Abstract
Natural killer T (NKT) cells develop from common CD4+ CD8+ thymocyte precursors. Transcriptional programs that regulate the development of NKT cells in the thymus development remain to be fully delineated. Here we demonstrate a cell-intrinsic requirement for transcription factors TCF1 and LEF1 for the development of all subsets of NKT cells. Conditional deletion of TCF1 alone results in a substantial reduction in NKT cells. The remaining NKT cells are eliminated when TCF1 and LEF1 are both deleted. These data reveal an essential role for TCF1 and LEF1 in development of NKT cells.
Keywords: TCF1, LEF1, NKT cells
INTRODUCTION
Natural killer T (NKT) cells develop from CD4+ CD8+ double positive (DP) thymocytes that express a T-cell receptor α (TCRα) chain, Vα14-Jα18, paired with Vβ8, Vβ7 or Vβ2 in mice to generate a limited TCR repertoire that recognizes glycolipid antigens presented by the MHC class I-like molecule CD1d. TCR-dependent recognition of lipid antigens presented by CD1d expressed on DP thymocytes signals commitment to the NKT lineage (Bendelac, 1995, Gapin, et al., 2001, Godfrey, et al., 2010, Hu, et al., 2011). Whereas the transcriptional programs that regulate development of conventional T cells from DP precursors have been defined, the transcriptional control of NKT cell generation remain to be delineated.
Several transcription factors are known to play an active role in the DP-to-NKT transition and the generation of NKT0 immature precursor cells that will subsequently mature into their effector phenotypes. Deletion of E protein HEB results in absence of NKT cells due to inability to generate Vα-Jα rearrangements (D'Cruz, et al., 2010). HEB also affects expression of RORγt that together with Runx1 regulates the half-life of DP cells (Egawa, et al., 2005). Likewise, c-Myb plays a role on the lifetime of DP cells and is also required for the expression of CD1d and signaling molecules SLAMF1, SLAMF6 and SAP (Hu, et al., 2010). Bcl11b deletion blocks early NKT cell development at the DP-to-NKT transition (Albu, et al., 2011). Finally, c-Myc controls proliferation during the DP-to-NKT transition (Dose, et al., 2009). Together these observations point to a complex developmental program that leads to commitment to the NKT lineage.
Evolutionarily conserved transcription factor T Cell Factor (TCF)-1 has been shown to repress and activate gene expression (Schilham and Clevers, 1998, Klingel, et al., 2012). In mammals, TCF1 is encoded by the Tcf7 gene and is expressed exclusively in T cells. TCF1 expression is required for conventional T cell development in the thymus and mice with germline deletion of TCF1 have very few thymocytes that are impaired at multiple stages during development (Staal and Sen, 2008, Staal, et al., 2008, Staal and Clevers, 2005, Staal and Clevers, 2000). Recently, we showed that germline deletion of TCF1 leads to reduced lifetime of CD4+ CD8+ double positive (DP) thymocytes in vivo (Sharma, et al., 2014). Reduction in lifetime results in failure to rearrange the distal TCR Vα14-Jα18 and express TCR proteins required for development of NKT cells. However, conditional deletion of the TCF1 gene with CD4-Cre has at least 30% DP thymocytes with undeleted TCF1 (Steinke, et al., 2014), which permits expression of selecting proteins on DP thymocytes and may allow the DP thymocytes that rearrange the TCR Vα14-Jα18 to develop into NKT cells. Thus, the issue of cell-intrinsic requirement for TCF1 for NKT cell development remains unanswered. Furthermore, some functions of TCF1 during conventional T cell development have been shown to be redundant with a related transcription factor called Lymphocyte Enhancer-binding Factor (LEF)-1 (Okamura, et al., 1998, Yu, et al., 2012). Thus, the cell intrinsic requirement for TCF1 and LEF1 in the generation and differentiation of NKT cells in the thymus remains to be fully defined.
