Abstract
Time-of-flight secondary ion mass spectrometry (ToF-SIMS) three-dimensional (3D) depth profiling and a novel background subtraction method were used to localize polymeric nanoparticles within cells. Results showed that ToF-SIMS 3D depth profiling is capable of localizing polymer nanoparticles within HeLa cells. ToF-SIMS results compared well with optical images of cells incubated with fluorescently labeled polymer nanoparticles, with both imaging techniques demonstrating clustering of nanoparticles in punctate regions consistent with endosomal localization as anticipated based on the nanoparticle design.
I. INTRODUCTION
Polymeric nanoparticles have been widely explored for intracellular delivery of therapeutics.1–5 Through control of physicochemical properties (e.g., size, charge, and shape), nanoparticles can be used to facilitate intracellular uptake and controlled or triggered release of diverse drug cargo and can also be engineered with molecular specificity to improve cellular targeting. A major challenge in the design of polymeric nanoparticles, however, is controlling the intracellular trafficking to appropriate subcellular compartments, including the cytosol, nucleus, and mitochondria.5–8 Hence, characterizing the intracellular distribution of the polymers and cargo within cell is essential to evaluating the efficacy of polymeric drug delivery systems and to optimizing nanoparticle properties to achieve delivery to desired subcellular compartments. Uptake patterns of polymeric nanoparticles by cells can be visualized by various optical methods such as fluorescent imaging as long as the nanoparticles contain a suitable label. This enables facile localization of the particles in 2D; however, localization of the particles in 3D is more challenging and few methods enable localization combined with information specific to the chemical composition of both the particle and the surrounding milieu. Toward this end, we have employed the use of state-of-the-art ToF-SIMS 3D depth profiling to image a model polymeric nanoparticle in HeLa cells as ToF-SIMS has previously been shown to successfully image subcellular features in HeLa cells.9 The high spatial resolution and detailed chemical information provided by ToF-SIMS could enable not only spatial localization of particles, but also chemical characterization of the surrounding area and any chemical changes that occur throughout the cells. ToF-SIMS 3D imaging of cells has been demonstrated previously.9–12 Nanoparticle localization using SIMS has been demonstrated with inorganic species such as TiO2,13 and in one recent publication with polymer nanoparticles.14 However, those nanoparticles produced chemical signatures that were unique and could easily be visualized without further processing. That is not the case with the materials used in this report where significant overlap of nanoparticle and cell peaks occurred. Here, nanoparticles are micelles that are assembled using poly((trifluoroethyl methacrylate-co-poly(ethylene glycol) methacrylate)-block-dimethylacrylamide), p((tFEMA-co-PEGMA)-bl-DMA), an amphiphilic diblock polymer with a hydrophobic fluorine containing block, and a hydrophilic dimethylacrylamide (DMA) block. To enable optical localization of the nanoparticles a fluorescent tag was conjugated to an amine-reactive tetrafluorophenyl methacrylate (tFPMA) monomer doped into the second DMA block. The polymer nanoparticles used here lack endosomal escape functionality15,16 and, hence, are anticipated to be localized mostly within endosomal compartments. Therefore, we have used these nanoparticles as a control for endosomal localization for our ToF-SIMS investigations.
One potential complication of localizing nanoparticles in cells is the fact that the most polymeric nanoparticles are made predominantly of carbon, nitrogen and oxygen, which are the same elements that make up the components of cells. Furthermore, the structure of many polymers resembles the types of structures seen in biological systems. This can make it difficult to know whether the signal one observes with ToF-SIMS is due to the presence of nanoparticles or is intrinsic to the cell. Herein we propose a background subtraction method that enables isolation of signal from the polymer nanoparticles from signal associated with the cell.
