Abstract
Immunoglobulin variable region heavy chain (IgVH) somatic gene diversification is instrumental in the transformation process that characterizes hepatitis C virus (HCV)-related B cell lymphoproliferative disorders. However, the extent to which activation-induced cytidine deaminase (AID), an enzyme essential for IgV gene somatic hypermutation (SHM), is active in cryoglobulinaemic vasculitis (CV) remains unclear. AID mRNA expression in the peripheral blood of 102 chronically hepatitis C virus (HCV)-infected patients (58 with and 44 without CV) and 26 healthy subjects was investigated using real-time reverse transcription–polymerase chain reaction (RT–PCR). The features of activation-induced cytidine deaminase (AID) protein and mRNA transcripts were explored in liver tissue biopsies and portal tracts isolated using laser capture microdissection. In chronically HCV-infected patients, AID mRNA expression was almost threefold higher in those with than in those without CV and sevenfold higher than in healthy subjects (median-fold: 6·68 versus 2·54, P = 0·03 and versus 0·95, P = 0·0003). AID transcript levels were significantly higher in polyclonal than in clonally restricted B cell preparations in either CV or non-CV patients (median-fold, 15·0 versus 2·70, P = 0·009 and 3·46 versus 1·58, P = 0·02, respectively). AID gene expression was found to be related negatively to age and virological parameters. AID protein was found in portal tracts containing inflammatory cells that, in several instances, expressed AID mRNA transcripts. Our data indicate that the aberrant expression of AID may reflect continuous B cell activation and sustained survival signals in HCV-related CV patients.
Keywords: cryoglobulinaemia, HCV infection, immunoglobulin variable gene, lymphoproliferation, somatic hypermutation
Introduction
Mixed cryoglobulinaemia (MC) is the most common extrahepatic manifestation of chronic hepatitis C virus (HCV) infection 1. Although the condition is detected in 50–60% of HCV-positive patients 2, cryoglobulin-related illness, known as cryoglobulinaemic vasculitis (CV), appears in only a minority (10–15%) of patients and includes a large spectrum of symptoms, ranging in severity from mild sporadic purpura to life-threatening features 3.
B cell activation and clonal expansions are peculiar events in the humoral immune response of patients with HCV-related CV 4. Dominant B cell clonalities are likely to contribute to the formation of liver intraportal follicle-like structures 5. Analysis of immunoglobulin heavy chain (IgH) complementarity-determining region-3, whether from circulating or tissue-derived B cell-expanded clones, showed several variations in this immunoglobulin variable region heavy chain (IgVH) gene segment, supporting the notion that these cells are the result of an antigen-driven response 6. In CV patients, restriction in the use of the B cell V gene has been shown to have a direct clinical impact, in that it is associated with higher levels of rheumatoid factor (RF) activity and lymphoproliferative disorders 7.
The cryoprecipitating immune complexes (ICs) in HCV-related CV are the result of a host reaction involving primarily immunoglobulin (Ig)M molecules with RF activity, capable of activating the complement cascade through the C1q receptor 8. HCV particles and HCV-encoded core protein linked to the corresponding anti-HCV antibodies form circulating ICs that bind to a RF-like B cell receptor, resulting in B cell activation and clonal expansion 9.
The mechanisms of B cell activation in CV are still undefined. Thus, it is unclear whether B cell clonal expansions are related directly to cryoglobulin formation or are the consequence of pathogenetic noxae that impair the regulation of the host peripheral immune response. B cell activation is regulated directly by the interaction of HCV with B cells, in that HCV binds to the cell membrane, probably via incomplete sets of receptors, including CD81, scavenger receptor class B member 1 (SRBI), low-density lipoprotein receptor (LDL-R) and Niemann-Pick C1-like 1 (NPC1L1), which permit strict attachment of the virus to the cell surface 10,11. A lack of claudin and occludin was shown to preclude virus internalization and to account for the lower efficiency of HCV infection of human B cells than hepatocytes 12,13. None the less, human B cells may be infected with HCV, and infection may result in functional abnormalities 14–17.
Proliferating B cells are always accompanied by the expression of the gene encoding activation-induced cytidine deaminase (AID), a B cell-restricted enzyme required for somatic hypermutation (SHM) and class-switch recombination (CSR) 18,19. While CSR induces antibody diversity by permitting antigen recognition by different classes of antibodies, SHM incorporates point mutations in the recombined VDJ exon of the heavy- and light-chain-encoding genes, which in turn enhances antibody affinity to specific antigens, resulting in selective B cell enrichment and clonal expansion 20.
Maturational events of normal B cells are concentrated in the germinal centres (GCs) of lymph nodes and possibly in other locations as well 21. It is becoming increasingly apparent that SHM and CSR also occur at sites of chronic or recurrent antigen stimulation, suggesting that unique local mechanisms are able to enhance the development of aggregates into ectopic GCs, as is the case in rheumatoid arthritis 22.
There is unequivocal evidence that B cell stimulating HCV antigens lead to a dysregulated polyclonal and subsequently monoclonal B cell expansion that can, in some instances, result in frank malignancy 23. In addition, intrinsic hypermutable sites enhance the affinity maturation of recurring mutational clusters in IgVH alleles and generate effective mutated antibody repertoires against an invading pathogen 24.
The ability of HCV to induce factors involved in DNA SHM leads to the question of whether B cell clonal changes are AID-mediated. In this study, we analysed the AID gene expression profiles of peripheral blood and liver-derived B cells from chronically HCV-infected patients, with the aim of assessing the difference in gene expression between those with and without CV.