This study shows that, whereas conditional deletion of TCF1 (TCF1-cKO) did not lead to a reduction in thymocyte numbers, TCF1 deficiency in NKT-precursor DP thymocytes substantially reduced the numbers of NKT cells. The few remaining NKT cells were NKT0 and NKT1 cells in TCF1-cKO thymus. Residual NKT cells were further eliminated in mice with conditional deletion of both transcription factors, TCF1 and LEF1, in DP thymocytes. These data show that cell autonomous expression of TCF1 and LEF1 expression are required for effective development of NKT cells at the earliest stages of development.
MATERIALS AND METHODS
Mice
Mice with single conditional deletion of LEF1 (LEF1-cKO), TCF1 (TCF1-cKO), and conditional deletion of both transcription factors (TCF1/LEF1-cDKO) are described elsewhere (Steinke, et al., 2014). CD1d knockout (CD1d-KO) mice were obtained from the Jackson Laboratory and CD45.1+ C57BL/6.SJL mice were purchased from Taconic. All the mice used are on a C57BL/6 genetic background. CD45.1+2+ mice were generated by breeding C57BL/6.SJL mice with C57BL/6 mice. Age-matched (7-12 weeks old) littermate controls or C57BL/6 mice were used in all experiments. CD1d-KO mice used in experiment were 3-4 weeks of age. All mice were bred and maintained in the animal facility at the National Institute on Aging (NIA). The studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals (NRC 2010). The protocol was approved by the Animal Care and Use Committee of the NIA Intramural Research Program, NIH. This program is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (File 000401), registered by the United States Department of Agriculture (51-F-0016) and maintains an assurance with the Public Health Service (A4149-01).
Flow cytometry
Single-cell suspensions were prepared from thymus and spleen as per standard protocols. Hepatic lymphocytes were isolated from livers that were homogenized, filtered through nylon mesh and washed in PBS with 1% FBS. Cells were then resuspended in 44% Percoll (GE Healthcare Bio-Sciences AB, Uppsala, Sweden), underlaid with 66% Percoll, and centrifuged for 20 min at 2000 rpm. Cells at the interface were collected, washed, and counted. Cells were stained and acquired on a FACSCantoII (Becton Dickinson) and analyzed with FlowJo (Treestar). Dead cells were excluded using the Fixable Viability Dye eFluor®506 (eBioscience). The following antibodies and their isotype controls conjugated to FITC, PE, PerCP-Cy5.5, PE-Cy7, APC, APC-Cy7 or Pacific Blue (from BD Biosciences, eBioscience or BioLegend) were used for staining: anti-CD4 (GK1.5), anti-CD8α (53-6.7), anti-TCRβ (H57-597), anti-NK1.1 (PK136), anti-CD24 (M1/69). PE- or APC- conjugated mouse CD1d tetramers loaded with glycolipid PBS-57 (CD1d-tet) and an unloaded tetramer comprised of only the glycolipid PBS57 were obtained from the tetramer facility of the US National Institutes of Health. In brief, cells were incubated with FC block and stained with antibodies, and then fixed with 2% paraformaldehyde. For PLZF, T-bet, TCF-1 and LEF-1 intracellular staining, cells were permeabilized and stained accordingly with anti-PLZF (D-9) (Santa Cruz Biotechnology, Inc.) plus FITC anti-mouse (BD Biosciences), anti-T-bet (eBio4B10) (eBioscience), anti-TCF-1 (C63D9) and/or anti-LEF-1 (C18A7) (both from Cell Signaling) followed by goat anti-rabbit-Alexa647 or Alexa488 (Invitrogen), using the Foxp3 Staining Buffer kit (eBioscience).
Bone marrow chimeras
For BM transplantation experiments, the CD45.1+ recipient mice were lethally irradiated with 950 rads 24 h before receiving BM transfers. BM cells were harvested from the femurs and tibias of two donor mice and depleted of mature T cells, B cells, and MHC class II-positive lymphocytes by using a cocktail of antibodies containing anti-CD4, anti-CD8, anti-CD19 (1D3), and anti-MHC class II (M5/114) followed by complement-mediated lysis. For the BM mixed chimera experiments, BM cells from two different types of donor mice were mixed at 1:1 ratio. Each recipient mouse received 9 × 106 cells in 250 μl of PBS through i.v. injection. For the other BM chimera experiments, each recipient mouse received 2 × 106 whole bone marrow cells from a single donor in 400 μl of PBS through i.v. injection. In all experiments, CD45 congenic markers were used to distinguish cells derived from the different sources. All BM chimeras were reconstituted for at least 7 weeks before analysis.