II. EXPERIMENT
A. Polymer synthesis
Reversible addition-fragmentation chain-transfer (RAFT) copolymerization of poly(ethylene glycol) methacrylate (average Mn 360; PEGMA) of 2,2,2-trifuoroethyl methacrylate (tFEMA)was conducted under a nitrogen atmosphere in dioxane (10 wt. % monomer) at 70 °C for 24 h with 4-cyano-4-(phenylcarbonothioylthio)pentanoic acid and 4,4′-Azobis(4-cyanovaleric acid) (V-501) as the RAFT chain transfer agent (CTA) and initiator, respectively. An alumina column was used to remove inhibitors from all monomers. The initial molar ratio of PEGMA to tFEMA was 50:50, and the initial monomer ([M]o) to CTA ([CTA]o) to initiator ([I]o) ratio was 50:1:0.2. The resultant poly(PEGMA-co-tFEMA) macrochain transfer agent (mCTA) was dialyzed against three exchanges of acetone over 3 days followed by dialysis against water for 6 h. The mCTA was lyophilized and subsequently used for block extension with DMA or diblock copolymerization with DMA (90%) and tFPMA (10%). Monomers were added to the mCTA {([M]o/[mCTA]o = 300) dissolved in dioxane (40 wt. % monomer) along with V-501 at an mCTA to initiator ratio ([mCTA]o/[I]o} of 5. The polymerization was allowed to proceed under a nitrogen atmosphere for 3 h at 70 °C. The resultant diblock polymer was precipitated into cold ether, redissolved in acetone, and precipitated an additional three times. The purified product was dried in vacuo for 4 days.
To prepare fluorescently labeled conjugates, tFPMA groups were used to couple a cadaverine-functionalized fluorophore (AlexaFluor488; Life Technologies) to the polymer backbone. The coupling reaction was performed overnight in Dimethyl sulfoxide (DMSO) containing 3% (v/v) triethylamine. The resultant product was dissolved in water, gel filtration (PD-10 columns, GE Healthcare Life Sciences) was used to remove DMSO and unreacted dye, and the purified product was lyophilized. The degree of polymer labeling was quantified using UV–vis spectroscopy and determined to be ∼1 dye/polymer chain.
B. Polymer characterization
The composition of the purified mCTA and diblock copolymers was analyzed by 1H NMR (CDCl3) spectroscopy (Bruker AV 500). NMR spectra and peak assignments can be found in Fig. S1 (supplementary material).17 Gel permeation chromatography was used to determine the molecular weights and polydispersity of the mCTA and diblock copolymer.18,19 The molecular weight (Mn) of the first block was determined to be 15.1 kD with a polydispersity index (PDI) of 1.2 and the Mn and PDI of the diblock determined to be 23.7 kD and 1.1, respectively.
C. Nanoparticle formulation and characterization
Diblock copolymers were dissolved in water at 25 mg/ml and subsequently diluted into phosphate buffered saline (PBS) at 1 mg/ml. Particle size was measured using dynamic light scattering (Malvern Nanoseries Zetasizer).
D. Cell culture and sample preparation
HeLa cells, human cervical carcinoma cells (ATCC CCL-2) were maintained in minimal essential media (MEM) containing l-glutamine (Gibco), 1% penicillin–streptomycin (Gibco), and 10% fetal bovine serum (Gibco) at 37 °C and 5% CO2. Silicon substrates (approximately 1 × 1 cm2) were used as substrates for cell growth for ToF-SIMS imaging studies. Substrates were cleaned prior to use by a 5 min sonication, followed 2× rinses in dichloromethane, acetone, and methanol. Substrates were placed into wells of a 24-well tissue culture plate and 10 000 cells in 1 ml of MEM were added to substrates and cells allowed to adhere overnight. Cells were incubated with 0.1 mg/ml polymer for 24 h, noninternalized polymer was removed, and cells were washed 3× with PBS. The silicon substrate was then dipped in ammonium acetate for 1–2 s and subsequently incubated in 4% formaldehyde in PBS for 30 min. The samples were then immersed in ammonium acetate again for 30 s and allowed to air dry prior to ToF-SIMS analysis.
E. Fluorescent microscopy
HeLa cells (2500 cells/cm2) were seeded on microscope chamber slides (Thermo Scientific) in 500 μl of media and allowed to adhere overnight. AlexaFluor488-labeled polymer was added to cells to a final concentration of 100 μg/ml and incubated with cells for 24 h. Prior to imaging, nuclei were stained with Hoechst for 1 h, polymer was aspirated, and cells were washed 3× with Dulbecco's phosphate buffered saline (DPBS) and imaged using a Nikon Ti-E live-cell fluorescence microscope. Cells were imaged with a mercury lamp and a 40× objective using a 480/40 nm excitation wavelength and 535/50 nm emission wavelength filter set (49000 Series, Chroma Technology, Rockingham, VT).
