Abstract
The paper is focused on a tiSsue-Based Standardization Technique (SBST) of magnetic resonance (MR) brain images. Magnetic Resonance Imaging intensities have no fixed tissue-specific numeric meaning, even within the same MRI protocol, for the same body region, or even for images of the same patient obtained on the same scanner in different moments. This affects postprocessing tasks such as automatic segmentation or unsupervised/supervised classification methods, which strictly depend on the observed image intensities, compromising the accuracy and efficiency of many image analyses algorithms. A large number of MR images from public databases, belonging to healthy people and to patients with different degrees of neurodegenerative pathology, were employed together with synthetic MRIs. Combining both histogram and tissue-specific intensity information, a correspondence is obtained for each tissue across images. The novelty consists of computing three standardizing transformations for the three main brain tissues, for each tissue class separately. In order to create a continuous intensity mapping, spline smoothing of the overall slightly discontinuous piecewise-linear intensity transformation is performed. The robustness of the technique is assessed in a post hoc manner, by verifying that automatic segmentation of images before and after standardization gives a high overlapping (Dice index >0.9) for each tissue class, even across images coming from different sources. Furthermore, SBST efficacy is tested by evaluating if and how much it increases intertissue discrimination and by assessing gaussianity of tissue gray-level distributions before and after standardization. Some quantitative comparisons to already existing different approaches available in the literature are performed.
Keywords: General intensity scale, Magnetic Resonance Imaging, Nonlinear registration, Intensity standardization, Alzheimer’s Disease Neuroimaging Initiative
Introduction
Magnetic resonance images from different sites and scanners are used extensively in medical and clinical research. They bring interesting challenges for image analysis algorithms [1], as well as for diagnosis and development of strategies of various disease treatments [2].
However, many problems can affect the results especially in a large multisite clinical study [3], where differences in subject positioning between sites or a baseline and a later scan, or protocol [4, 5] can be found, making the interpretation difficult [3].
As highlighted by Jäger and Hornegger [6], the lack of a standard intensity scale has no direct impact on medical diagnosis by experts but, when sophisticated automatic segmentation and quantification methods are needed, standardization of the observed image intensities is of crucial importance.
Moreover, most of the supervised parametric lesion identification and tissue type segmentation approaches (both automatic and semiautomatic) applied to brain Magnetic Resonance Imaging (MRI) volumes rely, explicitly or implicitly, on strong assumptions regarding the shape of the underlying distribution of various tissue type intensities. These assumptions require images to have standardized intensity ranges in order not to compromise the accuracy and efficiency of many image analysis applications in the medical field [6–8].
Furthermore, currently, a new class of hybrid imaging systems combining MR and positron emission tomography (PET) is being developed, and, in order to increase the PET image quality, a standardized attenuation correction utilizing the MR data has to be performed. For this purpose, the MR intensities have to be mapped to attenuation coefficients which correlate to tissue classes [8].
In the literature, many attempts to achieve intensity standardization by image histogram adjustment were published. Interesting and extensive reviews were given by Madabhushi et al. [2], Jäger and Hornegger [6], and Shah et al. [8]. Other approaches are reported by Christensen [9], who uses even-ordered derivatives of the image histogram to determine a single global scaling factor between two images, or by Weisenfeld and Warfield [10], who propose the use of Kullback-Leibler divergence to match the intensity distribution of two images. Leung et al. [1] proposed a semiautomated segmentation technique that delineates CSF/WM/GM tissue components, for which they computed mean intensities. However, this technique yields a linear transformation, which does not completely address the problem, guaranteeing the standardization of spatially corresponding tissue intensities [11].
In the study of Jäger and Hornegger [6], the properties of all acquired images (e.g., T1- and T2-weighted images) are stored in multidimensional joint histograms. In order to normalize the probability density function of a newly acquired dataset, a nonrigid image registration is performed between the joint histogram of a reference and the joint histograms of the acquired images, avoiding any prior registration or segmentation of the datasets [6].
This paper belongs to the field of intensity standardization by image histogram adjustment. The aim is to give insight on MRI standardization described by Cataldo et al. [12], as a part of a research, in which brain template generation for Alzheimer’s disease, using clusterization methods is described.
Against that background, many aspects regarding image standardization are deeper explained and results are compared with other approaches available in the literature.
Standardization techniques that employ histograms are indeed largely used in the literature [11, 13–16], we attempted to improve them, enhancing robustness, by considering tissue-specific information. The main novelty consists of computing three piecewise-linear standardizing transformations for the three main brain tissues, for each tissue class separately. In order to create a continuous intensity mapping, the three transformations are combined and spline smoothing of the overall slightly discontinuous intensity transformation is performed.
In this way, we are able to obtain similar gray values for comparable tissue classes. Our standardization procedure is hereafter called tiSsue-Based Standardization Technique (SBST).
The technique robustness was before all assessed through two indicators: the Dice index, as a measure of the overlap between tissue masks segmented before and after standardization, and the mean absolute error (MAE).
MAE was calculated on different voxel sets of the single brain tissues and the corresponding template images, in case of SBST and with other standardization procedures available in the literature [11, 14].
Dice index gave a very high (over 0.9) score, even for images belonging to diseased subjects.
