Abstract
Rationale
Impaired degradation of misfolded proteins is associated with a large subset of heart diseases. Misfolded proteins are degraded primarily by the ubiquitin-proteasome system (UPS) but the ubiquitin ligases responsible for the degradation remain largely unidentified. The cullin deneddylation activity of the COP9 signalosome (CSN) requires all 8 CSN subunits (CSN1 through CSN8) and regulates cullin-RING ligases (CRLs), thereby controlling ubiquitination of a large number of proteins; however, neither CSN nor CRLs are known to regulate the degradation of cytosolic misfolded proteins.
Objective
We sought to investigate the role of CSN8/CSN in misfolded protein degradation and cardiac proteinopathy.
Methods and Results
Cardiac CSN8 knockout causes mouse premature death; hence, CSN8 haploinsufficiency (CSN8hypo) mice were used. Myocardial neddylated forms of cullins were markedly increased and myocardial capacity of degrading a surrogate misfolded protein was significantly reduced by CSN8hypo. When introduced into proteinopathic mice in which a bona fide misfolded protein CryABR120G is overexpressed in the heart, CSN8hypo aggravated CryABR120G-induced restrictive cardiomyopathy and shortened the lifespan of CryABR120G mice, which was associated with augmented accumulation of protein aggregates, increased neddylated proteins, and reduced levels of total ubiquitinated proteins and LC3-II in the heart. In cultured cardiomyocytes, both CSN8 knockdown and CRL inactivation suppressed the ubiquitination and degradation of CryABR120G but not native CryAB, resulting in accumulation of protein aggregates and exacerbation of CryABR120G cytotoxicity.
Conclusions
(1) CSN8/CSN promotes the ubiquitination and degradation of misfolded proteins and protects against cardiac proteotoxicity and (2) CRLs participate in degradation of cytosolic misfolded proteins.
Subject Terms: Cardiomyopathy, Heart Failure, Myocardial Biology, Genetically Altered and Transgenic Models
Keywords: COP9 signalosome, ubiquitin, autophagy, misfolded proteins, Cops8, desmin-related cardiomyopathy, ubiquitin-proteosome system genetics, proteasome, crystalin, proteotoxicity
INTRODUCTION
Protein quality control (PQC) functions to minimize the level and toxicity of misfolded proteins in the cell, pivotal to intracellular proteostasis and cell survival.1, 2 PQC is accomplished by intricate collaboration between molecular chaperones and targeted proteolysis. The latter is done primarily by the ubiquitin (Ub)-proteasome system (UPS) and, sometimes, the autophagic-lysosomal pathway (ALP). PQC inadequacy allows misfolded proteins to undergo aberrant aggregation which can further impair PQC via mechanisms including suppressing UPS function;3–5 hence aberrant protein aggregation is both a consequence and a further cause of PQC inadequacy. Striking aberrant protein aggregation in cardiomyocytes, as evidenced by the presence of intracellular pre-amyloid oligomers and congophilic fibrils,6, 7 occurs in a large subset of human heart failure (HF) resulting from idiopathic cardiomyopathies. This links PQC inadequacy to the pathogenesis of common forms of heart diseases. PQC suppression via either ablating a chaperone gene or inhibiting targeted proteolysis is sufficient to cause cardiomyopathy and HF or to facilitate maladaptive cardiac remodeling;8–11 conversely, PQC improvement via chaperone overexpression or enhancement of target proteolysis confers cardiac protection against proteotoxicity in experimental animals.12–15 These experimental demonstrations are corroborated by clinical observations that a significant portion of cancer patients receiving proteasome inhibitors in their chemotherapy develop cardiac dysfunction or even heart failure.16, 17 A significant role of UPS malfunction in cardiac pathogenesis is further underscored by the identification of dominant negative mutations in TRIM63, the gene encodes a Ub ligase (muscle ring finger 1), as a cause of human familial hypertrophic cardiomyopathy.18 Hence, a better understanding of the molecular underpinnings of cardiac PQC is of paramount significance to developing strategies to improve cardiac PQC and thereby more effectively to treat a large subset of heart diseases.
