Summary
Double-stranded DNA (dsDNA) cleavage by Cas9 is a hallmark of type II CRISPR-Cas immune systems. Cas9–guide RNA complexes recognize 20-base-pair sequences in DNA and generate a site-specific double-strand break, a robust activity harnessed for genome editing. DNA recognition by all studied Cas9 enzymes requires a protospacer adjacent motif (PAM) next to the target site. We show that Cas9 enzymes from evolutionarily divergent bacteria can recognize and cleave single-stranded DNA (ssDNA) by an RNA-guided, PAM-independent recognition mechanism. Comparative analysis shows that in contrast to the type II-A S. pyogenes Cas9 that is widely used for genome engineering, the smaller type II-C Cas9 proteins have limited dsDNA binding and unwinding activity and promiscuous guide-RNA specificity. These results indicate that inefficiency of type II-C Cas9 enzymes for genome editing results from a limited ability to cleave dsDNA, and suggest that ssDNA cleavage was an ancestral function of the Cas9 enzyme family.
Introduction
Bacteria and archaea use CRISPR (clustered regularly interspaced short palindromic repeats) systems composed of Cas (CRISPR-associated) proteins and short RNA guides to provide adaptive immunity against invasive nucleic acids (Doudna and Charpentier, 2014; Jiang and Marraffini, 2015; van der Oost et al., 2014). Cas9, a protein component of Type II CRISPR-Cas systems, is a programmable, RNA-guided DNA endonuclease whose specificity is determined by RNA-DNA base pairing (Gasiunas et al., 2012; Jinek et al., 2012). The Streptococcus pyogenes Cas9 (Spy Cas9) has been employed widely for genome engineering based on its ability to generate site-specific double-stranded DNA breaks at sequences abutting an NGG PAM sequence (Doudna and Charpentier, 2014; Sontheimer and Barrangou, 2015; Terns and Terns, 2015). In nature, a large and diverse collection of cas9 genes have been identified, and a few different Cas9 enzymes have been characterized (Jinek et al., 2012; Chylinski et al., 2014; Esvelt et al., 2013; Fonfara et al., 2014; Ran et al., 2015). In all cases studied so far, PAM binding plays a key role in double-stranded DNA (dsDNA) target sequence recognition, although PAM sequences can differ between Cas9 variants. The nucleic-acid cleavage activities of more evolutionarily divergent Cas9 proteins, and whether all Cas9 proteins employ the same DNA recognition and cleavage mechanism have not been determined.
To investigate the function of highly divergent Cas9 proteins, we studied the DNA binding and cleavage activities of seven different Cas9 proteins belonging to the type II-A and type II-C subclasses of the Cas9 superfamily. Surprisingly, all of these enzymes were found to possess PAM-independent single-stranded DNA (ssDNA) cleavage activity, and the type II-C Cas9 proteins could utilize a range of guide RNA constructs for productive ssDNA binding and cleavage. We show that the type II-C Cas9s possess little dsDNA unwinding capability and consequently do not have robust dsDNA cleavage activity. These results suggest that ssDNA cleavage is an intrinsic activity of the Cas9 enzyme family that involves a distinct mode of substrate binding and catalytic domain organization. This activity may have evolved to target particular kinds of bacterial pathogens, and may be useful for applications that involve ssDNA binding or cleavage due to the lack of target sequence constraints.
Results
Single-stranded DNA cleavage by highly divergent Cas9 enzymes
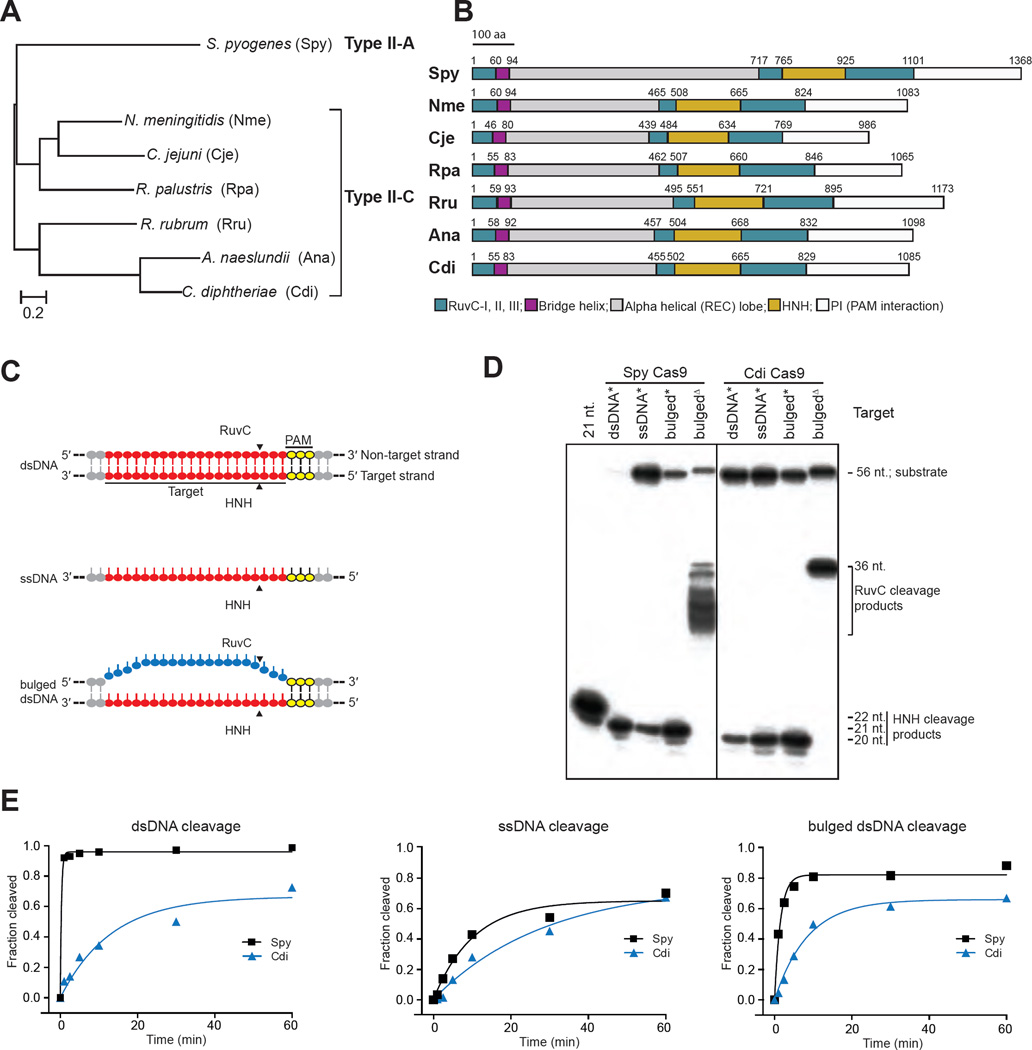
Phylogenetic analysis of the three established type II CRISPR-Cas9 subtypes, type II-A, -B and -C, suggests that type II-C is ancestral to the others (Chylinski et al., 2014; Fonfara et al., 2014). Of interest, Type II-C Cas9 proteins may harbor unique biochemical properties as they are diverse at the level of primary sequence and are generally smaller than type II-A and II-B Cas9 proteins (Chylinski et al., 2014; Fonfara et al., 2014). To determine whether biochemical activities of the smaller and ostensibly more ancient type II-C Cas9 proteins diverge from that of Spy Cas9, we selected six type II-C enzymes for analysis (Figure 1A). The type II-C Cas9 enzyme from N. meningitidis, as well as the type II-A enzyme from S. pyogenes, have been shown to function in eukaryotic cells for genome engineering applications, although Spy Cas9 is by far the most widely used to date (reviewed in Doudna and Charpentier, 2014).
