Abstract
In order to understand the mechanisms of drug–drug interaction (DDI), the study of pharmacokinetics (PK), pharmacodynamics (PD), and pharmacogenetics (PG) data are significant. In recent years, drug PK parameters, drug interaction parameters, and PG data have been unevenly collected in different databases and published extensively in literature. Also the lack of an appropriate PK ontology and a well-annotated PK corpus, which provide the background knowledge and the criteria of determining DDI, respectively, lead to the difficulty of developing DDI text mining tools for PK data collection from the literature and data integration from multiple databases.
To conquer the issues, we constructed a comprehensive pharmacokinetics ontology. It includes all aspects of in vitro pharmacokinetics experiments, in vivo pharmacokinetics studies, as well as drug metabolism and transportation enzymes. Using our pharmacokinetics ontology, a PK corpus was constructed to present four classes of pharmacokinetics abstracts: in vivo pharmacokinetics studies, in vivo pharmacogenetic studies, in vivo drug interaction studies, and in vitro drug interaction studies. A novel hierarchical three-level annotation scheme was proposed and implemented to tag key terms, drug interaction sentences, and drug interaction pairs. The utility of the pharmacokinetics ontology was demonstrated by annotating three pharmacokinetics studies; and the utility of the PK corpus was demonstrated by a drug interaction extraction text mining analysis.
The pharmacokinetics ontology annotates both in vitro pharmacokinetics experiments and in vivo pharmacokinetics studies. The PK corpus is a highly valuable resource for the text mining of pharmacokinetics parameters and drug interactions.
Keywords: Pharmacokinetics, Pharmacodynamics, Drug, drug interaction, Text mining, Corpus, Ontology, Relation extraction, Enzyme, Transporter
1 Introduction
Adverse drug reaction (ADR) is one of the major causes of morbidity and mortality occurring in clinical care every year. To investigate the crucial problem, the US Food and Drug Administration (FDA) found that more than 40 % of the US population is prescribed more than four medications at a single time, which makes them more susceptible to ADR [1]. A literature search in Medline and Embase database from 1990 to 2006 showed that drug–drug interactions (DDIs) were held responsible for 0.054 % of the emergency department (ED) visits, 0.57 % of the hospital admissions, and 0.12 % of the re-hospitalizations [2]. It is possible that drug interaction can be beneficial or detrimental. The use of multiple drugs might provide synergism such as increasing the efficacy of therapeutic effect, decreasing dosage but holding the same efficacy to avoid toxicity, or minimizing the drug resistance [3]. However, we have more interests in the investigation of negative interaction because pathological significance is often unexpected and hard to be diagnosed. To predispose DDI, the importance of high-risk factors like age, polypharmacy, and genetic polymorphisms should be carefully evaluated [4]. In the elder population, DDIs account for 4.8 % of the hospital admissions, which is much higher than the proportion of DDI victims within the total population. The reason is directed to the abatement of liver metabolism or kidney function [5, 6]. Genetic polymorphism has profound influence on enzyme function, which might result in increased drug metabolism and absence of drug response. Evidences [7] suggested that patients affected by genetic polymorphisms will experience severe toxicities upon drug intake.
For economic aspect, the problem of DDI effect or co-medication effect has scaled such heights that it has even led to withdrawing of drugs from the market after approval. The 1990s saw the withdrawal of more than 11 drugs as shown in ref. 8. In 2007, the biopharmaceutical industry invested roundabout $58.8 billion for the research and development as the withdrawing of drugs [9] is a major setback to the industry as the deployment of a single drug compound is estimated at $200 million.
1.1 Drug–Drug Interaction Mechanisms and In Vitro and In Vivo Drug Interaction Studies
DDI can result when a substance affects the activity of a drug or its metabolites when these two drugs are administrated at the same time. The simultaneous administration of two drugs, which causes synergistic or antagonistic effect, might lead to the alternation of medication effectiveness or some harming effects on patient body. Those potential influences on human body should be noticed to prevent from a high risk of multiple interactions because the number of approved drugs increases. To preclude the possibility of hazardous interaction, understanding the significant scientific principles or mechanisms of DDI is important.
Due to the continued growth in drug development and the insight into molecular biology, we come to realize that transporter and enzyme played an important role in drug elimination, which inspired a clue to dig the mechanisms surrounding DDI. In brief, there are two major molecular mechanisms of DDI, enzyme-based drug metabolism and transporter-based drug transportation [10]. If an enzyme that is responsible for the metabolism of one drug is induced or inhibited by another drug, then the clearance of original drug will be changed, which might result in being toxic or less effective. For transporter- based drug transportation, transporter is important to drug deposition. Drugs can be metabolized only after they are transported into liver cells. To understand how a transporter-mediated DDI happens, the knowledge of the transporter substrates and inhibitors can suggest potential DDIs [11].
There are two basic types of drug interaction, pharmacokinetics (PK) and pharmacodynamics (PD). In short, PK investigates the activity of drug combinations with drug absorption, disposition, metabolism, excretion, and transportation (ADMET), which describes how these five criteria influence drug level (concentration). Pharmacokinetically speaking, potentiative or reductive combinations are, respectively, correlated to positive or negative modulation of drug transport, permeation, distribution, localization, or metabolism. Potentiative modulation of drug transport will enhance drug absorption via the disruption of transport carrier, increase drug concentration in plasma by inhibiting metabolic process, and stimulate or inhibit the metabolism of drugs into active or inactive form. On the other hand, reductive modulation provides contrasting perspectives to potentiative modulation. The reductive modulation of drug transport typically blocks drug absorption, decreases drug concentration in plasma, and reduces drug metabolism activity [12]. Those information brings to systematically investigate the physiological and biochemical mechanisms of drug exposure in multiple tissue types, cells, animals, and human subjects [13], which links preclinical and clinical phase of drug development. If the PK can be interpreted as the dose– concentration relationship, pharmacodynamics (PD) can be defined as the mechanism of drug action and relationship between drug concentration and effect. A drug's pharmacodynamics effect ranges widely from the molecular signals (such as its targets or downstream biomarkers) to clinical symptoms (such as the efficacy or side effect endpoints). Classification of its therapeutic effects: It can be synergistic, additive, or antagonistic if the effect is greater than, equal to, or less than the summed effects of drug combinations [12].
As stated in the previous section, the complicated transporter– enzyme interplay in the deposition of drug leads to the difficulty for the identification of DDIs in drug administration and drug development. Thus, understanding the molecular mechanism underlying different types of drug interaction could facilitate the discovery of novel DDI. Recently, in vitro technologies can qualitatively provide an insight into the potential DDI based on the observation of enzyme kinetic parameters. Via ADME screening efforts as well as the assessment of CYP inhibition, the choice of test compound inhibiting the metabolism of one probe substrate for an enzyme in the in vitro experiment can be fulfilled to carry out the prediction of in vivo DDI. Wienkers and Heath [14] addressed the basic principles of in vitro inhibition prediction underlying the generation of in vitro drug metabolism data and suggested several factors that introduced error or uncertainty into a quantitative prediction of in vivo DDI based on in vitro-derived PK parameters. In ref. 15, three factors authors recommended for the ideal model to predict metabolic drug–drug interaction (M-DDI) should be an accurate measurement of the average increase in the area under the plasma concentration–time curve (AUC) of a victim drug following administration of a perpetrator drug, the plasma binding displacement interaction, and the impact of the concentration–time profile of the inhibitor. To evaluate the potential for M-DDI [15] developed an in silico software SIMCYP, which incorporates extensive data on demographics; disease states; anatomical, physiological, genetic, and biochemical variables; and input of information on in vitro drug metabolism and transport.
1.2 Computational Drug Interaction Prediction and Drug Interaction Text Mining
1.2.1 Overview of Computational Drug Interaction Prediction
The evaluation of the potential risk of DDI is of importance in patient safety since DDIs can raise the danger of patients and the cost of healthcare system. According to the guidance for industry from the Food and Drug Administration [16], study design, data analysis, and implication for dosing and labeling are suggested to deal with drug interaction studies. When studying DDI for a new drug, it usually begins with in vitro study to determine whether a drug is a substrate, inhibitor, or inducer of metabolizing enzymes. The consequence of in vitro investigations can serve as an evidence to screen out the candidate potential drug pairs for additional in vivo study. To conduct an in vivo DDI study for an investigating drug, a quantitative analysis to mathematically describe the kinetics of drug metabolism involved in ADME process is needed. The basic model for the initial assessment of DDI based on in vitro and in vivo studies can be achieved by physiologically based pharmacokinetics (PBPK) modeling. From published in vitro experiments and in vivo studies [17 – 24] had developed Bayesian models and computational algorithms to construct PBPK models for DDI prediction.
Another common way to explore novel DDI is literature-based discovery. The hidden knowledge among information embedded in publications can be dug out through finding connections between articles. To this end, many researchers took advantage of some commercial or public databases as resource, such as Metabolism and Transport Drug Interaction Database (DIDB) [25], PharGKB [26], and DrugBank [27] which provided extensive lists of DDI information published in articles, clinical files, or biomedical research reports. Gottlieb et al. [28] proposed a computational framework INDI to infer and explore DDI by calculating similarity measurement between drug pair via diverse feature measurements, i.e., chemical based, ligand based, side effect based, annotation based, and sequence based. However, the problem of data inconsistency arose when using different databases. Some significant scientific evidences associated with DDI are limited or lacking in some existing databases. This deficiency is hard to prevent because the tasks of data collections are manually accomplished by different research groups or professional experts. To conquer this problem, employing the technologies from information retrieval (IR) or natural language processing (NLP) can be a solution to help extract data more efficiently and consistently.