Materials and methods
Fifty-eight patients with CV [median age: 59·2 ± 11·7 years; male/female (M/F): 0·2] were selected according to recent validated criteria 25. The eligibility criteria were as follows: (a) no previous administration of interferons, steroids or immunosuppressive agents; (b) serum positivity for anti-HCV antibodies and circulating HCV RNA; (c) liver biopsies showing features of chronic active hepatitis; (d) serum negativity for hepatitis B virus surface antigen (HBsAg) and anti-HIV; and (e) exclusion of connective tissue diseases and malignant lymphoproliferative disorders. Forty-four additional patients (median age: 56·4 ± 13·1; M/F: 1·0) had a diagnosis of chronic active hepatitis and chronic HCV infection without CV. The control group consisted of 26 healthy blood donors (median age: 51·2 ± 13·5 years; M/F: 1·2). Local ethical committee approval was received for this study. Written informed consent was obtained from all participants in accordance with the Declaration of Helsinki. Activity score and degree of fibrosis in liver biopsies were evaluated according to the METAVIR scale 26. Serum cryoglobulins were isolated and purified as described elsewhere 27.
Laboratory parameters
Serum HCV antibodies were detected by third-generation enzyme-linked immunosorbent assay (ELISA) (Abbott Laboratories, Chicago, IL, USA). Serum HCV RNA was determined by reverse transcription–polymerase chain reaction (RT–PCR) (Roche Diagnostics System, Branchburg, NJ, USA) and quantified with the Versant HCV RNA quantitative 3·0 assay (Siemens Healthcare, Erlangen, Germany). HCV genotyping was performed with INNO-LiPA (Innogenetics NV, Ghent, Belgium).
Real-time RT–PCR for AID mRNA quantification in peripheral blood mononuclear cells (PBMCs)
PBMCs were isolated from blood samples using Histopaque-1077 (Sigma-Aldrich, St Louis, MO, USA). CD19+ B cells were counted by flow cytometry and normalized to 106 cells/sample. Frequency and distribution range of peripheral B lymphocytes in HCV-infected patients with and without CV and healthy individuals were 14·4 ± 12·1; 13·1 ± 7·6 and 16·6 ± 7·2, respectively. The groups had comparable total lymphocyte counts of 1336 ± 288 cells/µl in CV patients; 1400 ± 189 cells/µl in non-CV patients and 1290 ± 160 cells/µl in healthy individuals. Total RNA was extracted with the RNeasy mini kit (Qiagen, Hilden, Germany), and reverse-transcribed to complementary DNA (cDNA) using the iScript Select cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA, USA). AID mRNA was measured in patient samples by relative real-time RT–PCR using the RealTime Ready single assays for AID (ID: 137580) and β-actin (ID: 101125) (target and reference genes, respectively) (Roche Diagnostics, Mannheim, Germany) and the LC TaqMan Master kit (Roche Diagnostics) on a LightCycler 1·5 instrument (Roche Diagnostics).
B cell clonal expansions
Clonal IgH-chain gene rearrangements were analysed using the IgH somatic hypermutation assay version 2·0 (Invivoscribe, San Diego, CA, USA). Twenty μl of the reaction mixture was subjected to electrophoresis on a 4% certified low-range ultra agarose gel (Bio-Rad) in Tris–borate–ethylenediamine teraacetic acid (EDTA) (TBE) buffer, stained with GelRed (Biotium, Hayward, CA, USA). Control mixtures in the reaction were devoid of RNA or included RNA from a patient with chronic lymphocytic leukaemia.
Immunofluorescence assays
AID protein in liver biopsy samples was detected by an indirect immunofluorescence assay, using a rabbit anti-human AID antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and, as the secondary antibody, a swine anti-rabbit immunoglobulin/fluorescein isothiocyanate (FITC) (Dako, Glostrup, Denmark) at working concentrations of 4 and 20 µg/ml, respectively. As a positive control, human tonsil tissue sections were used. Further controls consisted of omission of the primary antibody and the use of an irrelevant antibody (anti-human chorionic gonadotrophin). The slides were viewed with a Leica DMBL research microscope (Leica Microsystems, Wetzlar, Germany), using the N-PLAN lens. Images were acquired using a Leica camera model DFC 490 (Leica Microsystems) and processed with Leica application suite version 2·4·0R1.
AID mRNA detection and analysis of B cell clonalities in portal tracts of MC patients
Portal tracts from liver biopsy specimens were isolated by means of laser capture microdissection (LCM), as described elsewhere 7,28. Briefly, the sections were placed on polyethylene naphthalate membrane slides (Leica Microsystems), fixed in acetone at −20°C for 5 min, and then analysed using a Leica AS LMD laser microdissection system. RNA was extracted using the RNeasy Micro kit (Qiagen) and reverse-transcribed. Five μl of cDNA was used in the PCR-based detection of AID mRNA (NM_020661.2) in a reaction containing the AmpliTaq Gold PCR master mix (Applied Biosystems, Foster City, CA, USA), and the following set of primers: sense 5′-CTGGGAAGGGCTGCATGAAA-3′ (position 562–581) and anti-sense 5′-AAAGTCCCAAAGTACGAAAT-3′ (position 674–655). The size of the final PCR product was 113 base pairs (bp) and all samples were analysed by electrophoresis on a 2% agarose gel. To confirm the validity of the PCR products, the cDNAs were sequenced using the BigDye Terminator version 1·1 cycle sequencing kit and the ABI Prism 310 genetic analyser (Applied Biosystems).
IgH gene rearrangement was characterized using 5 μl of the cDNA and the IgH SHM assay version 2·0 (Invivoscribe), as described previously 7,28. Each band was excised and loaded onto a DNA purification column (DNA gel extraction kit; Millipore, Billerica, MA, USA). The purified PCR product was cloned into a plasmid vector using the pGEM-T Easy vector system II (Promega, Madison, WI, USA). The recombinant vector was used to transform JM109 Escherichia coli competent cells (Promega). The transformants were plated onto duplicate Luria–Bertani (LB)/ampicillin/IPTG/X-Gal plates, incubated overnight at 37°C and processed for plasmid isolation. Five ml of LB-broth cultures of single colonies were grown overnight at 37°C. The plasmid DNA was purified using the QIAprep spin miniprep kit (Qiagen) and then solubilized in 100 μl of 10 mM Tris Cl (pH 8·5) buffer. The sequences of the cloned products were obtained with the BigDye Terminator version 1·1 cycle sequencing kit and ABI Prism 310 genetic analyser. All sequences were confirmed by sequencing in both directions, using the T7 and SP6 primers. Ten different clones were sequenced for each dominant band.