Figure 1. Diverse DNA cleavage activity of divergent Cas9 enzymes.
(A). Phylogenetic tree of the seven Cas9 proteins used in this study, generated using MAFFT (Katoh, 2002). (B). Secondary structures and sizes of divergent Cas9 enzymes in this study. Domains are colored and drawn proportionally to the full length of its protein sequence. (C). Schematic presentation of three DNA substrates used for the figures of D and E. Target sequence is presented in red and underlined. PAM (AGG) is shown in yellow. (D). In vitro cleavage of DNA substrates. Cleavages of single-stranded DNA (ssDNA), double-strand DNA (dsDNA) and bulged DNA (bulged) mediated by Spy and Cdi Cas9 proteins are shown. * denotes that the target strand is labeled; Δ indicates the non-target strand is labeled. Substrate and product sizes are labeled on the right. Vertical lines indicate the border between two separate gels (E). Quantification of DNA cleavage activity on dsDNA, ssDNA and bulged substrates with the 5′- end of the target strand radiolabeled. Cleavage assays were conducted in at least triplicate, as described in the Methods, and the quantified data were fitted with single-exponential decays to obtain pseudo first-order rate constants (kcleave) for each reaction. kcleave ± SD values for dsDNA cleavage by Spy and Cdi Cas9 are 3.5 ± 0.9 min−1 and 0.041 ± 0.003 min−1, respectively. kcleave ± SD values for ssDNA cleavage by Spy and Cdi Cas9 are 0.27 ± 0.09 min−1 and 0.16 ± 0.1 min−1, respectively. kcleave ± SD values for bulged dsDNA cleavage by Spy and Cdi Cas9 are 1.0 ± 0.5 min−1 and 0.59 ± 0.4 min−1, respectively. See also Figures S1, S2 and Table S1.
All of the selected Cas9 enzymes, found within evolutionarily divergent bacterial strains, share a common chassis consisting of the RuvC and HNH catalytic domains, the bridge helix, the alpha-helical domain (also known as the REC domain; Nishimasu et al., 2014), and the PAM-interaction domain (Figure 1B). However, the size of the alpha-helical domain and the primary sequence outside of the catalytic domains vary widely among the different Cas9 proteins tested (Figure S1A). After expressing and purifying these enzymes (Figure S2A), we tested their activities in DNA cleavage assays with various types of substrates. We focus here on the relative activities of the C. diphtheriae (Cdi) Cas9 enzyme, compared to the Spy Cas9 as a control. The Cdi Cas9 shares PAM recognition specificity with Spy Cas9, simplifying DNA substrate design, and its guide RNA has been validated (Ran et al., 2015). Conversely, in the absence of validated tracrRNA and PAM sequences, the Ana, Rpa and Rru Cas9 enzymes provided a useful benchmark of residual DNA-binding and cleavage abilities of Cas9 enzymes. Relevant biochemical data from Nme, Cje, Rpa, Rru and Ana Cas9 enzymes are shown in corresponding supplemental figures, where appropriate.
To explore the substrate selectivity of these Cas9 proteins, we examined cleavage of double-stranded and single-stranded DNA substrates, as well as a double-stranded DNA substrate containing a 20-base-pair mismatched segment along the length of the guide RNA recognition site (Figure 1C). Guide RNAs used in these experiments bear the single-guide RNA architecture (Jinek et al., 2012) consisting of CRISPR RNA (crRNA) and tracrRNA components derived from each bacterial host genome encoding the Cas9 in question(see Methods). Surprisingly, we found that in contrast to Spy Cas9, which has robust dsDNA cleavage activity but slow ssDNA cleavage activity, Cdi, as well as Nme and Cje Cas9 proteins demonstrated a preference for ssDNA cleavage (Figure 1D, Figure S2B). Unsurprisingly, Ana, Rpa and Rru Cas9s were unable to cleave dsDNA, due to a non-cognate guide RNA and PAM sequence in the substrate(Figure S2B). However, these enzymes were active for ssDNA cleavage (Figure S2B). Notably, the cleavage site in the ssDNA substrate was the same as that observed when the dsDNA substrate was used, implying that the mechanism of cleavage site selection does not require the PAM nucleotides, which are present in the non-target strand of the DNA (see Figure 1C). Although Spy Cas9 has been shown previously to cleave target ssRNA only in the presence of a PAM-containing DNA oligonucleotide (PAMmer) (O’Connell et al., 2014), we did not observe ssRNA cleavage (in the absence of a PAMmer) by Spy Cas9 or Cdi Cas9 (Figure S2D).
We next tested the ability of the RuvC and HNH domains to cleave DNA individually by conducting cleavage assays with the mismatched DNA substrate and only one strand of the substrate radiolabeled. Both the Spy and Cdi enzymes contain two active domains, but we did not detect catalytic activity of the RuvC domain in Ana Cas9 (Figure 1D, Figure S2B). We also noted that in contrast to the staggered RuvC-domain cleavage characteristic of the Spy Cas9 enzyme, the RuvC domain of Cdi Cas9 produced a single specific DNA cleavage product. Similar results were obtained for the other type II-C Cas9 enzymes tested in this study (Figure S2B).
Efficient dsDNA cleavage requires Cas9-mediated local DNA unwinding to promote the formation of an R-loop (Sternberg et al., 2014; Szczelkun et al., 2014). This DNA unwinding ability can be detected indirectly by assessing the dsDNA versus ssDNA cleavage activities of each Cas9 enzyme. Substrate cleavage kinetics, compared under single-turnover reaction conditions, quantify the marked difference in ssDNA versus dsDNA cleavage capabilities of the Spy versus Cdi Cas9 enzymes (Figure 1E). Whereas the Spy Cas9 rapidly cleaves a dsDNA substrate, only slow cleavage of this substrate is observed for Cdi Cas9 In contrast, when the same target DNA strand is presented as a ssDNA substrate, similar rates of cleavage are observed for both Cas9 enzymes. When the target DNA strand is presented in the context of a “bulged” dsDNA in which the 20-nucleotide target site is mismatched to the complementary DNA strand, Spy and Cdi Cas9 enzymes also show similar rates of cleavage (Figure 1E) Analogous results were obtained using a supercoiled plasmid DNA substrate (Figure S3A), and control experiments using a guide RNA with a scrambled target-recognition sequence did not support DNA cleavage (Figure S2C). These observations imply that although Cas9 enzymes share a fundamental RNA-targeted DNA cleavage activity, they have markedly different abilities to unwind a dsDNA substrate.