Then, we used the calculation of Jeffreys divergence to show how our standardization technique increases intertissue discrimination as compared to nonstandardized (NS) images, and to images standardized with the method described in [15], hereafter called L4.
Finally, SBST efficacy was tested by assessing gaussianity of tissue gray-level distributions before and after standardization.
According to our tests, SBST intensity standardization contributes to a better scale mapping of the various tissue types, in comparison with the NS and the L4 standardized images.
We processed images belonging to nondemented and demented older adults, characterized by clinical conditions ranging from good health state (normal) to probable dementia of AD type as well as with mild cognitive impairment (MCI), available from large, public datasets of MR brain images.
We also evaluated the technique on synthetic images with different amounts of noise.
Though SBST appears as a good standardizing method, some limits of the procedure are present. They are exposed in the “Discussion” section.
Materials and Methods
Materials
A consistent number (over 500) of MR brain images were employed to develop the procedure detailed in the next paragraphs.
They can be divided into the following:
MRIs of human brain, available from public databases, such as the Alzheimer’s Disease Neuroimaging Initiative (ADNI, Web site http://www.loni.ucla.edu/ADNI/) and the Open Access Series of Imaging Studies (OASIS, Web site http://www.oasis-brains.org/)
Synthetic MRIs, available from the Brainweb, McConnell Brain Imaging Centre (BIC) of the Montreal Neurological Institute, McGill University (Web site http://www.bic.mni.mcgill.ca/brainweb/
The ADNI initiative was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies, and nonprofit organizations, as a $60 million, 5-year public-private partnership. The primary goal of ADNI has been to test whether serial MRI, positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early Alzheimer’s disease (AD). Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials.
The principal investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California–San Francisco. ADNI is the result of efforts of many coinvestigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the USA and Canada. The initial goal of ADNI was to recruit 800 subjects, but ADNI has been followed by ADNI-GO and ADNI-2. To date, these three protocols have recruited over 1500 adults, ages 55 to 90, to participate in the research, consisting of cognitively normal older individuals, people with early or late MCI, and people with early AD. The follow-up duration of each group is specified in the protocols for ADNI-1, ADNI-2, and ADNI-GO. Subjects originally recruited for ADNI-1 and ADNI-GO had the option to be followed in ADNI-2 (for up-to-date information, see www.adni-info.org).
In this study, we used T1-weighted MP-RAGE protocol ADNI images belonged to men and women, aged 62 to 98 with a resolution of 256 × 256 × 166 and a slice thickness of 1 mm. Each image has undergone specific image preprocessing correction steps, including gradwarp, B1 and N3 correction (http://www.loni.ucla.edu/ADNI/).
The Open Access Series of Imaging Studies (OASIS) is a project aimed at making MRI datasets of the brain freely available to the scientific community. The aim is to facilitate future discoveries in basic and clinical neuroscience [17]. OASIS is made available by the Washington University Alzheimer’s Disease Research Center, Harvard University, the Neuroinformatics Research Group (NRG) at Washington University School of Medicine, and the Biomedical Informatics Research Network (BIRN) [17].
Longitudinal T1-weighted MRI data in nondemented and demented older adults were used. This set consists of a longitudinal collection of 150 subjects aged 60 to 96 with a resolution of 181 × 217 × 181 and a slice thickness of 1 mm. Each subject was scanned on two or more visits, separated by at least 1 year for a total of 373 imaging sessions (http://www.oasis-brains.org/). The subjects include both men and women, characterized by clinical conditions ranging from good health state (normal) to probable dementia of AD type as well as with MCI. The mini-mental state examination (MMSE) test was used to estimate the severity of the cognitive impairment.
From the OASIS database, we selected a subset comprising 30 subjects, with anonymized T1-weighted MP-RAGE protocol data. Subjects were scanned two different times, giving a total of 60 images.
Ten synthetic MRIs from the Brainweb (Web site http://www.bic.mni.mcgill.ca/brainweb/) were added to the employed dataset, allowing us to evaluate the performance of standardization techniques, including intersubject intensity variations and better focusing on interscanner differences.
Throughout this study, registration and segmentation tasks were performed with respect to two templates, i.e., the MNI152 and the COLIN27.
The MNI152 is the average of 152 normal MRI scans that have been matched to the MNI305 using a nine-parameter affine transform, and the COLIN27 is a high resolution (1-mm3 isotropic), high signal-to-noise average of 27 T1-weighted images of a single human brain. CSF, GM, and WM masks were available for both the templates.
Automatic registration and segmentation of the images, before and after standardization, was performed by using two popular software tools: FMRIB an open-source tool of the Oxford University Software Library (FSL, available at http://fsl.fmrib.ox.ac.uk/fsl) and the Statistical Parametric Mapping (SPM, Wellcome Dept. of Imaging Neuroscience, London, available at www.fil.ion.ucl.ac.uk/spm).
In this regard, we considered the results of Klein et al. [18] that evaluated 14 registration and segmentation methods, concluding that FSL and SPM-DARTEL Toolbox gave the most consistently high accuracy across subjects.
In particular, using the FSL package, FNIRT module registers the brain volume to the standard space (MNI152 or COLIN27), using a priori tissue probability maps. Then, segmentation is performed by the FMRIB’s Automated Segmentation Tool (FAST). The latter segments the brain volume into the three main tissue classes (GM, WM, CSF) while also correcting for spatial intensity variations.
