Abstract
Alterations in glycosylation of serum glycoproteins can provide unique and highly specific fingerprints of malignancy. Our previous mass spectrometric study revealed that the bifucosylation level of serum haptoglobin was distinctly increased in hepatocellular carcinoma (HCC) patients versus liver cirrhosis of all three major etiologies. We have thus developed a method for the analysis of large numbers of serum samples based on a 96-well plate platform for the evaluation of fucosylation changes of serum haptoglobin between HCC versus cirrhosis. Haptoglobin was isolated from the serum of individual patient samples based on an HPLC column immobilized with antihaptoglobin antibody via hydrazide immobilization chemistry. Only 10 μL of serum was required for glycan extraction and processing for MALDI-QIT mass spectrometry analysis using the 96-well plate format. The bifucosylation degrees of haptoglobin in individuals were calculated using a quantitative glycomics method. The MS data confirmed that the bifucosylated tetra-anntenary glycan was upregulated in HCC samples of all etiologies. This study provides a parallel method for processing glycan content for haptoglobin and evaluating detailed changes in glycan structures for a potentially large cohort of clinical serum samples.
Keywords: haptoglobin, glycan, hepatocellular carcinoma, 96-well plate format, HPLC, MALDI
INTRODUCTION
Aberrant protein glycosylation is highly associated with cancer development and progression. Besides the alterations in protein expression, characterization of unique changes in glycan structure on a target glycoprotein biomarker holds great promise to improve diagnosis specificity and sensitivity. AFP-L3 (alpha-fetoprotein with core fucosylation), a serum biomarker used for the detection of hepatocellular carcinoma (HCC), has been reported to be highly specific for HCC compared with AFP in clinical practice.1,2 AFP-L3 has also demonstrated increased clinical sensitivity in patients with early stage HCC.3 Therefore, it is essential to detect and monitor unique changes in glycan structures on target proteins associated with cancer processes to provide a highly specific fingerprint of malignancy.
Various efforts have been devoted to identifying glycan-based biomarkers, which require the isolation of a target glycoprotein from complex biological samples (i.e., blood serum/plasma) and subsequent glycan processing for accurate identification and quantitation of individual glycan structures. Tousi and coworkers developed an automated multidimensional HPLC platform for immunoaffinity-based isolation of clusterin from human plasma followed by quantitative analysis of the protein N-glycome.4 The three-column multidimensional platform, consisting of the removal of albumin and IgG (depletion column), affinity capture of clusterin (immunoaffinity column), and desalting (RP Trap column), was applied to evaluate clusterin glycosylation in a small group of renal cell carcinoma (RCC) patients;4 however, because clusterin forms complexes with numerous plasma proteins, there was still a significant amount of plasma proteins present in the eluted clusterin from the HPLC platform, and a complete isolation of the clusterin using 2DE gel separation was required.4 Goldman and coworkers alternatively developed a hemoglobin affinity column by coupling hemoglobin to CNBr-activated sepharose to purify haptoglobin from human plasma for site-specific glycoform analysis;5 however, a reverse-phase HPLC was subsequently performed for the isolation of the haptoglobin fraction.
Alternatively, a simple, high-throughput approach that can provide both high-purity isolation of a target glycoprotein from serum and accurate quantitation of glycans from the enriched glycoprotein in a large cohort of patients is needed. Jeong and coworkers recently developed a high-throughput N-glycan quantitative method that includes N-glycan release, purification, and derivatization on a 96-well plate platform for reliable absolute quantification of N-glycans from model glycoproteins (i.e., chicken ovalbumin and porcine thyroglobulin) using MALDI-QIT-TOF analysis.6 This approach, though, was applied only to glycoprotein standards using a large amount of material (>50 μg)6 but was not demonstrated on patient serum samples.
Serum haptoglobin (Hp) has attracted particular attention due to its potential as a reporter molecule for aberrant glycosylation in liver disease.5,7,8 Previously, we developed a mass-spectrometry-based approach to analyze the etiology-related alterations in fucosylation degree of serum haptoglobin in individual patients with hepatocellular carcinoma (HCC) and liver cirrhosis.9 Haptoglobin was extracted from IgG-depleted serum by immunoprecipitation using an antibody coupled to protein A/G agarose beads. A unique pattern of bifucosylated tetra-antennary glycan, with both core and antennary fucosylation, was identified in HCC patients.9 Notably, it was found that the elevated bifucosylated haptoglobin can discriminate early stage HCC patients from cirrhosis (AUC = 0.834, p < 0.0001), which outperforms serum AFP.9 The result suggests that serum bifucosylated haptoglobin may serve as a potential marker for early detection and prediction of HCC in patients with cirrhosis.9
Herein, we have developed an automated HPLC column immobilized with an antihaptoglobin antibody via hydrazide immobilization chemistry for high-purity, single-step isolation of haptoglobin from serum samples in combination with an improved high-throughput 96-well platform for evaluation of fucosylation changes of serum haptoglobin between HCC versus cirrhosis. In this method, serum haptoglobin was extracted from 20 μL of crude serum using the antibody-immobilized HPLC column, while half of the enriched haptoglobin, equal to that from 10 μL of serum, underwent N-glycan release, extraction and permethylation performed on a 96-well platform. Consequently, glycans of serum haptoglobin were analyzed on a MALDI-QIT-TOF mass spectrometer and the bifucosylation degrees were calculated using a quantitative glycomics method and compared with those obtained from our previous study9 using an immunoprecipitation method. The method provides a parallel method for processing glycan content for serum haptoglobin in a large number of samples and for evaluating detailed changes in glycan structures for a cohort of clinical serum samples. This work provides a strategy to identify glycan changes in target serum glycoproteins for biomarker discovery or validation.