1.2.2 Biomedical Text Mining
Text mining refers to the process of deriving high-quality information from text, which relies on NLP. To translate the text into computer-readable language, there are some basic steps of NLP [29], including sentence splitting, tokenization, part of speech, named entity recognition (NER), shallow parsing, and syntactic parsing. In this section, we do not go into the details of techniques for NLP tools. The attentions will be paid more on the tasks of corpus construction, IR, or information extraction (IE), which employs highly scalable statistics-based techniques to index and search large volume of text efficiently.
Extracting facts from texts is the goal of text-mining systems. The range of extraction tasks can be narrow from retrieving potentially relevant articles by sophisticated keyword search or classifying papers into different ontological types (IR), recognizing biological entities or concepts in text, and detecting relations between biological entities (IE) and broader to document summarization or question answering (beyond IE) [30]. To fulfill those tasks in biomedical domain, NER is an initial processing step because the significant knowledge is usually centered on the mechanism of biological activities which are described by nominalized verbs and nouns within sentences. Therefore, identifying text that satisfies various types of information needs is an important first step toward accurate text mining. But how to utilize the identified entities for improving text mining is challenging. One solution to this problem is an annotated corpus. The corpus annotated with such information allows real usage within text to be taken into account. The annotated sentence then can be represented in syntactic and semantic format, which shows the different levels of scientific characteristics. However, the strategy of constructing corpus is diverse. It differs with the purpose of text mining task and the methodology we used in extracting information. Kim et al. [31] introduces GENIA corpus with linguistically rich annotations for biomedical articles. The value of GENIA corpus comes from its annotations. All biologically meaningful terms are semantically annotated with descriptors from GENIA ontology. Wilbur et al. [32] suggest the basic guideline and criteria of corpus construction and annotation task for facilitating the training components of IE system by using machine learning method. Another value of annotated corpus is being a gold standard that facilitates the evaluation of approach. The success of practical applications crucially depends on the quality of extraction results, which is against the access of gold standard reference.
1.2.3 Relationship Extraction
Within IE methods, we are more interested in relationship extraction. The goal of relationship extraction is to detect the prespecified type of relation between a pair of entities of given types. A relation is typically represented as a pair of entities, linked by an arc that is either directed or undirected. The arc is given a label usually corresponding to a semantic type. In biology, the type of entities can be very specific such as gene, protein, or drug, while the type of relationship can be referred from some particular verbs, including transcribe, repress, or inhibit.
To effectively extract relationship, analysis of sentence structure is necessary. The use of semantic processing or deep parsing techniques that analyze both the syntactic and semantic structure of texts can benefit relation extraction. Several approaches had been reported in literature to extract the relation of interest. Generally, there are three main approaches for relationship extraction: co-occurrence-based, rule-based, and machine learning based approaches. Muller et al. [33] employ co-occurrence-based method, which is the simplest way to capture relationships relying on co-occurrence of two entities to derive a relation. Rule-based approaches [34, 35] are to take advantage of linguistic technology to grasp syntactic structure or semantic meaning for understanding the relationship from the unstructured text. Feldman et al. [34] employed an NP1–verb–NP2 template to identify the relation between two domain-specific entities. Fundel et al. [35] constructed a set of domain-specific rules and apply them to dependency parse tree to capture different forms of expressing a given relationship. Finally, classifiers using machine learning approaches such as support vector machines (SVM) [36] are often used for relation extraction. This method needs laborious efforts to define grammars or rules, and text in training dataset is manually tagged by a human expert. This text mining method uses the training data to automatically learn the “rules” so it can mine wanted information or identify the necessary knowledge [37–40].
The comparison among different methods is not easy because each method obtains its inherited pros and cons. Co-occurrence method provides the highest recall but poor precision among three. A large amount of false-positive relations are returned whenever the sentence is sophisticated with more than two entities or two key entities co-occurred in each single sentence but it does not state their relationship. Thus, co-occurrence method is more suitable to use as a simple baseline method for performance comparison. Rule-based method achieves better precision in extracting binary relationships due to the more precise rule conditions for defining relationship. But when it meets the complex sentence with various coordinates and relational clauses, the performance turns down obviously [41]. In general, machine learning-based method performs the best among methods. As an evidence in BioCreative challenge [42], the frameworks using supervised machine learning algorithm outperformed the existing methods in detecting protein–protein interaction (PPI). One important advantage is that system can predict categories for unseen samples. However, this advantage is heavily relying on annotated corpus [43]. Therefore, it can also be a big disadvantage because of the need for huge learning set.
1.2.4 Literature Review for Extracting Drug–Drug Interaction
Different approaches had been developed for extracting biomedical relationships such as PPI. From the experience of previous researches centered on PPI [36 – 40], few approaches have been proposed to the problem of detecting DDI. To promote the development of DDI extraction tools, DDIExtraction 2011, the first challenge task on DDI extraction, was held in 2011 at Spain. In this workshop, they provided evidence for the most effective methods available to solve specific problems and reveal the performance on these problems. In competition, most participants proposed systems using classifiers SVM or RLS. Their choices verified that machine learning can outperform other methods in relation extraction. Observed from results, approaches based on kernel methods achieved better performance than the classical feature-based methods [44]. Thus, the advantages of kernel-based method using machine learning classifier are spotlighted in this workshop.
In literature, some articles are outstanding in DDI extraction. The co-chairs of DDIExtraction 2011 [43] proposed a hybrid approach, which combines shallow parsing and pattern matching to extract relation between drugs based on annotated corpus. It utilizes the proposed syntactic patterns to split the sentence into clauses from which relations are extracted by matching patterns. The ability of dealing with complicated sentence is the advantage of this method. Complexity can be diminished by separating a long sentence into simplified clauses and by the detection of the apposition and coordinate structure. But there is one gap in the extraction of DDI information if used in pharmacokinetics or pharmacogenetics articles. Only exploring DDI based on literal denotation will lead to the missing detection of actual DDI information due to the lack of scientific knowledge. In ref. 45, DDIs are identified by aggregating gene–drug interactions which are extracted via rule-based method. The extracted interactions are then normalized and mapped into their standardized ontology to form the semantics network. The network could be useful to find potential DDIs. Differed from Percha et al. [45] who extracted DDI via the perspective of pharmacogenetics, Teri et al. [46] developed a method that combined text mining and automated reasoning to predict enzyme specific DDIs. In most situations, the extracted relations from the results of conventional relation extraction are not sufficient to derive DDI. By representing the general knowledge related to metabolism and interaction with the form of logic rules, DDI can be acquired in the reasoning phase.
2 Materials
For PK DDI text mining, the materials for the construction of PK ontology are prepared. A descriptor from specialized ontology can be used to describe the environment of PK experiments (in vivo and in vitro) and the nature of drug mechanisms (all drug metabolism and transportation enzymes.
For drug name, the dictionary is created using drug names from DrugBank 3.0 [27]. DrugBank consists of 6,829 drugs which can be grouped into different categories of FDA-approved, FDA-approved biotech, nutraceuticals, and experimental drugs. The drug names are mapped to generic names, brand names, and synonyms. The environment condition-specific in vitro PK experiment and their associated PK parameters are referred from [47 – 50]. The materials for in vivo study are summarized from two textbooks [13, 51]. The information of tissue-specific transporters and enzymes with all their probe inhibitors, inducers, and substrates were collected from industry standard (http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm064982.htm), reviewed in the top pharmacology journal [16].
3 Methods
To extract PK DDI by text-mining system, there are three noteworthy issues we should carefully deal with. (1) Recognition of drug name is one of the most salient issues in DDI text mining. Without satisfied performance in tagging drug name, false-positive or missing detection eliminates the accuracy of DDI results. Unlike gene's or protein's name, the representations of drug name are more sophisticated. The same drug may show in different documents with a number of ways, especially for metabolites of a compound [52]. The diversity of naming conventions perplexes the identification of drug names in pharmacokinetics articles. (2) Ontology is the main repository of formally represented knowledge for DDI text-mining system. The hierarchical repository provides a framework for knowledge integration and sharing, which give machine-readable descriptors of biomedical concepts and their relations. The challenge for ontology construction is to develop appropriate ontology resources and link them to adequate terminological lexicons [53]. (3) Corpus construction is essential to make text mining successful. It is not possible for a machine to capture useful information from text data written in natural language directly. To bridge the gap between text data and machine, corpus creates the accessibility for computer to read text data precisely [31, 54]. Another important issue within corpus is the scheme of biological annotation. The task of annotation can be regarded as identifying and classifying entities or sentences according to predefined categories. A well-defined scheme for annotation task is indispensable to corpus construction.
An ideal system for PK DDI extraction should provide not only a comprehensive list of DDIs in a cost-efficient manner but also the mechanism behind interactions. In current DDI extraction methods, most researches extract DDIs centering on exploring the semantics of sentence. Given a sentence with at least two drugs, they analyze sentence structure and identify drug entities and trigger words (e.g., verbs like inhibit or induce) to accomplish this task. However, in most situations, complete DDI information is presented in complicated ways with more than a single sentence. More concrete DDI conditions such as experimental measurements might be mentioned in those sentences which only have a single drug. For instance, the way to express DDI information in pharmacokinetics articles is quite different from that in pharmacodynamics articles. The sentence only with a single drug frequently mentions its corresponding PK parameters or other measurements, which show the practical conditions for drug metabolism. The merit of those parameters gives the clue to determine inhibition or induction of DDI as well as provides a criterion to exam the reliability of found DDIs.