Statistics
Statistical significance was determined by the Student’s t-test.
RESULTS
TCF1 controls the development of NKT cells
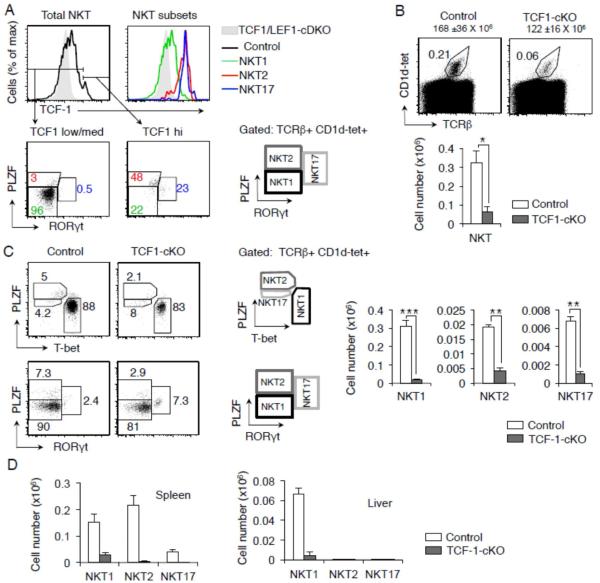
To study the role of TCF1 in NKT cell generation we analyzed the expression of TCF1 in NKT cells from C57BL/6 control mice. We found TCF1 was highly expressed in all NKT cells with highest level of expression in NKT2 and NKT17 subsets (Figure 1A). To determine a role for TCF1 in NKT cell generation, we analyzed thymocytes from mice with conditional deletion of TCF1 in thymocytes, Tcf7flox/flox CD4-Cre+ (TCF1-cKO) mice (Steinke, et al., 2014). Conditional deletion of TCF1 did not substantially alter the number of thymocytes compared to control mice (Figure 1B). By contrast, TCF1 deficiency resulted in greater than 80% reduction in NKT cell numbers from TCF1-cKO compared to control mice (Figure 1B). Detailed analysis showed that significantly fewer mature NKT1, NKT2 and NKT17 cells developed in the thymus of TCF1-cKO mice compared to control mice (Figure 1C). Finally, analysis of NKT cells in spleen and liver also showed a substantial reduction in all subsets of NKT cells in the peripheral organs (Figure 1D). These data show that when TCF1 is conditionally deleted in precursor DP thymocytes, NKT cell development was substantially blocked. We conclude that TCF1 plays an important role in specifying NKT cell lineage.
FIGURE 1. TCF1 is expressed in NKT cells and controls the development of NKT cells.
(A) Flow cytometry of TCF1 expression in total NKT cells and in NKT1, NKT2 and NKT17 subsets. Cells from TCF1/LEF1-cDKO are shown as a negative control in shaded grey. Data are representative of three mice. Flow cytometry from a representative experiment showing frequency of total NKT (B) and mature NKT1, NKT2 and NKT17 cells gated by two different strategies as shown (C) from control and TCF1-cKO mice. Graphs show cell numbers gated from each population from thymus, (B, C), spleen and liver (D) of control and TCF1-cKO mice (n ≥ 3, mean ± sem). Numbers over dot plots in (B) refer to total thymocyte cell numbers. * P<.05; ** P<.01; *** P<.001.