The proteasome and lysosomes can degrade two distinct repertoires of misfolded proteins, with the former degrading individual protein molecules and the latter removing protein aggregates; however, both often require the misfolded proteins to be covalently modified by Ub via a process known as ubiquitination.19 The specificity of ubiquitination is conferred by Ub ligases which recognize and bind a mature degron on the substrate proteins. In yeast, several Ub ligases such as Hrd1, San1, Ubr1 and Hul5 were identified to ubiquitinate misfolded proteins.20 Hrd1 was recently confirmed to be pivotal to endoplasmic reticulum (ER) associated degradation of ER misfolded proteins in mammalian cardiomyocytes.21 However, very little is known about the identity and regulation of Ub ligases responsible for ubiquitination and degradation of cytosolic misfolded proteins in mammals.22
The COP9 signalosome (CSN) is an evolutionarily conserved protein complex, playing an important role in regulating the catalytic dynamics of cullin-RING Ub ligases (CRLs).23 By estimate, CRLs are responsible for ~20% of Ub-dependent degradation of cellular proteins.24 CRLs are activated by covalent conjugation of a Ub-like protein NEDD8 (Neural precursor cell expressed developmentally down-regulated 8) to cullin proteins via a ubiquitination-like process known as neddylation which is catalyzed by the NEDD8 activating enzyme (NAE), conjugating enzyme, and ligases.25 CSN-mediated cullin deneddylation is essential to CRL catalytic dynamics in vivo, likely by promoting exchange of substrate receptors of CRLs.26 The deneddylation activity of CSN requires a holo-complex formed by all 8 subunits (CSN1 through CSN8). By regulating CRLs activities, CSN participates in the regulation of many cellular processes including cell cycle control, DNA repair, gene expression, apoptosis and signaling transduction.27 We have previously reported that conditional knockout of cops8, the gene encoding CSN8, in mouse hearts impairs cullin deneddylation and compromises myocardial UPS and ALP functions,28, 29 which leads to rapidly deteriorated HF and mouse premature death, preventing them from being used for studying the role of CSN8/CSN in a chronic setting. Hence, the Cops8 hypomorphic (CSN8hypo) mice were used here to address an unanswered question: does CSN8/CSN regulate the ubiquitination and degradation of misfolded proteins?
The pathogenic role of cardiac proteotoxicity is best illustrated by desmin-related cardiomyopathy (DRC) which is the cardiac manifestation of desmin-related myopathy (DRM). DRM is a heterogeneous group of myopathies caused by mutations in desmin or its partner proteins such as αB-crystallin (CryAB).30 DRC eventually progresses to HF and is the main cause of death in DRM. At cellular level, DRM is characterized by intrasarcoplasmic desmin-positive aberrant protein aggregates and disruption of the cytoskeletal network.30 Similar to other conformational disorders, protein misfolding and aggregation are identified as the proximal pathogenic factors to DRC.6, 14, 31–33 Human DRM-linked R120G missense mutation of CryAB (CryABR120G) has proven to be a bona fide misfolded cytosolic protein.34 Mice with cardiomyocyte-restricted transgenic (tg) expression of CryABR120G develop cardiomyopathy and HF, recapitulating human DRC.35 Hence, the CryABR120G tg mice represent a highly relevant animal model of HF,36 especially for studying cardiac PQC and proteotoxicity.6, 13, 14, 31–33
Here we report that CSN8hypo mouse hearts display decreased deneddylation and significantly reduced performance to degrade a surrogate misfolded protein. Myocardial CSN8 is significantly upregulated in CryABR120G tg mice. When introduced into the CryABR120G tg mice, CSN8hypo aggravated the CryABR120G-based DRC. Further experimentation reveals that CSN8 deficiency suppresses the ubiquitination and degradation of CryABR120G, resulting in accumulation of protein aggregates and exacerbation of CryABR120G cytotoxicity; similarly, inactivation of CRLs via inhibiting neddylation stabilizes CryABR120G proteins in cardiomyocytes. Our results demonstrate that CSN8/CSN promotes the ubiquitination and degradation of misfolded proteins and protects against cardiac proteotoxicity and that CRLs participate in degradation of cytosolic misfolded proteins.
METHODS
A detailed Methods section is provided in the Online Supplement.
Mouse models
Mice with Cops8 conditionally targeted alleles were previously described.37 Briefly, Cops8neoflox allele contains a neomycin resistant cassette in intron between exon 3 and 4; the CSN8 knockout allele (CSN8−) has a deletion of exon 4 to 6. The homozygous Cops8neoflox/neoflox mice were then mated with Cops8+/− mice to produce Cops8neoflox/− and Cops8neoflox/+ mice, which were in the FVB/N inbred background and used as CSN8hypo mice and control mice (CTL), respectively. The CryAB, CryABR120G, and GFPdgn tg mice were described previously.35, 38
Echocardiography
Trans-thoracic echocardiography was performed on mice using the VisualSonics Vevo 770 system and a 30-MHz probe as previously described.28
Neonatal rat ventricular cardiomyocytes (NRVMs) cultures and adenoviral delivery
Primary NRVMs culture and adenoviral delivery of CryABR120G were performed as reported.39
SiRNA transfection
To knock down the target gene expression, the Lipofectamine™-2000 transfection reagent (Invitrogen) was used for siRNA transfection following the manufacturer's protocol.39
Immunostaining and aggregate quantification
Immunofluorescence staining of mouse myocardial sections or cultured NRVMs were performed as described.29 Immunofluorescence images of CryAB positive aggregates were quantified using Image-Pro Plus as described.14
Statistical analyses
All continuous variables are expressed as mean±SD. Differences between groups were evaluated for significance using two-tailed Student’s t test for unpaired 2-group comparison or 1-way or 2-way analysis of variance (ANOVA) followed by the Scheffé test when appropriate. The probability value <0.05 is considered statistically significant.