Programmable and PAM-independent Cas9-catalyzed DNA cleavage
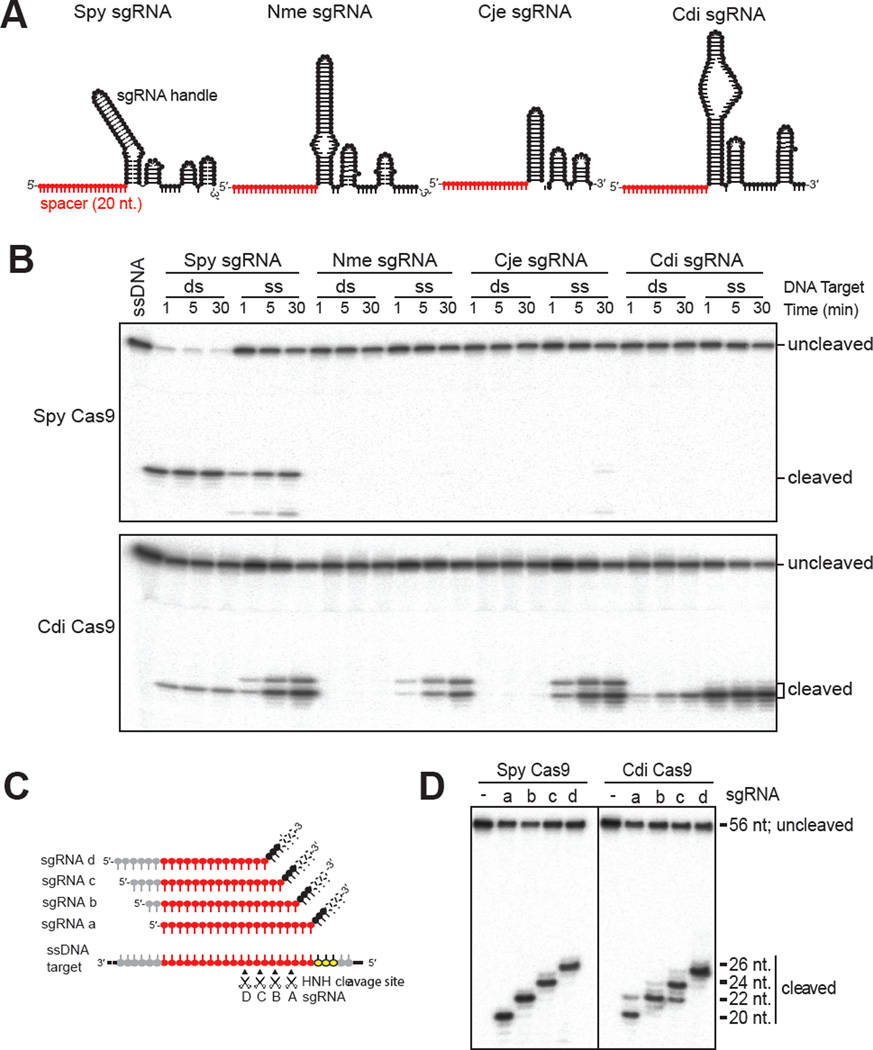
We next compared the guide RNA specificities of the Spy and Cdi Cas9 enzymes to support dsDNA versus ssDNA substrate cleavage (see Figure 2A for guide RNA schematic). As observed previously, we found that the Spy Cas9 enzyme is highly selective for its cognate guide RNA, and only supports dsDNA or ssDNA cleavage when its own guide RNA is used in the reaction ((Briner et al., 2014; Jinek et al., 2012); Figure 2B). In contrast, Cdi Cas9 can use a variety of guide RNAs corresponding to guide RNA sequences corresponding to five different Cas9 enzymes (Figure 2B). The Cdi enzyme catalyzed dsDNA cleavage in the presence of either its own guide RNA or the Spy guide RNA, but only ssDNA cleavage in the presence of the other three guide RNAs tested.
Figure 2. Programmable and PAM-independent Cas9-catalyzed DNA cutting.
(A). Schematic representation of the five sgRNAs used in this study. All of the sgRNAs were drawn based on their mfold predictions (Zuker, 2003) and contain the same 20 nt. ‘spacer’ sequence (shown in red), which is complementary to the target sequence in the DNA substrates. (B). In vitro cleavage assay of ssDNA (ss) and dsDNA (ds) using various combinations of Cas9 proteins and sgRNAs. The 5′-end of the target strand is radiolabeled. Cas9 proteins are shown on the left. (C). Schematic presentation of the four sgRNAs (sgRNAs A–D) used in D. All four sgRNAs have a same Spy sgRNA handle and contain different 20-nt. spacer sequence that is able to base pair to the target sequence in the DNA substrate. (D). Programmable ssDNA cleavage by divergent Cas9 enzymes. The product sizes are labeled on the right; solid lines indicate borders between separate gels. See also Figure S3.
We also observed some differences in DNA cleavage site specificity as a function of the guide RNA construct used in the reaction. With some guide RNAs, the Cdi Cas9 generated two cleavage products corresponding to a site three base pairs from the end of the target sequence (the cognate cleavage site) and a site ~2 nucleotides downstream of this position (Figure 2B). These cleavage products were generated only in reactions containing the Spy, Nme or Cje guide RNAs. These results suggest that guide RNA positioning and/or HNH domain docking may differ depending on the length and/or structure of the guide RNA used by these proteins.
We next tested whether ssDNA cleavage is programmable by generating a set of Spy guide RNAs with a common architecture but different 20-nt. target recognition sequences designed to bind successive DNA target sites shifted by two nucleotides in each case (Figure 2C). Both Spy and Cdi Cas9 showed programmable DNA cleavage such that the primary product generated with each guide RNA corresponded to cleavage at the phosphodiester bond positioned three nucleotides downstream of the 3′ end of the guide RNA (Figure 2D, S3B). In addition, we found that Cdi Cas9 was somewhat less precise in the cleavage reaction, generating two or three cleavage products in each reaction. These results show that like Cas9-catalyzed dsDNA cleavage, ssDNA cleavage is site-specific and programmable. In contrast to Cas9-catalyzed dsDNA cleavage, however, ssDNA cleavage is PAM-independent and the cleavage site appears to be determined largely by measuring from the end of the RNA-DNA hybrid. There may also be sequence-context effects that influence the precision of cleavage, perhaps reflecting differences in docking of the HNH domain that is catalyzing the cleavage reaction in each case.