Statistical analysis
Descriptive statistics included the mean or median, as appropriate for continuous variables, and frequency (%) for categorical variables. In the univariate analysis, χ2 and Fisher's exact tests were used as appropriate to compare categorical variables, and the non-parametric Mann–Whitney test to compare continuous variables. The differences were considered significant at P < 0·05. A Spearman's rank correlation coefficient (r) test was used to evaluate the relationships between variables. All statistical analyses were performed using IBM spss statistics version 19·0.
Results
AID mRNA expression in peripheral blood B cells
Quantitative real time RT–PCR assay for specific AID mRNA gene expression, carried out on nucleic acids extracted from peripheral B cells (PBCs) normalized to 106 CD19+ B cells/sample, showed progressively higher median levels in patients with than in those without CV. Figure 1 shows the quantitative differences in specific AID mRNA, normalized to β-actin mRNA expression for each group. In CV patients, AID mRNA expression was almost threefold higher than in non-CV patients and sevenfold higher than in healthy subjects [median fold change and range; 6·68 (0·09–580·4) versus 2·54 (0·04–36·8), P = 0·03 and versus 0·95 (0·16–7·9), P = 0·0003]. Surprisingly, a negative correlation was recorded between peripheral AID activity and patients’ age, in that the levels of AID transcripts were found to be significantly lower in older CV patients. Spearman's r value expressing the correlation between age and AID mRNA transcripts was −0·3551 [95% confidence interval (CI) = −0·5743 to −0·0883; P = 0·008].
Figure 1.
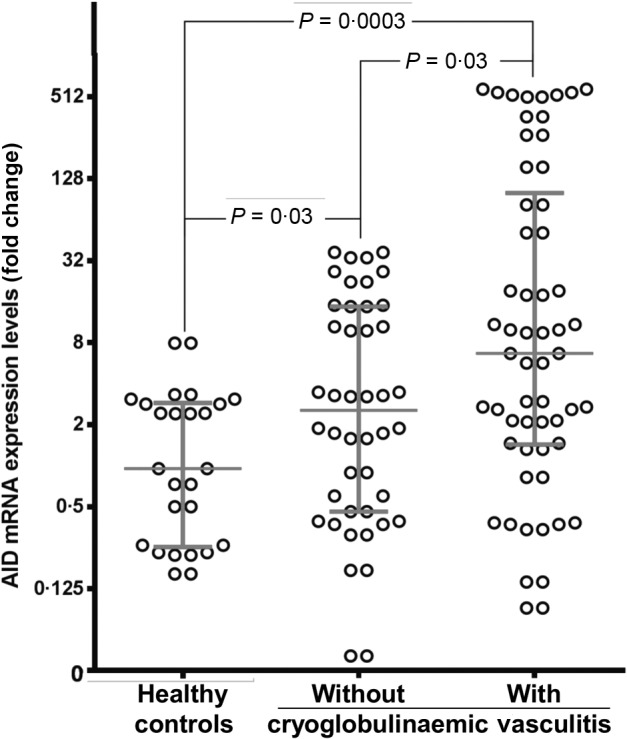
Activation-induced cytidine deaminase (AID) mRNA transcripts in peripheral blood B cells. Circulating cells from 58 patients with hepatitis C virus (HCV)-related cryoglobulinaemic vasculitis (CV), 44 with HCV infection without CV and 26 healthy blood donors were analysed with real-time reverse transcription–polymerase chain reaction (RT–PCR) for the level of AID transcript expression. Data are shown as the median with the interquartile range.
AID levels and B cell clonal expansions
Antigen-experienced memory cells frequently generate B cell clonal diversity in chronic HCV infection. As depicted in Fig. 2, analysis of the PBCs genotype showed clonal restriction in 34 of 58 (59%) and in seven of 44 (16%) patients with and without CV, respectively (P < 0·0001). Clonal PBCs restriction did not correspond to AID mRNA expression, as AID transcript levels were significantly higher in cells with polyclonal genotype than in those with clonal features in both CV and non-CV patients [median fold change and range; 15 (0·34–580·4) versus 2·7 (0·09–549), P = 0·009, and 3·46 (0·04–36·9) versus 1·58 (0·17–1·73), P = 0·02]. AID mRNA expression in polyclonal PBCs from CV patients was significantly higher than in the PBCs of non-CV patients (P = 0·02).
Figure 2.
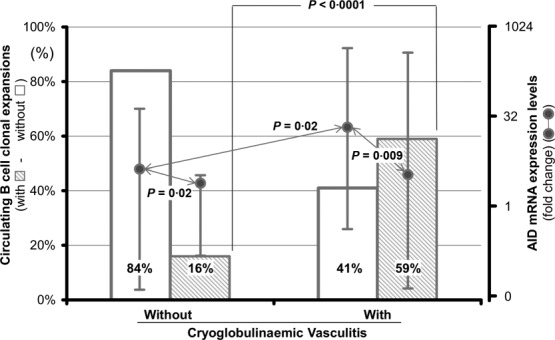
Activation-induced cytidine deaminase (AID) expression and circulating B cell clonal expansions. Relationship between B cell clonal restriction and AID mRNA expression in samples of peripheral blood from hepatitis C virus (HCV)-infected patients with and without cryoglobulinaemic vasculitis (CV).
