Abstract
Background
Argonaute 2 (AGO2), a central component of RNA-induced silencing complex, plays critical roles in cancer. We examined whether the single nucleotide polymorphisms (SNPs) of AGO2 were related to the risk of nasopharyngeal carcinoma (NPC).
Methods
Twenty-five tag SNPs within AGO2 were genotyped in Guangxi population consisting of 855 NPC patients and 1036 controls. The SNPs significantly associated with NPC were further replicated in Guangdong population consisting of 996 NPC patients and 972 controls. Functional experiments were conducted to examine the biologic roles of AGO2 in NPC.
Results
A significantly increased risk of advanced lymph node metastasis of NPC was identified for the AGO2 rs3928672 GA + AA genotype compared with GG genotype in both the Guangxi and Guangdong populations (combined odd ratio = 2.08, 95 % confidence interval = 1.44-3.01, P = 8.60 × 10−5). Moreover, the AGO2 protein expression levels of rs3928672 GA + AA genotype carriers were higher than the GG genotype carriers in the NPC tissues (P = 0.041), and AGO2 was significantly over-expressed in NPC tissues compared with non-cancerous nasopharyngeal tissues (P = 0.011). In addition, AGO2 knockdown reduced cell proliferation, induced apoptosis, and inhibited migration of NPC cells. Furthermore, gene expression microarray showed that genes altered following AGO2 knockdown were clustered in tumorigenesis and metastasis relevant pathways.
Conclusions
Our findings suggest that the genetic polymorphism in AGO2 may be a risk factor for the advanced lymph node metastasis of NPC in Chinese populations, and AGO2 acts as an oncogene in the development of NPC.
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-015-1895-4) contains supplementary material, which is available to authorized users.
Keywords: AGO2, Polymorphism, Nasopharyngeal carcinoma, Lymph node metastasis
Background
Nasopharyngeal carcinoma (NPC) is an epithelial malignancy with striking racial and geographic distribution differences. It is particularly prevalent among populations from southern China, Southeast Asia, northern Africa and Alaska. These incidence rates are approximately 100-fold higher than in the Caucasian populations [1]. Several environmental factors, including infection with the Epstein-Barr virus (EBV), long-term cigarette smoking, occupational exposure to formaldehyde, and various dietary factors, have been reported to confer the risk of developing NPC [2]. Furthermore, numerous genetic linkage and association studies have reported a few genes contributing to the risk of this malignancy [3, 4]. The identification of susceptibility genes contributing to NPC would assist in predicting individual and population risk of NPC development and would help to clarify the pathogenesis relevant to this disorder.
Argonaute 2 (AGO2), a member of the Argonaute protein family, can bind microRNAs (miRNAs) or short interfering RNAs (siRNAs) and mediate the repression of specific target RNAs either by degrading RNA or inhibiting translation [5]. Of the four human AGO proteins, AGO2 is the only member with intrinsic endoribonuclease activity and essential non-redundant slicer-independent function within the mammalian miRNA pathway [6]. Recently, alterations (genomic amplifications and/or over-expression) of the AGO2 gene have been extensively described in a variety of cancers [7–11], and these alterations have been shown to be linked with an increased metastasis [11–13] and poorer prognosis [12]. Further studies indicated that the AGO2 is involved in several steps of cancer development, including cell proliferation, differentiation, apoptosis, migration and invasion [8–10, 14, 15]. With regard to NPC, the elevated mRNA expressions of AGO2 were observed in tumor tissues and latent membrane protein 1 (LMP1)-positive tumors compared with normal adjacent nasopharyngeal epithelium tissues and LMP1-negative tumors, respectively [16]. Taken together, these studies suggest that the AGO2 may play crucial roles in the cancer development and progression.
Theoretically, the genetic variants such as single nucleotide polymorphisms (SNPs) within the AGO2 gene, which may alter the expression of AGO2 and then influence the miRNAs processing and function, could result in genotype-dependent differences in risk of cancers. Indeed, there is increasing evidence that the genetic variants in the AGO2 gene are associated with the risk or development of several cancers, including breast cancer [17, 18] and malignant peripheral nerve sheath tumor [19]. The role of genetic variants within AGO2 in NPC, however, has never been specifically investigated. In the present study, we examined whether the genetic variation in the AGO2 gene affect the risk or severity of NPC in the Chinese populations. We also evaluated the biologic roles of AGO2 in the development of NPC by functional assays.
Methods
Study population
This study consisted of two populations of patients with NPC and control subjects resided in Guangxi and Guangdong province, respectively, both of which were well-known high-risk regions for NPC located in southern China (Additional file 1: Table S1). The Guangxi population, which contained 855 incident patients with NPC and 1036 controls, has been described in detail previously [20]. Briefly, all subjects were unrelated ethnic adult Chinese and residents in Nanning city (Nanning, China) and the surrounding regions. All patients were newly diagnosed and pathologically confirmed, and were consecutively recruited between September 2003 and January 2008 at the Guangxi Cancer Hospital (Nanning, China). Patients that received chemotherapy or radiotherapy prior to surgery or had other type of cancer were excluded from this study. Tumor staging was performed according to the tumor-node-metastasis (TNM) classification by the 1997 American Joint Committee on Cancer (AJCC) system. All TNM classifications were determined by senior pathologists of the hospital on the basis of the postoperative histopathologic examination. All the controls were recruited in the same regions during the same time that the NPC cases were collected. The selection criteria for the controls included no individual history of cancer and frequency matching to the cases on sex and age (± 5 years). The Guangdong population, which contained 997 NPC patients and 972 controls, has been described in detail previously [3]. Briefly, all subjects were unrelated ethnic adult Chinese and residents in Guangdong province. All patients were consecutively recruited between October 2005 and October 2007 from Sun Yat-sen University Cancer Center (SYSUCC) in Guangzhou city (Guangzhou, China). All patients were histopathologically diagnosed by at least two pathologists according to the World Health Organization (WHO) classification. During the same period, control subjects were recruited from physical examination centers of several large comprehensive hospitals in local communities in Guangdong and were frequency matched to the cases by age (± 5 years), sex, geographic region and ethnicity. All the subjects were interviewed for collection of personal information on demographic factors, medical history, cigarette smoking and alcohol use via structured questionnaire. This study was approved by the Ethics Committee of Beijing Institute of Radiation Medicine (Beijing, China). At recruitment, the written informed consent was obtained from all the participants involved in this study.
From the Guangxi population of 855 patients with incident NPC, 37 patients who had undergone resection before receiving any further treatment at Guangxi Cancer Hospital were selected, and primary NPC biopsies were collected from them (Additional file 1: Table S2). The histological type of all tumor tissues was poorly differentiated squamous cell carcinoma (SCC). Histological non-cancerous nasopharyngeal epithelium tissues were collected from 18 of the 1036 control subjects (Additional file 1: Table S2). All the tissues were fixed in paraformaldehyde, embedded in paraffin wax, and prepared for subsequent immunohistochemical staining.