DNA unwinding capability differs among Cas9 variants
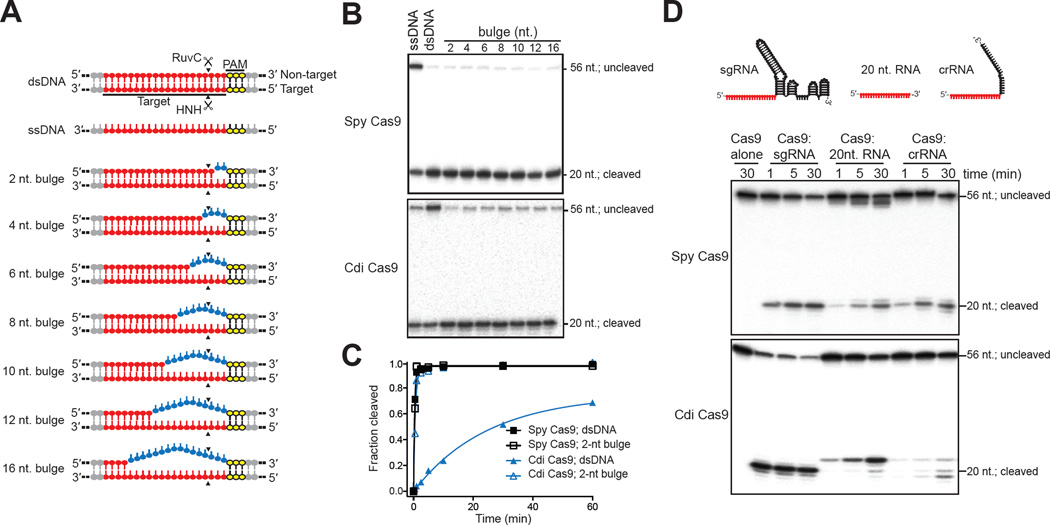
The different relative activities of Spy Cas9 compared to Type IIC Cas9 enzymes for cleavage of dsDNA versus ssDNA substrates implied a difference in DNA unwinding capabilities of these different proteins. To examine this, we tested the ability of each enzyme to cleave dsDNA substrates containing either a completely base-paired target site or varying numbers of mismatched base pairs (2–16) extending from the PAM-proximal end of the target site (Figure 3A). Importantly, in these experiments, the radiolabeled target strand of the DNA does not change, only the unlabeled strand to which it is annealed to generate the dsDNA substrate. Remarkably, a two base mismatch in the dsDNA target is required to greatly enhance dsDNA cleavage by Cdi Cas9, unlike Spy Cas9, which cleaves all of the tested substrates robustly (Figures 3B and 3C). These results show that the Cdi Cas9 have limited ability to cleave dsDNA unless the nucleotides located in the target sequence adjacent to the PAM are not base paired. Kinetic assays showed that Cdi Cas9 cleaves dsDNA at least 50-fold slower than Spy Cas9 unless the DNA substrate contains the two-base-pair bulge (Figures 3B, 3C and S4A). To ensure this observation wasn’t an artifact caused by the use of an artificial Cdi sgRNA, we repeated ssDNA vs. dsDNA cleavage experiments using the native Cdi dual crRNA:tracrRNA guide and saw a similar effect (Figures S4B, S4C). Thus, the type II-C proteins lack the robust DNA unwinding capacity observed for the type II-A Spy Cas9 enzyme.
Figure 3. Substrate recognition varies among divergent Cas9 enzymes.
(A). Schematic representation of DNA substrates used in this experiment. Target sequence is same in all of the substrates (red). For bulged dsDNAs, the bulge sizes are given on the left and mismatches are colored in blue. The PAM is indicated in yellow. (B). In vitro cleavage of bulged DNA substrates. Bulged substrates are indicated by the number of mismatches (2 to 16). Cas9 proteins are labeled on the left of each panel, and sizes of the substrate and cleaved products are labeled on the right. (C). Kinetic analysis of 2-nt. bulged substrate versus perfectly matched dsDNA. Cleavage assays were conducted in at least triplicate, as described in the Methods, and the quantified data were fitted with single-exponential decays to obtain pseudo first-order rate constants (kcleave) for each reaction. kcleave ± SD values for dsDNA cleavage by Spy and Cdi Cas9 are 2.9 ± 0.3 min−1 and 0.041 ± 0.002 min−1, respectively. kcleave ± SD values for 2-nt bulge dsDNA cleavage by Spy and Cdi Cas9 are 2.3 ± 0.1 min−1 and 0.99 ± 0.6 min−1, respectively. (D). DNA cleavage by Cas9 proteins can be guided by short RNAs. Guide RNAs (sgRNA, 20-nt. and crRNA) used in this study are represented schematically above each lane. Cas9 proteins are labeled on the left of each panel, and sizes of the substrate and cleaved products are labeled on the right. See also Figure S4 and Table S2.
These findings along with the weak selectivity for cognate guide RNA molecules suggested that extremely minimal guide RNA constructs might be sufficient for ssDNA target recognition. We compared ssDNA cleavage by Spy and Cdi Cas9 using the Spy single-guide RNA, the cognate crRNA or a 20 nucleotide RNA complementary to the ssDNA target sequence (Figure 3D). Spy Cas9 can use all three of these RNAs for sequence-specific ssDNA cleavage, although at similar concentrations the sgRNA supports faster cleavage kinetics (Figure 3D). In contrast, Cdi Cas9 only worked well with the full-length sgRNA; with the 20-nt guide RNA, a slightly longer cleavage product was slowly generated, and with the crRNA only trace reaction products appeared. We also noted that the ssDNA cleavage site for Cdi Cas9 changed to a 2-nt longer product when the shorter guide RNAs were used. These results suggest that an RNA-DNA hybrid provides some of the DNA cleavage specificity in these reactions, independent of PAM binding or the presence of the full-length guide RNA.