FAST requires as input a skull-stripped version of the image (running BET tool from the FSL package) and is based on a hidden Markov random field model with an associated expectation-maximization algorithm. The whole process is fully automated, producing a probabilistic and/or partial volume tissue segmentation.
The SPM-DARTEL Toolbox allows both registration and segmentation. It is based on a paper by Ashburner [19], starting from the idea of nonlinearly registering images by computing a “flow field” which can then be “exponentiated” to generate both forward and backward deformations. This procedure is repeated a number of times, writing out rigidly transformed versions of the tissue class images, such that they are in as close alignment as possible with the tissue probability maps.
Both tools are quite fast. Currently, they take less than 10 min to segment a 256 × 256 × 166 volume on the most common personal computer (Intel G640@2.80GHz).
The whole SBST procedure is currently developed under MATLAB, a high-level technical-computing language (http://www.mathworks.com/products/matlab).
Intensity Standardization
The presented SBST standardization technique starts from the gray-level standardization method addressed by Nyúl and Udupa, Nyúl et al., and Ge et al. works [13, 16], with some important differences discussed hereafter.
The original method is well known [13–16], and we briefly recall that it is a two-step approach. The first step (“training step”) involves finding the parameters of the standardizing transform from a set of images, by defining a set of landmarks in the image histograms. Thus, a continuous, piecewise-linear intensity mapping to a standard scale is achieved. The second step (“transformation step”) applies the learnt transformation to the intensity of each training set image and of any new image into the standardized grayscale: each image is standardized by projecting its landmarks onto the standard ones, while the gray levels between the landmarks are linearly interpolated.
In the original paper by Nyúl and Udupa [18], the landmarks were mode-based, i.e., the local maxima of the histogram were used. In their subsequent work, they chose a set of population percentiles instead, in order to make the method more robust and avoid incorrect standard scales. In fact, as pointed out by the authors, it might happen that a particular mode corresponded, in two images A and B, to different matters (e.g., WM in image A, GM in image B). In this case, the mode should not be used as a landmark, because it would lead to tissue mixing, as different tissues would be projected to the same “standard” levels. The consequence of training with such landmarks would be to obtain a meaningless standard scale.
In the decile formulation of the standardization method, deciles are chosen as the histogram landmarks, giving the intensity-landmark configuration CL as follows:
where plow = 1 and phigh = 99, each mi, i = {10, 20, …, 90} denotes the ith percentile of the histogram associated with the foreground part of the image with mode m.
A graphical illustration, directly derived from the literature [13–16], is provided in Fig. 1a.
Fig. 1.
A graphical illustration of the L4 method, directly derived from the literature (a). Histogram landmarks for the three tissues (CSF, GM, WM) in the tissue-based (SBST) and L4 standardization (b). A spline fitting of the SBST curve is shown (600 × 194 mm (300 × 300 DPI))
Our technique consists of these main steps:
Choice of a training set of images, from the original (hereafter called NS) MRI scans
Segmentation of GM, WM, and CSF tissue images for each member of the training set.
Calculation of the gray-level histogram for each tissue and for each training image.
Computing of three standardizing transformations for the three main brain tissues, for the training set images, similarly to [13–16], but for each tissue class separately. In order to create a continuous intensity mapping, as the three transformations do not exactly overlap in the two gray-value ranges shared by different brain tissues, spline smoothing of the overall slightly discontinuous piecewise-linear intensity transformation is performed.
Application of the standardizing transformation to each member of the training set and to other NS images, giving as output the SBST-standardized images.
The first issue regards how to construct the training set of images. In fact, the images are significantly different anatomically, and there is a big variance in the localization of the three main tissues. The major part of them belong to large, public datasets and encompass subjects characterized by clinical conditions ranging from good health state (normal) to probable dementia of AD type as well as with MCI. Before signal intensity standardization, the proper set of training histograms has to be chosen, checking that they are as representative of population variability as possible [12, 16]. In this regard, we took into account the considerations addressed by Cataldo et al. [12], highlighting that the number of images able to describe the population is lower for patients with homogeneous clinical conditions than with mixed degrees of neuropathology.
In the second step, the segmentation task is performed automatically, by the proper module available in SPM-DARTEL [19].
The gray-level standardization step represents the first and most important difference with respect to Nyúl and Udupa, Nyúl et al., and Ge et al. works [13–16], their standardization method being hereafter called L4, as in [15].
In L4, gray-level standardization is obtained by selecting for each image of a training set some histogram landmarks, averaging them to obtain a list of reference mean landmarks to be used as a standard scale. Each training set image is then standardized, by projecting its landmarks onto the standard ones, while the gray levels between the landmarks are linearly interpolated. Thus, a continuous, piecewise-linear intensity mapping to a standard scale is achieved. Unfortunately, as observed by Cataldo et al. [12], by this procedure, tissue “mixing” could happen.
In order to reduce this possibility, we propose a variant in which three independent standardizing transformations are calculated, after segmenting the training images into WM, GM, and CSF tissues.