SNP selection and genotyping
Twenty-five SNPs in the AGO2 gene were selected for genotyping in our study (Table 1). These SNPs were chosen on the basis of previous reports of their association with the risk for cancer, or with potential function, and a comprehensive tag SNP approach. Two SNPs (rs2292779 and rs7005286) significantly associated with cancer in previous studies [18, 19] were directly selected in our study. Two SNPs (rs4961280 and rs11996715) located in the promoter region (−1809A/C and −1686 A/C) of AGO2, were selected as they could alter the expression of AGO2. Tag SNPs of the AGO2 gene were selected from the Han Chinese in Beijing, China (CHB) HapMap database (HapMap release 27, February 2009) using HaploView (pairwise r2 > 0.8) [21]. To ensure enough statistical power, a value of 0.10 of minor allele frequency (MAF) was set as the threshold value of inclusion in this study. Finally, 25 tag SNPs were selected to capture the AGO2 gene.
Table 1.
Positions and frequencies of SNPs within AGO2 gene
| No. | SNPsa | Alleles (Minor/Major) | Frequenciesb | Positionsc | Regions |
|---|---|---|---|---|---|
| 1 | rs4961280 | C/A | 0.111 | −1809 | Promoter |
| 2 | rs11996715 | C/A | 0.430 | −1686 | Promoter |
| 3 | rs4961226 | G/T | 0.400 | 3050 | Intron 1 |
| 4 | rs10088596 | C/T | 0.453 | 5311 | Intron 1 |
| 5 | rs883596 | G/C | 0.105 | 9957 | Intron 1 |
| 6 | rs13261055 | A/G | 0.386 | 16208 | Intron 1 |
| 7 | rs7001653 | G/A | 0.444 | 24567 | Intron 1 |
| 8 | rs7819727 | A/G | 0.452 | 48394 | Intron 1 |
| 9 | rs7009635 | T/C | 0.367 | 50724 | Intron 2 |
| 10 | rs7005286 | C/T | 0.337 | 51145 | Intron 2 |
| 11 | rs11776034 | C/G | 0.211 | 53712 | Intron 2 |
| 12 | rs3735805 | C/T | 0.356 | 68083 | Intron 3 |
| 13 | rs2977490 | A/G | 0.453 | 72247 | Intron 3 |
| 14 | rs2292773 | C/T | 0.278 | 73107 | Intron 4 |
| 15 | rs3928672 | G/A | 0.222 | 73438 | Intron 4 |
| 16 | rs2271735 | G/T | 0.178 | 76295 | Intron 6 |
| 17 | rs2977481 | G/C | 0.183 | 83748 | Intron 10 |
| 18 | rs2292779 | G/C | 0.244 | 84394 | Intron 11 |
| 19 | rs2944764 | C/T | 0.239 | 84576 | Intron 11 |
| 20 | rs11166983 | G/A | 0.278 | 87519 | Intron 11 |
| 21 | rs12542354 | C/T | 0.239 | 89462 | Intron 13 |
| 22 | rs13252337 | A/G | 0.186 | 96222 | Intron 16 |
| 23 | rs2977469 | G/A | 0.465 | 96314 | Intron 16 |
| 24 | rs2977464 | C/T | 0.333 | 100412 | Intron 16 |
| 25 | rs2977462 | T/C | 0.467 | 102467 | Intron 16 |
SNPs in bold were not in Hardy-Weinberg equilibrium (HWE) (rs883596 and rs2977490, both P < 0.05) or failed genotyping (rs2977481) in the Guangxi population
adbSNP ID number
bMinor allele frequencies in HapMap-HCB (HCB, unrelated Han Chinese in Beijing, China)
cThe position of the SNPs is relative to the first nucleotide of the open reading frame of the AGO2 gene, referred to as accession number NT_008046.15
All SNPs were genotyped using GenomeLab SNPstream Genotyping System according to the manufacturer’s instructions (Beckman Coulter, USA) as previously described [22]. This platform uses a single base pair extension reaction to incorporate two-color fluorescence terminal nucleotides which are detected by a specialized imager. Details of the primer sequences are listed in Additional file 1: Table S3. The genotype data were analyzed by GenomeLab SNPstream version 2.2 software (Beckman Coulter, USA). To ensure genotyping quality, we genotyped 20 randomly selected duplicate samples and 4 blanks in each 384-well plate and obtained a concordance rate of >99 %.
Immunohistochemistry in NPC tissues and non-cancerous nasopharyngeal tissues
The paraformaldehyde-fixed and paraffin-embedded NPC tissues (n = 37) and non-cancerous nasopharyngeal tissues (n = 18) (Additional file 1: Table S2) were analyzed for protein expression of AGO2. Two slides from each biopsy were stained with hematoxylin and eosin for routine histological evaluation. Histologic slides with tissue sections were subjected to immunohistochemistry (IHC) as previously described [20, 22], using the primary antibody raised against AGO2 (diluted 1:200; ab57113, Abcam, UK). Negative controls and positive controls were performed at the same time. Photographs were taken (BX51 microscopic/Digital Camera System; Olympus) for study comparison. The IHC signals were scored as previously described [20, 22] by two pathologists (Wu J and Huang W) who did not have knowledge of ligand-binding assay results or patient outcome. Briefly, a proportion score was assigned representing the estimated proportion of positive staining tumor cells (0, none; 1, < 1/100; 2, 1/100 to < 1/10; 3, 1/10 to < 1/3; 4, 1/3 - 2/3; 5, > 2/3). Average estimated intensity of staining in positive cells was assigned an intensity score (0, none; 1, weak; 2, intermediate; 3, strong). The two parameters were combined and resulting in an overall score (0 or 2–8). The scores were classified into three groups: group 1 (score 0, negative expression), group 2 (score 2–4, low expression) and group 3 (score 5–8, high expression). A total of five fields per slide were selected, counted, and averaged.
AGO2 knockdown in NPC cell line
The human nasopharyngeal carcinoma cell line CNE2Z was obtained from the Peking Union Medical College (Chinese Academy of Medical Science, China) and maintained in RPMI 1640 (Gibco-BRL, USA) supplemented with 10 % fetal bovine serum (HyClone, USA) and 1 % penicillin/streptomycin (Gibco-BRL, USA) at 37 °C under a 5 % CO2 atmosphere. The AGO2 specific shRNA expression vectors and the scrambled ineffective shRNA cassette (as the negative control) in the pGPU6/GFP/Neo plasmid were purchased from GenePharma (China). The sequences of the three shRNAs directed against AGO2 are as follows: sh1: 5’-GCAAGGATCGCATCTTCAAGG-3’; sh2: 5’-GGTCTAAAGGTGGAGATAAGG-3’; and sh3: 5’-GCCTGAAGATCAACGTCAAGC-3’. The sequence of control shRNA is 5’-GTTCTCCGAACGTGTCACGT-3’. A mixture of constructs including three AGO2 shRNAs or the control shRNA construct was transfected into CNE2Z cells using Lipofectamine 2000 (Invitrogen, USA). The efficiency of RNA interfering was assessed by western blot analysis.