To clarify the possible interplay between AID activity and B cell clonal features over time, levels of AID transcripts and molecular profiles of PBCs clonal expansions were stratified over life decades. As summarized in Fig. 3a, AID expression diminished progressively over time [median fold change and range; 142·5 (0·37–580·4); 14·5 (1·32–155·5); 6·1 (0·34–549·0); 0·14 (0·09–0·38) in the fifth, sixth, seventh and eighth decades of life, respectively]. In sharp contrast, the percentage of B cell oligo/monoclonal profiles in CV patients increased over time, from 33·3% (four of 12) in the fifth decade to 50% (four of eight), 62·5% (10 of 16), and 71·4% (10 of 14) in the sixth, seventh and eighth decades, respectively. A similar trend of AID activity was demonstrated in the non-CV group. Although not statistically significant, median AID mRNA levels decreased progressively over the age [median fold change and range; 3·2 (0·31–33·6); 2·5 (0·04–22·3); 0·9 (0·17–15·0); 0·7 (0·1–3·5) in the fifth, sixth, seventh and eighth decades, respectively]. Conversely, no difference regarding the frequency of B cell clonalities was found over time. B cell clonal expansions were demonstrated in 12·5% (one of eight) in the fifth, 14·3% (two of 14) in the sixth, 14·3% (two of 14) in the seventh and 25% (two of eight) in the eighth decades (Fig. 3b).
Figure 3.
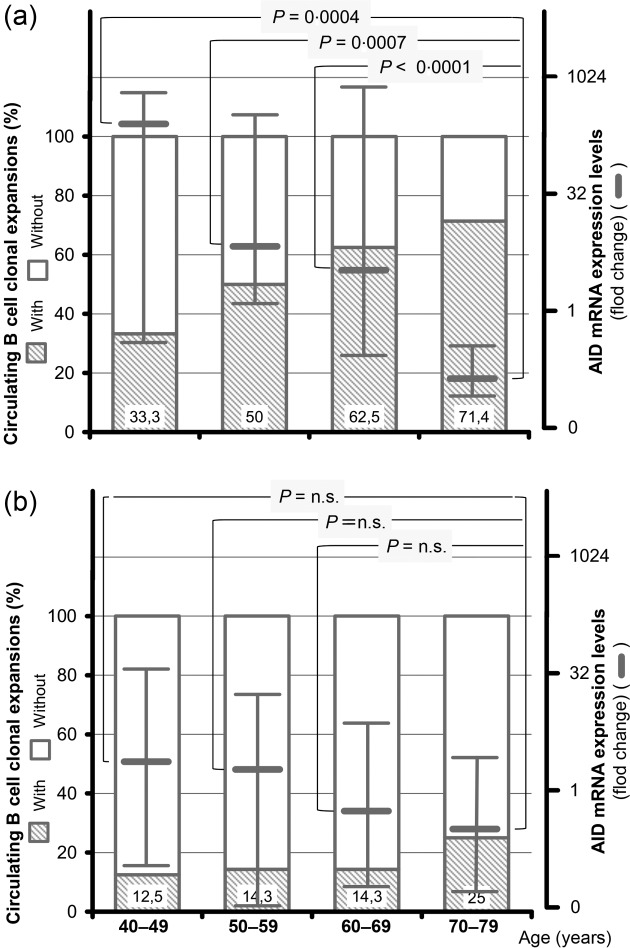
Age-related profile of activation-induced cytidine deaminase (AID) mRNA expression and frequency of circulating B cell clonal expansions in hepatitis C virus (HCV)-infected patients with (a) and without (b) cryoglobulinaemic vasculitis (CV). Expression of AID transcripts and the frequency of peripheral B cell clonal expansions of patients stratified for decades of age.
Virus-related AID expression
PBCs AID transcripts were measured and correlated with the circulating viral load (Fig. 4). The median concentrations of HCV RNA were found to increase progressively over the age in CV patients. This observation helped to establish a negative correlation between lower AID transcription levels and higher concentrations of HCV RNA (r = −0·4049; 95% CI = −0·6320 to −0·1137, P = 0·006). Increased HCV RNA levels were maintained with ageing, in that they were detected to a significant extent in the seventh [median and range; 5·9 log IU/ml (5·8–6·6)] and eighth decades [6·1 log IU/ml (5·8–6·4)], compared with levels found in the sixth decade [5·6 log IU/ml (4·5–6·5)] (P = 0·003 and P = 0·01, respectively). Conversely, no progressive increment of viral load over age was demonstrated in the non-CV group and no significant differences between decades were noticed [median levels and range of HCV RNA: 6·0 log IU/ml (5·5–6·3); 6·3 log IU/ml (5·7–6·9); 6·0 log IU/ml (4·8–7·3) and 5·8 log IU/ml (4·7–6·6) in the fifth, sixth, seventh and eighth decades, respectively].
Figure 4.
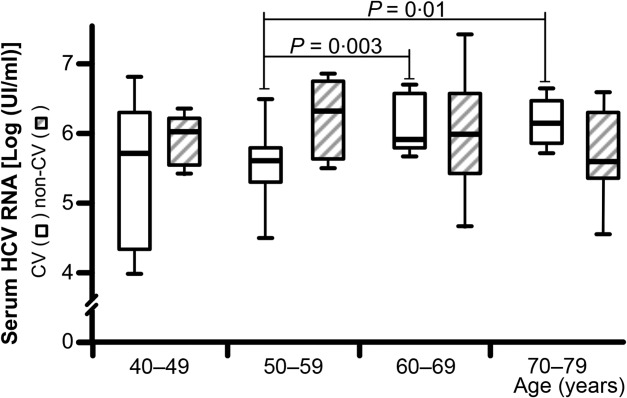
Age-related hepatitis C virus (HCV) RNA levels in patients with and without cryoglobulinaemic vasculitis (CV). Median levels and range of serum HCV RNA concentrations in patients stratified for decades of age.