TCF1 and LEF1 control the earliest stages of NKT cell development
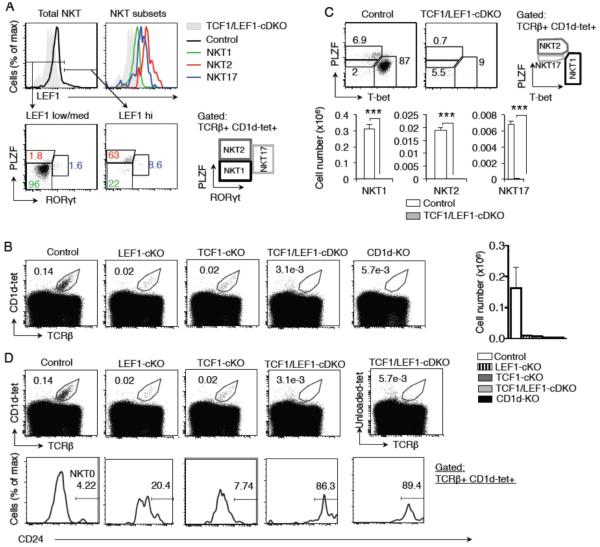
To determine if the residual NKT cells that develop in TCF1-cKO mice result from a redundant role of LEF1 in NKT cell development, we analyzed mice with conditional deletion of both transcription factors in DP thymocytes in TCF1/LEF1-cDKO mice (Steinke, et al., 2014). First, we noted that LEF1 was expressed in all NKT cells with highest level of expression in NKT2 and NKT17 cells (Figure 2A). Total number of thymocytes has been shown to be substantially comparable in mice with conditional deletion of TCF1 and/or LEF1 using Cd4-Cre compared to control mice (Steinke, et al., 2014). However, single deficiency of either LEF1 or TCF1 both showed lower NKT frequency and numbers, but did not completely abolish the NKT population (Figure 2B). Furthermore, the frequency of NKT cells was significantly (p=0.01) reduced in TCF1/LEF1-cDKO compared to control mice and was comparable to CD1d-KO mice (Figure 2B). Importantly, all subsets of NKT cells were significantly diminished TCF1/LEF1-cDKO compared to control mice (Figure 2C). We conclude that TCF1 and LEF1 both contribute to NKT cells maturation and are not compensatory in their function.
FIGURE 2. LEF1 is expressed in NKT cells and participates in the development of NKT cells.
(A) Flow cytometry of LEF1 expression in total NKT cells and in NKT1, NKT2 and NKT17 subsets. Cells from TCF1/LEF1-cDKO are shown as a negative control in shaded grey. Data are representative of three mice. (B) A representative flow plot showing frequency of total NKT cells from control (n=3), LEF1-cKO (n=2), TCF1-cKO (n=2), TCF1/LEF1-cDKO (n=5) and CD1d-KO (n=3) mice. Graph shows total thymic NKT cell numbers from each experimental group (mean ± sem). (C) Flow cytometry plot showing frequency of mature NKT cell types from control and TCF1/LEF1-cDKO mice. Graphs below show cell numbers of various NKT cell populations from thymus (n ≥ 3, mean ± sem). (D) Representative plots showing frequency of NKT cells (top) and NKT0 cells (CD1d-tet+TCRβ+CD24+) (bottom) from control, LEF1-cKO, TCF1-cKO and TCF1/LEF1-cDKO mice. Also shown is a representative plot of TCF1/LEF1-cDKO mouse stained with unloaded-tetramer as isotype control for CD1d-tet. Frequency numbers presented in NKT0 plots represent percentage of NKT cells. * P<.05; ** P<.01; *** P<.001.
Further analysis of NKT cells in TCF1-cKO and LEF1-cKO single mutant mice reveal a higher percentage CD24 expressing NKT cells, identified as immature NKT0 cells, compared to control mice (Figure 2D). Indeed, the few cells that were gated as NKT cells in TCF1/LEF1-cDKO showed high levels of CD24 expression (Figure 2D). These data demonstrate that TCF1 and LEF1 play a largely non-redundant role in the development of NKT cells. The few NKT cells that developed in TCF1-cKO mice were further eliminated in TCF1/LEF1-cDKO mice. The final analysis of the very few remaining cells showed that they expressed CD24 suggesting that some NKT0 cells may be generated in TCF1/LEF1-cDKO mice (Figure 2D). However, staining of CD1d-KO thymocytes with loaded and unloaded-tetramer mice showed similar staining pattern with anti-CD24 antibody (Figure 2D). These results revealed that when NKT cells were substantially reduced CD24 expression on other thymocytes was noted. In light of these observations we conclude that TCF1 and LEF1 both contribute to NKT cell development such that single mutant mice show significantly reduced NKT cell development with only few NKT0 cells remaining. The remaining NKT0 cells were also eliminated when both transcription factors were deleted.