RESULTS
Characterization of CSN8 hypomorphic mice
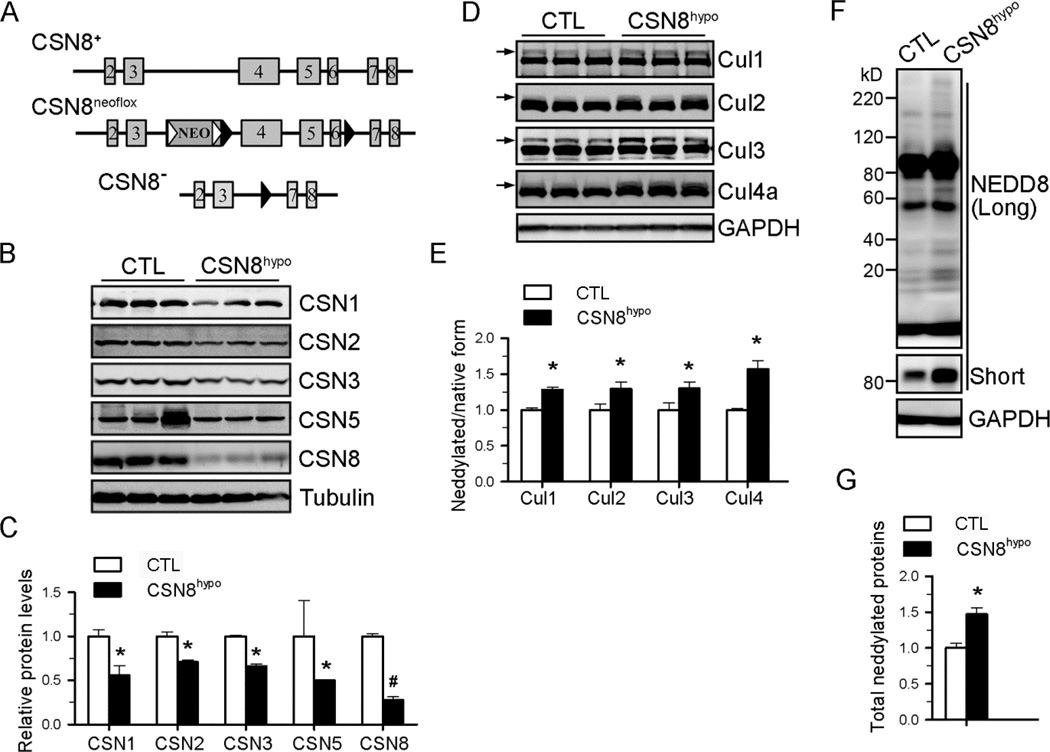
The early postnatal lethality of cardiomyocyte-restricted knockout of Cops8 in mice prevents the use of these mice to study the role of CSN in the degradation of misfolded proteins in adult hearts. To circumvent this problem, we used mice with different Cops8 targeted alleles (Figure 1A) to generate CSN8neoflox/− (CSN8hypo) and littermate CSN8neoflox/+ control (CTL) mice. CSN8hypo mice are viable, fertile and morphologically indistinguishable from wild-type mice and the CTL mice. Western blot analyses revealed that the CSN8hypo mice displayed ~80% reduction of CSN8 proteins in the heart (Figure 1B, 1C). In contrast, the CTL mice showed comparable CSN8 expression to wild-type mice (data not shown). Under the baseline condition, echocardiography showed no alteration in cardiac function, morphology or growth during the first 6 months of life of these CSN8hypo mice, compared to the CTL (Online Table I). Neither increased mortality nor gross abnormality was discerned in CSN8hypo mice by 1-year-of-age, the longest time monitored.
Figure 1. Decreased deneddylation activity in CSN8neoflox/− (CSN8hypo) mouse hearts.