To meet the abovementioned issues, this chapter tries to propose a system to detect DDI information not only from narrative sentences but also in those sentences with a single drug, which contains possible DDI candidate. Besides detecting DDI pairs from sentence structure, considering PK parameter as an evidence to determine DDI is important in our strategy. In the following sections, we carefully discuss how the task of drug name mapping works, the construction of an integrated pharmacokinetics ontology and corpus for text-mining system, and finally how to apply them in the text-mining system.
3.1 Drug Name Mapping
To detect the name by using NER, the performance of DDI extraction matters if the accuracy of drug name identification is not satisfied [52].
Drug names were created using the drug names from DrugBank 3.0 [27]. DrugBank consists of 6,829 chemicals with unique DrugBank ID which can be grouped into different categories of FDA-approved, FDA-approved biotech, nutraceuticals, and experimental drugs. The chemicals are mapped to generic names, brand names, and synonyms which results in 36,433 unique DrugBank ID–name pairs. 315 names in DrugBank have less than 4 letters such as chloramphenicol, DB0046 has a synonym CAP, and cholecalciferol, DB00169 has a synonym CC. The words with less than four letters may cause bad NER; therefore, they were removed.
In addition, drug metabolites were also tagged, because they are important in in vitro studies. The metabolites were judged by either prefix or suffix: oxi, hydroxyl, methyl, acetyl, N-dealkyl, N-demethyl, nor, dihydroxy, O-dealkyl, and sulfo. These prefixes and suffixes are due to the reactions due to phase I metabolism (oxidation, reduction, hydrolysis) and phase II metabolism (methylation, sulfation, acetylation, glucuronidation) [55].
3.2 PK Ontology Construction
The motivation for ontology in biomedical text mining is to make sense of raw text. According to the defined concepts, properties, relationships, instances, and axioms for a given domain, raw text can be interpreted by the descriptors of ontology with a standardized format and organized into hierarchical structure. Such advantages allow complex text to be represented with semantic and consistent manner [56].
The process of building ontology is a complex and tedious process. Various domain-specific resources and lexicons are required to satisfy the needs of a text-mining system using in a specific scope. According to the introduction of DDI mechanism we mentioned in Subheading 1.1, the domain of PK DDI is concerned with the process of drug disposition within the organism, the response of drug level, and the kinetics of drug exposure to different tissue types. Even in different experimental studies, DDI is defined with distinct measurements. However, no single system is currently capable of covering a complete domain for all aspects. For this reason, we introduce an integrated PK ontology which is composed of several components: experiment, metabolism enzyme, transporter, drug, and subject. In this work, the primary contribution is the ontology development for the PK experiment and integration of the PK experiment ontology with other PK-related ontologies.
Experiment specifies in vitro and in vivo PK studies and their associated PK parameters. The definitions and units for both in vitro or in vivo PK parameters and their corresponding experiment conditions should be included.
Within different types of in vitro PK experiments, different in vitro PK parameters are employed.
Single-drug metabolism experiment includes Michaelis–Menten constant (Km), maximum velocity of the enzyme activity (Vmax), intrinsic clearance (CLint), metabolic ratio, and fraction of metabolism by an enzyme (fmenzyme) [47].
Single-drug transporter experiment: PK parameters include apparent permeability (Papp), ratio of the basolateral to apical permeability and apical to basolateral permeability (Re), radio-activity, and uptake volume [57].
Drug interaction experiment: IC50 is the inhibition concentration that inhibits to 50 % enzyme activity; it is substrate dependent;and it does not imply the inhibition mechanism. Ki is the inhibition rate constant for competitive inhibition, noncompetitive inhibition, and uncompetitive inhibition. It represents the inhibition concentration that inhibits to 50 % enzyme activity, and it is substrate concentration independent. Kdeg is the degradation rate constant for the enzyme. KI is the concentration of inhibitor associated with half maximal inactivation in the mechanism-based inhibition; and Kinact is the maximum degradation rate constant in the presence of a high concentration of inhibitor in the mechanism-based inhibition. Emax is the maximum induction rate, and EC50 is the concentration of inducer that is associated with the half maximal induction [15].
Type of drug interaction: There are multiple drug interaction mechanisms, including competitive inhibition, noncompetitive inhibition, uncompetitive inhibition, mechanism-based inhibition, and induction [15].
For in vitro experiment conditions, metabolism enzyme, transporter, and some other factors should be considered.
Metabolism enzyme experiment conditions include buffer, NADPH sources, and protein sources. In particular, protein sources include recombinant enzymes, microsomes, and hepatocytes. Sometimes, genotype information is available for the microsome or the hepatocyte samples.
Transporter experiment conditions include bidirectional transporter, uptake/efflux, and ATPase.
Other factors of in vitro experiments include preincubation time, incubation time, quantification methods, sample size, and data analysis methods.
All these information can be found in the FDA website (http://www.abclabs.com/Portals/0/FDAGuidance_DraftDrug InteractionStudies2006.pdf).
Differed from in vitro study, in vivo refers to experimentation using a whole, living organism such that its experiment condition and parameters are quite different. Within in vivo study, in vivo PK parameters, pharmacokinetics models, study designs, and quantification methods are the key components to investigate an in vivo experiment.
All of the information for in vivo PK parameters is summarized from two text books [13, 51]. There are several main classes of PK parameters. Area under the concentration curve parameters are AUCinf, AUCSS, AUCt, and AUMC; drug clearance parameters are CL, CLb, CLu, CLH, CLR, CLpo, CLIV, CLint, and CL12; drug concentration parameters are Cmax and CSS; extraction ratio and bioavailability parameters are E, EH, F, FG, FH, FR, fe, and fm; rate constants include elimination rate constant k, absorption rate constant ka, urinary excretion rate constant ke, Michaelis–Menten constant Km, distribution rate constants (k12, k21), and two rate constants in the two-compartment model (λ1, λ2); blood flow rate (Q, QH); time parameters (tmax, t1/2); volume distribution parameters (V, Vb, V1, V2, Vss); maximum rate of metabolism, Vmax; and ratios of PK parameters that present the extent of the drug interaction (AUCR, CL ratio, Cmax ratio, Css ratio, t1/2 ratio).
Two types of pharmacokinetics models are usually presented in the literature: non-compartment model and one- or two-compartment models.
The design strategies are very diverse: single arm or multiple arms, crossover or fixed-order design, with or without randomization, with or without stratification, pre-screening or no pre-screening based on genetic information, prospective or retrospective studies, and case reports or cohort studies. The sample size includes the number of subjects and the number of plasma or urine samples per subject. The time points include sampling time points and dosing time points. The sample type includes blood, plasma, and urine. The hypotheses include the effect of bioequivalence, drug interaction, pharmacogenetics, and disease conditions on a drug's PK.
The drug quantification methods include HPLC/UV, LC/MS/MS, LC/MS, and radiographic.
Metabolism enzyme
The cytochrome P450 (officially abbreviated as CYP) enzymes predominantly exist in the gut wall and liver. The CYP450 super family is a large and diverse group of enzymes that catalyze the oxidation of organic substances. The substrates of CYP enzymes include metabolic intermediates such as lipids and steroidal hormones as well as xenobiotic substances such as drugs and other toxic chemicals. CYPs are the major enzymes involved in drug metabolism and bioactivation, accounting for about 75 % of the total number of different metabolic reactions [58]. CYP enzyme names and genetic variants were mapped from the Human Cytochrome P450 (CYP) Allele Nomenclature Database (http://www.cypalleles.ki.se/). This site contains the CYP450 genetic mutation effect on the protein sequence and enzyme activity with associated references.
In the pharmacology research, probe drug is another important concept. An enzyme's probe substrate means that this substrate is primarily metabolized or transported by this enzyme. In order to experimentally prove whether a new drug inhibits or induces an enzyme, its probe substrate is always utilized to demonstrate this enzyme's activity before and after inhibition or induction. An enzyme's probe inhibitor or inducer means that it inhibits or induces this enzyme primarily. Similarly, an enzyme's probe inhibitor needs to be utilized if we investigate whether a drug is metabolized by this enzyme. Due to its importance, all the probe inhibitors, inducers, and substrates of CYP enzymes are also included in our PK ontology. All this information was collected from industry standard (http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm064982.htm), reviewed in the top pharmacology journal [16].
Transporters are tissue specific. With different aliases, their tissue-specific transports and corresponding functions are different. Transport proteins are proteins which serve the function of moving other materials within an organism. Transport proteins are vital to the growth and life of all living things. Transport proteins are involved in the movement of ions, small molecules, or macromolecules, such as another protein, across a biological membrane. They are integral membrane proteins; that is, they exist within and span the membrane across which they transport substances. Their names and genetic variants were mapped from the Transporter Classification Database (http://www.tcdb.org). In addition, we also added the probe substrates and probe inhibitors and inducers to each one of the metabolism and transportation enzymes [16].