Western blot
Cells were lysed in Laemmli sample buffer (Bio-Rad, USA) with an EDTA-free, protease inhibitor cocktail (Roche, USA). Proteins at the same amount were separated by SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, USA). After probing with primary and secondary antibody, antigen-antibody complex was visualized by enhanced chemiluminescence-plus reagent (Pierce Biotechnologies, USA). For AGO2, mouse anti-AGO2 monoclonal antibody (diluted 1:1000; ab57113, Abcam, UK) was used. As an internal control, mouse anti-β-actin monoclonal antibody (diluted 1:1000; sc-130301, Snata cruz, USA) was used.
Cell proliferation, apoptosis and migration assays
Cell proliferation was evaluated by measuring cell viability with Cell Counting Kit-8 (CCK-8) assay (Beyotime Inst Biotech, China) according to the manufacturer’s instructions. Cells (2 × 103 cells per well) were plated in 96-well plates in triplicate. CCK-8 was added to each well at a final concentration of 10 % at different time points (24, 48, 72 and 96 h) and incubation continued at 37 °C for 50 min. Subsequently, the absorbance of the samples was measured at 450 nm using a Multiskan MK3 microplate reader (Thermo Labsystems, USA) to calculate the numbers of viable cells in each well.
Apoptosis was detected by flow cytometry using the Annexin V-APC/7-AAD Apoptosis Detection Kit (KeyGEN, China). Briefly, cells were harvested, washed, resuspended in the staining buffer, and doubly stained with annexin-V and 7-amino-actinomycin D (7-AAD). For each experiment, 5 × 104 cells were analyzed using FACSCalibur and CellQuest software (BD Biosciences, USA). The Annexin V-positive cells were regarded as apoptotic cells.
Ability of cell migration was evaluated by Transwell assay (Corning Inc, USA) according to the manufacturer’s instructions. Cells (1.5 × 105 cells per well) in 300 μl serum-free medium were placed in the upper chamber of the transwell, whereas the lower chamber was loaded with 500 μl medium containing 20 % fetal bovine serum. After 16 h of incubation, cells that migrated to the lower chamber were fixed and stained with crystal violet. The number of cells was counted in five random microscopic fields (magnification 400 ×).
Genome-wide expression microarray and pathway analysis
Total RNA was isolated for microarray analysis from three biological replicates of cells transfected with AGO2 shRNAs or control shRNA using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions. All samples with an RNA integrity number (RIN) ≥ 8.0 and A260/A280 ratio 1.8-2.1 were considered suitable for microarray analysis. The Affymetrix GeneChip® Human Gene U133 Plus 2.0 Arrays were used for genome-wide expression profiling assay, which was performed by CapitalBio Corporation (Beijing, China) according to the manufacturer’s instructions (Affymetrix, USA). Raw feature data from microarrays were subsequently corrected for background and normalized, and the log2 intensity expression summary values for each probe set were calculated using Robust Multiplechip Average (RMA). Significantly altered genes following AGO2 knockdown were analyzed by significance analysis of microarray (SAM, version 3.01), and the P values of the t test were calculated for each gene. Multiple hypothesis testing was performed to calculate the false discovery rates (FDRs) through the Bioconductor package Qvalue (http://www.bioconductor.org). Significantly altered genes following AGO2 knockdown were investigated for biological processes and signaling pathways using the cytoscape plug-in of Reactome FI [23].
Statistical analysis
Genotype and allele frequencies of AGO2 polymorphisms were determined by direct counting, and departures from Hardy-Weinberg equilibrium (HWE) were tested using the random-permutation procedure implemented in the Arlequin package (http://cmpg.unibe.ch/software/arlequin3/). Multivariate logistic regression analysis was done to evaluate whether there were genetic associations between the AGO2 polymorphisms and risk and severity of NPC. The P values, odds ratios (ORs), and 95 % confidence intervals (CIs) were calculated and adjusted for sex, age, status of smoking and drinking, smoking level, family history and nationality where it was appropriate. To assess the probability of a spurious association due to multiple comparisons, the online software SNPSpD (http://gump.qimr.edu.au/general/daleN/SNPSpD) was used to calculate the correction factor for multiple testing in gene unit [24]. A P value of 0.0036 (0.05/14.01; correction factor = 14.01) was used as the criterion of statistical significance in Guangxi population. Association analysis was performed by SNPStats (http://bioinfo.iconcologia.net/snpstats/start.htm). The power for our genetic association study was calculated using the Power for Genetic Association Analyses (PGA) [25]. Differences of the protein expression levels detected by IHC between the NPC tissues and non-cancerous nasopharyngeal tissues, and between the rs3928672 GG genotype and A allele (GA + AA genotype) carriers were assessed by a Wilcoxon signed-ranks test and logistic regression analysis, respectively. Unpaired t test was used to analyze the results of cell proliferation, apoptosis and migration assays. A P value of 0.05 was used as the criterion of statistical significance, and all statistical tests were two sided. These analyses were performed using SPSS software (Version 17.0; SPSS Inc., USA).
Results
Characteristics of the study population
For the Guangxi population, the selected demographic and clinical characteristics were described in detail previously (Additional file 1: Table S1) [20]. In the Guangdong population, there was no significant difference between the patients and control subjects in terms of mean age, sex, status of smoking and drinking; while more heavier smokers (≥ 24 pack-years) were present in the cases compared with controls (χ2 = 13.42, P = 2.48 × 10−4). According to the TNM systems, 3.6 %, 14.9 %, 50.5 % and 25.4 % of patients had stage I, II, III, and IV disease, respectively (Additional file 1: Table S1).
Genetic association between SNPs of the AGO2 gene and NPC
The 25 SNPs were initially genotyped in the Guangxi population. We succeeded in genotyping 24 (with the exception of rs2977481) out of 25 SNPs using the SNPstream assay. Of these 24 SNPs, 2 (rs883596 and rs2977490) were not in HWE (both P < 0.05). We therefore selected the remaining 22 SNPs for the subsequent analyses.
The genotyping results for the 22 SNPs are shown in Additional file 1: Table S4. On the basis of logistic regression analysis with adjustment for sex, age, status of smoking and drinking, smoking level, family history and nationality, only one SNP, i.e. rs12542354, was significantly associated with the susceptibility to NPC in the Guangxi population after correction for multiple comparisons (OR = 1.32, 95 % CI = 1.13-1.54, P = 0.00040; Additional file 1: Table S4). Then, the rs12542354 was further genotyped in the Guangdong population. However, the initial association between the rs12542354 and risk of NPC in the Guangxi population was not validated in the Guangdong population (Additional file 1: Table S4).