Age-matched littermate CSN8neoflox/+ mice were used as controls (CTL). (A) Csn8-targeted alleles described in this study. The numbered light grey rectangles denote the exons. In the neoflox allele (CSN8neoflox), a neomycin phosphotransferase II gene (NEO) flanked by Frt sites (empty triangles) are inserted between exon 3 and 4; and exons 4 through 6 are flanked by LoxP sites (solid triangles). In CSN8 knockout allele (CSN8−), the NEO cassette and exons 4 through 6 are deleted by FLP- and Cre- mediated recombination. (B, C) Western blot analyses of indicated proteins. Representative images (B) and pooled densitometry data (C) are shown. GAPDH serves as a loading control. (D, E) Representative images (D) and pooled densitometry data (E) from western blot analyses of indicated cullin proteins in mouse hearts. Arrows mark the neddylated form of cullins. (F, G) Representative images (F) and pooled densitometry data (G) from western blot analyses of neddylated proteins in mouse hearts. Total neddylated proteins (revealed by long-time exposure) and neddylated cullin (revealed by short-time exposure) are increased in CSN8hypo hearts. N=3 mice/group; *p<0.05, #p<0.01 vs. CTL; t-test.
Impaired deneddylation activity in CSN8hypo mice
In CSN8hypo hearts, the reduction of CSN8 proteins was sufficient to discernibly reduce the protein levels of several other CSN subunits tested, including CSN1, CSN2, CSN3 and CSN5 (Figure 1B and 1C), confirming the essential role of CSN8 in the integrity and stability of the CSN holocomplex in cardiomyocytes. Consistent with the notion that the intact CSN complex is required for CSN deneddylation activity, we found that CSN8hypo increased neddylated forms of cullin (Cul) 1, 2, 3, and 4a (Figure 1D and 1E). Notably, CSN8hyop also increased the neddylated forms of many other non-cullin proteins (Figure 1F and 1G), indicating that CSN has a broad range of deneddylation substrates. Together, these data demonstrate that CSN8hypo impairs CSN deneddylation activity in the heart.
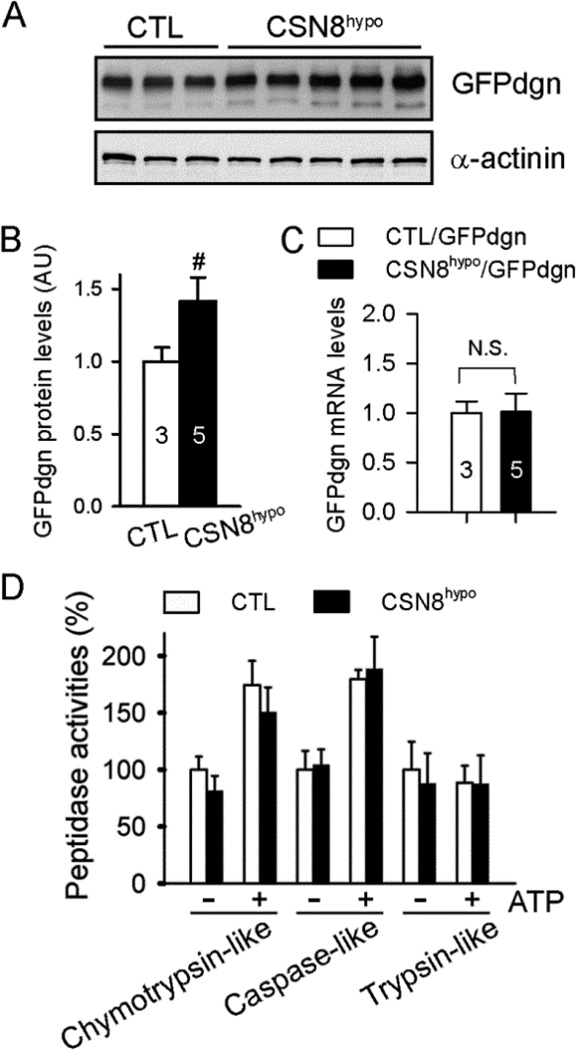
CSN8 hypomorphism impairs degradation of a surrogate misfolded protein in the heart
A proven UPS substrate protein GFPdgn was created by carboxyl fusion of a green fluorescence protein (GFP) with degron CL1, a 17-amino-acid sequence with surface exposure of a stretch of hydrophobic residues that mimics the signature conformation of misfolded proteins and is capable of triggering ubiquitination of its fusion protein by a pathway known to target misfolded proteins;25 hence, GFPdgn is considered a surrogate misfolded protein.40 To assess whether CSN8hypo affects UPS-mediated degradation of misfolded proteins in vivo, we employed GFPdgn tg mice, in which GFPdgn is ubiquitously expressed.38 By cross-breeding tg GFPdgn into the CTL or CSN8hypo mice, we found myocardial GFPdgn protein levels were significantly accumulated in CSN8hypo mouse hearts (Figure 2A and 2B) in absence of changes in GFPdgn mRNA levels (Figure 2C), suggesting that CSN8 haploinsufficiency impairs UPS degradation of GFPdgn. The defect does not appear to arise from alterations in proteasome activities because all three proteasome peptidase activities were comparable between CTL and CSN8hypo hearts (Figure 2D).