F. ToF-SIMS depth profiling
ToF-SIMS dual beam depth profiling was performed using an IonTof ToF-SIMS 5 spectrometer using a 25 kV Bi3+ cluster ion source in the pulsed mode for imaging, and a 20 keV C60++ beam for sputtering.20 The images were acquired in negative ion mode over a mass range of m/z = 0 to 800 using a 100 × 100 μm area at a pixel density of 256 × 256. The images were acquired using the high spatial resolution mode with unit mass resolution spectra. The ion source was operated with at a current at the surface of 0.07 pA, and the primary ion dose per “slice” of the 3D profiles was ∼3 × 1011 ions/cm2. Secondary ions of a given polarity were extracted and detected using a reflectron time-of-flight mass analyzer. The negative ion spectra were mass calibrated using the CH−, OH−, and C2H− peaks. Sputtering was done over a 500 × 500 μm area with a current of 0.7 nA for 6 s, providing a sputter dose of ∼1 × 1013 ions/cm2. Sputtering through the cells took 70 cycles (image/sputter). Imaging and sputtering settings were kept the same between the control and nanoparticle exposed cells. The dose ratio of Bi to C60 was kept low to minimize issues with accumulated damage of the Bi beam.20 ToF-SIMS depth profiling data were reconstructed using the zcorrectorgui (Ref. 11) in the nbtoolbox (Dan Graham Ph.D., NESAC/BIO, University of Washington) in matlab (Mathworks, Natick, MA).
G. Background subtraction for ToF-SIMS 3D images
Since the chemical composition and structure of many polymers is similar in elemental composition to that found in cells and tissues, it can be challenging to isolate signal from polymer nano- or microparticles in biological systems. The overlap in chemical structures results in the same peaks being present in both the particles and the cells or tissues. This presents the challenge of determining if the signals seen in a ToF-SIMS data set are truly from the added particles or due to “background” from the cells or tissues. To overcome this problem, we have employed a background subtraction method to remove the background signal from the cells and leave any signal from the particles. This works under the hypothesis that the presence of the particles will cause an increase in the relative intensity of any “nanoparticle” peaks also present in the cells. Therefore, after subtracting the average background signal, one should be left with signal predominantly from the particles. It is likely that there will be some error in this assumption; however, we believe this error should be minimal since we are subtracting the average signal from the untreated cells that will overestimate the amount of background in some cases and underestimate it in others.
For the 3D depth profiles, we employed the following methodology. First, the 3D depth profiles were carried out on cells with nanoparticles and cells without nanoparticles using the same analysis and sputtering conditions. Second, each respective data set was normalized and scaled so that the image stack data for each given peak ranged from 1 to 255 counts. This enabled working on equal intensity scales between data sets. Finally, the average signal from the unexposed sample for a given peak, across all slices of the depth profile, was subtracted pixel by pixel across all slices for the same peak in the nanoparticle exposed sample. Using a peak by peak subtraction should help account for signals that vary between various cellular compartments since these variations should be consistent cell to cell. All resulting negative intensities were set to zero based on the assumption that any signal below the average would not be from nanoparticles since the presence of the nanoparticles should increase the relative intensity of the given peak(s). The result of this process is a data set with signal that should be representative of the nanoparticles. It is noted that this methodology assumes a constant sputter rate throughout the cells, which has been demonstrated previously with HeLa cells.9
III. RESULTS AND DISCUSSION
Here, ToF-SIMS depth profiling was utilized to enable facile localization of the polymeric nanoparticle in three dimensions via imaging and reconstruction of cells in three dimensions. The process starts from sequentially collecting an image, and then, sputtering away a layer of material until the entire cell has been ablated. The data can then be visualized by stacking the sequential images and reconstructing a 3D image. To reconstruct the data properly, one must take into account the fact that ToF-SIMS creates images that are a 2D projection of a 3D object. This reconstruction process can be done using the zcorrectorgui (Ref. 11) and has also been described by others.12 Once the data have been reconstructed, distribution of the target of interest in 3D can be determined by visualizing the peaks associated with the target.