EXPERIMENTAL METHODS
Materials
N-Glycosidase F (PNGase F) was purchased from New England Biolabs (Ipswich, MA). Neuraminidase, sodium hydroxide, methyl iodide, β-mercaptoethanol, chloroform, dimethyl sulfoxide (DMSO), HPLC-grade acetonitrile (ACN), and water and the 96-well vacuum manifold were purchased from Sigma-Aldrich (St. Louis, MO). The MALDI matrix, 2,5-dihydroxybenzoic acid (2,5-DHB), the UltraLink hydrazide resin, the Zeba Spin Desalting column, and the SPE 96-well plate packed with 100% porous graphitic carbon (PGC) were purchased from Thermo Scientific (Rockford, IL). The 96-well SpinColumn plate was purchased from Harvard Apparatus (Holliston, MA). Antihuman haptoglobin antibody (CatLog No. ab13429) and a human haptoglobin standard were purchased from Abcam (Cambridge, MA). The empty PEEK column was purchased from MicroSolv Technology Corp. (Eatontown, NJ).
Serum Samples
Serum samples from patients with hepatocellular carcinoma (HCC) and liver cirrhosis were provided by the University Hospital, Ann Arbor, Michigan according to IRB approval, which includes 15 HCC cases (5 HBV-, 5 HCV-, and 5 ALC-related, respectively) and 15 cirrhosis cases (5 HBV-, 5 HCV-, and 5 ALC-related, respectively). The clinical features of patients with HCC and cirrhosis are summarized in Table 1. Samples were aliquoted and stored at −80 °C until further use.
Table 1.
Summary of Patient Characteristics
| disease diagnosisa | HCC | cirrhosis |
|---|---|---|
| number | 15 | 15 |
| etiology (HBV/HCV/ALC)b | 5/5/5 | 5/5/5 |
| gender % (M/F) | 56/44 | 65/35 |
| age (mean ± SD) | 56.8 ± 12 | 56.3 ± 9 |
| AFP level (median), ng/mLc | 97.40 | 3.92 |
| MELD scored | 7.9 ± 2.8 | 8.9 ± 2.8 |
| TNM stage % (I/II/III/IV) | 40/47/13/0 | NA |
| BCLC stage % (A/B/C/D) | 40/40/20/0 | NA |
Samples were provided by Division of Gastroenterology, University of Michigan.
HBV: hepatitis B virus; HCV: hepatitis C virus; ALC: alcohol consumption.
AFP level was provided by Division of Gastroenterology, University of Michigan.
MELD: Model for end stage liver disease.
Anti-Haptoglobin Immobilized HPLC Column
We have developed an HPLC-based hydrazide column that was covalently immobilized with antihaptoglobin antibody via hydrazide chemistry to purify haptoglobin from patient serum. The immobilization of antihaptoglobin antibody to the UltraLink hydrazide resin was performed according to the manufacturer’s instruction (Thermo Scientific, Rockford, IL). In brief, 500 μg of antihaptoglobin antibody was diluted with 1 mL of coupling buffer (0.1 M sodium phosphate buffer, pH 7.0) and then incubated with sodium meta-periodate (at a final concentration of 10 mM) at room temperature for 30 min. The oxidized antibody was then desalted using the Zeba Spin Desalting Column (Thermo Scientific, Rockford, IL), followed by coupling to the UltraLink hydrazide resin. One milliliter of hydrazide resin (average size 50–80 μm) was packed on a gravity-flow column and then was washed and equilibrated with 5 resin-bed volumes of coupling buffer. Subsequently, the oxidized antibody was added to the resin and incubated at room temperature for 6 h with gentle end-over-end mixing. After incubation, the antibody-coupled resin was washed with 1 mL of coupling buffer, 5 resin-bed volumes of 1 M NaCl, and 5 washes of PBS containing 0.05% sodium azide, respectively. The antihaptoglobin immobilized resin was subsequently packed into a PEEK column (4.6 mm × 50 mm).
HPLC Immunoaffinity Enrichment of Haptoglobin
Haptoglobin was purified from 20 μL of serum using the antibody-immobilized HPLC column on a Beckman Coulter ProteomeLab PPS system (Fullerton, CA) with a flow rate of 0.5 mL/min. The serum sample was diluted with 200 μL of 1× dilution buffer (10 mM TBS, 150 mM NaCl, pH 7.4) and filtered through a 0.45 μm-pore-size Costar Spin-X centrifuge tube filter (Corning) at 10 000g for 1 min to remove any particulates prior to loading onto the HPLC column. The immunoaffinity enrichment of haptoglobin was achieved within 40 min. A representative chromatogram is shown in Supplemental Figure S2a. The bound materials were eluted with five volumes of stripping buffer (0.1 M Glycine, pH 2.5). Then the column was neutralized with 3 volumes of neutralization buffer (0.1 M Tris-HCl, pH 8.0) and equilibrated with 1× dilution buffer. The eluted fraction between 19 and 26 min (~3.5 mL) was collected and immediately neutralized with the neutralization buffer. The fraction was desalted using a 4 mL YM-3 centrifugal device (Millipore, Billerica, MA) by buffer exchange three times with deionized water. The desalted eluent was aliquoted into two tubes and then dried down using a SpeedVac concentrator (Labconco). Only half of the enriched haptoglobin was applied for the following N-glycan extraction and processing.
The purity of the eluted haptoglobin was further evaluated by SDS-PAGE, followed by silver staining using ProteoSilver Plus Silver Stain Kit (Sigma) following the manufacturer’s instruction. 1/10 of haptoglobin eluent was checked on a 4–20% SDS-PAGE gel (Bio-Rad, Hercules, CA). In comparison, 1/400 volume of the unbound fractions and 0.1 μL of crude serum were also loaded on the gel. The result of gel analysis is shown in Supplemental Figure S2b.