AID mRNA and laboratory/clinical parameters
In addition, Spearman's rank correlation analysis was used to assess the strength of the association between AID mRNA expression and laboratory parameters (Table1). No difference in the estimated disease duration was found between patients with (9·9 ± 5·8 years) and without CV (10·7 ± 5·4 years). No linear regression was provided between AID expression and duration of disease in CV patients (r = 0·2267, P = 0·09). Conversely, we found a positive correlation between AID mRNA levels and circulating C3 (P = 0·004) and C4 (P = 0·01) complement fractions. No significant differences were found between AID transcripts and cryocrit percentages, circulating RF activity, IgM, IgG and IgA concentrations, and alanine aminotransferase activity.
Table 1.
Spearman's rank correlation analysis between activation-induced cytidine deaminase (AID) mRNA expression levels in circulating peripheral B cells (PBCs) and clinical/laboratory parameters in 58 hepatitis C virus (HCV)-related cryoglobulinaemic vasculitis (CV) patients
| Parameters | r Coefficient | 95% Confidence interval | P-value (P < 0.05) |
|---|---|---|---|
| Epidemiology | |||
| Disease duration (years) | 0·2267 | −0·0465 to 0·4684 | 0·09 |
| Laboratory | |||
| Cryocrit (%) | −0·1370 | −0·3885 to 0·1335 | 0·3 |
| Rheumatoid factor (IU/ml) | 0·2332 | −0·0854 to 0·5085 | 0·14 |
| IgM (mg/dl) | 0·2826 | −0·0506 to 0·5592 | 0·08 |
| IgG (mg/dl) | 0·1368 | −0·2007 to 0·4453 | 0·4 |
| IgA (mg/dl) | 0·0947 | −0·2413 to 0·4105 | 0·6 |
| C3 (mg/dl) | 0·4400 | 0·1395 to 0·6663 | 0·004 |
| C4 (mg/dl) | 0·3772 | 0·0734 to 0·6169 | 0·01 |
| ALT (IU/l) | 0·1201 | −0·1849 to 0·4041 | 0·43 |
Ig = immunoglobulin; ALT = alanine transaminase.
An overview of the symptomatology observed at the time of study admission, related to the measured levels of AID transcripts, is shown in Table2. Of 58 patients, signs of full-blown CV included purpura in 39 (67%), torpid leg ulcers in 19 (33%), nephropathy in 12 (21%) and peripheral neuropathy in 19 (33%). The AID transcript levels were largely comparable in patients with and without signs of active vasculitis.
Table 2.
Comparison of activation-induced cytidine deaminase (AID) mRNA transcript levels in 58 hepatitis C virus (HCV)-positive patients with or without cryoglobulinaemic vasculitis (CV) symptomatology
| Categories | AID mRNA transcripts (fold change) | ||
|---|---|---|---|
| With | Without | P | |
| Palpable purpura, n (%) | 39 (67) | 19 (33) | 0·41 |
| Median | 4·18 | 2·95 | |
| Range | (0·14–526·7) | (0·09–51·3) | |
| Cutaneous ulcers n (%) | 19 (33) | 39 (67) | 0·99 |
| Median | 2·95 | 4·18 | |
| Range | (0·14–364·8) | (0·09–526·7) | |
| Nephropathy n (%) | 12 (21) | 46 (79) | 0·24 |
| Median | 19·2 | 2·8 | |
| Range | (1·3–364·8) | (0·09–526·7) | |
| Neuropathy n (%) | 19 (33) | 39 (67) | 0·76 |
| Median | 2·70 | 4·3 | |
| Range | (0·09–364·8) | (0·14–526·7) | |
Detection of AID protein and mRNA transcripts in liver tissue
It has been repeatedly shown that chronically HCV-infected inflamed livers contain lymphoid aggregates that share many of the structural and functional features of secondary lymphoid tissues. Thus, we sought to detect the distribution of AID protein and mRNA transcripts in liver tissue biopsies. As shown in Fig. 5, immunoreactive signal was demonstrated in 10 of 25 (40%) and seven of 30 (23%) liver biopsies from patients with and without CV, respectively. AID immune reactants were found mainly in the inflammatory cells of portal tracts (Fig. 5a,b) and sometimes as single elements in sinusoidal spaces (Fig. 5c). There were no differences in terms of the liver distribution of AID-positive cells between patients with and without CV. Histopathology evaluation showed features of chronic active hepatitis with fibrosis and necro-inflammatory activity scores (METAVIR units) ranging from F1 to F3 and from A0 to A3, respectively. The occurrence of AID protein in situ was not related to the severity of liver damage (data not shown).
Figure 5.