Then, we assessed the effects of the AGO2 polymorphisms on the severity of NPC (as measured by TNM staging system) in the Guangxi population. After adjustment for sex, age, status of smoking and drinking, smoking level, family history and nationality, logistic regression analysis revealed that significant association with the advanced local tumor invasion of NPC was observed with the rs2271735 (Additional file 1: Table S5). In the meantime, the rs2977469 and rs3928672 were significantly associated with the advanced lymph node involvement of NPC (Additional file 1: Table S6). The associations remained significant even after correction for multiple comparisons. To confirm the initial associations of these three SNPs (rs2271735, rs2779249 and rs3928672) with the NPC severity, the Guangdong population, composed of 997 patients with NPC, were also genotyped (Additional file 1: Table S5 and Additional file 1: Table S6). Only rs3928672 was validated in the Guangdong population. In the Guangxi population, the patients bearing rs3928672 A allele (GA + AA genotype) had a significantly increased frequent of involvement of lymph node compared with ones bearing GG genotype (N3 vs. N0 + N1 + N2; OR = 2.47, 95 % CI = 1.47-4.13, P = 0.00030; Table 2). Consistently, patients with the rs3928672 A allele (GA + AA genotype) had an advanced lymph node metastasis (N3 vs. N0 + N1 + N2; OR = 1.75, 95 % CI = 1.03-2.98, P = 0.034; Table 2) compared to ones with the GG genotype in the Guangdong population. The distribution of rs3928672 genotype frequencies is not different between the four N classification sub-populations. Therefore, we pooled the two case-control studies from the Guangxi and Guangdong populations, and found that the association of rs3928672 with the susceptibility to advanced lymph node metastasis was stronger (N3 vs. N0 + N1 + N2; OR = 2.08, 95 % CI = 1.44-3.01, P = 8.60 × 10−5; Table 2). A meta-analysis also confirmed the strong evidence of association of rs3928672 with the advanced lymph node metastasis (N3 vs. N0 + N1 + N2; OR = 2.09, 95 % CI = 1.44-3.03, P = 9.64 × 10−5; Table 2). There was little evidence of heterogeneity among sample sets (Pheterogeneity = 0.36).
Table 2.
Genetic associations of rs3928672 with the lymph node metastasis of NPC
| Populations | Genotypes | N classification, n (%) | OR (95 % CI)a | P valuea | |||
|---|---|---|---|---|---|---|---|
| N0 | N1 | N2 | N3 | ||||
| Guangxi | (n = 177) | (n = 406) | (n = 180) | (n = 78) | |||
| GG | 91 (51.4) | 187 (46.1) | 95 (52.8) | 22 (28.2) | 1.00 (reference) | ||
| GA | 72 (40.7) | 177 (43.6) | 69 (38.3) | 47 (60.3) | 2.50 (1.47-4.25) | 0.0010 | |
| AA | 14 (7.9) | 42 (10.3) | 16 (8.9) | 9 (11.5) | 2.31 (1.01-5.27) | 0.046 | |
| GA + AA | 86 (48.6) | 219 (53.9) | 85 (47.2) | 56 (71.8) | 2.47 (1.47-4.13) | 0.00030 | |
| Guangdong | (n = 226) | (n = 372) | (n = 260) | (n = 65) | |||
| GG | 123 (54.4) | 184 (49.5) | 134 (51.5) | 25 (38.5) | 1.00 (reference) | ||
| GA | 87 (38.5) | 152 (40.8) | 105 (40.4) | 37 (56.9) | 1.95 (1.14-3.34) | 0.015 | |
| AA | 16 (7.1) | 36 (9.7) | 21 (8.1) | 3 (4.6) | 0.79 (0.23-2.72) | 0.71 | |
| GA + AA | 103 (45.6) | 188 (50.5) | 126 (48.5) | 40 (61.5) | 1.75 (1.03-2.98) | 0.034 | |
| Pooled | (n = 403) | (n = 778) | (n = 440) | (n = 143) | |||
| GG | 214 (53.1) | 371 (47.7) | 229 (52.0) | 47 (32.9) | 1.00 (reference) | ||
| GA | 159 (39.5) | 329 (42.3) | 174 (39.5) | 84 (58.7) | 2.22 (1.51-3.20) | 4.07 × 10−5 | |
| AA | 30 (7.4) | 78 (10.0) | 37 (8.5) | 12 (8.4) | 1.54 (0.79-2.99) | 0.21 | |
| GA + AA | 189 (46.9) | 407 (52.3) | 211 (48.0) | 96 (67.1) | 2.08 (1.44-3.01) | 8.60 × 10−5 | |
| Overallb | (n = 403) | (n = 778) | (n = 440) | (n = 143) | |||
| GG | 214 (53.1) | 371 (47.7) | 229 (52.0) | 47 (32.9) | 1.00 (reference) | ||
| GA | 159 (39.5) | 329 (42.3) | 174 (39.5) | 84 (58.7) | 2.21 (1.52-3.23) | 3.82 × 10−5 | |
| AA | 30 (7.4) | 78 (10.0) | 37 (8.5) | 12 (8.4) | 1.66 (0.83-3.29) | 0.15 | |
| GA + AA | 189 (46.9) | 407 (52.3) | 211 (48.0) | 96 (67.1) | 2.09 (1.44-3.03) | 9.64 × 10−5 | |
Note: Guangxi and Guangdong populations consist of 855 and 962 patients with known lymph node involvement (N classification) stage, respectively. Due to DNA quality and/or quantity, the actual sample sizes were 841 and 923 patients in the Guangxi and Guangdong population, respectively
Abbreviations: N lymph node involvement, OR odds ratio, CI confidence interval
aIn the Guangxi population, the ORs, 95 % CIs and P values were calculated for N3 vs. N0 + N1 + N2 and adjusted for sex, age, status of smoking and drinking, smoking level, family history and nationality. In the Guangdong population, the ORs, 95 % CIs and P values were calculated for N3 vs. N0 + N1 + N2 and adjusted for sex, age, status of smoking and drinking, and smoking level. In the pooled population, the ORs, 95 % CIs and P values were calculated for N3 vs. N0 + N1 + N2 and adjusted for sex, age, status of smoking and drinking, and smoking level. Another confounding factor, native place, was also adjusted in the pooled population
bA meta-analysis combining two independent case–control studies. The P values for heterogeneity among sample sets are 0.52 for GA vs. GG group, 0.18 for AA vs. GG and 0.36 for GA + AA vs. GG group, respectively
The association between the rs3928672 A allele (GA + AA genotype) and advanced lymph node metastasis of NPC were further examined with stratification by age, sex, status of smoking and drinking, smoking level, family history and nationality (Additional file 1: Table S7). A significantly increased risk of lymph node metastasis of NPC was found in the heavier smokers (≥ 24 pack-years) compared with the lighter smokers (P = 0.022, test for homogeneity). However, the smoking level had no modification effect on the severity of NPC related to the rs3928672 in the Guangdong population.
The power for this association study was calculated using the PGA program [25], based on our sample sizes in the Guangxi population (78 patients with N3 stage vs. 763 patients with N0 + N1 + N2 stage), the minor A allele frequency of 31.3 %, and an OR of 2.47. By this calculation, the power of our initial association study in the Guangxi population to detect rs3928672 was estimated to be 44.0 %.
Effect of the rs3928672 on AGO2 protein expression
We further investigated the protein expression level of AGO2 in NPC tissues and non-cancerous nasopharyngeal tissues by IHC assay. Consistent with a previous report [12], the AGO2 protein was mainly accumulated in the cytoplasm of malignant cells. We detected high expression of AGO2 in the 32 of 37 NPC tissues (86.5 %), but in the 10 of 18 non-cancerous nasopharyngeal tissues (55.5 %) (χ2 = 6.42, P = 0.011; Fig. 1 and Additional file 1: Table S8). Furthermore, there was significant association between the rs3928672 genotypes and expression level of AGO2 protein in the NPC tissues, with the A allele (GA + AA genotype) carriers having higher AGO2 expression than the GG genotype carriers (P = 0.041; Additional file 1: Table S8).