Divergent Cas9 proteins have varying affinities for guide RNAs and substrates
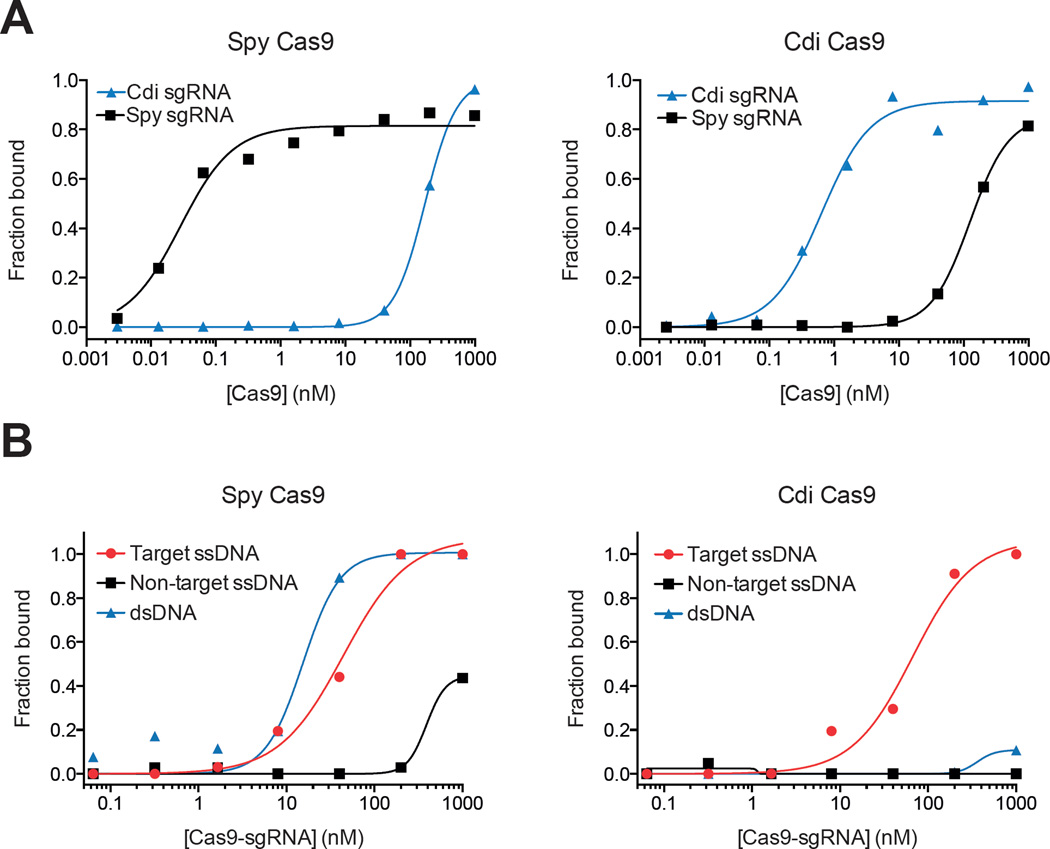
It is possible that the different cleavage activities and guide RNA specificities of the Spy and Cdi Cas9 enzymes reflect differences in binding affinities of these complexes. To test this, we first used filter-binding assays to test Cas9 protein affinity for cognate versus non-cognate guide RNA constructs. We found that Spy Cas9 binds to its cognate guide RNA with an apparent Kd of ~30 pM, whereas binding affinities for non-cognate guide RNAs were ~10,000-fold weaker (Figure 4A). The Cdi Cas9 has a binding affinity of ~1 nM for its cognate guide RNA, whereas binding to a non-cognate guide RNA is >100-fold weaker. Based on these results, binding affinities alone do not account for the differences observed for DNA cleavage activity among the Spy, and Cdi Cas9 enzymes, since cleavage assays were conducted under saturating or near-saturating conditions for the different guide RNAs with the Spy and Cdi enzymes. Instead, we suspect that the large Cas9 protein architecture can accommodate RNAs in distinct binding sites that may differ in their capacity to induce formation of a functional protein-RNA complex. Evidence for alternate guide RNA binding sites within Spy Cas9 was obtained previously (Jinek et al., 2012).
Figure 4.
Binding affinity of Cas9 proteins for sgRNA and DNA. (A). Binding affinity of Cas9 proteins for cognate and non-cognate guides as determined by filter binding assays. Measurements were made in at least triplicate to determine KD and a representative replicate is shown. Data were fit to a binding isotherm. (KD ± SD for Spy Cas9 to Spy sgRNA, 28 pM ± 6 pM; Spy Cas9 to Cdi sgRNA, 146 ± 11nM; Cdi Cas9 to Spy sgRNA, 115 ± 51nM; Cdi Cas9 to Cdi sgRNA, 0.56 ± 0.13nM) (B). Binding affinity of Cas9 proteins for dsDNA, target ssDNA and non-target ssDNA as measured by electrophoretic mobility shift assay (EMSA). Each Cas9 protein was incubated with its cognate guide. Measurements were made in at least triplicate to determine KD and a representative replicate is shown. Data were fit to a binding isotherm. (KD ± SD [where appropriate] for Spy Cas9 to dsDNA, 25 ± 11 nM; Spy Cas9 to target ssDNA, 40 ± 11 nM; Spy Cas9 to non-target ssDNA, >1000 nM; Cdi Cas9 to dsDNA, >1000 nM; Cdi Cas9 to target ssDNA, 58 ± 32 nM; Cdi Cas9 for non-target ssDNA, >1000 nM. See also Figure S5 and Table S2
We next used gel mobility shift assays to test the affinity of pre-formed Cas9-guide RNA complexes for binding to a dsDNA substrate and to each ssDNA strand of this substrate individually (target and non-target) (Figure 4B and S5A). Both Spy and Cdi Cas9-guide RNA complexes had a binding affinity of ~10–60 nM for the target-strand ssDNA substrate. However, they had markedly different affinities for the dsDNA and non-target ssDNA molecules. Whereas the Spy Cas9-guide RNA complex bound to dsDNA with a similar ~10 nM affinity, binding to the non-target ssDNA was >1000 nM under these conditions. By contrast, the Cdi Cas9-guide RNA complex bound to dsDNA with a Kd of >1000 nM, and binding to non-target ssDNA was not detectable (Figure 4B). These data show that only Spy Cas9 possesses a robust dsDNA binding capability. Consistent with DNA cleavage behavior for the Spy and Cdi Cas9 proteins, this may reflect the superior ability of Spy Cas9 to unwind a target dsDNA to enable guide RNA strand annealing.
Cas9 conformational changes reveal differences in nucleic acid binding modes
Previous studies of Spy Cas9 revealed that the protein undergoes a substantial conformational rearrangement upon binding to guide-RNA and subsequently target DNA that can be detected by susceptibility to proteolytic cleavage (Jiang et al., 2015; Jinek et al., 2014; Sternberg et al., 2014). We used trypsin digestion to assess structural changes that might accompany Cdi Cas9 assembly with guide RNA and DNA substrates. In control experiments we observed that Spy Cas9 alone is rapidly cleaved by trypsin, but in the presence of guide RNA the protein is largely protected (Figure 5A; Jiang et al., 2015; Jinek et al., 2014). Similarly, the Cdi Cas9 enzyme is cleaved rapidly by trypsin in the absence of guide RNA but becomes protected upon binding to guide RNA. We also noted that although the addition of non-target-strand ssDNA to Cdi Cas9: guide-RNA results in only small changes in protease sensitivity, Cdi Cas9 becomes more susceptible to proteolysis in the presence of target strand ssDNA (Figure 5A). Therefore, there is likely an additional conformational change occurring upon guide RNA-DNA hybridization, perhaps undocking of the mobile HNH domain and subsequent interaction with the substrate. Furthermore, the distinct cleavage patterns of Cdi Cas9 in the presence of target or non-target ssDNA indicate that in contrast to Spy Cas9, Cdi Cas9 maintains its ssDNA sequence specificity even at these elevated concentrations (15µM DNA). By contrast, the Ana Cas9 enzyme did not exhibit much difference in proteolytic digestion patterns in the absence or presence of (non-cognate) guide RNA and DNA (Figure S5B). Together, these results suggest that like Spy Cas9, Cdi Cas9 undergoes structural changes as it assembles into a functional RNA-guided DNA-bound complex. The lack of conformational change detected for Ana Cas9, even under conditions that favor guide RNA-DNA hybrid binding, may reflect an alternate mode of nucleic acid recognition in the absence of a cognate guide RNA.