The binding efficiency of the antibody-immobilized HPLC column was compared with traditional immunoprecipitation (IP) using the Cross-link IP kit (Pierce Scientific, Rockford, IL) which enables antigen immunoprecipitation by covalently cross-linking antibody onto protein A/G resin. Twenty μL of serum was applied for immunoprecipitation according to the procedure previously described.9 One-tenth of the haptoglobin eluted from the HPLC column and immunoprecipitation, respectively, was run on a 4–20% SDS-PAGE gel (Bio-Rad, Hercules, CA) and visualized by silver staining. The result of the comparison is shown in Supplemental Figure S3.
Deglycosylation and Desialylation of Haptoglobin
Half of the enriched haptoglobin, which is equal to that from 10 μL of serum, was applied for the following N-glycan processing. Haptoglobin denaturation, deglycosylation, and desialylation were performed successively in a single 96-well plate, which can greatly reduce the sample loss during multiple procedures. Haptoglobin purified from patient serum with HCC and liver cirrhosis was dissolved in 8 μL of deionized water and then denatured on a 96-well plate (Axygen, Union City, CA) by adding 2 μL of denaturing solution (0.1% SDS, 50 mM 2-mercaptoethanol) and incubated at 65 °C in a GC oven (PerkinElmer) for 30 min. After cooling at room temperature, ammonium bicarbonate solution was then added to make a final concentration of 15 mM. Two units of N-glycosidase F (PNGase F) were added and incubated with the sample at 37 °C for 16 h. The action of PNGase F was quenched by heating the reaction mixture to 95 °C in a GC oven for 10 min. Subsequently, the mixture was dried and reconstituted in 20 mM ammonium acetate, followed by desialylation with neuraminidase (40 mU) (Sigma-Aldrich, St. Louis, MO) at 37 °C for 18 h. The mixture of desialylated N-glycans and the protein was dried in a SpeedVac concentrator (Labconco).
N-Glycan Extraction
The desialylated N-glycans were extracted using a PGC containing 96-well plate by centrifugation to remove the salts and other contaminants (Thermo Scientific, Rockford, IL). In brief, the PGC-containing well was washed with 200 μL of 50% acetonitrile (ACN)/water 3 times and then was equilibrated with 200 μL of 0.1% TFA in water. The N-glycan and protein mixture, which was constituted in 100 μL of water with 0.1% TFA, was loaded onto the well and then recycled through the PGC column for at least 5 times. After three washes with 200 μL of water with 0.1% TFA, the N-glycans were eluted into the 96-well collection plate with 100 μL of 10% acetonitrile (with 0.1% TFA), followed by 100 μL of 25% acetonitrile (with 0.1% TFA). All centrifugation steps were performed at 1300 rpm for 1 min in a Thermo Scientific centrifuge with a 96-well plate rotator. The collected glycan eluent was then dried in a SpeedVac concentrator (Labconco).
Solid-Phase Permethylation
Subsequently the N-glycans were permethylated using a solid-phase permethylation method10 on a 96-well SpinColumn plate6 with 5 μm frit (Harvard Apparatus, Holliston, MA). In brief, NaOH beads (20–40 mesh) that were prepared in ACN were packed into the 96-well SpinColumn plate to ~1 cm depth.10 The well was then washed three times with 200 μL of DMSO and kept in DMSO prior to applying the samples. The sample was suspended in 70.8 μL of DMSO, 26.4 μL of iodomethane (CH3I), and 2.8 μL of water, where the ratios were optimized according to the procedure of Kang.10 After discarding the DMSO from the NaOH packed 96-well SpinColumns, the sample was applied through the NaOH beads and recycled for at least 8 times to minimize the sample loss. Subsequently, the permethylated sample was eluted into a 96-well collection plate by adding 100 μL of ACN. The eluent was collected by suction using a 96-well vacuum manifold (Sigma) and then transferred to an eppendorf tube. The permethylated glycans were extracted with 200 μL of chloroform and washed with 200 μL of water for four times. Finally, the permethylated N-glycans were dried under a SpeedVac concentrator (Labconco).
MALDI-QIT-TOF Analysis
The permethylated N-glycans were redissolved in 2 μL of 20% acetonitrile for mass spectrometric analysis using an Axima MALDI quadrupole ion trap-TOF instrument (Shimadzu Biotech, Manchester, U.K.).9 Ionization was performed with a pulsed N2 laser (337 nm) at 5 Hz. The glycan sample (0.5 μL) was manually spotted on the MALDI plate and allowed to air-dry. Then, DHB matrix (0.5 μL, 10 mg/mL DHB in 50% ACN with 1 mM sodium acetate) was deposited on the dried sample layer. The TOF detector was calibrated with 1 nmol/μL peptide mixtures of Angiotensin II (m/z 1046.54), Angiotensin I (m/z 1296.68), Substance P (m/z 1347.74), Bombesin (m/z 1619.82), ACTH 1–17 (m/z 2093.09), ACTH 18–39 (m/z 2465.20), and Somatostatin 28 (m/z 3147.47) prior to data acquisition. The mass accuracy with calibration was 30 ppm. All glycans were sodiated and analyzed in positive ion mode, and a total 500 laser shots were acquired for each sample. Glycomod tool (http://www.expasy.org/tools/glycomod) was utilized to predict the glycan compositions, which were further confirmed by collision-induced dissociation (CID) MS/MS analysis on the MALDI-QIT-TOF mass spectrometer. Additional settings for the MS analysis included: (i) mass range 500–5000; (ii) mass window for precursor ion isolation 2 Da; (iii) one precursor was selected and stored in the quadrupole ion trap for fragmentation when performing MS/MS analysis; and (iv) argon was used as the collision gas with a collision energy manually adjusted between 120 and 200 V (low-energy CID) to achieve an optimum degree of fragmentation.