Drug names were created using the drug names from DrugBank 3.0 [27]. DrugBank consists of 6,829 drugs which can be grouped into different categories of FDA-approved, FDA-approved biotech, nutraceuticals, and experimental drugs. The drug names are mapped to generic names, brand names, and synonyms.
Subject included the existing ontologies for human disease ontology (DOID), suggested Ontology for Pharmacogenomics (SOPHARM), and mammalian phenotype (MP) from http://bioportal.bioontology.org.
The PK ontology was implemented with Protégé [59] and uploaded to the BioPortal ontology platform.
3.3 PK Corpus
Corpus is the key component to make NLP technologies successfully applied to text. To materialize text into computer-readable format, two types of annotations are needed, biological annotation and linguistic annotation [54]. Biological annotation belongs to event annotation, which identifies the location of biological information in the article. The scope of biological annotation can be narrowed down at single biological terms or broadened to include a whole sentence, which describes a biological event. Practically, event annotations are more complicated than term annotations. Term annotation only needs the terms to be annotated and hierarchically organized into categories. Unlike term annotation, an event has its own internal structure and it also involves biological entities (from term annotation) as its participants. Therefore, well-defined conditions to call biological events are required. On the other hand, linguistic annotation gives linguistic parsing such as POS or syntactic trees to know the type and role of term in natural language. The main purpose of linguistic annotation is to use it in the study of language through analysis of natural-occurring data. It involves computational methods and tools for analyzing linguistic pattern IR based on annotated corpora.
Most existing DDI extraction methods are designed to capture pairs of drugs that have the relation of interaction via semantic interpretation. There is one gap if we continue to use the same method for extracting DDI information from a pharmacokinetics perspective. The gap comes from the lack of knowledge to define a PK DDI. Pharmacokinetics parameters and knowledge from in vitro and in vivo DDI experimental designs, especially the selection of enzyme-specific probe substrates and inhibitors, should be considered. For instance, important pharmacokinetic parameters such as Ki, IC50, and AUCR have not been included in the existing text mining approaches to DDI. This kind of pharmacokinetic information may be particularly relevant when seeking evidence of causal mechanisms behind DDIs and as a complement to DDI text mining of patient records.
3.3.1 Corpus Construction
A PK abstract corpus was constructed to cover four primary classes of PK studies: clinical PK studies (n = 56); clinical pharmacogenetic studies (n = 57); in vivo DDI studies (n = 218); and in vitro drug interaction studies (n = 210). The PK corpus construction is a manual process. The abstracts of clinical PK studies related to the most popular CYP3A substrate, midazolam, were investigated [60]. The clinical pharmacogenetic abstracts were selected based on the most polymorphic CYP enzyme, CYP2D6. We think that these two selection strategies represent very well all the in vivo PK and PG studies. In searching for the drug interaction studies, the abstracts were randomly selected from a PubMed query, which used probe substrates/inhibitors/inducers for metabolism enzymes.
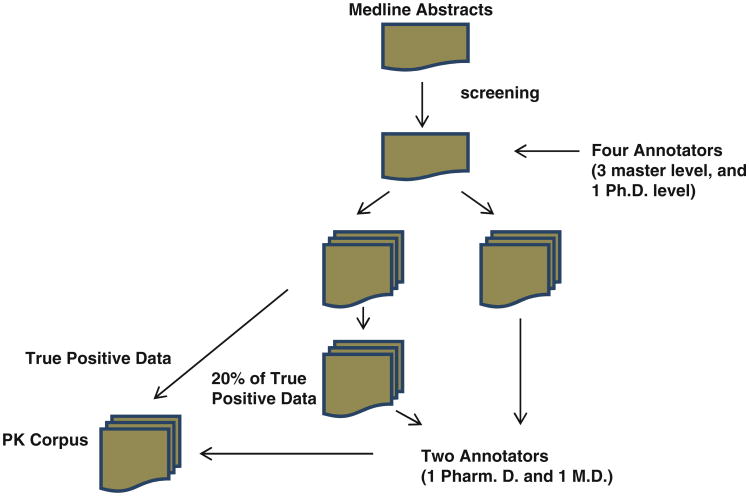
Once the abstracts have been identified in four classes, their annotation is a manual process (Fig. 1). The annotation was firstly carried out by three master-level annotators (Shreyas Karnik, Abhinita Subhadarshini, and Xu Han) and one Ph.D. annotator (Lang Li). They have different training backgrounds: computational science, biological science, and pharmacology. Any differentially annotated terms were further checked by Sara K. Quinney and David A. Flockhart, one Pharm D. scientist and one M.D. scientist with extensive pharmacology training background. Among the disagreed annotations between these two annotators, a group review was conducted (Drs Quinney, Flockhart, and Li) to reach the final agreed annotations. In addition a random subset of 20 % of the abstracts that had consistent annotations among four annotators (three masters and one Ph.D.) were double checked by two Ph.D.-level scientists.
Fig. 1. PK corpus annotation flow chart.
3.3.2 DDI Annotation Scheme
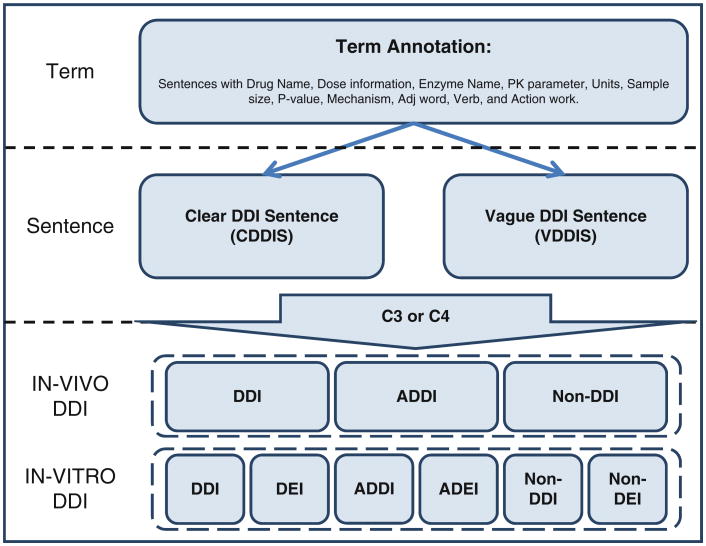
A structured annotation scheme was implemented to annotate three layers of pharmacokinetics information: key terms, DDI sentences, and DDI pairs (Fig. 2). DDI sentence annotation scheme depends on the key terms; and DDI annotations depend on the key terms and DDI sentences. Their annotation schemes are described as follows.
Fig. 2. A three-level hierarchical PK and DDI annotation scheme.
Term-level annotation
Key terms include drug names, enzyme names, PK parameters, numbers, mechanisms, and change. The boundaries of these terms among different annotators were judged by the following standard.
Drug names were defined mainly on DrugBank 3.0 [27]. In addition, drug metabolites were also tagged, because they are important in in vitro studies. The metabolites were judged by either prefix or suffix: oxi, hydroxyl, methyl, acetyl, N-dealkyl, N-demethyl, nor, dihydroxy, O-dealkyl, and sulfo. These prefixes and suffixes are due to the reactions due to phase I metabolism (oxidation, reduction, hydrolysis) and phase II metabolism (methylation, sulfation, acetylation, glucuronidation) [55].
Enzyme names covered all the CYP450 enzymes. Their names are defined in the Human Cytochrome P450 Allele Nomenclature Database, http://www.cypalleles.ki.se/. The variations of the enzyme or the gene names were considered.
PK parameters were annotated based on the defined in vitro and in vivo PK parameter ontology. In addition, some PK parameters have different names, such as CL = clearance, t1/2 = half-life, AUC = area under the concentration curve, and AUCR = area under the concentration curve ratio. Those terms need to be handled carefully because their formats are varied.
Numbers such as dose, sample size, values of PK parameters, and p-values were all annotated. If presented, their units were also covered in the annotations.
Mechanisms denote the drug metabolism and interaction mechanisms. Linguistic realization of those terms is usually presented in various contexts. The nominalization of the following terms, inhibit, catalyze, correlate, metabolize, induce, form, stimulate, activate, and suppress, is annotated with regular expression patterns.
Change describes the change of PK parameters. The following words and its nominalizations were annotated in the corpus to denote the change: strong, moderate, high, slight, significant, obvious, marked, great, pronounced, modest, probably, may, might minor, little negligible, doesn't interact, affect, reduce, and increase.
Sentence-level annotation
The middle-level annotation focused on the drug interaction sentences. Because two interaction drugs were not necessary all presented in the sentence, sentences were categorized into two classes:
Clear DDI sentence (CDDIS): Two drug names (or drug–enzyme pair in the in vitro study) are in the sentence with a clear interaction statement, i.e., either “interaction” or “non-interaction”, or ambiguous statement (i.e., such as “possible interaction” or “might interact”).
Vague DDI sentence (VDDIS): One drug or enzyme name is missed in the DDI sentence, but it can be inferred from the context. Clear interaction statement also is required.