Here, p((tFEMA-co-PEGMA)-bl-DMA), an amphiphilic diblock polymer with a hydrophobic fluorine-containing block and a hydrophilic DMA block, was utilized to form micelles (∼30 nm) in aqueous solution (Fig. S2, supplementary material17). Peak characteristics of the polymer nanoparticles were determined by acquiring high mass resolution spectra from a sample of just the polymer deposited onto a silicon wafer. Table I lists the negative secondary ion peaks determined to be characteristic of the polymer. Characteristic peaks were chosen as peaks that showed a high relative intensity in the control polymer and did not have other significant peaks within the same unit mass, as these peaks could provide false signals when doing high spatial resolution imaging with unit mass resolution. As seen in the table, the characteristic polymer peaks consisted mostly of fragments of the ethylene glycol portions of the polymers used in the nanoparticles and a possible nitrogen containing fragment that could be from the dimethylacrylamide.
Table I.
Characteristic peaks for polymer nanoparticles.
| Nanoparticle peaks (m/z) | Possible identity |
|---|---|
| 45.99 | CHO2 |
| 59.01 | C2H3O2 |
| 69.00 | C3HO2 or CF3 |
| 85.04 | C3H5N2O possible |
| 99.01 | Unknown |
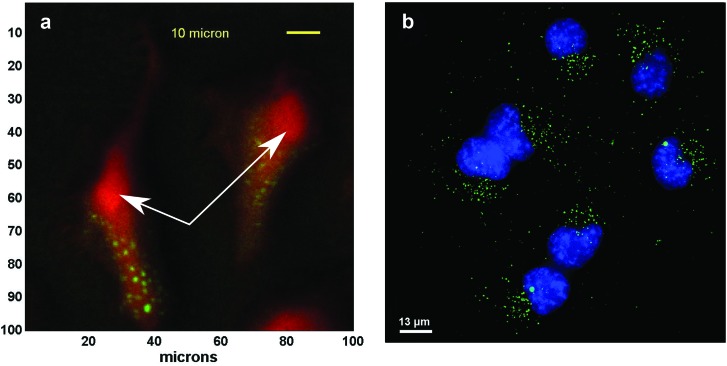
We demonstrate that ToF-SIMS depth profiling can generate cell images that are similar to fluorescence images. Figure 1 shows the top view of the summed signal from a ToF-SIMS 3D depth profile of a cell containing nanoparticles compared with a fluorescence image of similar cells. As seen in the figure, the ToF-SIMS data clearly show punctate spots (green) of polymer signal (summed signal from peaks in Table I) in the area around the nucleus of the cell (rounded area indicated by white arrows in the figure). The ToF-SIMS image is very similar to the fluorescence image, which shows the nanoparticle clusters (green) located around the cell nuclei (blue). It should be noted that the cell nuclei are not directly shown in the ToF-SIMS image and the cell membrane is not seen in the fluorescence image.
Fig. 1.
ToF-SIMS 3D and fluorescence images of cells with nanoparticles. Top view of summed signal from a ToF-SIMS 3D depth profile (a) and fluorescence image (b). Nanoparticle clusters (green—polymer peaks) can be seen within the cells (red—CN− peak in ToF-SIMS image). The particles are located outside of the cell nuclei (blue in fluorescence image, and red rounded areas in ToF-SIMS image). The white arrows indicate the location of the nuclei in the ToF-SIMS images.
The appearance of bright punctate spots in the fluorescence image is consistent with polymers localized within endosomal compartments [additional fluorescence images are provided in Fig. S3 (Ref. 17)]. These polymers lack any membrane-destabilzing activity and therefore are almost exclusively localized within endosomes with minimal delivery to the cytosol.21–23 For ToF-SIMS, the cell nuclei were identified as the thickest area of the cell where the phosphate signals (m/z 63 and 79) typically had a higher relative intensity in the summed depth profile, and where nanoparticle signals were not seen since the polymers used have been shown to become trapped in endosomes.
It is noted that though areas showing nanoparticle signal are seen in the summed depth profile image (Fig. 1), it is not clear that all of the signals from those peaks originates from the nanoparticles as some signal may also come from the cell. This overlapping signal can be seen by the slight yellow tones around the nanoparticle clusters indicating overlap between polymer and cell signals. It is also noted that due to the spatial resolution used to acquire the ToF-SIMS images (∼1 μm), it is likely that only clusters of nanoparticles are detected. However, since the nanoparticles are designed to be trafficked into endosomes, they majority of the polymer should be in relatively large clusters.