Figure 2. Effect of CSN8hypo on myocardial UPS function.
Mice were created via cross-breeding between CSN8neoflox/neoflox and CSN8+/−::GFPdgn mice. The resultant 8-week-old CTL::GFPdgn mice and CSN8hypo::GFPdgn mice were used. (A, B) Images (A) and pooled densitometry data (B) from western blot analyses for myocardial GFPdgn. Alpha-actinin was probed as a loading control. n= 3 or 5 mice per group, #p < 0.01 vs. CTL. (C) GFPdgn mRNA levels are assessed by real time RT-PCR using total RNA extracted from mouse ventricular myocardium. (D) Proteasomal peptidase activity assays of ventricular myocardium. n=4~6 per group, t-test.
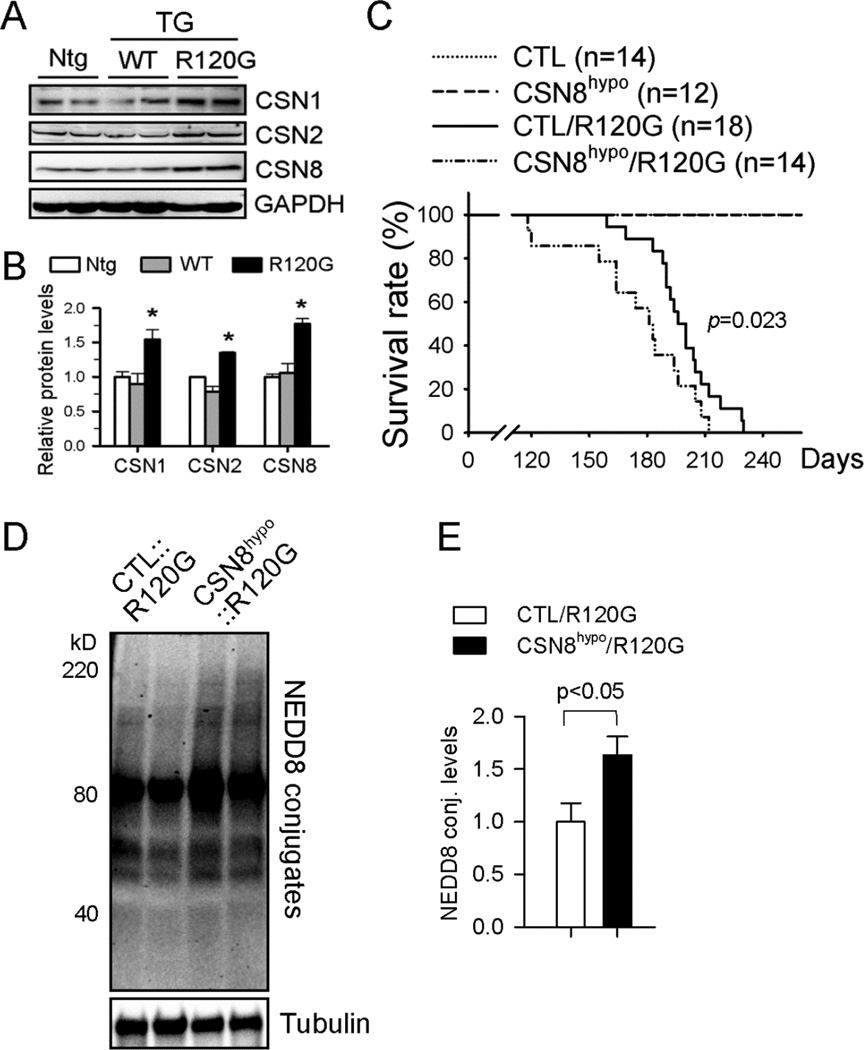
CSN8 hypomorphism exacerbates DRC in mice
Our examination of protein expression of representative CSN subunits (CSN1, CSN2, CSN8) revealed that CSN abundance was significantly increased in CryABR120G (line 134), but not wild type CryAB (line 11), tg mouse hearts (Figure 3A and 3B) although previous studies have shown that CryAB mRNA and protein overexpression in line 11 is greater than in line 134.35 To determine the role of CSN8/CSN upregulation in DRC mice, we crossbred the CSN8hypo mice with CryABR120G tg mice, a bona fide model of cardiac proteinopathy with defined disease progression.14, 32, 35 We obtained a cohort of mice with a genotype of CTL, CSN8hypo, CTL::CryABR120G, or CSN8hypo::CryABR120G and performed a Kaplan-Meier survival analysis which revealed that CSN8hypo significantly accelerated the premature death of the DRC mice (Figure 3C). Transthoracic echocardiography was performed on these animals at 12 weeks of age. Compared with CTL mice, CTL::CryABR120G mice displayed a cardiac functional phenotype characteristic of compensatory restrictive cardiomyopathy, as evidenced by marked decreases in left ventricular (LV) internal diameters and volumes at the end of diastole. These abnormalities were further augmented in CSN8hypo::CryABR120G mice (Table 1).