Fig. 1.
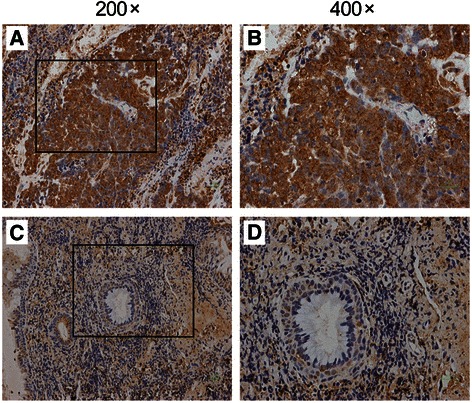
Protein expression level of AGO2 by immunohistochemical staining in representative NPC tissues and non-cancerous nasopharyngeal tissues. Panels a and b NPC tissues; Panels c and d non-cancerous nasopharyngeal tissues. Images in the box (left, magnification 200 ×) were enlarged and shown in the right (magnification 400 ×). The AGO2 protein was found to locate in the cytoplasm of malignant cells
Effect of AGO2 knockdown on cell proliferation, apoptosis and migration in vitro
Given that the genetic variant of AGO2 is associated with NPC progression, we speculated that AGO2, which was over-expressed in NPC tissues, might influences the tumorigenic properties of NPC cells. We used three AGO2 specific shRNAs to knock down AGO2 in a human nasopharyngeal carcinoma cell line CNE2Z, and western blots assay confirmed that the protein expression level of AGO2 was remarkably decreased in CNE2Z cells after AGO2 knockdown (Fig. 2a). The CCK-8 assay was carried out to examine whether AGO2 knockdown affected NPC cell proliferation. As shown in Fig. 2b, CNE2Z cells transfected with AGO2 shRNAs had a significant viability reduction over time compared with controls (P < 0.0001), suggesting that AGO2 knockdown significantly inhibited proliferation of NPC cells. Moreover, the percentages of apoptotic cells in CNE2Z cells transfected with AGO2 shRNAs was increased 14.4 % compared with controls (P < 0.0001; Fig. 2c), indicating AGO2 knockdown induced apoptosis of NPC cells. Furthermore, migratory cells in CNE2Z cells transfected with AGO2 shRNAs was 36 ± 3, approximately 7.8-fold lower (P < 0.0001) than those in CNE2Z cells transfected with control shRNA (282 ± 34), suggesting AGO2 knockdown reduced migration ability of NPC cells (Fig. 2d).
Fig. 2.
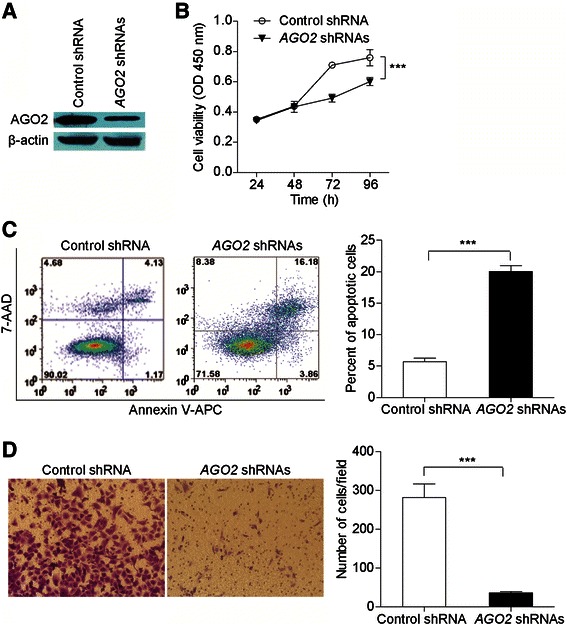
The effect of AGO2 knockdown on proliferation, apoptosis and migration of NPC cells. a Confirmation of AGO2 knockdown in CNE2Z cells transfected with AGO2 shRNAs or control shRNA by western blot assay. b Cell proliferation was evaluated by CCK-8 assay in CNE2Z cells transfected with AGO2 shRNAs or control shRNA at different time points (24, 48, 72 and 96 h). The experiments were repeated at least three times, and the points represent the mean values of triplicate tests (mean ± SD). ***P < 0.0001, compared with the controls (ANOVA test). c Apoptosis was assessed by flow cytometric analysis of annexin V-APC/7-AAD staining in CNE2Z cells transfected with AGO2 shRNAs or control shRNA. Representative flow cytometric analysis are shown (left panel). The Annexin V-positive cells were regarded as apoptotic cells. The experiments were repeated at least three times, and the histogram (right panel) represents the mean values of apoptotic cells from triplicate tests (mean ± SD). ***P < 0.0001, compared with the controls (Unpaired t test). d Cell migration was evaluated by transwell assay in CNE2Z cells transfected with AGO2 shRNAs or control shRNA. Representative results are shown (left panel). Magnification: 400 ×. The experiments were repeated at least three times, and the histogram (right panel) represents the mean numbers of transferred cells from triplicate tests (mean ± SD). ***P < 0.0001, compared with the controls (Unpaired t test). CCK-8, Cell Counting Kit-8; SD, standard deviation
Altered gene expression in response to AGO2 knockdown
To elucidate the comprehensive regulation in transcriptional profile induced by AGO2, whole-genome expression microarray analysis was conducted between the CNE2Z cells transfected with AGO2 shRNAs and control shRNA. A total of 1160 genes were altered significantly, including 767 genes were down-regulated and 393 genes were up-regulated, in CNE2Z cells after AGO2 knockdown compared with controls (P < 0.05; Additional file 2: Table S9).
To determine whether specific biological processes or signaling pathways were involved in the AGO2 knockdown-induced anticancer effects, we conducted biological network analysis of these 1160 deferentially expressed genes using the cytoscape plug-in of Reactome FI [23]. Figure 3 shows some of the most significant biological processes or signaling pathways (full list in Additional file 3: Table S10). Remarkably, the majority of the enriched biological processes were related to the cell cycle, apoptosis and cell adhesion, which were hallmarks during the development of cancers. Furthermore, signaling pathways related to the tumor metastasis, namely “Signaling by Rho GTPases” [26], “Notch signaling pathway” [27] and “Wnt signaling pathway” [28] were also significantly enriched. All together, these observations indicate that AGO2 is associated with the development of NPC by regulating expression of numerous cancer related genes.
Fig. 3.