Gray-level standardization is performed after taking into account that, because of signal intensity outliers, it is not advisable to use the full intensity range, but only the range up to the 99.8 % intensity percentile. Moreover, deciles are chosen as the histogram landmarks, so as to have a smooth map function (Fig. 1b).
In this way, we consider and solve the problems highlighted in [6], i.e., tissue classes with a small number of voxels could not be correctly transformed. Consequently, it is no longer possible to find a plausible global transformation of the intensity [6]. One straightforward solution to this is to split the datasets into smaller subvolumes, represented in our case by the different tissues, and intensity-standardize separately [6].
Due to the independent standardization of the tissues, intensity discontinuities can occur at the two common gray-value ranges.
Smoothing of the piecewise-linear intensity transformations, achieved by a spline function, gives a fitting that closely follows the transformation shape, just avoiding discontinuities.
A large set of landmarks composed by deciles chosen in the histograms of each of the three tissues, as said above, permits to achieve a “standard scale” used to standardize each image in the training dataset and other images.
Figure 1b gives an example of a SBST standardizing transformation, in comparison with the transformation calculated with the L4 procedure.
Results
This paragraph consists of four subsections which explain the assessment of the results and the metric applied for quantitative evaluation of the performance of the proposed technique.
Comparison with the L4 Procedure
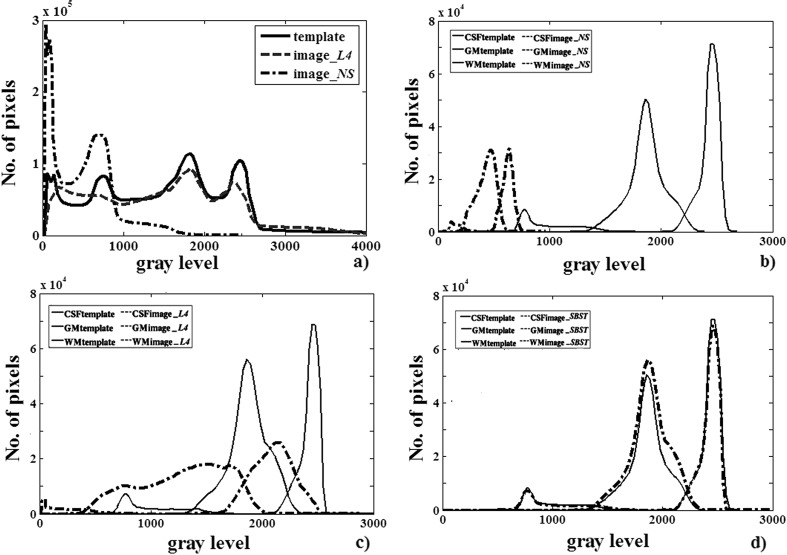
Some histograms before and after standardization, with both the L4 and SBST procedures, with respect to the COLIN27 template, are shown in Fig. 2.
Fig. 2.
An example of image standardization. Histograms of the template and the whole L4-standardized image (a). Histograms of the template and the three tissues (CSF, GM, WM): nonstandardized (NS) (b), L4-standardized (c), and SBST-standardized (d) histograms, according to the transformation in Fig. 1b (189 x 123 mm (300 x 300 DPI))
This figure shows (Fig. 2a) that the chosen image was apparently standardized with success by L4. However, once the fat, bone, and background are removed and the images are segmented in the three fundamental tissues, a different situation is represented (Fig. 2b–d).
In particular, Fig. 2b compares the template and the image histograms before any standardization: they look quite different in shape and actually need intensity standardization. In Fig. 2c, the three matters are shown after L4 standardization (this is the clean equivalent of Fig. 2a): no correspondence exists, even if in Fig. 2a standardization looked satisfying. Finally, Fig. 2d shows that SBST correctly and cleanly standardized each tissue.
This confirms, at the histogram level, the robustness of the SBST approach, in which three standardizing transformations are separately calculated on the GM, WM, and CSF images, and then fused, giving histograms with similar shape for the same tissues.
Thus, as already shown in [12], the possibility of tissue “mixing” is reduced.
Comparison Between NS and SBST-Standardized Images
We now demonstrate the robustness of the SBST technique, by comparing WM/GM/CSF segmentations of SBST-standardized images with the corresponding segmentations of the NS segmented images, taken as a gold standard. This is a fundamental issue, as solely pure histogram comparisons of SBST with previously published methods, already presented in this section, could be by no means informative for the robustness of the technique in terms of brain tissue type segmentation. So, spatially specific comparison of transformed images with a gold standard is needed.
It could be happen, in fact, that anatomical match of resulting images with a gold standard anatomy could be low because negatively affected by the smoothed piecewise-linear intensity transformations employed in standardization.
The Dice overlap metric was used to measure the similarity of the segmented results across the two image types, i.e., between the NS segmented images, considered as a gold standard, and the SBST segmented ones. This cross-validation is largely used in the literature, providing a simple yet effective way to compare the consistency of the segmentation especially between images segmented with different algorithms.
Furthermore, considering that the used MRI scans contained pathological images belonging to MCI and AD subjects too, the Dice overlap is evaluated for each different pathology, in order to assess if the SBST technique respects or negatively affects lesion morphology.
Dice index between tissue masks was measured for the whole dataset, after automatic segmentation of the tissues, before and after SBST standardization, with the proper tools available in FSL and in SPM-DARTEL packages.