Data Analysis
The MALDI MS data were acquired and processed in Launchpad software (Karatos, Manchester, U.K.). The glycan masses were searched in Glycomod using the following parameters: (1) mass values are monoisotopic; (2) mass tolerance 0.2 Da; (3) positive ion mode; (4) ion adducts Na+; (5) N-linked Free/PNGase released oligosaccharides; and (6) monosaccharide residues permethylated. The m/z values and intensities were exported as ASCII files, where the first column corresponds to the m/z and the second column corresponds to the intensity. Glycan peak area integration was performed with Matlab (Natick, MA) using the script described in our previous study.11 The abundance of each glycan was normalized by the sum of all glycan abundances identified in each sample. The bifucosylation degree of serum haptoglobin was calculated using the equation previously described,9 which is the ratio of the sum of abundances of bifucosylated glycans to all glycan abundances. For data visualization, a scatter plot of the calculated bifucosylation degree was generated with GraphPad Prism 6 (La Jolla, CA). The p value was generated by Student’s t test between HCC and cirrhosis groups, reflecting the statistical significance of the difference in bifucosylation level of serum haptoglobin.
RESULTS AND DISCUSSION
Strategy for N-Glycan Analysis
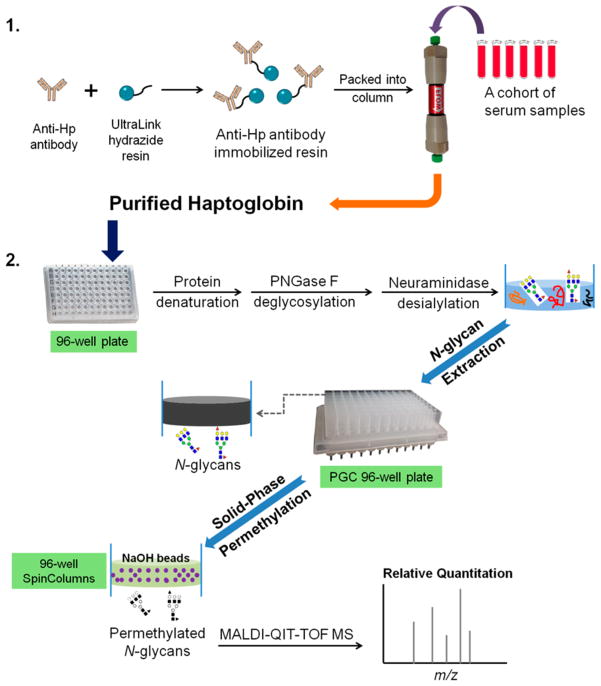
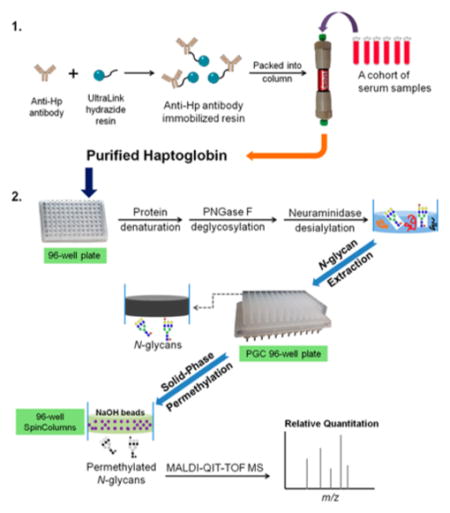
In this work, a strategy was developed to monitor fucosylation alterations of serum haptoglobin by integrating an antibody-immobilized HPLC column, a 96-well plate platform, and MALDI-QIT-MS glycan structural analysis. The schematic of the method used for N-glycan analysis for serum haptoglobin is shown in Figure 1. In this scheme, serum haptoglobin was extracted from serum using an automated HPLC column immobilized with antihuman haptoglobin antibody via hydrazide chemistry, followed by N-glycan release, desialylation, extraction, and permethylation performed on a 96-well plate platform. Notably, protein denaturation, deglycosylation, and desialylation were performed sequentially in a single plate, which can greatly reduce the sample loss during the multiple procedures. The removal of sialic acids can eliminate the complicated heterogeneity of sialic acids in N-glycans and then improve the peak sensitivity so that we can focus on the bifucosylated haptoglobin, which was found as a potential marker in HCC patients in our previous study.9 The released N-glycans were extracted by a PGC containing 96-well plate to remove the salts and other contaminants. Subsequently, the N-glycans were permethylated using a solid-phase permethylation method on a microspin column 96-well plate filled with NaOH beads. These two steps, glycan extraction and permethylation, were performed within 2 h, and up to 96 samples can be handled at the same time. The permethylated glycans were analyzed on a MALDI-QIT-TOF mass spectrometer. MS data were integrated and the bifucosylation degree of haptoglobin in each serum sample was calculated by using a quantitative glycomics method described in our previous study and compared with previous results.9
Figure 1.
Workflow scheme of the purification of haptoglobin from serum samples using an antibody-immobilized HPLC column followed by N-glycan analysis on a 96-well plate format. Photograph courtesy of Jianhui Zhu. Copyright 2015.
Anti-Hp Antibody HPLC Column
An HPLC-based hydrazide column, which was covalently immobilized with a monoclonal antihaptoglobin antibody (Abcam, Cambridge, MA), was developed for the purification of haptoglobin from patient serum. The UltraLink hydrazide resin (Thermo Scientific, Rockford, IL) was used for the immobilization of the antihaptoglobin antibody where the glycol groups of the sugar moieties on the Fc portion can be oxidized by sodium meta-periodate to generate aldehydes that can react with hydrazide groups on the resin to form stable hydrazone bonds. The antibody is coupled to the resin through the Fc portion only, leaving its antigen-binding sites unobstructed, which offers optimal purification capability. Any antibodies (glycosylated proteins) that contain carbohydrate groups can be immobilized on the hydrazide-activated resin. The binding efficiency of the antibody-immobilized HPLC column was measured by a human haptoglobin standard with different loading amounts, that is, 10, 20, 30, and 50 μg, respectively. The chromatogram is shown in Supplemental Figure S1. The efficiency of the anti-Hp HPLC column was calculated by the ratio of peak area of the bound fraction to the sum of peak areas of bound and unbound fractions. As listed in Table S1, the column efficiency for the enrichment of haptoglobin is 66.0, 61.1, 62.5, and 64.1%, respectively, when loading 10, 20, 30, and 50 μg haptoglobin.