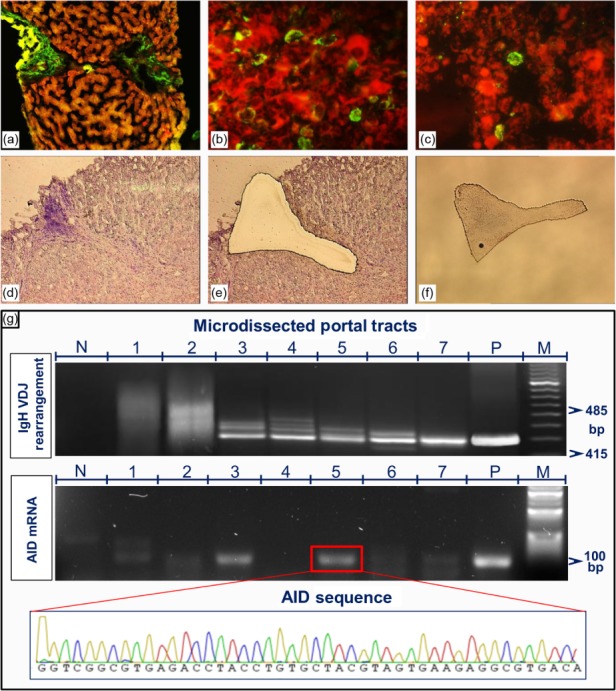
Detection of activation-induced cytidine deaminase (AID) protein and mRNA transcripts in liver tissue. (a,b) AID immune reactants are present in portal tracts containing inflammatory cells (low and high magnification, ×5 and ×20/0·40, respectively). (c) A single AID-positive cell is seen in the sinusoid (magnification, ×20/0·40). Example of laser capture microdissection (LCM)-based analysis: (d) section of liver biopsy showing enlarged portal tract with heavy inflammatory infiltrate and lymphoid aggregate, with appearance of a lymphoid follicle; (e) cleared area remaining in the context of tissue section after complete dissection of the portal structure; (f) dissected portal tract in the cap tube containing lysis buffer for molecular procedure. (g) Comparison of the polymerase chain reaction (PCR) products of immunoglobulin variable region heavy chain (IgVH) rearrangement (upper panel, M = molecular-weight size marker; N = no RNA; P = positive sample from a patient with chronic lymphocytic leukaemia) and AID transcripts (middle panel, M = molecular-weight size marker; N = no RNA; P = AID mRNA reverse transcription–polymerase chain reaction (RT–PCR) product cloned into a plasmid vector) determined in seven dissected portal tracts derived from a single liver biopsy section. The chromatogram indicates the specificity of the AID RT–PCR product (lower panel).
To determine whether the tissue expression of AID protein paralleled AID transcription and PBCs clonal expansions, molecular analyses were carried out on nucleic acids extracted from portal structures isolated by the laser capture microdissection (LCM) technique. Laser pulses were used to obtain dissected samples of comparable areas. An example of a portal tract obtained using LCM is shown in Fig. 5d–f. In each sample, the integrity of the template was verified by the amplification of β-actin gene sequences. Analyses were conducted on liver biopsy sections from 15 patients with and 15 without CV. For each biopsy, seven or more portal tracts were isolated. The results provided evidence of the presence of AID mRNA transcripts in the portal tracts of all 15 patients with and in 12 of the 15 without CV. AID expression was usually associated with SHM and clonally expanded local B cells. However, a notable discordance between B cell clonal expansions and AID expression was recorded among different portal tracts of the same liver biopsy, as shown in the example provided in Fig. 5g. B cell clonal expansion was detected clearly in portal tracts, but AID expression was not (lines 4, 6 and 7). Additional molecular features were portal tracts negative for both AID mRNA expression and B cell clonal expansions (lines 1 and 2), whereas in lines 3 and 5 both AID RNA transcript and B cell clonal expansions were demonstrated. The heterogeneous molecular profiles of B cell-containing portal structures in the same liver support the contention of a peculiar immunological competence of each intraportal lymphoid aggregate.
Discussion
This study shows that AID mRNA is expressed in the PBCs of CV patients at levels significantly higher than those determined in the cells of HCV-infected patients without CV, whereas expression in the cells of healthy controls was very low or absent. These results emphasize the profound dysregulation of B cell function and the striking heterogeneity of AID transcripts in HCV infection, indicating biological differences between individuals. Surprisingly, Spearman's rank correlation analysis demonstrated a negative link between AID expression and higher levels of HCV RNA.
Conversely, the frequency of B cell clonal expansions was found to be increased significantly over time. In addition, our results indicate that a significantly higher frequency of B cell clonal expansions occurs in CV than in non-CV patients, and that in both groups unmutated B cells exhibit remarkable differences in AID transcript expression. The median levels of AID transcripts in HCV-positive patients were much higher in polyclonal B cell preparations, suggesting that B cell clonal expansion was highly regulated in low AID-expressing cells. Thus, the present data define a subset of patients with ‘non-mutated’ B cells with high AID transcript levels, who should be differentiated from patients with ‘mutated’ B cells with lower AID expression and a tendentially older age.
Another finding of our study is that ectopic lymphoid structures in the liver, the major target organ of HCV infection, are potentially functional, given that AID expression was maintained within these lymphoid aggregates. As reported previously, these tertiary lymphoid structures can lead to self-perpetuating autoimmunity 5,29,30 and may critically impact the capacity of B cell-depleting biologicals to modulate chronic inflammation and autoimmunity in CV patients 31. Molecular analyses of the B cell-containing microdissected lymphoid structures of liver tissue demonstrated that they are hypermutated and clonally restricted, suggesting that an antigen-driven B cell response is ongoing in the liver microenvironment 4–7. The persistence of ectopic lymphoid structures in the liver from chronically HCV-infected patients with or without CV probably reflects the sustained over-expression of genes regulating ectopic lymphoneogenesis 32,33.
Aberrant expression of AID serves as a link between the cellular editing machinery and a high mutation frequency, leading to lymphoid and non-lymphoid malignancies 34. The present findings are consistent with those found in chronic HIV-1 infection, in which basal up-regulation of AID transcripts was seen to be associated with high risk of development of malignant lymphoproliferative disorders 35. Thus, this work outlines the potential importance of patients with ‘unmutated’ B cells and over-expression of AID that may reflect the existence of continuous stimulation of BCR able to sustain survival signals, with subsequent emergence of autonomous B cell clone.