Figure 3. DRC exacerbation by CSN8hypo.
(A, B) Western blot analysis for the indicated CSN subunits in mouse hearts with tg overexpression of wild type CryAB (WT) or CryABR120G (R120G). Representative images (A) and pooled densitometry data (B) are shown. n=4 mice/group; *p<0.05 vs. Ntg or WT; ANOVA followed by the Scheffé test. (C) Kaplan-Meier survival analysis of a cohort of mixed-sex littermate CTL, CSN8hypo, CTL::CryABR120G, and CSN8hypo::CryABR120G mice. Log-rank test. (D, E) Representative image (D) and pooled densitometry data (E) of western blot analysis for myocardial NEDD8 conjugates (conj.) of mice of indicated genotypes at 8 weeks (n=4 mice/group); t-test.
Table 1.
Echocardiographic Measurements at 12 Weeks of Age
| CTL n=6 |
CSN8hypo n=8 |
CTL::CryABR120G n=10 |
CSN8hypo::CryABR120G n=8 |
|
|---|---|---|---|---|
| Body weight (g) | 30.4±2.1 | 27.2±3.5 | 28.2±3.7 | 28.8±4.2 |
| Heart rate (bpm) | 471±24 | 487±42 | 432±43 | 472±43 |
| LVIDd (mm) | 4.37±0.35 | 4.09±0.14 | 3.90±0.26† | 3.54±0.28†‡ |
| LVPWd (mm) | 0.70±0.13 | 0.73±0.13 | 0.93±0.15† | 0.88±0.06* |
| LVIDs (mm) | 3.17±0.26 | 3.12±0.21 | 2.45±0.22† | 1.89±0.22†‡ |
| LVPWs (mm) | 1.13±0.12 | 1.09±0.24 | 1.45±0.27† | 1.44±0.14† |
| LVAWd (mm) | 0.71±0.08 | 0.67±0.12 | 1.00±0.20† | 0.97±0.19† |
| LVAWs (mm) | 1.15±0.12 | 1.03±0.27 | 1.52±0.25† | 1.55±0.21† |
| LV FS (%) | 30.02±2.48 | 28.59±2.00 | 37.84±5.93* | 45.07±4.11†‡ |
| LV EF (%) | 53.83±5.68 | 58.55±5.52 | 65.19±8.95* | 77.25±4.20†‡ |
| LVVd (mm3) | 87.58±15.20 | 77.33±2.40 | 66.20±10.92† | 49.59±11.02†‡ |
| LVVs (mm3) | 40.28±8.18 | 41.29±6.18 | 22.28±9.46† | 11.31±3.41†‡ |
| SV (µl) | 47.3±10.3 | 35.2±6.2 | 43.9±6.7 | 38.3±8.6 |
| CO (ml/min) | 22.3±4.9 | 17.2±3.9 | 19.0±4.3 | 18.1±4.2 |
LVIDd, end-diastolic left ventricular (LV) internal diameter; LVIDs, end-systolic LVID; LVPWd, end-diastolic LV posterior wall thickness; LVPWs, end-systolic LVPW; LVAWd, end-diastolic LV anterior wall thickness; LVAWs, end-systolic LVAW; FS, fractional shortening; EF, ejection fraction; LVVd, end-diastolic LV volume; LVVs, end-systolic LVV; SV, stroke volume; CO, cardiac output.
p<0.05,
p<0.01 vs. CTL;
p<0.05 vs. CTL::CryABR120G; ANOVA followed by the Scheffé test.