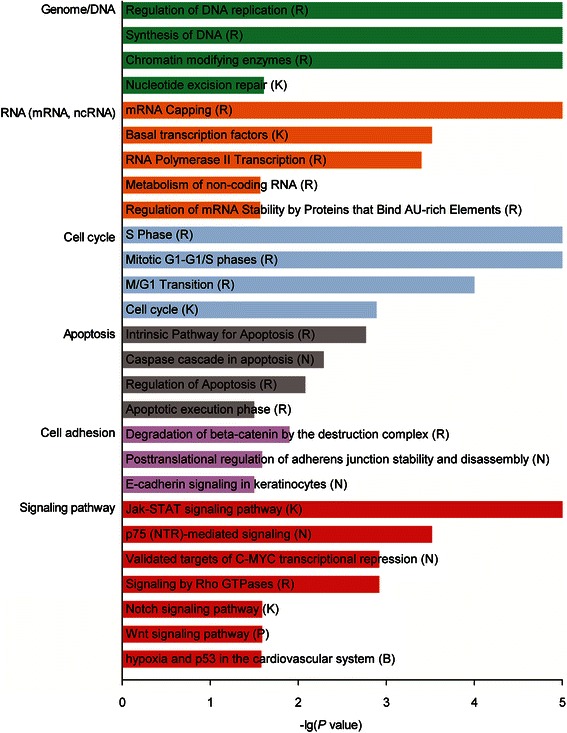
The most enriched biological processes and signaling pathways of the 1160 genes with significantly altered expression following AGO2 knockdown in CNE2Z cells by cytoscape plug-in of Reactome FI analysis. “G” represents “GO biological process”, “K” represents “KEGG pathway”, “N” represents “NCI PID”, “R” represents “Reactome”, “P” represents “Reactome Pathway” and “B” represents “BioCarta”. The detailed results were shown in Additional file 3: Table S10
Discussion
In the present study, we systematically evaluated the effect of SNPs in the AGO2 gene on the risk of occurrence or progression of NPC in two case–control populations of Chinese ancestry. We found that the AGO2 rs3928672 was significantly associated with the advanced lymph node metastasis of NPC. The functional experiments demonstrated that the genotype of rs3928672 was significantly associated with the expression of AGO2 in NPC tissues, and AGO2 can play an oncogenic role in the development of NPC by regulating genes related to the tumorigenesis and metastasis. To the best of our knowledge, this is the first report that the genetic polymorphisms of AGO2 may be risk factor for the progression of NPC, and AGO2 acts as an oncogene in NPC.
The polymorphisms in the AGO2 gene have been used to search for susceptibility alleles of a wide spectrum of cancers (Additional file 1: Table S11). For instance, three SNPs, i.e. rs3864659, rs2292779 and rs11786030, in the AGO2 gene were shown to be associated with breast cancer in Korea population [17, 18]. Unfortunately, on the basis of analysis of 855 NPC cases and 1036 controls in the Guangxi population, we did not find a statistically significant association between these SNPs and the susceptibility to NPC. The inconsistent results may be attributed to different molecular mechanisms of carcinogenesis among cancers, small sample size, marginal statistical significance and different ethnicities of study populations. However, by using the tag SNP approach, we identified a novel tag SNP (i.e. rs3928672) that was associated with the risk of lymph node metastasis of NPC in two Chinese subpopulations. Together, our results support the AGO2 gene as a susceptibility gene for cancers.
In both study populations, the subjects bearing the rs3928672 A allele (GA + AA genotype) had a significantly increased frequency of involvement of lymph node metastasis of NPC compared with ones bearing GG genotype. Moreover, the rs3928672 A allele carriers having higher AGO2 protein expression than the GG genotype carriers in NPC tissues. Given the role of AGO2 in the development of NPC, one might expect individuals who carry the rs3928672 A allele, and thus have increased expression of AGO2 over a lifetime, may have a higher risk of developing lymph node metastasis of NPC after establishment of this malignancy. However, the rs3928672, which is a polymorphism in intron 4 of AGO2, may not have a functional consequence. It is plausible that some other functional polymorphisms in linkage disequilibrium (LD) with the rs3928672 should be responsible for the allele-specific expression. However, the rs3928672 is in low linkage disequilibrium with other SNPs in both the patients and comparison subjects in the Guangxi population as well as in the HapMap CHB (r2 < 0.80), indicating that the association of rs3928672 with NPC severity is likely independent. Alternatively, we cannot exclude the possibility that the rs3928672 itself is a functional variant directly affecting the AGO2 production. Introns in eukaryotes fulfill a broad spectrum of functions, such as acting as transposable elements, and are involved in virtually every step of mRNA processing [29]. Indeed, by computer analysis (F-SNP database, http://compbio.cs.queensu.ca/F-SNP) we found that the polymorphism rs3928672 maybe influence binding of transcription factor RUNX1 (runt-related transcription factor 1, also called AML-1) and CAP1 [CAP, adenylate cyclase-associated protein 1 (yeast)], since the sequences flanking rs3928672 A allele (5’-AGTGGT-3’) as a potential binding site for the RUNX1, and the sequences flanking G allele (5’-TCACCGCT-3’) as a potential binding site for the CAP1, respectively. Therefore, one mechanism by which this could occur is if the risky rs3928672 A allele can influence binding of transcription factor in the intron 4 of AGO2. Further studies are needed to clarify which polymorphism(s) may possess functional consequence(s) for AGO2, and in turn to provide mechanistic plausibility for the observed association between rs3928672 and involvement of lymph node metastasis of NPC.
The mechanism of how AGO2 SNPs regulates human susceptibility to cancer is unknown. However, AGO2 has significant roles controlling the tumorigenesis and progression of several cancers, including tumor invasion and metastasis [8–10, 14, 15]. This processes appears to be through two different molecular mechanisms, RNAi-dependent gene silencing [8, 30] and RNAi-independent ways such as stabilizing insulator-independent looping [31], facilitating DNA double-strand break repair [32], targeting the intragenic long interspersed nuclear element-1 (LINE-1) transcription complexes [33], and directly regulating downstream gene expression [9]. Consistent with the previous study, we showed that the AGO2 protein was significantly over-expressed in NPC tissues compared with non-cancerous nasopharyngeal epithelium tissues in the present study. Moreover, functional experiments illustrate that AGO2 knockdown reduced cell proliferation, induced apoptosis, and inhibited migration of NPC cells. These effects of AGO2 knockdown were further corroborated by genes with altered expression following AGO2 knockdown, which were functionally clustered in biological processes related with cell cycle, apoptosis and cell migration. Furthermore, several tumorigenesis and metastasis associated genes with altered expression following AGO2 knockdown were targets of miRNAs (e.g., TPBG [34], JUP [35], CDKN1A [36] and S100A11 [37] were experimentally validated targets of miR-155 [38, 39]). Therefore, AGO2 may promote tumorigenesis and metastasis by regulating miRNAs and their targets in NPC. Taken together, our findings indicated that AGO2 over-expression can contribute to NPC malignant behaviors.
In reviewing the results of this study, one must also keep several potential limits in mind. First, as a hospital-based study, our NPC cases were recruited from the hospital, while the controls were selected from the community population; inherent selection bias cannot be completely excluded. However, by further adjustment and stratification in data analyses, the potential confounding effects might have been minimized. Second, a number of association studies have addressed identifying the genes that may relate to the susceptibility to NPC [40–42]. Most of the results, however, could not be replicated in subsequent studies in other populations. Although the highly significant association between the AGO2 and lymph node metastasis of NPC was strengthened by our two independent case–control studies, our initial findings should be independently verified in other populations with high incidence rate of NPC, such as other Southern Chinese, Singaporeans, and Taiwanese.
Conclusions
In conclusion, we have shown that the AGO2 polymorphism may be a genetic risk factor for NPC in Chinese population. If confirmed by other studies, knowledge of genetic factors contributing to the pathogenesis of the NPC as presented here may have implications for the cancer screening and treatment of this disorder.