Purification of Serum Haptoglobin
Serum haptoglobin was purified from 20 μL of patient serum using the anti-Hp HPLC column, and the immunoaffinity enrichment was achieved within 40 min. A total of 15 HCC cases (5 HBV-, 5 HCV-, and 5 ALC-related, respectively) and 15 cirrhosis cases (5 HBV-, 5 HCV-, and 5 ALC-related, respectively) were investigated in this study. A representative chromatogram and 1D gel analysis of all fractions are shown in Supplemental Figure S2, where the fraction C represents the eluted haptoglobin.
We compared the enriched Hp from an HCC serum sample with a human Hp standard by running the two samples on the same gel followed by silver staining (Figure 2). The gel analysis showed that only haptoglobin was observed in the eluted fraction, which included Hp β chain (~42 kDa), α-2 chain (~18 kDa), and α-1 chain (~13 kDa). No contamination from other proteins was observed in the eluted fraction. The bands corresponding to Hp β chain migrated in the same position (~42 kDa) in the HPLC eluent and the Hp standard. The bands corresponding to α-2 chain were also in the same position (~18 kDa), while the α-1 chain varied in these two samples. This is consistent with Hp being characterized by a genetic polymorphism that arises from differences in α chains, while β chains are identical in all Hp types.12 The four potential N-glycosylation sites of Hp are located on its β chain.12 The gel analysis demonstrated that the antibody-immobilized HPLC column resulted in a high-purity enrichment of haptoglobin (100%). The total yield of Hp β chain was estimated to be around 4–6 μg per 20 μL of serum, which recovers 40–50% of the Hp from serum samples. The binding efficiency of the antibody-immobilized column was further compared with the traditional immunoprecipitation (IP), which showed that the yield of haptoglobin using the HPLC antibody column was more than twice that from immunoprecipitation (Supplemental Figure S3).
Figure 2.
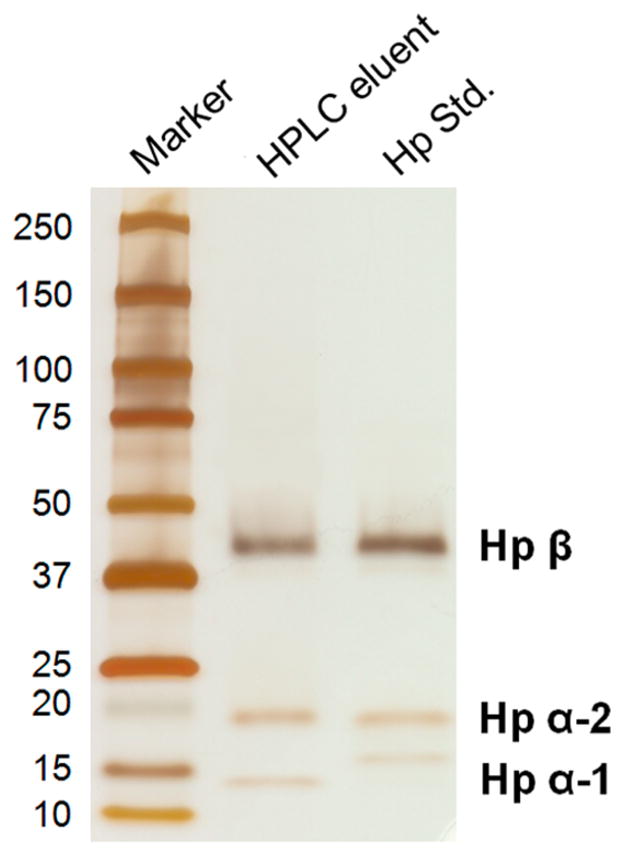
SDS-PAGE analysis of the eluted haptoglobin from an HCC serum sample using the antibody-immobilized HPLC column, with a comparison of a human haptoglobin standard. One-tenth of the eluent and 0.5 μg of haptoglobin standard were subjected to 4–20% SDS-PAGE, followed by silver staining. Only haptoglobin was observed in the eluted fraction, including Hp β chain, α-2 chain, and α-1 chain. No contamination from other proteins was observed in the eluted haptoglobin. The bands corresponding to Hp β and α-2 chains migrated in the same position in the eluted Hp and the Hp standard, while the α-1 chain varied in these two samples. This is consistent with the fact that Hp is a genetic polymorphism with differences in α chains, while β chains are identical in all Hp types.
Immunoprecipitation (IP) and immunoaffinity chromatography (IAC) approaches are generally used for the enrichment of target proteins. The purity of the enriched glycoprotein is essential for the subsequent glycan structural analysis. Even trace quantities of coextracted glycoprotein impurities will interfere with the quantitative oligosaccharide profiling of haptoglobin. In some HPLC methods4 where the column was packed with anticlusterin ligand immobilized on agarose-based media, an extra step of depletion of the two most abundant proteins (i.e., albumin and IgG) is required prior to HPLC enrichment to reduce the complexity of serum and decrease the chance of nonspecific binding. The issue also exists in the immunoprecipitation method, where protein A/G beads are used to immobilize the antibody, and it is therefore required to remove IgG before incubation with the serum sample. Notably, the HPLC-based column we developed here was covalently immobilized with a monoclonal antihaptoglobin antibody via hydrazide immobilization chemistry, resulting in high-purity enrichment of haptoglobin from crude serum. This single-step HPLC enrichment method does not require an IgG depletion step.
This automated platform enables affinity purification of serum haptoglobin in a single step with minimal sample handling in a short amount of time (40 min), while the traditional immunoprecipitation usually takes 2 days to complete. The column can be reused for more than 100 times without significant loss in binding capacity. This platform is versatile and can be adapted to immunoenrichment of any target protein from human serum or plasma, and the single-step HPLC immunoaffinity platform is useful for handling large numbers of samples required for biomarker discovery/ validation.