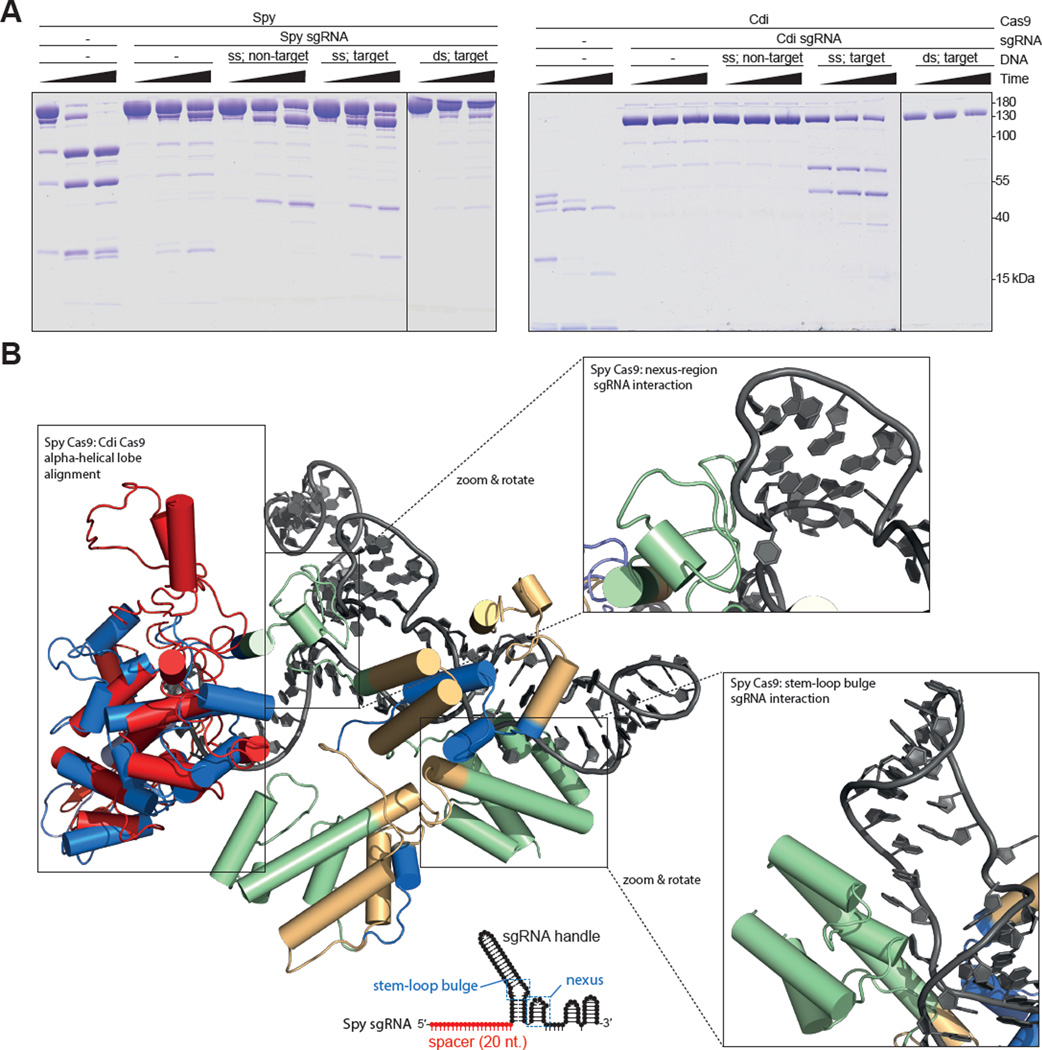
Figure 5.
Limited proteolysis and structural comparison of Spy and Cdi Cas9 proteins. (A). Each Cas9 (5 µM) was incubated with its preferred guide RNA before the addition of DNA. Trypsin was added to the preformed complexes and allowed to digest the protein for 5, 15 or 30min. Degradation products were analyzed on 10% SDS-PAGE gels. ds: dsDNA; ss: ssDNA. Vertical lines indicate the border between two separate gels and approximate molecular weights are indicated on the right. (B). Structural alignment between Spy Cas9 alpha-helical (REC) lobe (PDB ID: 4ZT9) and the homology model of the alpha-helical lobe of Cdi Cas9 (based on the crystal structure of Ana Cas9 PDB ID: 4OGE); all atom RMSD: 3.9Å. The alpha-helical lobe of Cdi Cas9 is shown in red, sgRNA in shown in grey, and Spy Cas9 is colored according to primary sequence alignment with Cdi Cas9: green represents regions not present in the Cdi Cas9 protein sequence, blue represents regions present and conserved in the Cdi Cas9 sequence, and straw represents regions that are loosely conserved in the Cdi protein sequence. Spy sgRNA schematic is shown below with regions of interest boxed and labeled in blue. See also Figure S5.
Similarities between the Cdi and Ana Cas9 protein sizes (1085 and 1098 amino acids, respectively) and sequences (45.4% sequence identity) enabled creation of a homology model of Cdi Cas9 based on the crystal structure (Jinek et al., 2014) of Ana Cas9 (Figure 5B). Comparison to the Spy Cas9 protein structure in the presence of guide RNA (PDB ID: 4ZT9), suggest that the alpha-helical (REC) lobe contributes both to guide RNA recognition specificity (Briner et al., 2014) and possibly to DNA unwinding activity (Figure 5B). This analysis suggests that ancestral Cas9 proteins like Cdi may have lacked robust activity against dsDNA due to a more compact alpha-helical lobe structure that doesn’t appear to include additional regions of the alpha-helical lobe found in Spy Cas9 that bind to the stem-loop bulge and nexus regions of the sgRNA, and contribute to guide-RNA specificity (Figure 5B) and DNA cleavage efficiency (Briner et al., 2014). Evolutionary pressure may have led to enhanced dsDNA binding and cleavage capability due to acquisition of new protein architectures and consequent guide-RNA specificity.