DDI-level annotation
Once DDI sentences were labeled, the DDI pairs in the sentences were further annotated. Because of the fundamental difference between in vivo DDI studies and in vitro DDI studies, their DDI relationships were defined differently. In in vivo studies, three types of DDI relationships were defined (Table 1): DDI, ambiguous DDI (ADDI), and non-DDI (NDDI). Four conditions are specified to determine these DDI relationships. Condition 1 (C1) requires that at least one drug or enzyme name has to be contained in the sentence; condition 2 (C2) requires that the other interaction drug or enzyme name can be found from the context if it is not from the same sentence; condition 3 (C3) specifies numeric rules to define the DDI relationships based on the PK parameter changes; and condition 4 (C4) specifies the language expression patterns for DDI relationships. Using the rules summarized in Table 1, DDI, ADDI, and NDDI can be defined by C1 ∧ C2 ∧ (C3 ∨ C4). The priority rank of in vivo PK parameters is AUC > CL > t1/2 > Cmax. In in vitro studies, six types of DDI relationships were defined (Table 1). DDI, ADDI, and NDDI were similar to in vivo DDIs, but three more drug–enzyme relationships were further defined: DEI, ambiguous DEI (ADEI), and non-DDI (NDEI). C1, C2, and C4 remained the same for in vitro DDIs. The main difference is in C3, in which either Ki or IC50 (inhibition) or EC50 (induction) was used to defined DDI relationship quantitatively. The priority rank of in vitro PK parameters is Ki > IC50. Table 2 presents eight examples of how DDIs or DEIs were determined in the sentences.
Table 1. DDI definitions in corpus.
| DDI relationship | C1 | C2 | C3b | C4b |
|---|---|---|---|---|
| In vivo study | ||||
|
| ||||
| DDI | Yes | Yes | The PK parameter with the highest prioritya must satisfy p-value < 0.05 and FC > 1.50 or FC < 0.67 | Significant, obviously, markedly, greatly, pronouncedly, etc. |
| Ambiguous DDI (ADDI) | The PK parameter with the highest prioritya in the conditions of p-value < 0.05 but 0.67 <FC < 1.50; or FC>1.50 or FC < 0.67, but p-value >0.05 | Modestly, moderately, probably, may, might, etc. | ||
| Non-DDI (NDDI) | The PK parameter with the highest prioritya is in the condition of p-value > 0.05 and 0.67 < FC < 1.50 | Minor significance, slightly, little or negligible effect, does not interact, etc. | ||
|
| ||||
| In vitro study | ||||
|
| ||||
| DDI DEI | Yes | Yes | (0 < Ki< 10 or 0 < EC50 < 10 μM, and p-value < 0.05) | Significant, obviously, markedly, greatly, pronouncedly, etc. |
| ADDI Ambiguous DEI (ADEI) | (10<Ki<100 or 10<EC50<100 μM, and p-value <0.05 or vice versa) | Modestly, moderately, probably, may, might, etc. | ||
| NDDI Non-DEI (NDEI) | (Ki>100 μM or EC50>100 μM, and p-value >0.05) | Minor significance, slightly, little or negligible effect, does not interact, etc. | ||
Note:
C1: At least one drug or enzyme name has to be contained in the sentence
C2: Need to label the drug name if it is not from the same sentence
C3: PK parameter and value dependent
C4: Significance statement
For the priority of PK parameters: AUC > CL > t1/2 > Cmax; the priority of in vitro PK parameters: Ki>IC50.
Priority issue: When C3 and C4 occur and conflict, C3 dominates the sentence
Table 2. Examples of DDI definitions.
| PMID | DDI sentence | Relationship and commend |
|---|---|---|
| 20012601 | The pharmacokinetic parameters of verapamil were significantly altered by the co-administration of lovastatin compared to the control | Because of the words, “significantly,” (verapamil, lovastatin) is a DDI |
| 20209646 | The clearance of mitoxantrone and etoposide was decreased by 64 and 60 %, respectively, when combined with valspodar | Because the fold changes were less than 0.67 (mitoxantrone, valspodar) and (etoposide, valspodar) are DDIs |
| 20012601 | The (AUC (0-infinity)) of norverapamil and the terminal half-life of verapamil did not significantly changed with lovastatin co-administration | Because of the words, “not significantly changed,” (verapamil, lovastatin) is an NDDI |
| 17304149 | Compared with placebo, itraconazole treatment significantly increases the peak plasma concentration (Cmax) of paroxetine by 1.3-fold (6.7 ±2.5 versus 9.0 ±3.3 ng/mL, p≤ 0.05) and the area under the plasma concentration–time curve from zero to 48 h (AUC(0–48)) of paroxetine by 1.5-fold (137 ±73 versus 199 ±91 ng×*h/mL, p≤0.01) | AUC has a higher rank than Cmax, and it had a 1.5-fold change and less than 0.05 p-value; thus, (itraconazole, paroxetine) is a DDI |
| 13129991 | The mean (SD) urinary ratio of dextromethorphan to its metabolite was 0.006 (0.010) at baseline and 0.014 (0.025) after St John's wort administration (p= 0.26) | The change in PK parameter is more than 1.5-fold but p-value is >0.05. Thus, (dextromethorphan, St John's wort) is an ADDI |
| 19904008 | The obtained results show that perazine at its therapeutic concentrations is a potent inhibitor of human CϒP1A2 | Because of the word, “potent inhibitor,” (perazine, CYP1A2) is a DEI |
| 19230594 | After human hepatocytes were exposed to 10 μM ϒM758, microsomal activity and mRNA level for CϒP1A2 were not induced while those for CϒP3A4 were slightly induced | Because of the words, “not induced” and “slightly induced,” (YM758, CYP1A2) and (YM758, CYP1A2) are NDEIs |
| 19960413 | From these results, DPT was characterized to be a competitive inhibitor of CϒP2C9 and CϒP3A4, with K(i) values of 3.5 and 10.8 μM in HLM and 24.9 and 3.5 μM in baculovirus–insect cell-expressed human CYPs, respectively | Because Kwzs larger than 10 μM, (DPT, CYP2C9) and (DPT, CYP3A4) are ADEIs |
Corpus evaluation
Agreement measurement is one of the important steps in corpus construction, which carries out the assessment of reference standard quality. If there is little agreements among annotators, that means that the task of annotation is not reliable and the quality of reference standard is suspected. In this work, Krippendorff's alpha [61] was calculated to evaluate the reliability of annotations from four annotators. The frequencies of key terms, DDI sentences, and DDI pairs are presented in Table 3. Their Krippendorff's alphas are 0.953, 0.921, and 0.905, respectively. Please note that the total DDI pairs refer to the total pairs of drugs within a DDI sentence from all DDI sentences.
Table 3. Annotation performance evaluation.
| Key terms | Annotation categories | Frequencies | Krippendorff's alpha |
|---|---|---|---|
| Drug | 8,633 | 0.953 | |
| CYP | 3,801 | ||
| PK parameter | 1,508 | ||
| Number | 3,042 | ||
| Mechanism | 2,732 | ||
| Change | 1,828 | ||
| Total words | 97,291 | ||
|
| |||
| DDI sentences | CDDI sentences | 1,191 | 0.921 |
| VDDI sentences | 120 | ||
| Total sentences | 4,724 | ||
|
| |||
| DDI pairs | DDI | 1,239 | 0.905 |
| ADDI | 300 | ||
| NDDI | 294 | ||
| DEI | 565 | ||
| ADEI | 95 | ||
| NDEI | 181 | ||
| Total drug pairs | 12,399 | ||
The PK corpus was constructed by the following process. Raw abstracts were downloaded from PubMed in XML format. Then XML files were converted into GENIA corpus format following the gpml.dtd from the GENIA corpus [31]. The sentence detection in this step is accomplished by using the Perl module Lingua∷ EN∷Sentence, which was downloaded from the Comprehensive Perl Archive Network (CPAN, www.cpan.org). GENIA corpus files were then tagged with the prescribed three levels of PK and DDI annotations. Finally, a cascading style sheet (CSS) was implemented to differentiate colors for the entities in the corpus. This feature allows the users to visualize annotated entities. We would like to acknowledge that a DDI corpus was recently published as part of a text-mining competition DDIExtraction 2011 (http://labda.inf.uc3m.es/DDIExtraction2011/dataset.html). Their DDIs were clinical outcome oriented, not PK oriented. They were extracted from DrugBank, not from PubMed abstracts. Our PK corpus complements to their corpus very well.
3.4 DDI Text Mining
We implemented the approach described by [37] for the DDI extraction. Prior to performing DDI extraction, the testing and validation DDI abstracts in our corpus were preprocessed and converted into the unified XML format [37]. The following steps were conducted:
Drugs were tagged in each of the sentences using dictionary based on DrugBank. This step revised our prescribed drug name annotations in the corpus. One purpose is to reduce the redundant synonymous drug names. The other purpose is only to keep the parent drugs and remove the drug metabolites from the tagged drug names from our initial corpus, because parent drugs and their metabolites rarely interact. In addition, enzymes (i.e., CYPs) were also tagged as drugs, since enzyme–drug interactions have been extensively studied and published. The regular expression of enzyme names in our corpus was used to remove the redundant synonymous gene names.
Each of the sentences was subjected to tokenization, POS tags, and dependency tree generation using the Stanford parser [62].
C2n drug pairs from the tagged drugs in a sentence were generated automatically, and they were assigned with default labels as no-drug interaction. Please note that if a sentence had only one drug name, this sentence did not have a DDI. This setup limited us to consider only CDDI sentence in our corpus.