Glycan Extraction and Analysis
Half of the enriched haptoglobin, which is equal to that from 10 μL of serum, was applied for the N-glycan extraction and analysis. The glycan processing for the enriched serum haptoglobin, including protein denaturation, N-glycan release, desialylation, extraction, and permethylation, was performed on a 96-well plate format. Notably, protein denaturation, deglycosylation, and desialylation were performed sequentially in a single 96-well plate, which can greatly reduce the sample loss during the multiple procedures. The protein denaturation was performed at 65 °C for 30 min, and deglycosylation and desialylation were performed at 37 °C overnight, respectively. The released N-glycans were extracted by a PGC containing 96-well plate to remove the salts and other contaminants. Subsequently the N-glycans were permethylated using a solid-phase permethylation method6 on a 96-well SpinColumn plate filled with NaOH beads. These two steps, glycan extraction and permethylation, were carried out within 2 h, respectively. All of the procedures from glycan release to MALDI MS analysis were carried out within 2 days, demonstrating the capability for simultaneous analysis of a number of samples, which will be essential for marker discovery and validation.
In Jeong’s N-glycan preparation method using a stack of four 96-well plates (i.e., sample plate, PVDF, PGC, and 96-well SpinColumn plates),6 an extra PVDF plate was used to collect the released N-glycans from the PVDF membrane plate. Jeong’s method can be used for N-glycan analysis of standard glycoproteins with an amount of >50 μg. While handling limited materials enriched from serum samples, it is important to minimize sample loss during the multiple steps. Compared with Jeong’s method, our 96-well plate format contains three 96-well plates (i.e., sample plate, PGC, and NaOH plates), where protein denaturation, glycan release, and desialylation were performed sequentially in a single 96-well sample plate, which can greatly reduce the sample loss during the multiple procedures.
MALDI Analysis
The permethylated N-glycans were deposited onto a MALDI plate for mass spectrometric analysis using an Axima MALDI quadrupole ion trap-TOF instrument (Shimadzu Biotech). The N-glycan MS data were acquired and processed in Launchpad software where the Glycomod tool is utilized to predict the glycan composition. In total, eight glycan structures were identified as listed in the Supplemental Table S2. The glycan compositions were further confirmed by MS/MS analysis. CID MS/MS spectra of the bifucosylated glycans of haptoglobin are shown in the Supporting Information and Supplemental Figure S4. Glycan peak area integration was performed using the Matlab for quantification as previously described.9 The abundance of each glycan was normalized by the sum of all glycan abundances identified in each sample. The fucosylation/ bifucosylation degree is calculated by the ratio of the sum of abundances of fucosylated/bifucosylated glycans to all glycans according to our previous work.9 In this work, we focused on bifucosylation of haptoglobin because our previous work uncovered that the bifucosylation level may serve as a promising predictor for early detection of HCC in patients with cirrhosis.9
Validation of Reproducibility of the Method
The performance of glycan processing on the 96-well platform was optimized using a human haptoglobin standard. A typical desialylated N-glycan profile of human haptoglobin standard is shown in Supplemental Figure S5, including nonfucosylated bi-, tri-, and tetra-antennary glycans (m/z 2070.07, 2519.28, and 2968.49, respectively) and monofucosylated bi-, tri-, and tetra-antennary glycans (m/z 2244.13, 2693.40, and 3142.69, respectively). To measure the detection range of this 96-well format, we performed a test on a human haptoglobin standard (Abcam, Cambridge, MA) in sequential aliquots of 0.5, 1, 2, 5, 10, and 20 μg. The limit of detection (LOD) was found to be 1 μg. As shown in the Supplemental Figure S5, no significant differences in glycan distribution were observed in the five spectra from 1 to 20 μg. Although the total intensity of the spectrum increased with the sample amount, the fucosylation degree is consistent for the five sequential aliquots, and the relative standard deviation (RSD) is 10.2%.
We also investigated the reproducibility of the 96-well format by processing and analysis of N-glycans of haptoglobin enriched from an ALC-related HCC serum sample. The enriched haptoglobin from 40 μL of serum was separated into four aliquots and applied onto the 96-well plate format. The glycan spectra from these four replicates are presented in Supplemental Figure S6, which shows that all eight glycans of haptoglobin in HCC patient were observed, including bifucosylated tri- (m/z 2867.48) and tetra-antennary (m/z 3316.69) glycans. The RSD for the bifucosylation degree is 9.6% for the four replicates. The result demonstrated the consistency of evaluation of bifucosylation degree in serum haptoglobin for a given patient.
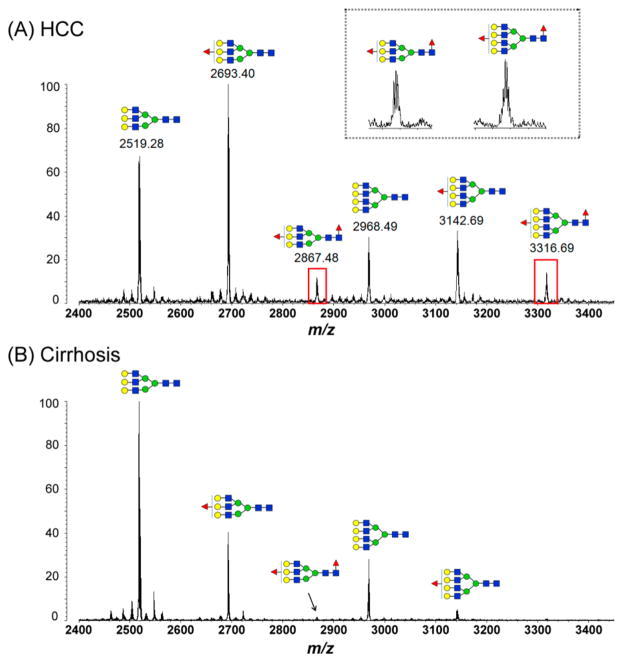
We then applied this platform for processing of N-glycans from haptoglobin, which was from 10 μL of serum, for all 30 samples. All samples were analyzed in the same manner on the 96-well plate to reduce inconsistency. The representative spectra are shown in Figure 3, which represents a comparison of tri- and tetra-antennary structures of serum Hp in HCC and cirrhosis. The enlarged insert in Figure 3 shows two peaks of the bifucosylated tri- (m/z 2867.48) and tetra-antennary (m/z 3316.69) N-glycans that are highly associated with HCC. The elevated intensity of the peak (m/z 3316.69) corresponding to bifucosylated tetra-antennary glycan was observed in all HCCs compared with liver cirrhosis in this sample set.