The function of TCF1 and LEF1 in NKT cell development is cell autonomous
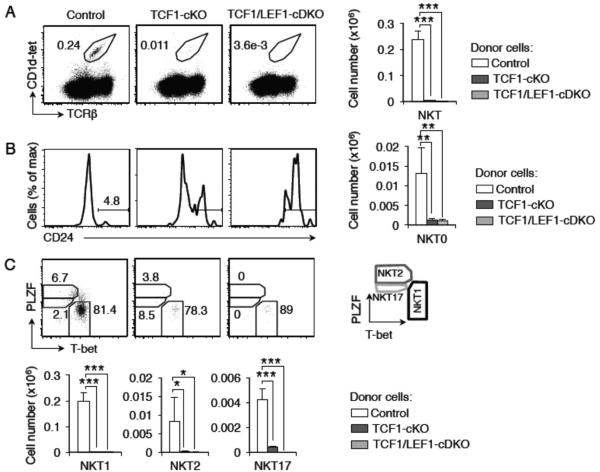
To determine if TCF1 controls NKT cell development in a cell-intrinsic manner, we transferred TCF1-cKO bone marrow cells into irradiated CD45.1+ recipients. Thirteen weeks after transplant, we observed that the recipients of TCF1-cKO bone marrow were devoid of nearly all NKT cells (Figure 3A). We also tested if the function of both TCF1 and LEF1 was cell intrinsic and found that recipients of TCF1/LEF1-cDKO bone marrow also lacked NKT cells (Figure 3A). The few NKT cells that developed in TCF1-cKO showed a preference towards NKT0 stage compared to controls (Figure 3B). These data were further substantiated by the observation that essentially no mature NKT cells developed in transplanted mice (Figure 3C) [Number of donor-derived mature NKT cells in the thymus of recipient mice: NKT1: control 0.199±0.03 × 106 cells, TCF1-cKO 0.0037±0.00016 × 106 cells and TCF1/LEF1-cDKO 0.0012±0.0004 × 106 cells; NKT2: control 0.0083±0.006 × 106 cells, TCF1-cKO 0.00027±0.00015 × 106 cells and TCF1/LEF1-cDKO 0.00001±0.00001 × 106 cells; NKT17: control 0.004±0.0008 × 106 cells, TCF1-cKO 0.00039±0.00001 × 106 cells and TCF1/LEF1-cDKO 0±0x 106 cells]. As shown in figure 2B, TCF1/LEF1-cDKO thymocytes stained with unloaded-tet showed staining for CD24 expressing cells that were not NKT cells. This indicates that the apparent high frequency of NKT0 cells in transplanted mice may be a reflection of non-NKT thymocytes that express CD24 (Figure 3B). These data demonstrate a cell- autonomous requirement for TCF1 and LEF1 in the generation of NKT cells. Thus we conclude that transcription factors TCF1 and LEF1 are required for NKT cell development at the earliest stages of development in a cell-intrinsic manner.
FIGURE 3. TCF1 and LEF1 control the development of NKT cells in a cell-intrinsic manner.
Flow cytometry of a bone-marrow chimera experiment analyzed 13 weeks post-transplantation. CD45.1+ recipient C57BL/6.SJL irradiated mice were transplanted with CD45.2+ cells from control, TCF1-cKO or TCF1/LEF1-cDKO mice. Thymic cells gated on CD45.2+ cells (donor) were further analyzed. Representative experiment shows frequency of total NKT (A), NKT0 (B) and mature NKT subsets (C) from control, TCF1-cKO and TCF1/LEF1-cDKO donor cells. Graphs show cell numbers gated from each population from thymus of recipient that received donor control (n=2), TCF1-cKO (n=2) or TCF1/LEF1-cDKO mice (n = 3) (mean ± st dev). * P<.05; ** P<.01; *** P<.001.