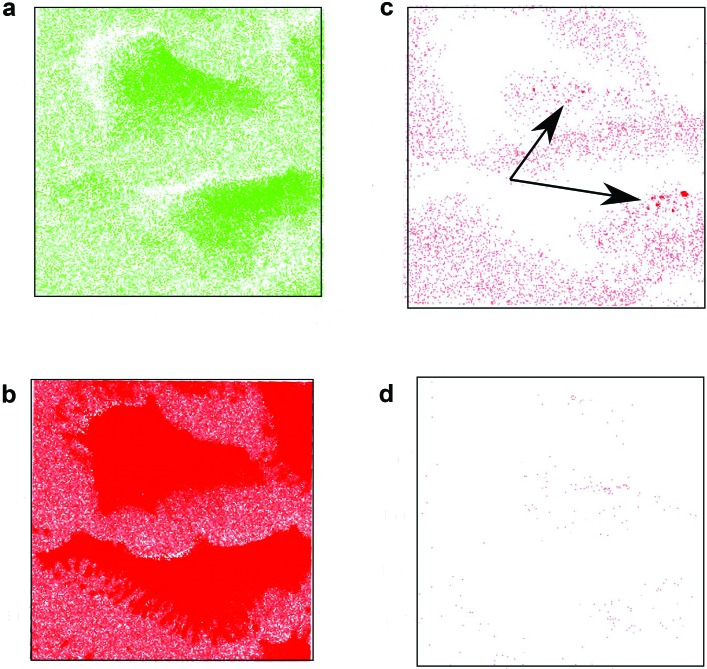
To distinguish signal specifically associated with the polymeric nanoparticles from the signal from the cell, a background subtraction routine was implemented. This involved subtracting the average signal for a given peak from a control cell without nanoparticles from each voxel of the 3D data from a cell with nanoparticles. Figure 2 shows a series of images from a 3D depth profile of cells treated with nanoparticle before and after carrying out the background subtraction. As can be seen in the figure, after background subtraction the signals characteristic of cellular components disappear [Fig. 2(d)], while characteristic nanoparticle peaks show several punctate spots along with some diffuse signal inside and outside the cells [Fig. 2(c)]. This suggests that the background subtraction method is working properly. The peaks corresponding to cellular components are present in both cells in Fig. 2, and therefore, after background subtraction, little to no signal remains [Fig. 2(d)]. In contrast, peaks corresponding with the polymer nanoparticles still show signal in areas where the particles are present after background subtraction [Fig. 2(c)]. This is due to the fact that the relative intensity of these peaks is higher in areas with nanoparticles. The polymer signal outside the cells is presumably from nanoparticles that have adsorbed to the silicon substrate. Figure S4 (Ref. 17) shows the same background subtraction using two untreated cells. As expected, after background subtraction, almost all signals are gone from both the polymer and cell related peaks.
Fig. 2.
ToF-SIMS data before and after background subtraction. Images from before [(a) and (b)] and after [(c) and (d)] for polymer [(a) and (c)] and cell [(b) and (d)]. The arrows highlight the location of the punctate spots in the polymer image after background subtraction. The polymer signal is the sum signal from the peaks in Table I. The cell signal is m/z 26 (CN). Similar results are obtained for other cell related peaks.