Discussion
CRISPR-Cas9 proteins share a common domain composition, but are highly diverse in overall size, primary sequence and guide RNA and PAM specificity (Jinek et al., 2012; Esvelt et al., 2013; Chylinski et al., 2014; Fonfara et al., 2014; Ran et al., 2015). We show here that evolutionarily divergent Cas9 enzymes from the type II-C Cas9 subclass preferentially cleave ssDNA, as opposed to the dsDNA activity typified by the type II-A Spy Cas9. Since dsDNA cleavage requires guide-RNA strand invasion and local DNA unwinding to form an R-loop (Szczelkun et al., 2014), these findings imply that the smaller and perhaps ancestral type II-C Cas9 proteins have limited DNA unwinding capability.
Distinct Cas9 cleavage activities correlate with protein size and architecture. Structural and biochemical studies indicate that the type II-A Spy Cas9 undergoes a large conformational rearrangement upon guide RNA binding, with additional changes upon dsDNA target recognition (Jiang et al., 2015; Jinek et al., 2014). Similarly, we found that the type II-C Cdi Cas9 undergoes substantial structural change upon binding to guide RNA, as detected by partial proteolysis (Figure 5A). By contrast, the similar proteolytic digestion patterns detected for Ana Cas9 in the absence or presence of guide RNA or DNA suggest a similar or highly dynamic structure under all conditions tested, an observation likely related to the minimal dsDNA-cleavage activity of this enzyme without its cognate guide-RNA and PAM present.
We also found that the type II-C enzymes can utilize a variety of guide RNAs to recognize and cleave ssDNA substrates, and type II-A and II-C Cas9 proteins can employ the crRNA or even a simple 20-nt RNA for residual site-specific ssDNA cleavage. The ability of type II-C enzymes to use non-cognate full-length guide RNAs is markedly different from the activity of Spy Cas9, which is highly selective for its cognate guide RNA (Briner et al., 2014; Jinek et al., 2012). Structural differences between Spy and Cdi Cas9 proteins suggest an interesting connection between recognition of the guide RNA in the region of the nexus (Briner et al., 2014; Figure 5B) and DNA binding capability. The lack of guide RNA binding specificity in the type II-C Cas9s may also relate to their limited ability to unwind dsDNA.
The observations that type II-C Cas9 enzymes are more promiscuous for guide RNA binding and have limited dsDNA unwinding and cleavage activity have important implications for genome editing applications. Because guide RNA binding is less specific, the potential for spurious guide RNA association may increase the likelihood of unintended editing events. In addition, the lack of robust dsDNA binding and cleavage activity imply inhibited ability to generate dsDNA breaks in cells. This finding may explain why type II-C Cas9 enzymes have reduced or in some cases undetectable genome editing activity relative to the type II-A Spy Cas9 (Hou et al., 2013; Ran et al., 2015).
Based on previous phylogenetic analysis (Chylinski et al., 2014; Fonfara et al., 2014), it is possible that the properties of the type II-C proteins represent ancestral activities of the Cas9 enzyme family. Although the physiological functions of ssDNA cleavage by type II-C Cas9 enzymes have not been determined, these enzymes may have evolved to recognize substrates such as ssDNA bacteriophage. However, bacteriophage ssDNA is generally stably bound and protected by ssDNA-binding proteins (Chase and Williams, 1986) and ssDNA cleavage by type II-C Cas9 enzymes would therefore require either transient exposure of naked ssDNA during replication or the ability to displace cellular ssDNA-binding proteins. In addition, Cas9 proteins could potentially silence active gene loci in a manner analogous to the ssDNA targeting that occurs during transcription in vivo by type III-A CRISPR/Cas systems (Deng et al., 2013; Samai et al., 2015). In all cases, type II-C Cas9 enzymes might require additional cellular co-factors to promote efficient dsDNA cleavage. Persistent evolutionary pressure to protect bacteria from dsDNA phage or plasmids might have driven the emergence of larger variants of Cas9 with insertions in the alpha-helical (REC) lobe that confer both guide RNA recognition specificity and the ability to unwind dsDNA target sites. We also envision that despite limited efficiency for genome editing applications requiring dsDNA cleavage, it may be possible to harness the intrinsic type II-C Cas9 activity for ssDNA target recognition in vitro or possibly in cells.
Methods
Cloning and purification of Cas9 proteins
In this study, seven Cas9 proteins have been used. The sources and molecular weights of these Cas9s are summarized in Table S1. To produce the Cas9 proteins for in vitro studies, we amplified each ORF from genomic DNA via PCR with primers tagged with a common sequence for ligase-independent cloning (LIC), we then cloned the PCR products into the 2CT expression vector (His6-MBP-N10-tev-ORF, from UC Berkeley MacroLab). All Cas9 proteins were expressed in Rosetta cells (Novagen), purified with Ni2+ resin, cleaved with TEV protease to remove the MBP tag, and further purified using Superdex 200 size-exclusion column (Figure S1A). The Cas9 proteins were stored at −80 °C in Cas9 storage buffer (20 mM HEPES (pH 7.5), 0.2 M KCl and 5% Glycerol).
Construction of single guide RNA (sgRNA)
Guide RNAs were designed according to the single-guide RNA construct structure established previously (Jinek et al 2012) in which crRNA and tracrRNA sequences were linked by an RNA tetraloop. We used predictions based on type II CRISPR system and Cas9 protein phylogenies (Chylinski et al., 2014; Fonfara et al., 2014) to generate crRNA and tracrRNA sequences; the Cdi guide RNA was based on an experimentally validated construct (Ran et al., 2015). sgRNA-encoding DNAs were amplified by PCR to add a T7 promoter fragment (TAATACGACTCACTATAGG). The T7 fragment-tagged DNAs were then used as templates for the in vitro transcription of sgRNAs as described The transcribed sgRNAs were purified by denaturing PAGE, ethanol precipitated and folded according to (Lin et al., 2014). All other RNA oligos (and all DNA oligos) were synthesized by Integrated DNA Technologies, Inc. (Coralville, Iowa). DNA and RNA Oligo sequences as well as sgRNA sequences used in this study are listed in Table S2.
In vitro DNA cleavage assays
In vitro cleavage reactions were carried out in a total volume of 10 µl of Cas9 cleavage buffer (150 mM KCl, 20 mM HEPES (pH7.5), 1% glycerol, 5 mM MgCl2 and 1 mM DTT) in the presence of 30 nM Cas9, 30 nM sgRNA and ~2 nM 5’-end 32P-labeled target DNA. Reactions were incubated at 37°C for one hour (unless otherwise stated). The reactions were stopped by the addition of 12 ul of 2× loading buffer (2× loading buffer: 0.2 mg/ml of bromophenol blue, 0.2 mg/ml of xylene cyanol, 40 mM EDTA and 95% of formamide) and incubated at 70°C for 10 min. Cleavage products were analyzed by 10% denaturing PAGE, visualized by phosphorimaging and quantified with ImageQuant TL (GE Healthcare). The fraction of RNA cleaved at each time point was plotted as a function of time, and these data were fit with a single exponential decay curve using Prism 6 (GraphPad Software, Inc., La Jolla, USA), according to the equation: Fraction cleaved =A × (1 – exp(−k × t)), where A is the amplitude of the curve, k is the first-order rate constant, and t is time. All experiments were carried out at least in triplicate, with representative replicates shown in the figure panels. Pseudo first-order rate constants (kcleave) and associated errors are reported in the figure legends.