DISCUSSION
In this study we demonstrate that TCF1 deficiency in NKT-cell precursor DP thymocytes blocks the development of all NKT cells. The few NKT cells that remained in TCF1-cKO mice were eliminated when TCF1 and LEF1 were both deleted in the NKT-precursor cells. Thus, in mice with TCF1 and LEF1 conditional double-deletion in DP thymocytes we found very few NKT cells. We posit that TCF1-dependent gene expression is required to promote NKT cell development and LEF1 failed to compensate for TCF1 deficiency.
Parameters that regulate NKT cell development include rearrangement of TCR Vα14-Jα18 gene to pair with Vβ8, Vβ7 or Vβ2 to generate the restricted TCR selected by CD1d on DP thymocytes. TCR Vα14-Jα18 gene rearrangement has been shown to be a distal event (Guo, et al., 2002). Therefore, the recent observation showing that germline deletion of TCF1, in which all DP thymocytes lack TCF1 expression, failed to undergo this rearrangement, provided one role for TCF1 expression in the generation of NKT cells (Sharma, et al., 2014). Enforced expression of BclXL rescued the lifetime of TCF1-deficient DP thymocytes and TCR Vα14-Jα18 gene rearrangements but did not rescue NKT cell development (Sharma, et al., 2014). Likewise, enforced expression of rearranged Vα14-Jα18 transgene failed to rescue NKT cell development in TCF1-deficient background (Sharma, et al., 2014). Along these lines, even though mice with CD4-Cre-dependent conditional deletion of TCF1 have approximately 20-30% DP thymocytes that retain expression of TCF1 (Steinke, et al., 2014), we found that these mice had a profound defect in NKT cell development. These data suggest a more extensive role for this transcription factor in NKT cell commitment and development. In this report we show that TCF1 is required at multiple stages of NKT cell development. First, TCF1-deficiency partially impairs maturation of NKT0 cells. At this stage of development LEF1 contributes in a partially redundant manner. Furthermore, the NKT cells that develop in TCF1-deficient mice are largely of NKT1 subset with greater impairment in the development of NKT2 and NKT17 cells. Molecular mechanisms downstream of TCF1-dependent gene expression remain to be defined.
CONCLUSIONS
In conclusion, this report indicates a cell-intrinsic role for TCF1 in the development of NKT cells beyond the requirement of this transcription factor in maintaining the required lifetime of DP thymocytes for rearrangement of TCR Vα14-Jα18 gene to pair with Vβ8, Vβ7 or Vβ2 to generate the restricted TCR for selection by CD1d on DP thymocytes. The data also supports a role for TCF1 in the differentiation and/or expansion of NKT2 and NKT17 cells.
Highlights.
Cell-intrinsic requirement for transcription factor TCF1 for the development of NKT cells.
TCF1 differentially regulates differentiation and/or survival of NKT cell subsets, NKT1, 2 and 17.
ACKNOWLEDGEMENTS
We thank NIA animal facility and genotyping facility for animal husbandry and genotyping and the Tetramer Facility of the US National Institutes of Health for providing PE- and APC-conjugated mouse CD1d tetramers loaded with glycolipid PBS-57. This research was supported by the Intramural Research Program of the National Institute on Aging at the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors declare no conflict of interest.