In addition to showing the polymeric nanoparticles inside of the cells in 2D, the ToF-SIMS depth profiles can also show the nanoparticles inside of the cells in 3D. Though other methods can image in 3D, ToF-SIMS brings the additional capability of chemical imaging. This could enable not only localization of nano- or microparticles in 3D, but also determination of the chemical changes occurring around the particles. Furthermore, since a full mass spectrum is obtained during a depth profile, one can do retrospective analysis to determine other chemical changes occurring throughout the cell. Figure 3 shows 3D renders of the cells with nanoparticles produced using blender (www.blender.org). The 3D cell data were processed and corrected using the nbtoolbox zcorrectorgui (available at http://www.nb.uw.edu/mvsa/multivariate-surface-analysis-homepage). The data were then exported as individual images for each slice as .jpg files that were imported into blender and used as the texture channel to produce volumetric renders of the cells and nanoparticles. To produce images in Fig. 3, the nonbackground subtracted cell peak data were overlaid with the background subtracted nanoparticle data. The nonbackground subtracted data for m/z 26 (CN−) were used due to the fact that after the background subtraction all cell related peaks show little to no signal as discussed above. As can be seen in the 3D data sets (Fig. 3), the nanoparticles appear to be localized outside of the cell nuclei but within the cells. The punctate spots seen are most likely agglomerated nanoparticles within endosomal compartments consistent with the fate of endocytosed nanoparticles that lack endosomal escape properties.21,23–25 The yellowish tint around the particles is now gone since the overlapping polymer and cell signals have been removed. It should be noted that in order to accurately reconstruct the data in 3D one should account for any sputter rate differences between the polymer and cell. We did not notice any direct signs of a sputter rate difference, such as increased height in reconstructed regions where the nanoparticles are located so we did not explore the sputter rates of the nanoparticles and cells in this study. However, even if there was a difference in sputter rate, it would not change the conclusions about the polymer localization, and it would only change the relative position of the voxels containing nanoparticle signal. Furthermore, correcting for this type of sputter rate difference in 3D is nontrivial.
Fig. 3.
Three-dimensional reconstructed ToF-SIMS data of nanoparticles in cells. Red indicates cell, and green indicates nanoparticles. (a) Side view, (b) side view cut halfway through cells, and (c) top view. From previous data using similar sputter conditions, the sputter rate through similar cells was around 10.8 nm/1.25 × 1013 ions/cm2 (Ref. 11). This gives an estimated cell thickness of ∼0.6 μm.
IV. SUMMARY AND CONCLUSIONS
The high spatial resolution imaging and depth profiling capabilities of ToF-SIMS presents great promise for localization of nano- and microparticles within cells. The ability to track these particles can aid in understanding how these particles traffic within cells and could provide further insight into the local chemical changes occurring due to the presence of the particles. In this study, we have demonstrated a background subtraction method that enables isolation of signal corresponding to polymeric nanoparticles and overcomes the challenge of signal overlap with cellular components. Successful localization of clusters of nanoparticles within HeLa cells was also demonstrated and shown to corroborate with fluorescence images of the nanoparticles within cells. As expected, nanoparticles appeared to be localized within endosomal compartments within the cells. Future work will focus on use of ToF-SIMS to compare the intracellular distribution of nanoparticles with and without endosomal escape capabilities.
The background subtraction method presented in this study represents a simple, easy to implement method that effectively highlights the location of the nanoparticle signals. This method should be applicable to any system where the molecules of interest produce peaks that overlap with the host system or substrate. Other possible methods to deal with this type of overlap could include the use of multivariate statistics, or a combination of background subtraction and multivariate analysis. A comparison with these other methods is planned for future work.
ACKNOWLEDGMENTS
The authors would like to thank Geoffrey Berguig (University of Washington) for assistance with fluorescent microscopy and Max Jacobson (Vanderbilt University) for assistance with dynamic light scattering data acquisition. The authors are grateful to the Irvington Institute Fellowship Program of the Cancer Research Institute (J.T.W.) for funding this research. The authors gratefully acknowledge support of the research by NIH Grant No. EB-002027 to NESAC/BIO.