Filter binding assays
Filter binding was carried out in RNA Binding Buffer (20 mM Tris, pH 7.5, 150 mM KCl, 5 mM MgCl2, 1 mM DTT, 5% glycerol, 0.01% Igepal CA-630, 10µg/ml yeast tRNA and 10 µg/ml BSA). Cas9 was incubated with radiolabeled RNA (<0.02nM) for 1hr at room temperature. Tufryn, Protran and Hybond-N+ were assembled onto a dot-blot apparatus in the order listed above. The membranes were washed with 50µL Equilibration Buffer (20 mM Tris, pH7.5, 150 mM KCl, 5 mM MgCl2, 1 mM DTT, 5% glycerol) twice before the sample was applied to the membranes. Membranes were again washed twice with 50 µL Equilibration Buffer, dried and visualized by phosphorimaging. Data were quantified with ImageQuant TL Software (GE Healthcare) and fit to a binding isotherm using Prism (GraphPad Software). All experiments were carried out at least in triplicate, with representative replicates shown in the figure panels Dissociation constants (KD) and associated errors are reported in the figure legends.
Electrophoretic mobility-shift assay
Assays were done in Binding Buffer (20 mM Tris, pH 7.5, 150 mM KCl, 5 mM EDTA, 1 mM DTT, 5% (v/v) glycerol, 50 µg/mL heparin, 0.01% Tween 20, and 100 µg/mL BSA). Cas9-sgRNA complexes were preformed by incubating 1µM of Cas9 and sgRNA for 30min at room temperature. Cas9-sgRNA complexes were diluted and DNA (<0.05nM) was added and allowed to incubate for another 30min at room temperature. Samples were then analyzed by 8% polyacrylamide gel containing 0.5× TBE. Gels were imaged by phosphorimaging, quantified using ImageQuant TL Software (GE Healthcare) and fit to a binding isotherm using Prism (GraphPad Software). All experiments were carried out at least in triplicate, with representative replicates shown in the figure panels. Dissociation constants (KD) and associated errors are reported in the figure legends.
Limited proteolysis of Cas9 complexes
Limited proteolysis was conducted in a 10µL reaction using 5 µM Cas9 with or without guide (1:1.5 molar ratio) and/or DNA (1:3 molar ratio). The Cas9 and guide mixture was pre-incubated for 30min at room temperature in 1× Trypsin Buffer (100mM Tris, pH 7.5, 150 mM KCl, 2% glycerol, 1 mM EDTA and 1 mM DTT). DNA was added and the reaction was incubated for 5 to 30 min before the addition of 0.2µg trypsin per 10µL of Cas9 complex. The reaction was quenched by addition of 5× loading dye (5% β-Mercaptoethanol, 0.02% bromophenol blue, 30% glycerol, 10% sodium dodecyl sulfate, 250 mM Tris-Cl, pH 6.8) to a final concentration of 1× and the products were analyzed on 10% PAGE gels. All experiments were carried out at least in triplicate, with representative replicates shown in the figure panels.
Supplementary Material
Acknowledgements
We thank Doudna Lab members and Rodolphe Barrangou for helpful discussions. This work was supported by the Center for RNA Systems Biology (NIH P50, J. Cate, P.I.) and the Howard Hughes Medical Institute. L.H. is supported by a graduate fellowship from the National Science Foundation. J.A.D. is a Howard Hughes Medical Institute Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
E.M., L.H., M.O. and J.A.D. designed the experiments; E.M., L.H. and K.Z. conducted the experiments; E.M., L.H., M.O. and J.A.D. analyzed the data; E.M., L.H., M.O. and J.A.D. wrote the manuscript.
References
- Briner AE, Donohoue PD, Gomaa AA, Selle K, Slorach EM, Nye CH, Haurwitz RE, Beisel CL, May AP, Barrangou R. Guide RNA Functional Modules Direct Cas9 Activity and Orthogonality. Mol. Cell. 2014;56:333–339. doi: 10.1016/j.molcel.2014.09.019. [DOI] [PubMed] [Google Scholar]
- Chase JW, Williams KR. Single-stranded DNA binding proteins required for DNA replication. Annu. Rev. Biochem. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- Chylinski K, Makarova KS, Charpentier E, Koonin EV. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:6091–6105. doi: 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Garrett RA, Shah SA, Peng X, She Q. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol. Microbiol. 2013;87:1088–1099. doi: 10.1111/mmi.12152. [DOI] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Sci. 2014;346 doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Meth. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, Le Rhun A, Chylinski K, Makarova KS, Lécrivain A-L, Bzdrenga J, Koonin EV, Charpentier E. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:2577–2590. doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Zhang Y, Propson NE, Howden SE, Chu L-F, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl. Acad. Sci. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Marraffini La. CRISPR-Cas: New Tools for Genetic Manipulations from Bacterial Immunity Systems. Annu. Rev. Microbiol. 2015;69 doi: 10.1146/annurev-micro-091014-104441. 150724172101001. [DOI] [PubMed] [Google Scholar]
- Jiang F, Zhou K, Ma L, Gressel S, Doudna Ja. A Cas9-guide RNA complex preorganized for target DNA recognition. Science (80-.) 2015;348:1477–1481. doi: 10.1126/science.aab1452. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Sci. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, et al. Structures of Cas9 endonucleases reveal RNAmediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Staahl B, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516:263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samai P, Pyenson N, Jiang W, Goldberg GW, Hatoum-Aslan A, Marraffini LA. Co-transcriptional DNA and RNA Cleavage during Type III CRISPR-Cas Immunity. Cell. 2015;161:1164–1174. doi: 10.1016/j.cell.2015.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer E, Barrangou R. The Bacterial Origins of the CRISPR Genome Editing Revolution. Hum. Gene Ther. 2015 doi: 10.1089/hum.2015.091. [DOI] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna Ja. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczelkun MD, Tikhomirova MS, Sinkunas T, Gasiunas G, Karvelis T, Pschera P, Siksnys V, Seidel R. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc. Natl. Acad. Sci. U. S. A. 2014;111:9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns RM, Terns MP. CRISPR-based technologies: prokaryotic defense weapons repurposed. Trends Genet. 2015;30:111–118. doi: 10.1016/j.tig.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.