REFERENCES
- Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. 1995;182:2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nature immunology. 2001;2:971–978. doi: 10.1038/ni710. [DOI] [PubMed] [Google Scholar]
- Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nature immunology. 2010;11:197–206. doi: 10.1038/ni.1841. [DOI] [PubMed] [Google Scholar]
- Hu T, Gimferrer I, Alberola-Ila J. Control of early stages in invariant natural killer T-cell development. Immunology. 2011;134:1–7. doi: 10.1111/j.1365-2567.2011.03463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Cruz LM, Knell J, Fujimoto JK, Goldrath AW. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nature immunology. 2010;11:240–249. doi: 10.1038/ni.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa T, Eberl G, Taniuchi I, Benlagha K, Geissmann F, Hennighausen L, Bendelac A, Littman DR. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity. 2005;22:705–716. doi: 10.1016/j.immuni.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Hu T, Simmons A, Yuan J, Bender TP, Alberola-Ila J. The transcription factor c-Myb primes CD4+CD8+ immature thymocytes for selection into the iNKT lineage. Nat Immunol. 2010;11:435–441. doi: 10.1038/ni.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albu DI, VanValkenburgh J, Morin N, Califano D, Jenkins NA, Copeland NG, Liu P, Avram D. Transcription factor Bcl11b controls selection of invariant natural killer T-cells by regulating glycolipid presentation in double-positive thymocytes. Proc Natl Acad Sci U S A. 2011;108:6211–6216. doi: 10.1073/pnas.1014304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dose M, Sleckman BP, Han J, Bredemeyer AL, Bendelac A, Gounari F. Intrathymic proliferation wave essential for Valpha14+ natural killer T cell development depends on c-Myc. Proc Natl Acad Sci U S A. 2009;106:8641–8646. doi: 10.1073/pnas.0812255106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilham MW, Clevers H. HMG box containing transcription factors in lymphocyte differentiation. Semin Immunol. 1998;10:127–132. doi: 10.1006/smim.1998.0114. [DOI] [PubMed] [Google Scholar]
- Klingel S, Morath I, Strietz J, Menzel K, Holstein TW, Gradl D. Subfunctionalization and neofunctionalization of vertebrate Lef/Tcf transcription factors. Dev Biol. 2012;368:44–53. doi: 10.1016/j.ydbio.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Staal FJ, Sen JM. The canonical Wnt signaling pathway plays an important role in lymphopoiesis and hematopoiesis. Eur J Immunol. 2008;38:1788–1794. doi: 10.1002/eji.200738118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staal FJ, Luis TC, Tiemessen MM. WNT signalling in the immune system: WNT is spreading its wings. Nat Rev Immunol. 2008;8:581–593. doi: 10.1038/nri2360. [DOI] [PubMed] [Google Scholar]
- Staal FJ, Clevers HC. WNT signalling and haematopoiesis: a WNT-WNT situation. Nat Rev Immunol. 2005;5:21–30. doi: 10.1038/nri1529. [DOI] [PubMed] [Google Scholar]
- Staal FJ, Clevers H. Tcf/Lef transcription factors during T-cell development: unique and overlapping functions. Hematol J. 2000;1:3–6. doi: 10.1038/sj.thj.6200001. [DOI] [PubMed] [Google Scholar]
- Sharma A, Berga-Bolanos R, Sen JM. T cell factor-1 controls the lifetime of CD4+ CD8+ thymocytes in vivo and distal T cell receptor alpha-chain rearrangement required for NKT cell development. PloS one. 2014;9:e115803. doi: 10.1371/journal.pone.0115803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinke FC, Yu S, Zhou X, He B, Yang W, Zhou B, Kawamoto H, Zhu J, Tan K, Xue HH. TCF-1 and LEF-1 act upstream of Th-POK to promote the CD4(+) T cell fate and interact with Runx3 to silence Cd4 in CD8(+) T cells. Nature immunology. 2014;15:646–656. doi: 10.1038/ni.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura RM, Sigvardsson M, Galceran J, Verbeek S, Clevers H, Grosschedl R. Redundant regulation of T cell differentiation and TCRalpha gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 1998;8:11–20. doi: 10.1016/s1074-7613(00)80454-9. [DOI] [PubMed] [Google Scholar]
- Yu S, Zhou X, Steinke FC, Liu C, Chen SC, Zagorodna O, Jing X, Yokota Y, Meyerholz DK, Mullighan CG, Knudson CM, Zhao DM, Xue HH. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity. 2012;37:813–826. doi: 10.1016/j.immuni.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Hawwari A, Li H, Sun Z, Mahanta SK, Littman DR, Krangel MS, He YW. Regulation of the TCRalpha repertoire by the survival window of CD4(+)CD8(+) thymocytes. Nature immunology. 2002;3:469–476. doi: 10.1038/ni791. [DOI] [PubMed] [Google Scholar]