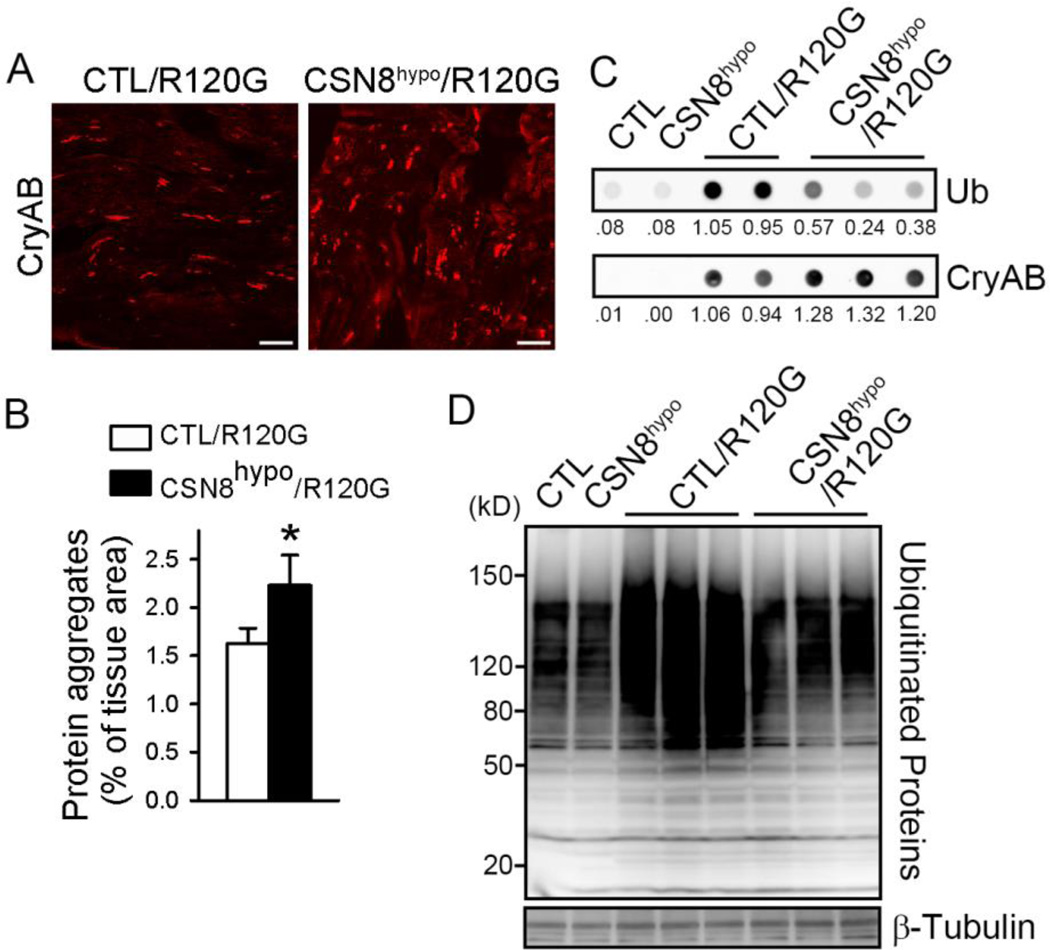
CSN8 hypomorphism increases NEDD8 conjugates and aberrant protein aggregation in DRC mouse hearts
Confirming that the deneddylation activity in DRC hearts was decreased by CSN8hypo, western blot analyses showed that myocardial total NEDD8 conjugate levels were increased by over 60% (p<0.05) in CSN8hypo::CryABR120G mice, compared with the CTL::CryABR120G mice (Figure 3D and 3E). Since protein aggregation is a causative pathogenic factor of DRC, we then sought to determine if the exacerbation of DRC in CSN8hypo mice was associated with altered protein aggregation. Immunofluorescence confocal microscopy revealed that CryAB-positive protein aggregates were significantly increased by CSN8hypo in DRC mouse hearts (Figure 4A and 4B). Consistently, filter-trap assays also showed a substantially increase in detergent-resistant CryAB-positive aggregates in the myocardium from CSN8hypo::CryABR120G mice, compared with those from CTL::CryABR120G mice (Figure 4C). These data compellingly demonstrate that CSN8hypo accumulates protein aggregates in DRC hearts. Surprisingly, the increased protein aggregates in CSN8hypo::CryABR120G hearts were accompanied by significantly reduced Ub conjugates in both detergent-resistant myocardial fraction (Figure 4C) and total myocardial protein extract (Figure 4D),suggesting that CSN8 is required for the ubiquitination of misfolded proteins in DRC hearts.