References
- 1. Hubbell J. A. and Langer R., Nat. Mater. 12, 963 (2013). 10.1038/nmat3788 [DOI] [PubMed] [Google Scholar]
- 2. Cheng C. J., Tietjen G. T., Saucier-Sawyer J. K., and Saltzman W. M., Nat. Rev. Drug Discovery 14, 239 (2015). 10.1038/nrd4503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Petros R. A. and DeSimone J. M., Nat. Rev. Drug Discovery 9, 615 (2010). 10.1038/nrd2591 [DOI] [PubMed] [Google Scholar]
- 4. Davis M. E., Chen Z., and Shin D. M., Nat. Rev. Drug Discovery 7, 771 (2008). 10.1038/nrd2614 [DOI] [PubMed] [Google Scholar]
- 5. Pack D. W., Hoffman A. S., Pun S., and Stayton P. S., Nat. Rev. Drug Discovery 4, 581 (2005). 10.1038/nrd1775 [DOI] [PubMed] [Google Scholar]
- 6. Malhi S. S. and Murthy R. S. R., Expert Opin. Drug Delivery 9, 909 (2012). 10.1517/17425247.2012.694864 [DOI] [PubMed] [Google Scholar]
- 7. Ge Z. S. and Liu S. Y., Chem. Soc. Rev. 42, 7289 (2013). 10.1039/c3cs60048c [DOI] [PubMed] [Google Scholar]
- 8. Fu A. L., Tang R., Hardie J., Farkas M. E., and Rotello V. M., Bioconjugate Chem. 25, 1602 (2014). 10.1021/bc500320j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brison J., Robinson M. A., Benoit D. S. W., Muramoto S., Stayton P. S., and Castner D. G., Anal. Chem. 85, 10869 (2013). 10.1021/ac402288d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fletcher J. S., Lockyer N. P., Vaidyanathan S., and Vickerman J. C., Anal. Chem. 79, 2199 (2007). 10.1021/ac061370u [DOI] [PubMed] [Google Scholar]
- 11. Robinson M. A., Graham D. J., and Castner D. G., Anal. Chem. 84, 4880 (2012). 10.1021/ac300480g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Breitenstein D., Rommel C. E., Stolwijk J., Wegener J., and Hagenhoff B., Appl. Surf. Sci. 255, 1249 (2008). 10.1016/j.apsusc.2008.05.289 [DOI] [Google Scholar]
- 13. Angerer T. B. and Fletcher J. S., Surf. Interface Anal. 46, 198 (2014). 10.1002/sia.5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kokesch-Himmelreich J., Woltmann B., Torger B., Rohnke M., Arnhold S., Hempel U., Muller M., and Janek J., Anal. Bioanal. Chem. 407, 4555 (2015). 10.1007/s00216-015-8647-9 [DOI] [PubMed] [Google Scholar]
- 15. Shete H. K., Prabhu R. H., and Patravale V. B., J. Nanosci. Nanotechnol. 14, 460 (2014). 10.1166/jnn.2014.9082 [DOI] [PubMed] [Google Scholar]
- 16. Convertine A. J., Benoit D. S. W., Duvall C. L., Hoffman A. S., and Stayton P. S., J. Controlled Release 133, 221 (2009). 10.1016/j.jconrel.2008.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.See supplementary material at http://dx.doi.org/10.1116/1.4934795E-BJIOBN-11-304602 for NMR Spectra and peak assignments, additional fluorescense images and background subtraction examples.
- 18. Wilson J. T., Keller S., Manganiello M. J., Cheng C., Lee C. C., Opara C., Convertine A., and Stayton P. S., ACS Nano 7, 3912 (2013). 10.1021/nn305466z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Keller S., Wilson J. T., Patilea G. I., Kern H. B., Convertine A. J., and Stayton P. S., J. Controlled Release 191, 24 (2014). 10.1016/j.jconrel.2014.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brison J., Muramoto S., and Castner D. G., J. Phys. Chem. C 114, 5565 (2010). 10.1021/jp9066179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Duvall C. L., Convertine A. J., Benoit D. S. W., Hoffman A. S., and Stayton P. S., Mol. Pharmaceutics 7, 468 (2010). 10.1021/mp9002267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berguig G. Y., Convertine A. J., Shi J., Palanca-Wessels M. C., Duvall C. L., Pun S. H., Press O. W., and Stayton P. S., Mol. Pharmaceutics 9, 3506 (2012). 10.1021/mp300338s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Convertine A. J., Diab C., Prieve M., Paschal A., Hoffman A. S., Johnson P. H., and Stayton P. S., Biomacromolecules 11, 2904 (2010). 10.1021/bm100652w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shi B. and Abrams M., J. Histochem. Cytochem. 61, 407 (2013). 10.1369/0022155413484152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sahay G. et al. , Nat. Biotechnol. 31, 653 (2013). 10.1038/nbt.2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1116/1.4934795E-BJIOBN-11-304602 for NMR Spectra and peak assignments, additional fluorescense images and background subtraction examples.