A typical box plot of the Dice index is drawn in Fig. 3, evaluated for 250 ADNI MRIs, segmented with the FAST. The index is very high (over 0.9) for each tissue class, and more important, it gives high values even for pathological images belonging to MCI and AD subjects.
Fig. 3.
Box plot of the Dice index for 250 ADNI MRIs with different degree of neurodegenerative pathology, segmented with the FMRIB’s Automated Segmentation Tool (FAST) (414 x 190mm (300 x 300 DPI))
The result is independent on the employed segmentation tool, and small differences in the Dice index may depend on the peculiarities of the segmentation algorithm [20, 21].
Dice index was then measured between the NS segmented images and the L4-standardized segmented ones, obtaining best values around 0.6.
However, for every degree of neurodegenerative pathology, we observed that the CSF tissue class exhibits a greater spread, probably since CSF accounts for a small portion of voxels in total brain matter; thus, even slight variations in the CSF can yield large overlap errors [21].
In Fig. 4, a sample image without standardization and with the SBST and the L4 intensity standardization, and their histograms, are given.
Fig. 4.
A brain image, non-standardized (NS), SBST, and L4 intensity standardized, and their histograms (339 x 388 mm (300 x 300 DPI))
Comparison by Using the MAE
Here, the SBST results are compared with those obtained by its parent standardization technique, i.e., L4, and another tissue-based standardization technique called STandardization of Intensities (STI) [11]. The measure employed is the voxel-wise MAE [11], computed on different voxel sets.
The MAE for each image is defined by
| 1 |
where N is the number of voxels in the considered regions (e.g., CSF, WM…), and Io,v and Is,v are intensity values for the template and the nonlinearly registered images (NS, SBST, or L4-standardized), respectively, at voxel v. MAE can be expressed in percentage [11].
The STI technique uses spatial correspondence between an input image and a standard one, determined via global linear and nonlinear registration. Registration allows thus the use of joint histograms to determine intensity correspondence in each tissue, defined within voxel masks [11].
As regards the standardizing transformation in STI, first the mode, i.e., maximum, in the joint histogram [11] is found, and then, a histogram landmark pair corresponding to the input-to-standard intensity mapping for each tissues is determined. So, in order to create the standardized image, the authors added an experimentally determined heuristic to their algorithm. This resulted in estimating the background (BKG) first, then the WM and the GM, removing overlap between BKG/GM and GM/WM, by using the standard image masks for these tissues. They considered CSF is mostly similar to BKG, and they found that it was more robust to indirectly correct the former through BKG standardization [11].
In Fig. 5, we give MAE values for a subset of 250 MR images (a mixture of ADNI and OASIS images randomly selected), for the CSF, WM, and GM tissue classes and the corresponding templates, in case of NS images or standardized by the L4 and SBST procedures.
Fig. 5.
Box plots (left to right, top to bottom) of CSF, GM, WM, and BRAIN mean absolute errors for 250 MR images (a mixture of ADNI and OASIS images randomly selected), non-standardized (NS), L4 and SBST standardized
Also the whole brain, obtained by combining the CSF, WM, and GM images, labeled as BRAIN in the figure, is shown.
It represents a typical situation, observed on the whole dataset, i.e., MAE values in SBST outperformed always L4 in a significant manner with respect to each considered tissue, giving values reduced over 50 %. t Test evaluation of the statistical significance of MAE differences between L4 and SBST gave P(T<=t) two tails <10−6.
On the contrary, Robitaille et al. [11] observed a less homogeneous behavior of STI with respect to L4. In their case, compared to NS images, both L4 and STI exhibited better MAE, but STI significantly outperformed L4 for WM, with no difference for GM.
L4 was superior for foreground (FRG), corresponding to the set of voxels for which the intensity is higher than or equal to the mean intensity computed over the whole image and lower than the intensity corresponding to the percentile value 99.8 obtained for the whole image.
Obviously, the conclusion in [11] that standardization techniques should not be aimed solely at matching histograms and that spatial information should also be incorporated is valid for SBST and STI approaches.
Evaluation on Synthetic Images
For this experiment, we used only the Brainweb synthetic datasets [22], available from the McConnell BIC of the Montreal Neurological Institute, McGill University.
The simulated datasets had a resolution of 181 × 217 × 181 and a slice thickness of 1 mm. The advantage of using the Brainweb dataset in the comparison was the availability of ground truth for the tissue classes (CSF, GM, and WM) from which the digital phantoms were created.
The Brainweb dataset, while consisting of a single digital phantom, comes with different simulation options pertaining to the amount of noise and the amount of RF inhomogeneity in the simulated image. We chose T1-weighted images with a noise level from 0 to 9 % and no signal intensity inhomogeneities.
We standardized these images with the same scale obtained for the other employed images coming from the other public datasets. Standardization quality assessment was performed evaluating the Dice overlap measures.
Interestingly enough, the best results (Dice index >0.99) with both segmentation tools (FSL and SPM-DARTEL) were obtained when using simulated images with a moderate amount of Rician noise, as opposed to images without any noise (Dice index around 0.8). This is also noted by Ferreira da Silva [23], and bodes well for real datasets since imaging noise is an inevitable part of image acquisition. The observation that the CSF tissue class exhibits a greater spread is confirmed also for the simulated images, probably for the same reasons addressed in the previous subsection.