Figure 3.
MALDI-QIT-TOF MS spectra show the difference of fucosylation in tri- and tetra-antennary N-glycans of haptoglobin between HCC (A) and cirrhosis (B). The elevated presence of bifucosylated tri- and tetra-antennary glycans in HCC samples compared with cirrhosis is highlighted with a red rectangle. The enlarged inset shows two peaks of the bifucosylated tri- (m/z 2867.48) and tetra-antennary (m/z 3316.69) N-glycans, which are highly associated with HCC.
Relative Quantitation of Bifucosylation Degree
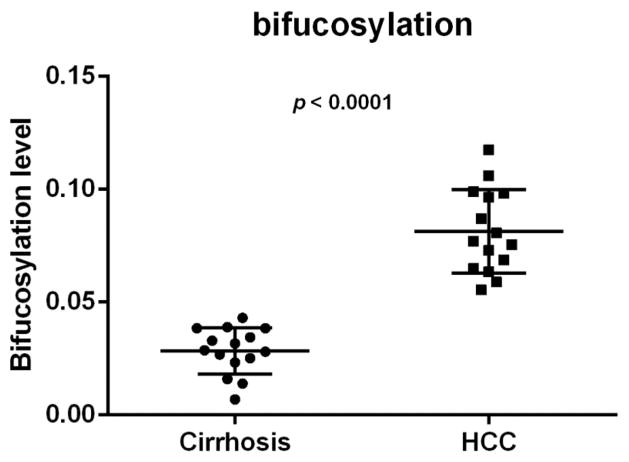
The bifucosylation degrees of haptoglobin enriched from individual serum samples are listed in Supplemental Table S3, within a range between 0.007 and 0.12 for all cirrhosis and HCC samples. The cirrhosis groups have low bifucosylation degrees 0.028 ± 0.010 (mean ± SD); however, bifucosylation degrees are highly elevated in HCCs of all the three major etiologies, with a mean of 0.081. Figure 4 shows a dot plot of bifucosylation degrees of serum haptoglobin in patients with HCC and cirrhosis, respectively. As shown in Figure 4, all HCC patients investigated in this study have a distinctly increased bifucosylation degree of haptoglobin as compared with cirrhosis patients (p < 0.0001). The result was consistent with those reported in our previous study,9 which confirmed that the bifucosylated haptoglobin may serve as a potential marker for HCC.
Figure 4.
Scatter plot of bifucosylation level of haptoglobin N-glycans in cirrhosis and HCC patients, which shows significantly elevated bifucosylation level in all HCCs compared with cirrhosis patients in this sample set (p < 0.0001). Each spot represents an individual patient.
The core-fucosylation degree was also evaluated by the ratio of the sum of abundances of core-fucosylated glycans to all glycans (Supplemental Table S3), which showed slightly elevated core-fucosylation level between HCCs and cirrhosis samples in this sample set (p < 0.05).
CONCLUSIONS
In this study, we have developed a strategy to monitor fucosylation alterations of serum haptoglobin between HCC and liver cirrhosis patients by integrating an antibody-immobilized HPLC column, a 96-well plate platform, and MALDI-QIT-MS glycan structural analysis. The HPLC column immobilized with antihaptoglobin antibody via hydrazide chemistry enables high-purity, single-step enrichment of haptoglobin from patient serum within 40 min, without an extra step of serum IgG depletion required. This single-step HPLC immunoaffinity platform can be adapted to immunoenrichment of any target protein from human serum/plasma and is useful for handling large numbers of samples required for biomarker discovery. The bifucosylated tetra-antennary N-glycan derived from serum haptoglobin is a minor peak, where 10 μL of serum was required in this assay for accurate detection of this signal as a potential biomarker. The MS result confirmed that the bifucosylated haptoglobin may serve as a potential marker for HCC. Although lectin blots8 or lectin-ELISAs7,13 can be used to detect the core-fucosylation level, these tools cannot detect differences between mono- and bifucosylated structures. The assay developed here combined a 96-well plate format for N-glycan processing and MALDI-QIT MS for N-glycan relative quantitation, which is highly specific and able to detect both alterations in glycan expression and detailed changes in glycan structure. This method may be useful for not only processing glycans from the target glycoprotein but also detecting detailed changes in glycan structures for a potentially large cohort of clinical serum samples.
Supplementary Material
Acknowledgments
We acknowledge the support of this work from the National Cancer Institute under grants 1R01 CA160254 (D.M.L.) and 1R01 CA154455 (D.M.L.) and from the National Institutes of Health through grant R01 GM 49500 (D.M.L.). We thank Dr. Hee-Jin Jeong (Tokyo Institute of Technology) for her valuable advice on the permethylation step on the 96-well plate format.
Footnotes
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.5b00662.