This raises the hypothesis that B cell malignant transformation does not arise from memory B cells. Given that the present study was designed to determine the AID levels in B cells from HCV-infected patients with and without CV, it was obviously beyond its scope to perform a longitudinal survey aimed to identify individuals with high AID phenotype cells who eventually progress to overt lymphoproliferative disorders. The relationship between chronic HCV infection and B cell non-Hodgkin's lymphoma (B-NHL) may provide a model for the kinetics of progression. Approximately 15% of HCV patients show disease progression to B-NHL after a median of 15 years 36. The degree of somatic hypermutation in the IgH chain gene and the levels of AID expression by B cells may represent powerful prognostic factors. In the cases assessed, the high-AID phenotype cells have demonstrated a low frequency of B cell clonalities, whereas abnormal clonal B cell populations, rather than monoclonal response, show low AID levels of transcripts, supporting the notion that there are two biologically distinct cell populations and, possibly, two different clinical entities. The identification of B cell phenotype in HCV chronically infected individuals has many implications for understanding the developing of malignant disease. The cells are readily recognizable, and can be isolated to examine gene expression profiles in relation to normal B lymphocytes to identify the potential early stages of lymphomagenesis and to clarify the mechanism(s) of disease progression. Prospective studies in a larger series of patients are needed to achieve conclusive evidence to our results.
Acknowledgments
This work was supported by Foundation ‘Cassa di Risparmio di Puglia’, Bari, Italy (grant: 510201104001’); Italian Foundation for Cancer Research (FIRC) (fellow: 15166), Milan, Italy; University of Bari (grant: 00474712Ricat); Associazione Italiana per la Ricerca sul Cancro (AIRC) (investigator grant (no. 14095) and Special Program Molecular Clinical Oncology 5 per 1000 (n. 9965), Milan, Italy.
Glossary
- BCR
B cell receptor
- CAH
chronic active hepatitis
- CSR
class switch recombination
- CV
cryoglobulinemic vasculitis
- GC
germinal center
- ICs
immune complexes
- LCM
laser capture microdissection
- MC
mixed cryoglobulinemia
- PBC
peripheral blood CD19+ B cells
- PBMCs
peripheral blood mononuclear cells
- RF
rheumatoid factor
- SHM
somatic hypermutation
Author contributions
All authors were involved in drafting the paper and revising it critically for important intellectual content, and all authors approved the final version to be published. D. S. and F. D. had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study conception and design: D. S. and F. D. Acquisition of data: S. R., S. S. and F. P. Analysis and interpretation of data: D. S., S. R. and F. D.
Disclosure
The authors declare that they have no conflicts of interest with respect to this paper.
References
- Agnello V. The etiology and pathophysiology of mixed cryoglobulinemia secondary to hepatitis C virus infection. Springer Semin Immunopathol. 1997;19:111–29. doi: 10.1007/BF00945029. [DOI] [PubMed] [Google Scholar]
- El-Serag HB, Hampel H, Yeh C, Rabeneck L. Extrahepatic manifestations of hepatitis C among United States male veterans. Hepatology. 2002;36:1439–45. doi: 10.1053/jhep.2002.37191. [DOI] [PubMed] [Google Scholar]
- Sansonno D, Dammacco F. Hepatitis C virus, cryoglobulinaemia, and vasculitis: immune complex relations. Lancet Infect Dis. 2005;5:227–36. doi: 10.1016/S1473-3099(05)70053-0. [DOI] [PubMed] [Google Scholar]
- Sansonno D, De Vita S, Iacobelli AR, Cornacchiulo V, Boiocchi M, Dammacco F. Clonal analysis of intrahepatic B cells from HCV-infected patients with and without mixed cryoglobulinemia. J Immunol. 1998;160:3594–601. [PubMed] [Google Scholar]
- Racanelli V, Sansonno D, Piccoli C, D'Amore FP, Tucci FA, Dammacco F. Molecular characterization of B cell clonal expansions in the liver of chronically hepatitis C virus-infected patients. J Immunol. 2001;167:21–9. doi: 10.4049/jimmunol.167.1.21. [DOI] [PubMed] [Google Scholar]
- Ivanovski M, Silvestri F, Pozzato G, et al. Somatic hypermutation, clonal diversity, and preferential expression of the VH 51p1/VL kv325 immunoglobulin gene combination in hepatitis C virus-associated immunocytomas. Blood. 1998;91:2433–42. [PubMed] [Google Scholar]
- Sansonno D, Lauletta G, De Re V, et al. Intrahepatic B cell clonal expansions and extrahepatic manifestations of chronic HCV infection. Eur J Immunol. 2004;34:126–36. doi: 10.1002/eji.200324328. [DOI] [PubMed] [Google Scholar]
- Sansonno D, Tucci FA, Ghebrehiwet B, et al. Role of the receptor for the globular domain of C1q protein in the pathogenesis of hepatitis C virus-related cryoglobulin vascular damage. J Immunol. 2009;183:6013–20. doi: 10.4049/jimmunol.0902038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrier B, Cacoub P. Cryoglobulinemia vasculitis: an update. Curr Opin Rheumatol. 2013;25:10–18. doi: 10.1097/BOR.0b013e32835b15f7. [DOI] [PubMed] [Google Scholar]
- Bartosch B, Vitelli A, Granier C, et al. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J Biol Chem. 2003;278:41624–30. doi: 10.1074/jbc.M305289200. [DOI] [PubMed] [Google Scholar]
- Hsu M, Zhang J, Flint M, et al. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc Natl Acad Sci USA. 2003;100:7271–7276. doi: 10.1073/pnas.0832180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, von Hahn T, Tscherne DM, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–5. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- Dustin LB, Charles ED. Primary, post-primary and non-specific immunoglobulin M responses in HCV infection. Antivir Ther. 2012;17:1449–52. doi: 10.3851/IMP2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai M, Seya T, Matsumoto M, Shimotohno K, Sakamoto N, Aly HH. The J6JFH1 strain of hepatitis C virus infects human B-cells with low replication efficacy. Viral Immunol. 2014;27:285–94. doi: 10.1089/vim.2013.0140. [DOI] [PubMed] [Google Scholar]