Tissue Divergences
This evaluation was performed on the whole MRI dataset, including the synthetic Brainweb images, with the aim to reproduce as much as possible MRIs coming from heterogeneous sources, including scanners from different manufacturers as well as different scanner models from the same manufacturer.
The rationale is in considering that MRI intensities of the different brain tissues follow normal distributions that can be depicted at least as mixtures of Gaussians, a basis utilized by the approaches motivated by the Gaussian mixture model (GMM) based tissue analysis and segmentation procedures [8 and references therein].
In this way, we should be able to quantify the proximity of voxel intensities to a Gaussian distribution as a result of the intensity standardization [8].
Consider what happens if we assume that the data distribution obtained from the data histogram of each tissue (GM, WM, CSF) does not differ significantly from the Gaussian distribution over the tissue mean and variance that is supposed to generate the samples. If this hypothesis holds, we should not find any significant advantage toward data modeling as a result of intensity standardization.
The Jeffreys divergence (JD) is the metric used to evaluate how much the data distributions differ from the Gaussian models. JD is a symmetric measure of similarity between two distributions, giving low values when there is a small difference between them.
We first compute for the NS MRIs, the mean and the variance of gray-level values from each tissue, and then, we generate samples of Gaussian distributions parameterized by those means and variances.
Next, for each tissue, we generate histograms from the data over each tissue taking into account 98 data percentiles (upper and lower 1 percentile data left out as noise and outliers) [8]. This gives an account of the actual model of the data for the given tissue type, calculated by means of the JD measure.
We are especially interested in showing how much standardization increases per-tissue similarity to Gaussian distribution, and intertissue discrimination, for L4- and SBST-standardized MRIs with respect to NS images.
For this purpose, starting from NS images, we obtain the histograms of the voxel intensities for each tissue and the corresponding Gaussian models. Then, the JDs are calculated between each histogram and its corresponding Gaussian model (i.e., per-tissue) and between histograms corresponding to couples of different tissues (i.e., intertissue).
Next, we do the same for both the SBST- and L4-standardized images.
If the intensity standardization improves tissue contrast and gaussianity, then we should see an increase in the intertissue and a decrease in the per-tissue divergence measures, on the standardized images.
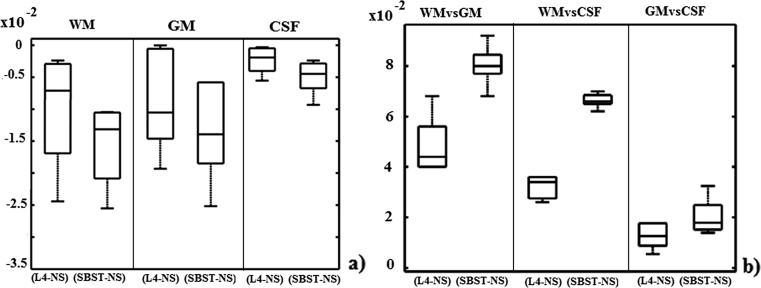
Figure 6a shows the per-tissue JD variation distribution (across images), calculated by subtracting (for each MR image) JD values between the histogram of each tissue and its Gaussian model, SBST or L4 standardized, with respect to NS. For each tissue, the statistical significance of the results is in 95 % interval of confidence.
Fig. 6.
Variation in per-tissue Jeffreys divergences, between the same tissue, SBST and L4 standardized, with respect to the NS corresponding one (a). Variation in intertissue Jeffreys divergences (b)
From the above, if standardization leads to better Gaussian compliance of the various tissue types, variation values are expected to be lower than 0, and from Fig. 6, it is evident that the SBST standardization results in improved tissue type separation in intensity space.
Figure 6b shows the intertissue JD variation, calculated by subtracting JD values between two tissues in NS images, from JD values for the corresponding SBST or L4 standardized images. Higher values represent better discrimination. Noteworthy, SBST appears to outperform L4 in discriminating brain tissues.
Discussion
A questionable point of the paper could regard the assessment of the robustness of the SBST technique through the Dice index, as a measure of the overlap between tissue masks segmented before and after standardization.
This could be considered an apparently circular argument, in a sense that segmentation is used to define tissue classes; then, intensities are standardized, and subsequent segmentation is applied again to show that segmentation remains largely unchanged. As such, it could be considered that the result of a high Dice index between segmentation before and after SBST is not surprising and does not demonstrate the advantage of the method.
To overcome such a potential limitation, we quantitatively investigated how representative the voxel intensities from various tissue types are of a Gaussian model built around a Gaussian distribution centered at the considered tissue mean and with equal variance.
As shown in the “Tissue Divergences” section, SBST standardization results in improved tissue type separation in intensity space and better discrimination of tissue types.
As regards the effect of the SBST intensity standardization on tasks that foresee for example the unsupervised learning of the tissue classes/clusters in different MRIs, Cataldo et al. [12] demonstrated, especially in Fig. 5 of the reference, that automated classifiers may work more reliably on the SBST-standardized images than in NS ones.
Furthermore, in that procedure for generating set of templates for the hippocampal region, it was assessed that the “minimum” number of templates is largely independent on the clusterization method and on the number of the MR images [12].
