1007/978-3-319-04843-7_10. [DOI] [PubMed] [Google Scholar]
- 17.Clucas J, Valderrama F. ERM proteins in cancer progression. Journal of cell science. 2014;127:267–75. doi: 10.1242/jcs.133108. [DOI] [PubMed] [Google Scholar]
- 18.Ren L, Hong SH, Cassavaugh J, Osborne T, Chou AJ, Kim SY, et al. The actin-cytoskeleton linker protein ezrin is regulated during osteosarcoma metastasis by PKC. Oncogene. 2009;28:792–802. doi: 10.1038/onc.2008.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulut G, Hong SH, Chen K, Beauchamp EM, Rahim S, Kosturko GW, et al. Small molecule inhibitors of ezrin inhibit the invasive phenotype of osteosarcoma cells. Oncogene. 2012;31:269–81. doi: 10.1038/onc.2011.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaur K, Jain M, Reddy RP, Jain R. Quinolines and structurally related heterocycles as antimalarials. European journal of medicinal chemistry. 2010;45:3245–64. doi: 10.1016/j.ejmech.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Gamo FJ, Sanz LM, Vidal J, de Cozar C, Alvarez E, Lavandera JL, et al. Thousands of chemical starting points for antimalarial lead identification. Nature. 2010;465:305–10. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
- 22.Guiguemde WA, Shelat AA, Bouck D, Duffy S, Crowther GJ, Davis PH, et al. Chemical genetics of Plasmodium falciparum. Nature. 2010;465:311–5. doi: 10.1038/nature09099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meister S, Plouffe DM, Kuhen KL, Bonamy GM, Wu T, Barnes SW, et al. Imaging of Plasmodium liver stages to drive next-generation antimalarial drug discovery. Science. 2011;334:1372–7. doi: 10.1126/science.1211936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spangenberg T, Burrows JN, Kowalczyk P, McDonald S, Wells TN, Willis P. The open access malaria box: a drug discovery catalyst for neglected diseases. PloS one. 2013;8:e62906. doi: 10.1371/journal.pone.0062906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reczek D, Berryman M, Bretscher A. Identification of EBP50: A PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family. The Journal of cell biology. 1997;139:169–79. doi: 10.1083/jcb.139.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bosch J, Paige MH, Vaidya AB, Bergman LW, Hol WG. Crystal structure of GAP50, the anchor of the invasion machinery in the inner membrane complex of Plasmodium falciparum. Journal of structural biology. 2012;178:61–73. doi: 10.1016/j.jsb.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beauchamp EM, Ringer L, Bulut G, Sajwan KP, Hall MD, Lee YC, et al. Arsenic trioxide inhibits human cancer cell growth and tumor development in mice by blocking Hedgehog/GLI pathway. The Journal of clinical investigation. 2011;121:148–60. doi: 10.1172/JCI42874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paige M, Kosturko G, Bulut G, Miessau M, Rahim S, Toretsky JA, et al. Design, synthesis and biological evaluation of ezrin inhibitors targeting metastatic osteosarcoma. Bioorganic & medicinal chemistry. 2014;22:478–87. doi: 10.1016/j.bmc.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Westerfield M. The Zebrafish Book; A Guide for the Laboratoy Use of Zebrafish (Brachydanio rerio) University of Oregon Press; Eugene: 1993. [Google Scholar]
- 30.Link V, Carvalho L, Castanon I, Stockinger P, Shevchenko A, Heisenberg CP. Identification of regulators of germ layer morphogenesis using proteomics in zebrafish. Journal of cell science. 2006;119:2073–83. doi: 10.1242/jcs.02928. [DOI] [PubMed] [Google Scholar]
- 31.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Developmental dynamics : an official publication of the American Association of Anatomists. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 32.Mendoza A, Hong SH, Osborne T, Khan MA, Campbell K, Briggs J, et al. Modeling metastasis biology and therapy in real time in the mouse lung. The Journal of clinical investigation. 2010;120:2979–88. doi: 10.1172/JCI40252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced drug delivery reviews. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 34.Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. Journal of medicinal chemistry. 2002;45:2615–23. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 35.Marina N, Gebhardt M, Teot L, Gorlick R. Biology and therapeutic advances for pediatric osteosarcoma. The oncologist. 2004;9:422–41. doi: 10.1634/theoncologist.9-4-422. [DOI] [PubMed] [Google Scholar]
- 36.Hawkins PC, Skillman AG, Nicholls A. Comparison of shape-matching and docking as virtual screening tools. Journal of medicinal chemistry. 2007;50:74–82. doi: 10.1021/jm0603365. [DOI] [PubMed] [Google Scholar]
- 37.Christofori G. New signals from the invasive front. Nature. 2006;441:444–50. doi: 10.1038/nature04872. [DOI] [PubMed] [Google Scholar]
- 38.Steeg PS, Theodorescu D. Metastasis: a therapeutic target for cancer. Nature clinical practice Oncology. 2008;5:206–19. doi: 10.1038/ncponc1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iiizumi M, Liu W, Pai SK, Furuta E, Watabe K. Drug development against metastasis-related genes and their pathways: a rationale for cancer therapy. Biochimica et biophysica acta. 2008;1786:87–104. doi: 10.1016/j.bbcan.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kais B, Schneider KE, Keiter S, Henn K, Ackermann C, Braunbeck T. DMSO modifies the permeability of the zebrafish (Danio rerio) chorion-implications for the fish embryo test (FET) Aquatic toxicology. 2013;140-141:229–38. doi: 10.1016/j.aquatox.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 41.Zuccala ES, Baum J. Cytoskeletal and membrane remodelling during malaria parasite invasion of the human erythrocyte. British journal of haematology. 2011;154:680–9. doi: 10.1111/j.1365-2141.2011.08766.x. [DOI] [PubMed] [Google Scholar]
- 42.Parish LA, Mai DW, Jones ML, Kitson EL, Rayner JC. A member of the Plasmodium falciparum PHIST family binds to the erythrocyte cytoskeleton component band 4.1. Malaria journal. 2013;12:160. doi: 10.1186/1475-2875-12-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gould KL, Bretscher A, Esch FS, Hunter T. cDNA cloning and sequencing of the protein-tyrosine kinase substrate, ezrin, reveals homology to band 4.1. The EMBO journal. 1989;8:4133–42. doi: 10.1002/j.1460-2075.1989.tb08598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panichakul T, Ponnikorn S, Roytrakul S, Paemanee A, Kittisenachai S, Hongeng S, et al. Plasmodium vivax inhibits erythroid cell growth through altered phosphorylation of the cytoskeletal protein ezrin. Malaria journal. 2015;14:138. doi: 10.1186/s12936-015-0648-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.