Supplemental Figure S1. Chromatogram shows the binding efficiency of the antibody-immobilized HPLC column measured by a human haptoglobin standard with different loading amounts, that is, 10, 20, 30, and 50 μg, respectively. Supplemental Figure S2. Representative chromatogram of haptoglobin enrichment from patient serum using an antibody-immobilized HPLC platform. 1D gel analysis of unbound fractions A and B and the eluted fraction C after HPLC enrichment. Supplemental Figure S3. Comparison of the yield of haptoglobin enriched from patient serum using the antibody-immobilized HPLC platform versus the conventional immunoprecipitation method. Supplemental Figure S4. Representative MALDI-QIT-TOF MS/MS spectra of glycans at (A) m/z 2244.13, (B) m/z 2693.40, and (C) m/z 2867.48 for glycan composition and core/antennary fucosylation assignment. Supplemental Figure S5. MALDI-QIT-TOF MS spectra of N-glycans from a human haptoglobin standard processed on the 96-well plate format in sequential aliquots of 1, 2, 5, 10, and 20 μg, respectively. Supplemental Figure S6. MALDI-QIT-TOF MS analysis of four replicates of haptoglobin derived from 10 μL of serum of an ALC-related HCC patient followed by N-glycan processing on the 96-well plate format. Supplemental Table S1. The binding efficiency of the anti-Hp HPLC column measured by a human haptoglobin standard, which was calculated by the ratio of peak area of the bound fraction to the sum of peak areas of bound and unbound fractions. Supplemental Table S2. Eight N-glycans identified in serum haptoglobin after desialylation and permethylation. Supplemental Table S3. The bifucosylation and core-fucosylation degrees of haptoglobin enriched from individual serum samples including 15 HCC and 15 cirrhosis cases. (PDF)
References
- 1.Oka H, Saito A, Ito K, Kumada T, Satomura S, Kasugai H, Osaki Y, Seki T, Kudo M, Tanaka M. Multicenter prospective analysis of newly diagnosed hepatocellular carcinoma with respect to the percentage of Lens culinaris agglutinin-reactive alpha-fetoprotein. J Gastroenterol Hepatol. 2001;16:1378–1383. doi: 10.1046/j.1440-1746.2001.02643.x. [DOI] [PubMed] [Google Scholar]
- 2.Kumada T, Toyoda H, Tada T, Kiriyama S, Tanikawa M, Hisanaga Y, Kanamori A, Tanaka J, Kagebayashi C, Satomura S. High-sensitivity Lens culinaris agglutinin-reactive alpha-fetoprotein assay predicts early detection of hepatocellular carcinoma. J Gastroenterol. 2014;49:555–563. doi: 10.1007/s00535-013-0883-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oda K, Ido A, Tamai T, Matsushita M, Kumagai K, Mawatari S, Saishoji A, Kure T, Ohno K, Toyokura E, Imanaka D, Moriuchi A, Uto H, Oketani M, Hashiguchi T, Tsubouchi H. Highly sensitive lens culinaris agglutinin-reactive alpha-fetoprotein is useful for early detection of hepatocellular carcinoma in patients with chronic liver disease. Oncol Rep. 2011;26:1227–1233. doi: 10.3892/or.2011.1425. [DOI] [PubMed] [Google Scholar]
- 4.Tousi F, Bones J, Iliopoulos O, Hancock WS, Hincapie M. Multidimensional liquid chromatography platform for profiling alterations of clusterin N-glycosylation in the plasma of patients with renal cell carcinoma. J Chromatogr A. 2012;1256:121–128. doi: 10.1016/j.chroma.2012.07.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pompach P, Brnakova Z, Sanda M, Wu J, Edwards N, Goldman R. Site-specific glycoforms of haptoglobin in liver cirrhosis and hepatocellular carcinoma. Mol Cell Proteomics. 2013;12:1281–1293. doi: 10.1074/mcp.M112.023259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeong HJ, Kim YG, Yang YH, Kim BG. High-throughput quantitative analysis of total N-glycans by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Chem. 2012;84:3453–3460. doi: 10.1021/ac203440c. [DOI] [PubMed] [Google Scholar]
- 7.Ang IL, Poon TC, Lai PB, Chan AT, Ngai SM, Hui AY, Johnson PJ, Sung JJ. Study of serum haptoglobin and its glycoforms in the diagnosis of hepatocellular carcinoma: a glycoproteomic approach. J Proteome Res. 2006;5:2691–2700. doi: 10.1021/pr060109r. [DOI] [PubMed] [Google Scholar]
- 8.Zhang S, Shu H, Luo K, Kang X, Zhang Y, Lu H, Liu Y. N-linked glycan changes of serum haptoglobin beta chain in liver disease patients. Mol BioSyst. 2011;7:1621–1628. doi: 10.1039/c1mb05020f. [DOI] [PubMed] [Google Scholar]
- 9.Zhu J, Lin Z, Wu J, Yin H, Dai J, Feng Z, Marrero J, Lubman DM. Analysis of serum haptoglobin fucosylation in hepatocellular carcinoma and liver cirrhosis of different etiologies. J Proteome Res. 2014;13:2986–2997. doi: 10.1021/pr500128t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang P, Mechref Y, Novotny MV. High-throughput solid-phase permethylation of glycans prior to mass spectrometry. Rapid Commun Mass Spectrom. 2008;22:721–734. doi: 10.1002/rcm.3395. [DOI] [PubMed] [Google Scholar]
- 11.Lin Z, Lubman DM. Permethylated N-glycan analysis with mass spectrometry. Methods Mol Biol. 2013;1007:289–300. doi: 10.1007/978-1-62703-392-3_12. [DOI] [PubMed] [Google Scholar]
- 12.Kurosky A, Barnett DR, Lee TH, Touchstone B, Hay RE, Arnott MS, Bowman BH, Fitch WM. Covalent structure of human haptoglobin: a serine protease homolog. Proc Natl Acad Sci U S A. 1980;77:3388–3392. doi: 10.1073/pnas.77.6.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu J, Zhu J, Yin H, Buckanovich RJ, Lubman DM. Analysis of glycan variation on glycoproteins from serum by the reverse lectin-based ELISA assay. J Proteome Res. 2014;13:2197–2204. doi: 10.1021/pr401061c. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.