So that, the best strategy to be used when nonhomogeneous populations are considered, strictly depends on the features and characteristics we want to emphasize better [12].
This means that information about tissue classes/clusters are very robust with regard to the signal intensity changes made by the SBST technique.
A limitation of the technique regards the fact that it is reliant on WM/GM segmentation and therefore cannot be applied to pathologies that take up significant part of the brain, e.g., glioblastoma, or partial field of view acquisitions (such as for example in physiological sequences), or when other physiological conditions, such as administered contrast agent, may prevent segmentation and therefore the technique from being effective.
Finally, it is to be considered that segmentation of the MR images is performed after whole head, fat, bone, and background are removed. This fact implies that the technique cannot be applied in cases when adipose or osseous tissues may be important.
Apart from these considerations about the technique limits, it is to be mentioned that SBST standardization could be applied across image sets acquired with different modalities, e.g., similarly to what is described in Shah et al. [8] in which T1- and T2-weighted or proton density images are L4 standardized.
Conclusions
The paper details a standardization technique for brain MR images, called SBST, able to obtain similar gray values for comparable tissue classes, so that automatic segmentation of images before and after standardization gives high overlap for each tissue class.
By using both histogram and tissue-specific intensity information, piecewise-linear intensity transformations between GM, WM, and CSF images are separately calculated; then, a single smoothed transformation is applied to the images. The technique was evaluated on large, public datasets of MR brain images belonging to older adults, characterized by clinical conditions ranging from good health state to probable dementia of AD type as well as with MIC. We evaluated the technique also on synthetic MR images with different amounts of noise.
First of all, the technique proved to be effective in reducing the possibility of tissue “mixing”.
Then, the robustness of this technique was assessed in two ways: (a) the nonstandardized and standardized images were segmented into WM, GM, and CSF, and segmentation masks were compared by the Dice index, with the aim of checking if the information contained in the images was somewhat corrupted by the procedure, and (b) MAE was calculated between a (single) standardization template and each standardized image (after coregistration).
As to test (a), the results showed that the Dice index between standardized and nonstandardized images was very high (over 0.9) for each tissue class and independently on the clinical conditions. It is important to highlight that the number of images for the training set has to describe the population as possible, and it is lower for patients with homogeneous clinical conditions than with mixed degrees of neuropathology. As to test (b), MAE was smaller for SBST than for two other standardization techniques, L4 and STI.
Furthermore, we assessed how much standardization increases intertissue discrimination, considering NS versus L4-standardized and versus SBST-standardized MRIs, respectively, by using the variation in JD, before and after standardization. The efficacy of the SBST technique was finally tested by assessing gaussianity of gray-level distributions of each tissue before and after standardization. In all the cases, SBST performed better.
In conclusion, the technique shows very promising results even compared with other approaches available in the literature or over calibration techniques, since it does not require a reference material of known MRI property for calibration and does not require explicit manual sampling of different tissue regions. Intensity standardization results in a usable modified image in which all tissues have standardized intensities, up to the accuracy of the technique.
The technique could be applied to other MRI protocol, such as T2-weighted or proton density images and can be used to correct for intrapatient/interpatient, intrascanner/interscanner, and intrasite/intersite MR image intensity variations.
Acknowledgments
First of all, we warmly thank our anonymous reviewers for their pertinent comments and useful suggestions. We thank the Alzheimer’s Disease Neuroimaging Initiative. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129 and K01 AG030514. We thank the Open Access Structural Imaging Series (OASIS), a project dedicated to making brain imaging data openly available to the public (grant numbers P50 G05681, P01 AG03991, R01 AG021910, P20 MH071616, and U24 RR021382). This work is inserted in the framework of the “Programma Operativo Nazionale (PON) 254/Ric- Ricerca e competitività 2007-2013” of the Italian Ministry of Education, University, and Research (upgrading of the “Centro ricerche per la salute dell'uomo e dell'ambiente” PONa3_00334). It is also supported by the Italian “Istituto Nazionale di Fisica Nucleare” (INFN).