- Sansonno D, Tucci FA, Lauletta G, et al. Hepatitis C virus productive infection in mononuclear cells from patients with cryoglobulinaemia. Clin Exp Immunol. 2007;147:241–48. doi: 10.1111/j.1365-2249.2006.03272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskus T, Operskalski EA, Radkowski M, et al. Negative-strand hepatitis C virus (HCV) RNA in peripheral blood mononuclear cells from anti-HCV-positive/HIV-infected women. J Infect Dis. 2007;195:124–33. doi: 10.1086/509897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliviero B, Cerino A, Varchetta S, et al. Enhanced B-cell differentiation and reduced proliferative capacity in chronic hepatitis C and chronic hepatitis B virus infections. J Hepatol. 2011;55:53–60. doi: 10.1016/j.jhep.2010.10.016. [DOI] [PubMed] [Google Scholar]
- Muto T, Okazaki IM, Yamada S, et al. Negative regulation of activation-induced cytidine deaminase in B cells. Proc Natl Acad Sci USA. 2006;103:2752–7. doi: 10.1073/pnas.0510970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Sankaranand VS, Anant S, et al. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–6. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- Keim C, Kazadi D, Rothschild G, Basu U. Regulation of AID, the B-cell genome mutator. Genes Dev. 2013;27:1–17. doi: 10.1101/gad.200014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch EE, Li Z, Takizawa M, et al. Regulation of AID expression in the immune response. J Exp Med. 2007;204:1145–56. doi: 10.1084/jem.20061952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humby F, Bombardieri M, Manzo A, et al. Ectopic lymphoid structures support ongoing production of class-switched autoantibodies in rheumatoid synovium. PLOS Med. 2009;6:e1. doi: 10.1371/journal.pmed.0060001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Saadoun D, Calabrese LH, Cacoub P. The pathophysiology of HCV induced B-cell clonal disorders. Autoimmun Rev. 2007;6:581–7. doi: 10.1016/j.autrev.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Laskov R, Yahud V, Hamo R, Steinitz M. Preferential targeting of somatic hypermutation to hotspot motifs and hypermutable sites and generation of mutational clusters in the IgVH alleles of a rheumatoid factor producing lymphoblastoid cell line. Mol Immunol. 2011;48:733–45. doi: 10.1016/j.molimm.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Quartuccio L, Isola M, Corazza L, et al. Validation of the classification criteria for cryoglobulinaemic vasculitis. Rheumatology (Oxf) 2014;53:2209–13. doi: 10.1093/rheumatology/keu271. [DOI] [PubMed] [Google Scholar]
- The French METAVIR Study Group. Intraobserver and interobserver variations in liver biopsy interpretation in patients with chronic hepatitis C. Hepatology. 1994;20:15–20. [PubMed] [Google Scholar]
- Dammacco F, Sansonno D, Piccoli C, Tucci FA, Racanelli V. The cryoglobulins: an overview. Eur J Clin Invest. 2001;31:628–38. doi: 10.1046/j.1365-2362.2001.00824.x. [DOI] [PubMed] [Google Scholar]
- Sansonno D, Tucci FA, De Re V, et al. HCV-associated B cell clonalities in the liver do not carry the t(14;18) chromosomal translocation. Hepatology. 2005;42:1019–27. doi: 10.1002/hep.20887. [DOI] [PubMed] [Google Scholar]
- Dammacco F, Sansonno D, Piccoli C, Racanelli V, D'Amore FP, Lauletta G. The lymphoid system in hepatitis C virus infection: autoimmunity, mixed cryoglobulinemia, and Overt B-cell malignancy. Semin Liver Dis. 2000;20:143–57. doi: 10.1055/s-2000-9613. [DOI] [PubMed] [Google Scholar]
- Ferri C, Zignego AL. Relation between infection and autoimmunity in mixed cryoglobulinemia. Curr Opin Rheumatol. 2000;12:53–60. doi: 10.1097/00002281-200001000-00009. [DOI] [PubMed] [Google Scholar]
- Dammacco F, Tucci FA, Lauletta G, et al. Pegylated interferon-alpha, ribavirin, and rituximab combined therapy of hepatitis C virus-related mixed cryoglobulinemia: a long-term study. Blood. 2010;116:343–53. doi: 10.1182/blood-2009-10-245878. [DOI] [PubMed] [Google Scholar]
- Sansonno D, Tucci FA, Troiani L, et al. Increased serum levels of the chemokine CXCL13 and up-regulation of its gene expression are distinctive features of HCV-related cryoglobulinemia and correlate with active cutaneous vasculitis. Blood. 2008;112:1620–7. doi: 10.1182/blood-2008-02-137455. [DOI] [PubMed] [Google Scholar]
- Manzo A, Paoletti S, Carulli M, et al. Systematic microanatomical analysis of CXCL13 and CCL21 in situ production and progressive lymphoid organization in rheumatoid synovitis. Eur J Immunol. 2005;35:1347–59. doi: 10.1002/eji.200425830. [DOI] [PubMed] [Google Scholar]
- Okazaki IM, Hiai H, Kakazu N, et al. Constitutive expression of AID leads to tumorigenesis. J Exp Med. 2003;197:1173–81. doi: 10.1084/jem.20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epeldegui M, Breen EC, Hung YP, Boscardin WJ, Detels R, Martinez-Maza O. Elevated expression of activation induced cytidine deaminase in peripheral blood mononuclear cells precedes AIDS-NHL diagnosis. Aids. 2007;21:2265–70. doi: 10.1097/QAD.0b013e3282ef9f59. [DOI] [PubMed] [Google Scholar]
- Lauletta G, Russi S, Conteduca V, Sansonno L, Dammacco F, Sansonno D. Impact of cryoglobulinemic syndrome on the outcome of chronic hepatitis C virus infection: a 15-year prospective study. Medicine (Balt) 2013;92:245–56. doi: 10.1097/MD.0b013e31829d2abc. [DOI] [PMC free article] [PubMed] [Google Scholar]