The drug interaction labels were then manually flipped based on their true drug interaction annotations from the corpus. Please note that our corpus had annotated DDIs, ADDIs, NDDIs, DEIs, ADEIs, and NDEIs. Here only DDIs and DEIs were labeled as true DDIs. The other ADDIs, NDDIs, DEIs, and ADEIs were all categorized into the no-drug interactions.
Then sentences were represented with dependency graphs using interacting components (drugs) (Fig. 3). The graph representation of the sentence was composed of two items: (1) one dependency graph structure of the sentence and (2) a sequence of POS tags (which was transformed to a linear order “graph” by connecting the tags with a constant edge weight). We used the Stanford parser [62] to generate the dependency graphs. Airola et al. proposed to combine these two graphs to one weighted, directed graph. This graph was fed into a SVM for DDI/non-DDI classification. More details about the all paths graph kernel algorithm can be found in [37].
Fig. 3. Sentence separation.
DDI extraction was implemented in the in vitro and in vivo DDI corpus separately. Table 4 presents the training sample size and testing sample size in both corpus sets. Then Table 5 presents the DDI extraction performance. In extracting in vivo DDI pairs, the precision, recall, and F-measure in the testing set are 0.67, 0.79, and 0.73, respectively. In the in vitro DDI extraction analysis, the precision, recall, and F-measure are 0.47, 0.58, and 0.52, respectively, in the in vitro testing set. In our early DDI research published in the DDIExtract 2011 Challenge [63], we used the same algorithm to extract both in vitro and in vivo DDIs at the same time, and the reported F-measure was 0.66. This number is in the middle of our current in vivo DDI extraction F-measure 0.73 and in vitro DDI extraction F-measure 0.52.
Table 4. DDI data description.
| Datasets | Abstracts | Sentences | DDI pairs | True DDI pairs |
|---|---|---|---|---|
| In vivo DDI training | 174 | 2,112 | 2,024 | 359 |
| In vivo DDI testing | 44 | 545 | 574 | 45 |
| In vitro DDI training | 168 | 1,894 | 7,122 | 783 |
| In vitro DDI testing | 42 | 475 | 1,542 | 146 |
Table 5. DDI extraction performance.
| Datasets | Precision | Recall | F-measure |
|---|---|---|---|
| In vivo DDI training | 0.67 | 0.78 | 0.72 |
| In vivo DDI testing | 0.67 | 0.79 | 0.73 |
| In vitro DDI training | 0.51 | 0.59 | 0.55 |
| In vitro DDI testing | 0.47 | 0.58 | 0.52 |
Error analysis was performed in testing samples. Table 6 summarizes the results. Among the known reasons for the false positives and false negatives, the most frequent one is that there are multiple drugs in the sentence or the sentence is long. The other reasons include that there is no direct DDI relationship between two drugs, but the presence of some words, such as dose and increase, may lead to a false-positive prediction; or DDI is presented in an indirect way; or some NDDIs are inferred due to some adjectives (little, minor, negligible).
Table 6. DDI extraction error analysis from testing DDI sets.
| No. | Error categories | Error type | Frequency | Examples | |
|---|---|---|---|---|---|
|
| |||||
| In vivo | In vitro | ||||
| 1 | There are multiple drugs and PK parameters in the sentence, and the sentence is long | FP | 6 | 34 | PMID: 12426514: In three subjects with measurable concentrations in the single-dose study, rifampin significantly decreased the mean maximum plasma concentration (C(max)) and area under the plasma concentration-time curve from 0 to 24 h (AUC(0–24)) of praziquantel by 81%(p<0.05) and 85 % (p<0.01), respectively, whereas rifampin significantly decreased the mean C(max) and AUC(0–24) of praziquantel by 74 % (p<0.05) and 80%(p<0.01), respectively, in five subjects with measurable concentrations in the multiple-dose study |
| FN | 2 | 17 | PMID: 10608481: Erythromycin and ketoconazole showed a clear inhibitory effect on the 3-hydroxylation of lidocaine at 5 μM of lidocaine (IC50 9.9 μM and 13.9 μM, respectively) but did not show a consistent effect at 800 μM of lidocaine (IC50 > 250 μM and 75.0 μM, respectively) | ||
|
| |||||
| 2 | There is no direct DDI relationship between two drugs, but the presence of some words, such as dose and increase, may lead to a false-positive prediction | FP | 6 | 14 | PMID: 17192504: A significant fraction of patients to be treated with HMR1766 is expected to be maintained on warfarin |
|
| |||||
| 3 | DDI is presented in an indirect way | FN | 2 | 19 | PMID: 11994058: In CYP2D6 poor metabolizers, systemic exposure was greater after chlorpheniramine alone than in extensive metabolizers, and administration of quinidine resulted in a slight increase in CLoral |
|
| |||||
| 4 | Design issue: Some NDDIs are inferred due to some adjectives (little, minor, negligible) | FP | 1 | 3 | PMID: 10223772: In contrast, the effect of ranitidine or ebrotidine on CYP3A activity in vivo seems to have little clinical significance |
|
| |||||
| 5 | Unknown | FP | 5 | 44 | PMID: 10383922: CYP1A2, CYP2A6, and CYP2E1 activities were not significantly inhibited by azelastine and the two metabolites |
| FN | 6 | 26 | PMID: 10681383: However, the most unusual result was the interaction between testosterone and nifedipine | ||
4 Notes (Challenges and Possible Solutions)
As we have seen, there had been a number of approaches for DDI extraction research. Nonetheless, there are significant unsolved problems or difficulties when we apply those approaches in PK DDI text mining. According to our annotation scheme, the three-level annotation is designed to identify key terms, DDI sentences, and DDI pairs. From our DDI extraction error analysis, we found that major errors come from the challenges of annotations. Most missing detections result from the issue of drug name mapping in term annotation level. The reason to cause the errors classified into the third category is that the approach we use to extract DDI lacks the ability of co-reference resolution. Due to the omission of VDDIS in DDI sentence level, these kinds of errors happen. Finally, a major part of failure resulted largely from the long sentences with multiple drugs and PK parameters. To meet these three issues, we discuss the problem of errors and try to explore their possible solution in the following three subsections.
4.1 Issues in Drug Name Mapping
In term annotation level, most biological terms can be annotated with satisfied performance by using NER, except for drug name. The representations of drug names are diverse in pharmacology articles. The main reason to this issue comes from the naming convention of different drug companies. Each drug with the same generic name might have multiple brand names or synonym name. Due to the different backgrounds of authors, the preferences of name adoption are quite different. Among drug names, some really confuse NER tools by its confliction with other terms. For example, one of ketoconazole's synonyms (DB01026) is “2 %” and a small molecule (db03951) is denominated with “16 g.” For the possible solution for this issue, we recommend to remove those terms from your dictionaries because few authors use those peculiar terms as drug names. Another issue in NER is to recognize acronym and abbreviation of drugs or other terms. There are no rules or exact patterns for the creation of acronym and abbreviation from their full form. To meet this problem, there are two possible solutions. First, parenthetical expression might be the solution to distinguish acronyms. By using Schwartz and Hearst's algorithm, it searches for parentheses in text and limits context around brackets as a mark of term, such as single or more words, e.g., nevi-rapine (NVP) or human liver microsomes (HLMs). Otherwise, using FDA-provided acronym and abbreviation database as another dictionary can be the second solution. This database can be downloaded at the following link: http://www.fda.gov/AboutFDA/FDAAcronymsAbbreviations/ucm070296.htm.
4.2 Vague DDI Sentence Problem
In most DDI extraction approaches, CDDIS are considered to be candidates for the analysis of DDI extraction. Nevertheless, we found that the number of VDDISs amounts to one-tenth of CDDISs' quantity in Table 3. If we omit investigating those sentences, it means that up to 10 % of true information is possibly missed. Although this problem also happens in many articles related to protein–protein or protein–gene interactions, it harms PK DDI articles more. It is because the interactions between proteins or genes are more often expressed with narrative ways while many of PK DDIs in text can be determined only with the measurements of ADME activities. Such omissions will highly increase the chance of missing detections, especially for the task of PK curation.
To retrieve VDDIS, human beings can easily recognize DDI information from VDDIS via the reference to other sentences. The process of determining the pronoun or the antecedent from its context is called co-reference resolution [64]. Some previous works [65, 66] had considered this problem on a pre-sentence basis and used it to explore neglected useful information in the same article. Grosz et al. [67] considered the feature of significant entities which are mentioned multiple times in context and its transitivity property to extract event–argument relations. But no one has yet considered using it to improve the performance of DDI extraction. Here we would like to choose one appropriate approach among published co-reference resolution method to transform the VDDIS into CDDIS.
Bridging references arise when a reference to a noun phrase that is not directly mentioned is made. For an example sentence in PMID-17518508 (example 1), it does not mention that which drug is the CYP3A4 inhibitor in the sentence, but readers can figure out that it is ketoconazole from few sentences before. Another type of VDDIS is more challenging to determine because the noun phrase and pronoun are not even mentioned in the sentence of PMID-17909805 (example 2). The pronoun or the antecedent for inhibitor drug even does not show in this sentence, and its argument is located in few sentences behind, which makes it more difficult to find its co-reference.
Example 1
Co-administration of a potent CϒP3A4 inhibitor moderately increased cinacalcet exposure in study subject.
Example 2
The plasma clearances of docetaxel and midazolam were reduced by 1.7-and 6-fold, respectively.