References
- 1.Leung KK, Clarkson MJ, Bartlett JW, Clegg S, Jack CR, Jr, Weiner MW, Fox NC, Ourselin S, Alzheimer's Disease Neuroimaging Initiative Robust atrophy rate measurement in Alzheimer's disease using multi-site serial MRI: Tissue-specific intensity standardization and parameter selection. Neuroimage. 2010;50:516–523. doi: 10.1016/j.neuroimage.2009.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Madabhushi A, Udupa JK, Moonis G. Comparing MR image intensity standardization against tissue characterizability of magnetization transfer ratio imaging. J Magn Reson Imaging. 2006;24:667–675. doi: 10.1002/jmri.20658. [DOI] [PubMed] [Google Scholar]
- 3.Stonnington CM, Tan G, Klöppel S, Chu C, Draganski B, Jack CR, Chen K, Ashburner J, Frackowiak RSJ. Interpreting scan data acquired from multiple scanners: a study with Alzheimer's disease. Neuroimage. 2008;39(3):1180–1185. doi: 10.1016/j.neuroimage.2007.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R, Kennedy D, Schmitt F, Brown G, Macfall J, Fischl B, Dale A. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. Neuroimage. 2006;30:436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- 5.Preboske GM, Gunter JL, Ward CP, Jack CR., Jr Common MRI acquisition non-idealities significantly impact the output of the boundary shift integral method of measuring brain atrophy on serial MRI. Neuroimage. 2006;30:1196–1202. doi: 10.1016/j.neuroimage.2005.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jäger F, Hornegger J. Nonrigid registration of joint histograms for intensity standardization in Magnetic Resonance Imaging. IEEE T Med Imaging. 2009;28(1):137–150. doi: 10.1109/TMI.2008.2004429. [DOI] [PubMed] [Google Scholar]
- 7.Madabhushi A, Udupa JK. Interplay of intensity standardization and inhomogeneity correction in MR image analysis. IEEE T Med Imaging. 2005;24:561–576. doi: 10.1109/TMI.2004.843256. [DOI] [PubMed] [Google Scholar]
- 8.Shah M, Xiao Y, Subbanna N, Francis S, Arnold DL, Collins DL, Arbel T. Evaluating intensity standardization on MRIs of human brain with multiple sclerosis. Med Image Anal. 2011;15:267–282. doi: 10.1016/j.media.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Christensen James D. Standardization of brain magnetic resonance images using histogram even-order derivative analysis. Magn Reson Imaging. 2003;21:817–820. doi: 10.1016/S0730-725X(03)00102-4. [DOI] [PubMed] [Google Scholar]
- 10.Weisenfeld N and Warfield S: Standardization of joint image-intensity statistics in MRI using the Kullback-Leibler divergence. In: I S Biomed Imaging. Arlington (VA), 2004
- 11.Robitaille N, Mouiha A, Burt Crépeault B, Valdivia F, Duchesne S, The Alzheimer’s Disease Neuroimaging Initiative Tissue-Based MRI Intensity Standardization: Application to Multi-Centric Datasets. Int J Biomed Imaging. 2012 doi: 10.1155/2012/347120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cataldo R, Agrusti A, De Nunzio G, Carlà A, De Mitri I, Favetta M, Quarta M, Monno L, Rei L. Fiorina E and Alzheimer’s Disease Neuroimaging Initiative (ADNI): Generating a minimal set of templates for the hippocampal region in MR neuroimages. J Neuroimaging. 2013;23(3):473–483. doi: 10.1111/j.1552-6569.2012.00713.x. [DOI] [PubMed] [Google Scholar]
- 13.Nyul LG, Udupa JK. On Standardizing the MR Image Intensity Scale. Magn Reson Med. 1999;42:1072–1081. doi: 10.1002/(SICI)1522-2594(199912)42:6<1072::AID-MRM11>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 14.Ge Y, Udupa JK, Nyúl LG, Wei L, Grossman RI. Numerical tissue characterization in MS via standardization of the MR image intensity scale. Magn Reson Med. 2000;12(5):715–721. doi: 10.1002/1522-2586(200011)12:5<715::aid-jmri8>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 15.Nyúl LG, Udupa JK, Zhang X. New variants of a method of MRI scale Standardization. IEEE T Med Imaging. 2000;19(2):143–150. doi: 10.1109/42.836373. [DOI] [PubMed] [Google Scholar]
- 16.Madabhushi A, Udupa JK. New methods of MR image intensity standardization via generalized scale. Med Phys. 2006;33(9):3426–3434. doi: 10.1118/1.2335487. [DOI] [PubMed] [Google Scholar]
- 17.Marcus DS, Wang TH, Parker, Csernansky JG, Morris JC, Buckner RL. Open Access Series of Imaging Studies (OASIS): Cross-sectional MRI Data in Young, Middle Aged, Nondemented, and Demented Older Adults. J Cogn Neurosci. 2007;19(9):1498–1507. doi: 10.1162/jocn.2007.19.9.1498. [DOI] [PubMed] [Google Scholar]
- 18.Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P, Song JH, Jenkinson M, Lepage C, Rueckert D, et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46:786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashburner J. A Fast Diffeomorphic Image Registration Algorithm. Neuroimage. 2007;38(1):95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Tsang O, Gholipour A, Kehtarnavaz, Gopinath K, Briggs R, Panahi H: Comparison of tissue segmentation algorithms in neuroimage analysis software tools. In: IEEE, 30th Annual International IEEE EMBS Conference, Vancouver, British Columbia, Canada,3924- 3928,2008 [DOI] [PubMed]
- 21.Ortiz A, Górriz JM, Ramírez J, Salas-Gonzalez D. Improving MRI segmentation with probabilistic GHSOM and multiobjective optimization. Neurocomputing. 2013;114(19)):118–131. doi: 10.1016/j.neucom.2012.08.047. [DOI] [Google Scholar]
- 22.Kwan RKS, Evans AC, Pike GB. MRI simulation based evaluation of image-processing and classification methods. IEEE T Med Imaging. 1999;18(11):1085–97. doi: 10.1109/42.816072. [DOI] [PubMed] [Google Scholar]
- 23.da Silva AR F. A Dirichlet Process Mixture Model for Brain MRI Tissue Classification. Med Image Anal. 2007;11(2):169–82. doi: 10.1016/j.media.2006.12.002. [DOI] [PubMed] [Google Scholar]