Availability of supporting data
Gene expression data have been submitted to Gene Expression Omnibus (GEO) database with the series accession number GSE74154, and these data are available free of charge via the Internet at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE74154.
Acknowledgments
We are grateful to the patients and their families for participating in this study. We also thank all of the clinicians, nurses and study coordinators for their contributions to the work. This project was supported by grants from Chinese National Natural Science Foundation (No. 30900819 and 81125017). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- AGO2
argonaute 2
- CI
confidence interval
- IHC
immunohistochemistry
- NPC
nasopharyngeal carcinoma
- OR
odds ratio
- SNP
single nucleotide polymorphism
- TNM classification
tumor-node-metastasis classification
Additional files
Selected characteristics of patients with NPC and controls in the Guangxi and Guangdong population. Table S2. Selected characteristics of patients with NPC and controls for the immunohistochemistry analysis. Table S3. Primers used for the genotyping assays. Table S4. Genotype frequencies in patients with NPC and controls in the Guangxi population. Table S5. Association between AGO2 polymorphisms genotypes and local tumor invasion of NPC. Table S6. Association between AGO2 polymorphisms genotypes and lymph node metastasis of NPC. Table S7. Risk of lymph node metastasis of NPC associated with the AGO2 rs3928672 by potential risk factors in the Guangxi and Guangdong population. Table S8. Correlation between protein expression levels of AGO2 and rs3928672 genotypes in NPC tissues and non-cancerous nasopharyngeal tissues by immunohistochemistry. Table S11. Summary of previous studies on the genetic associations between AGO2 and cancers. (DOC 525 kb)
Genes with significantly altered expression following AGO2 knockdown in CNE2Z cells. (XLS 208 kb)
Biological network analysis of genes with significantly altered expression following AGO2 knockdown in CNE2Z cells. (XLS 29 kb)
Footnotes
Peiyao Li and Jinfeng Meng contributed equally to this work.
Competing interests
The authors have declared that no competing interests exist.
Authors’ contributions
PL and JM conceived and designed the experiments. PL performed the genotyping, analyzed the data, and drafted the manuscript. JM performed the functional experiments, and analyzed the data. PC and XC analyzed the microarray data. HZ, LY, ZW, XZ, YZ, HC, YL and YL helped to the sample preparation and genotyping. YZ and YH helped to perform functional experiments. YC was responsible for recruitment of Guangxi subjects, phenotype collection and biological sample collection. JB and YZ were responsible for recruitment of Guangdong subjects, phenotype collection and biological sample collection. GZ and FH conceived and designed the experiments, and drafted and revised the manuscript. All authors read and approved the final manuscript.
Contributor Information
Peiyao Li, Email: lipeiyao214@gmail.com.
Jinfeng Meng, Email: zb.meng@163.com.
Yun Zhai, Email: zhaiyun2012@126.com.
Hongxing Zhang, Email: zhanghx08@126.com.
Lixia Yu, Email: yulixia_lili@163.com.
Zhifu Wang, Email: wangzhifu_1@163.com.
Xiaoai Zhang, Email: babylovehopi@163.com.
Pengbo Cao, Email: birchcpb@163.com.
Xi Chen, Email: arhelion@163.com.
Yuqing Han, Email: hanyuqing_1985@163.com.
Yang Zhang, Email: y_zh2001@hotmail.com.
Huipeng Chen, Email: chenhp0909@163.com.
Yan Ling, Email: lingny@me.com.
Yuxia Li, Email: liyuxia990306@126.com.
Ying Cui, Email: 13907863066@139.com.
Jin-Xin Bei, Email: beijinx@mail.sysu.edu.cn.
Yi-Xin Zeng, Email: zengyx@sysucc.org.cn.
Fuchu He, Phone: 86-10-68177417, Email: hefc@nic.bmi.ac.cn.
Gangqiao Zhou, Phone: 86-10-80705255, Email: zhougq114@126.com.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Wei WI, Sham JS. Nasopharyngeal carcinoma. Lancet. 2005;365(9476):2041–2054. doi: 10.1016/S0140-6736(05)66698-6. [DOI] [PubMed] [Google Scholar]
- 3.Bei JX, Li Y, Jia WH, Feng BJ, Zhou G, Chen LZ, et al. A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat Genet. 2010;42(7):599–603. doi: 10.1038/ng.601. [DOI] [PubMed] [Google Scholar]
- 4.Zhou G, Zhai Y, Cui Y, Zhang X, Dong X, Yang H, et al. MDM2 promoter SNP309 is associated with risk of occurrence and advanced lymph node metastasis of nasopharyngeal carcinoma in Chinese population. Clin cancer res : an official journal of the American Association for Cancer Research. 2007;13(9):2627–2633. doi: 10.1158/1078-0432.CCR-06-2281. [DOI] [PubMed] [Google Scholar]
- 5.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15(2):185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328(5986):1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoo NJ, Hur SY, Kim MS, Lee JY, Lee SH. Immunohistochemical analysis of RNA-induced silencing complex-related proteins AGO2 and TNRC6A in prostate and esophageal cancers. APMIS: acta pathol, microbiol, et immunol Scand. 2010;118(4):271–276. doi: 10.1111/j.1600-0463.2010.02588.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Y, Chen L, Barlogie B, Stephens O, Wu X, Williams DR, et al. High-risk myeloma is associated with global elevation of miRNAs and overexpression of EIF2C2/AGO2. Proc Natl Acad Sci U S A. 2010;107(17):7904–7909. doi: 10.1073/pnas.0908441107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng N, Li Y, Han ZG. Argonaute2 promotes tumor metastasis by way of up-regulating focal adhesion kinase expression in hepatocellular carcinoma. Hepatology. 2013;57(5):1906–1918. doi: 10.1002/hep.26202. [DOI] [PubMed] [Google Scholar]
- 10.Chang SS, Smith I, Glazer C, Hennessey P, Califano JA. EIF2C is overexpressed and amplified in head and neck squamous cell carcinoma. ORL; journal for oto-rhino-laryngology and its related specialties. 2010;72(6):337–343. doi: 10.1159/000320597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L, Yu C, Gao H, Li Y. Argonaute proteins: potential biomarkers for human colon cancer. BMC Cancer. 2010;10:38. doi: 10.1186/1471-2407-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang FQ, Huang JH, Liu M, Yang FP, Li W, Wang GC, et al. Argonaute 2 is up-regulated in tissues of urothelial carcinoma of bladder. Int J Clin Exp Pathol. 2014;7(1):340–347. [PMC free article] [PubMed] [Google Scholar]
- 13.Wang M, Ren D, Guo W, Wang Z, Huang S, Du H, et al. Loss of miR-100 enhances migration, invasion, epithelial-mesenchymal transition and stemness properties in prostate cancer cells through targeting Argonaute 2. Int J Oncol. 2014;45(1):362–372. doi: 10.3892/ijo.2014.2413. [DOI] [PubMed] [Google Scholar]