Centering theory [66, 68, 69] is a method to model the relations among focus of attention, choice of referring expression, and perceived coherence of utterances within a discourse segment. This approach should conquer the problem of example 1. As for the second example, it cannot be answered by only finding the co- reference of pronoun. Finding the relation between an event and its argument across co-reference relations will really help find the argument of events. To achieve the cross-sentence event–argument relation, some previous works [67, 70] had been capable of identifying the event for intra-sentence argument. To handle the challenges in both examples, we are eager to look for a method which reaches the best performance.
4.3 Multiple Drug Pairs and PK parameters
The purpose of DDI-level annotation is to label drug pairs and PK parameters in text and conduct the relationships for DDI pairs. From the experience of evaluating corpus and the error analysis for DDI results, there are two challenges when extracting DDI. (1) Long sentences with multiple drugs significantly complicated syntactic structure and led to the most frequent faults of first category in Table 6. In fact, such sentences often occurred in the articles related to in vivo and in vitro experiments. Authors try to compare the intensity of drug interactions among different drugs and place their PK parameters as well as dose conditions after. (2) How to take advantage of PK parameters for DDI extraction is another challenge. In the previous works, machine learning-based approach deals with the task of extracting relations by classifying pairs of drug with/without DDI categories, while rule-based or pattern-based method locates drug pairs as well as trigger words to build up a tree for determining their relationship. But, no one has yet considered using it to improve the performance of DDI extraction.
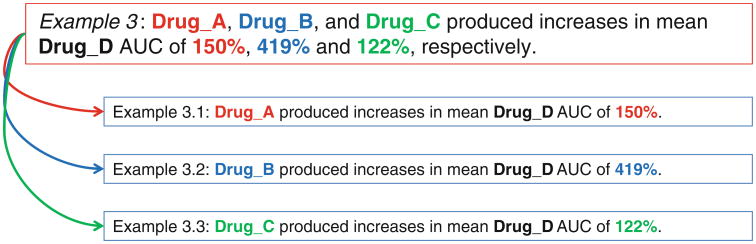
To overcome both the problem of multiple drugs and PK parameters and the challenge of utilizing PK parameters, simplifying sentence is an idea that came from Segura-Bedmar's method [43], which split the long sentences into clauses from which relations are extracted by a pattern matching algorithm. Such a simplification significantly improves the performance of dealing with long sentences. This inspires us to split a long sentence with different way. According to the characteristics of utterances in PK articles, the orders and locations of drug names and their corresponding PK parameters are parallelly located. The example in Fig. 3 shows that there are three different drugs interacting one drug followed by the corresponding fold change of AUC value. But when looking into its structure of dependent graph tree which is often used for machine learning- or rule-based pattern (Fig. 4), we found that both drugs and PK values are connected with conj_and edge. It is not possible to differentiate which PK value is belonging to which drug. Thus, splitting the sentence according to drugs and PK parameters before machine learning-or rule-based pattern matching is necessary. Using example 3 as an instance, we hope that the sentence can break down into three sentences (examples 3.1, 3.2, and 3.3 in Fig. 4). This separation greatly simplifies the sentences' complexity and resolves the problem of matching PK parameters.
Fig. 4. Dependency graph tree of example 3.
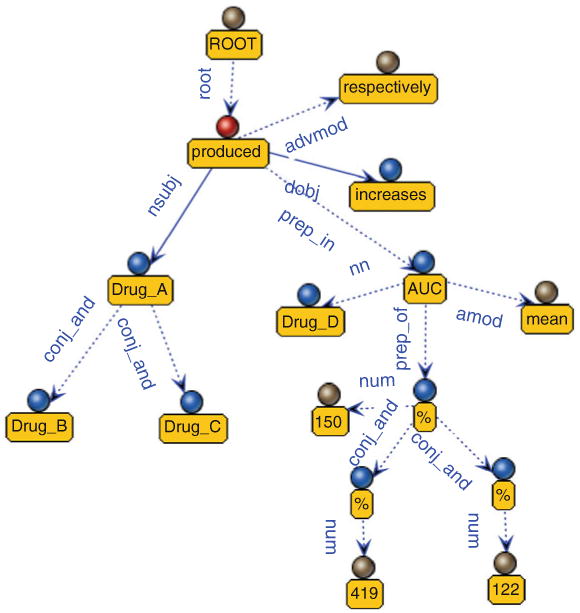
Example 3
Drug_A, Drug_B, and Drug_C produced increases in mean Drug_D AUC of 150, 419, and 122 %, respectively.
Acknowledgments
This work is supported by the US National Institutes of Health grant R01 GM74217 (Lang Li).
References
- 1.Second Annual Adverse Drug/Biologic Reaction Report. US Food and Drug Administration; 1987. [Google Scholar]
- 2.Becker ML, et al. Hospitalisations and emergency department visits due to drug–drug interactions: a literature review. Pharmacoepidemiol Drug Saf. 2007;16:641–651. doi: 10.1002/pds.1351. [DOI] [PubMed] [Google Scholar]
- 3.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58(3):621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 4.Magro L, Moretti U, Leone R. Epidemiology and characteristics of adverse drug reactions caused by drug–drug interactions. Expert Opin Drug Saf. 2012;11(1):83–94. doi: 10.1517/14740338.2012.631910. [DOI] [PubMed] [Google Scholar]
- 5.Juurlink DN, et al. Drug–drug interactions among elderly patients hospital ized for drug toxicity. JAMA. 2003;289(13):1652–1658. doi: 10.1001/jama.289.13.1652. [DOI] [PubMed] [Google Scholar]
- 6.Merle L, et al. Predicting and preventing adverse drug reactions in the very old. Drugs Aging. 2005;22(5):375–392. doi: 10.2165/00002512-200522050-00003. [DOI] [PubMed] [Google Scholar]
- 7.Johansson I, Ingelman-Sundberg M. Genetic polymorphism and toxicology: with emphasis on cytochrome p450. Toxicol Sci. 2011;120(1):1–13. doi: 10.1093/toxsci/kfq374. [DOI] [PubMed] [Google Scholar]
- 8.Ajayi FO, Sun H, Perry J. Adverse drug reactions: a review of relevant factors. J Clin Pharmacol. 2000;40(10):1093–1101. [PubMed] [Google Scholar]
- 9.DiMasi JA, Grabowski HG. The cost of biopharmaceutical R&D: is biotech different? Manage Decis Econ. 2007;28:469–479. [Google Scholar]
- 10.Pang KS, Rodrigues AD, Peter RM. Enzyme- and transporter-based drug–drug interactions. Vol. 746. Springer; New York: 2010. [Google Scholar]
- 11.The European Medicines Agency. Guideline on the investigation of drug interactions. The European Medicines Agency; London: 2012. [Google Scholar]
- 12.Jia J, et al. Mechanisms of drug combinations: interaction and network perspectives. Nat Rev Drug Discov. 2009;8(2):111–128. doi: 10.1038/nrd2683. [DOI] [PubMed] [Google Scholar]
- 13.Rowland M, Tozer TN. Clinical pharmacokinetics: concepts and applications. Lippincott Williams & Wilkins; London: 1995. [Google Scholar]
- 14.Wienkers LC, Heath TG. Predicting in vivo drug interactions from in vitro drug discovery data. Nat Rev Drug Discov. 2005;4(10):825–833. doi: 10.1038/nrd1851. [DOI] [PubMed] [Google Scholar]
- 15.Rostami-Hodjegan A, Tucker G. ‘In silico’ simulations to assess the ‘in vivo’ consequences of ‘in vitro’ metabolic drug– drug interactions. Drug Discov Today. 2004;1(4):441–448. doi: 10.1016/j.ddtec.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Huang SM, et al. Drug interaction studies: study design, data analysis, and implications for dosing and labeling. Clin Pharmacol Ther. 2007;81(2):298–304. doi: 10.1038/sj.clpt.6100054. [DOI] [PubMed] [Google Scholar]
- 17.Li L, Yu M, Chin R, Lucksiri A, Flockhart D, Hall S. Drug–drug interaction prediction: a Bayesian meta-analysis approach. Stat Med. 2007;26(20):3700–3721. doi: 10.1002/sim.2837. [DOI] [PubMed] [Google Scholar]
- 18.Yu M, et al. A Bayesian meta-analysis on published sample mean and variance pharmacokinetic data with application to drug–drug interaction prediction. J Biopharm Stat. 2008;18(6):1063–1083. doi: 10.1080/10543400802369004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou J, et al. A new probabilistic rule for drug–drug interaction prediction. J Pharmacokinet Pharmacodyn. 2009;36:1–18. doi: 10.1007/s10928-008-9107-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou J, Qin Z, Kim S, Wang Z, Hall DS, Li L. Drug–drug interaction prediction assessment. J Pharmacokinet Pharmacodyn. 2009;19:641–657. doi: 10.1080/10543400902964084. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z, Kim S, Quinney SK, Zhou J, Li L. Non-compartment model/compartment model transformation. BMC System Biol. 2010;4(1):S8. doi: 10.1186/1752-0509-4-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L. Discussion on parameter estimation for differential equations: a generalized smoothing approach. J Royal Stat Soc B. 2007;69:787–788. [Google Scholar]
- 23.Chien JY, Lucksiri A, Ernest CS, Gorski JC, Wrighton SA, Hall SD. Stochastic prediction of CYP3A-mediated inhibition of midazolam clearance by ketoconazole. Drug Metab Dispos. 2006;34(7):1208–1219. doi: 10.1124/dmd.105.008730. [DOI] [PubMed] [Google Scholar]
- 24.Quinney SK, Zhang X, Lucksiri A, Gorski JC, Li L, et al. Physiologically based pharma-cokinetic model of mechanism-based inhibition of CYP3A by clarithromycin. Drug Metab Dispos. 2010;38(2):241–248. doi: 10.1124/dmd.109.028746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hachad H, Ragueneau-Majlessi I, Levy RH. A useful tool for drug interaction evaluation: the University of Washington Metabolism and Transport Drug Interaction Database. Hum Genomics. 2010;5(1):61–72. doi: 10.1186/1479-7364-5-1-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hewett M, et al. PharmGKB: the pharmacogenetics knowledge base. Nucleic Acids Res. 2002;30(1):163–165. doi: 10.1093/nar/30.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knox C, et al. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 2011;39(Database issue):D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gottlieb A, et al. INDI: a computational framework for inferring drug interactions and their associated recommendations. Mol Syst Biol. 2012;8:592. doi: 10.1038/msb.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. J Am Med Inform Assoc. 2011;18:544–551. doi: 10.1136/amiajnl-2011-000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zweigenbaum P, et al. Frontiers of biomedical text mining: current progress. Brief Bioinform. 2007;8(5):358–375. doi: 10.1093/bib/bbm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JD, et al. GENIA corpus–semantically annotated corpus for bio-text mining. Bioinformatics. 2003;19(Suppl 1):i180–i182. doi: 10.1093/bioinformatics/btg1023. [DOI] [PubMed] [Google Scholar]
- 32.Wilbur WJ, Rzhetsky A, Shatkay H. New directions in biomedical text annotation: definitions, guidelines and corpus construction. BMC Bioinformatics. 2006;7:356. doi: 10.1186/1471-2105-7-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muller HM, Kenny EE, Sternberg PW. Textpresso: an ontology-based information retrieval and extraction system for biological literature. PLoS Biol. 2004;2(11):e309. doi: 10.1371/journal.pbio.0020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feldman R, et al. Mining biomedical literature using information extraction. Curr Drug Discov. 2002;2:19–23. [Google Scholar]
- 35.Fundel K, Küffner R, Zimmer R. RelEx: relation extraction using dependency parse trees. Bioinformatics. 2007;23:365–371. doi: 10.1093/bioinformatics/btl616. [DOI] [PubMed] [Google Scholar]
- 36.Qian L, Zhou G. Tree kernel-based protein–protein interaction extraction from biomedical literature. J Biomed Inform. 2012;45(3):535–543. doi: 10.1016/j.jbi.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Airola A, et al. All-paths graph kernel for protein–protein interaction extraction with evaluation of cross-corpus learning. BMC Bioinformatics. 2008;9(Suppl 11):S2. doi: 10.1186/1471-2105-9-S11-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pyysalo S, et al. Comparative analysis of five protein–protein interaction corpora. BMC Bioinformatics. 2008;9(Suppl 3):S6. doi: 10.1186/1471-2105-9-S3-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tikk D, et al. A comprehensive benchmark of kernel methods to extract protein– protein interactions from literature. PLoS Comput Biol. 2010;6:e1000837. doi: 10.1371/journal.pcbi.1000837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, Liu F, Manderick B. Normalizing interactor proteins and extracting interaction protein pairs using support vector machines. BioCreative II 5 Workshop 2009 on Digital Annotations 2009 [Google Scholar]
- 41.Zhou D, He Y. Extracting interactions between proteins from the literature. J Biomed Inform. 2008;41(2):393–407. doi: 10.1016/j.jbi.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 42.Krallinger M, Leitner F, Valencia A. The BioCreative II.5 challenge overview. Proceedings of the BioCreative II 5 Workshop 2009 on Digital Annotations 2009 [Google Scholar]
- 43.Segura-Bedmar I, Martinez P, de Pablo-Sanchez C. A linguistic rule-based approach to extract drug–drug interactions from pharmacological documents. BMC Bioinformatics. 2011;12(Suppl 2):S1. doi: 10.1186/1471-2105-12-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Segura-Bedmar I, Martinez P, Sanchez-Cisneros D. Proceedings of the 1st challenge task on drug–drug interaction extraction 2011. Spain: 2011. The 1st DDIExtraction-2011 challenge task: extraction of drug–drug interactions from biomedical texts. [Google Scholar]
- 45.Percha B, Garten Y, Altman RB. Discovery and explanation of drug–drug interactions via text mining. Pac Symp Biocomput. 2012:410–421. [PMC free article] [PubMed] [Google Scholar]
- 46.Tari L, et al. Discovering drug–drug interactions: a text-mining and reasoning approach based on properties of drug metabolism. Bioinformatics. 2010;26(18):i547–i553. doi: 10.1093/bioinformatics/btq382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Segel IH. Enzyme kinetics: behavior and analysis of rapid equilibrium and steady state enzyme systems. Wiley; New York: 1975. [Google Scholar]
- 48.Consortium IT et al. Membrane transporters in drug development. Nat Rev Drug Discov. 2010;9(3):215–236. doi: 10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rostami-Hodjegan A, Tucker G. In silico simulations to assess the in vivo consequences of in vitro metabolic drug–drug interactions. Drug Disc Today Technol. 2004;1:441–448. doi: 10.1016/j.ddtec.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 50.Lam YW, Alfaro CL, Ereshefsky L, Miller M. Pharmacokinetic and pharmacodynamic interactions of oral midazolam with ketoconazole, fluoxetine, fluvoxamine, and nefazodone. J Clin Pharmacol. 2003;43(11):1274–1282. doi: 10.1177/0091270003259216. [DOI] [PubMed] [Google Scholar]
- 51.Gibaldi M, Perrier D. Pharmacokinetics. 2nd. Marcel Dekker; New York: 1982. [Google Scholar]
- 52.Vazquez M, et al. Text mining for drugs and chemical compounds: methods, tools and applications. Mol Inform. 2011;30:506–519. doi: 10.1002/minf.201100005. [DOI] [PubMed] [Google Scholar]
- 53.Spasic I, et al. Text mining and ontologies in biomedicine: making sense of raw text. Brief Bioinform. 2005;6(3):239–251. doi: 10.1093/bib/6.3.239. [DOI] [PubMed] [Google Scholar]
- 54.Kim JD, Ohta T, Tsujii J. Corpus annotation for mining biomedical events from literature. BMC Bioinformatics. 2008;9:10. doi: 10.1186/1471-2105-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brunton LL, Chabner BA, Knollmann BC. Goodman & Gilman's the pharmacological basis of therapeutics. 12th. McGraw-Hill; New York: 2011. [Google Scholar]
- 56.Witte R, Kappler T, Baker CJO. Ontology design for biomedical text mining, in semantic Web: revolutionizing knowledge discovery in the life sciences. Springer; USA: 2007. pp. 281–313. [Google Scholar]
- 57.Giacomini KM, et al. Membrane transporters in drug development. Nat Rev Drug Discov. 2010;9(3):215–236. doi: 10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guengerich FP. Cytochrome p450 and chemical toxicology. Chem Res Toxicol. 2008;21(1):70–83. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- 59.Rubin DL, Noy NF, Musen MA. Protege: a tool for managing and using terminology in radiology applications. J Digit Imaging. 2007;20(Suppl 1):34–46. doi: 10.1007/s10278-007-9065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Z, et al. Literature mining on pharmacokinetics numerical data: a feasibility study. J Biomed Inform. 2009;42(4):726–735. doi: 10.1016/j.jbi.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krippendorff K. Content analysis: an introduction to its methodology. SAGE; Thousand Oaks, CA: 2004. [Google Scholar]
- 62.de Marneffe MC, MacCartney B, Manning CD. Generating typed dependency parses from phrase structure parses. LREC 2006 [Google Scholar]
- 63.Karnik S, et al. The 1st challenge task on drug–drug interaction extraction. Huelva, Spain: 2011. Extraction of drug–drug interactions using all paths graph kernel. [Google Scholar]
- 64.van Deemter K, Kibble R. On core ferring: coreference in muc and related annotation schemes. Comput Linguist. 2000;26(4):629–637. [Google Scholar]
- 65.Hobbs J. Resolving pronoun references Readings in natural language processing. Morgan Kaufmann Publishers Inc.; San Francisco, CA, USA: 1986. pp. 339–352. [Google Scholar]
- 66.Grosz BJ, Weinstein S, Joshi AK. Centering: a framework for modeling the local coherence of discourse. Comput Linguist. 1995;21(2):203–225. [Google Scholar]
- 67.Yoshikawa K, et al. Coreference based event-argument relation extraction on biomedical text. J Biomed Semantics. 2011;2(Suppl 5):S6. doi: 10.1186/2041-1480-2-S5-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brennan SE, Friedman MW, Pollard CJ. Proceedings of the 25th annual meeting on Association for Computational Linguistics. Morristown, NJ, USA: 1987. A centering approach to pronouns. [Google Scholar]
- 69.Elango P. Coreference resolution: a survey. University of Wisconsin; Madison, WI: 2005. [Google Scholar]
- 70.Lee H, et al. Deterministic coreference resolution based on entity-centric, precision-ranked rules. Comput Linguist. 2013;34(4):885–916. [Google Scholar]