- 14.Naoghare PK, Tak YK, Kim MJ, Han E, Song JM. Knock-down of argonaute 2 (AGO2) induces apoptosis in myeloid leukaemia cells and inhibits siRNA-mediated silencing of transfected oncogenes in HEK-293 cells. Basic Clin Pharmacol Toxicol. 2011;109(4):274–282. doi: 10.1111/j.1742-7843.2011.00716.x. [DOI] [PubMed] [Google Scholar]
- 15.Bian XJ, Zhang GM, Gu CY, Cai Y, Wang CF, Shen YJ, et al. Down-regulation of Dicer and Ago2 is associated with cell proliferation and apoptosis in prostate cancer. Tumour biol: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35(11):11571–11578. doi: 10.1007/s13277-014-2462-3. [DOI] [PubMed] [Google Scholar]
- 16.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131(6):1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 17.Sung H, Lee KM, Choi JY, Han S, Lee JY, Li L, et al. Common genetic polymorphisms of microRNA biogenesis pathway genes and risk of breast cancer: a case–control study in Korea. Breast Cancer Res Treat. 2011;130(3):939–951. doi: 10.1007/s10549-011-1656-2. [DOI] [PubMed] [Google Scholar]
- 18.Sung H, Jeon S, Lee KM, Han S, Song M, Choi JY, et al. Common genetic polymorphisms of microRNA biogenesis pathway genes and breast cancer survival. BMC Cancer. 2012;12:195. doi: 10.1186/1471-2407-12-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weng Y, Chen Y, Chen J, Liu Y, Bao T. Common genetic variants in the microRNA biogenesis pathway are associated with malignant peripheral nerve sheath tumor risk in a Chinese population. Cancer Epidemiol. 2013;37(6):913–916. doi: 10.1016/j.canep.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Zhang H, Zhai Y, Wang Z, Ma F, Wang H, et al. A functional tandem-repeats polymorphism in the downstream of TERT is associated with the risk of nasopharyngeal carcinoma in Chinese population. BMC Med. 2011;9:106. doi: 10.1186/1741-7015-9-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Zhai Y, Hu Z, Wu C, Qian J, Jia W, et al. Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers. Nat Genet. 2010;42(9):755–758. doi: 10.1038/ng.638. [DOI] [PubMed] [Google Scholar]
- 23.Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39(Database issue):D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74(4):765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menashe I, Rosenberg PS, Chen BE. PGA: power calculator for case–control genetic association analyses. BMC Genet. 2008;9:36. doi: 10.1186/1471-2156-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitz AA, Govek EE, Bottner B, Van Aelst L. Rho GTPases: signaling, migration, and invasion. Exp Cell Res. 2000;261(1):1–12. doi: 10.1006/excr.2000.5049. [DOI] [PubMed] [Google Scholar]
- 27.Hu YY, Zheng MH, Zhang R, Liang YM, Han H. Notch signaling pathway and cancer metastasis. Adv Exp Med Biol. 2012;727:186–198. doi: 10.1007/978-1-4614-0899-4_14. [DOI] [PubMed] [Google Scholar]
- 28.Xi Y, Chen Y. Wnt signaling pathway: Implications for therapy in lung cancer and bone metastasis. Cancer Lett. 2014;353(1):8–16. doi: 10.1016/j.canlet.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 29.Chorev M, Carmel L. The function of introns. Front genet. 2012;3:55. doi: 10.3389/fgene.2012.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang M, Haase AD, Huang FK, Coulis G, Rivera KD, Dickinson BC, et al. Dephosphorylation of tyrosine 393 in argonaute 2 by protein tyrosine phosphatase 1B regulates gene silencing in oncogenic RAS-induced senescence. Mol Cell. 2014;55(5):782–790. doi: 10.1016/j.molcel.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moshkovich N, Nisha P, Boyle PJ, Thompson BA, Dale RK, Lei EP. RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev. 2011;25(16):1686–1701. doi: 10.1101/gad.16651211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao M, Wei W, Li MM, Wu YS, Ba Z, Jin KX, et al. Ago2 facilitates Rad51 recruitment and DNA double-strand break repair by homologous recombination. Cell Res. 2014;24(5):532–541. doi: 10.1038/cr.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aporntewan C, Phokaew C, Piriyapongsa J, Ngamphiw C, Ittiwut C, Tongsima S, et al. Hypomethylation of intragenic LINE-1 represses transcription in cancer cells through AGO2. PLoS One. 2011;6(3) doi: 10.1371/journal.pone.0017934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elkord E, Dangoor A, Drury NL, Harrop R, Burt DJ, Drijfhout JW, et al. An MVA-based vaccine targeting the oncofetal antigen 5 T4 in patients undergoing surgical resection of colorectal cancer liver metastases. J Immunother. 2008;31(9):820–829. doi: 10.1097/CJI.0b013e3181876ab3. [DOI] [PubMed] [Google Scholar]
- 35.Kowalewska M, Radziszewski J, Goryca K, Bujko M, Oczko-Wojciechowska M, Jarzab M, et al. Estimation of groin recurrence risk in patients with squamous cell vulvar carcinoma by the assessment of marker gene expression in the lymph nodes. BMC Cancer. 2012;12:223. doi: 10.1186/1471-2407-12-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng X, Xia W, Yang JY, Hsu JL, Chou CK, Sun HL, et al. Activation of p21(CIP1/WAF1) in mammary epithelium accelerates mammary tumorigenesis and promotes lung metastasis. Biochem Biophys Res Commun. 2010;403(1):103–107. doi: 10.1016/j.bbrc.2010.10.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao MB, Jiang F, Ni WK, Chen BY, Lu CH, Li XY, et al. High expression of S100A11 in pancreatic adenocarcinoma is an unfavorable prognostic marker. Med Oncol. 2012;29(3):1886–1891. doi: 10.1007/s12032-011-0058-y. [DOI] [PubMed] [Google Scholar]
- 38.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 39.Xie Q, Chen X, Lu F, Zhang T, Hao M, Wang Y, et al. Aberrant expression of microRNA 155 may accelerate cell proliferation by targeting sex-determining region Y box 6 in hepatocellular carcinoma. Cancer. 2012;118(9):2431–2442. doi: 10.1002/cncr.26566. [DOI] [PubMed] [Google Scholar]
- 40.Cho EY, Hildesheim A, Chen CJ, Hsu MM, Chen IH, Mittl BF, et al. Nasopharyngeal carcinoma and genetic polymorphisms of DNA repair enzymes XRCC1 and hOGG1. Cancer epidemiol, biomarkers prev: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2003;12(10):1100–1104. [PubMed] [Google Scholar]
- 41.Tiwawech D, Srivatanakul P, Karalak A, Ishida T. Glutathione S-transferase M1 gene polymorphism in Thai nasopharyngeal carcinoma. Asian Pac j cancer prev: APJCP. 2005;6(3):270–275. [PubMed] [Google Scholar]
- 42.He Y, Zhou G, Zhai Y, Dong X, Lv L, He F, et al. Association of PLUNC gene polymorphisms with susceptibility to nasopharyngeal carcinoma in a Chinese population. J Med Genet. 2005;42(2):172–176. doi: 10.1136/jmg.2004.022616. [DOI] [PMC free article] [PubMed] [Google Scholar]