Figure 4. CSN8hypo aggravates CryABR120G-induced aberrant protein aggregation and decreases the ubiquitination in mouse hearts.
Littermate mice with the indicated genotypes were used at the age of 8 weeks. (A) Representative confocal fluorescent images of myocardial sections immunostained for CryAB (white). Scale bar = 20 µm. (B) Quantitative analysis of the relative abundance of the CryAB-positive protein aggregates in the heart. n=3 mice/group. *p<0.05 vs. CTL::CryABR120G, t-test. (C) Filter trapping assays for CryAB and ubiquitin (Ub) in ventricular myocardium of mice with indicated genotypes. The proteins retained on the filter were immunostained for CryAB or Ub. The relative density of each dot is shown below the blots. (D) Western blot analyses of myocardial total ubiquitinated proteins. β-tubulin was probed as a loading control.
CSN8/CSN regulates the stability of CryABR120G in cultured cardiomyocytes
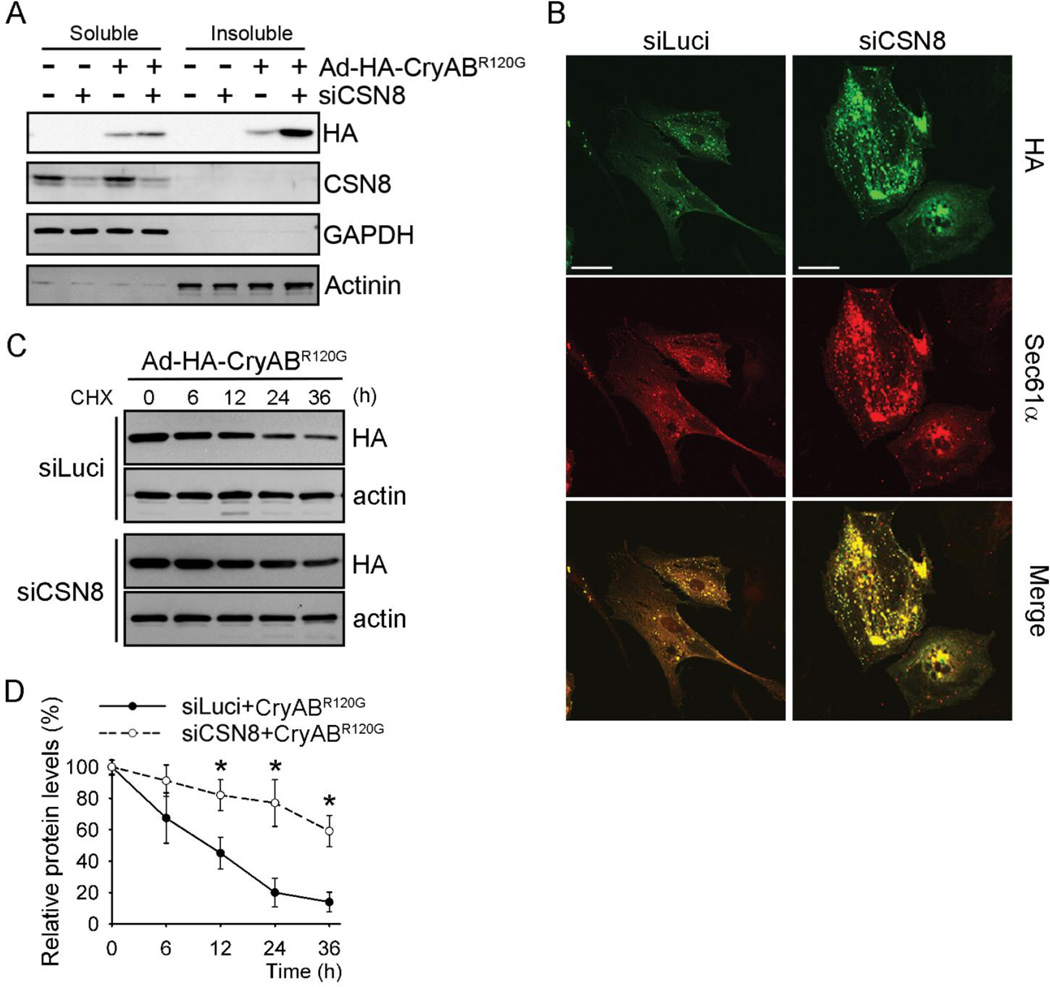
To further test if CSN8hypo-induced accumulation of protein aggregates in DRC mouse hearts is due to impaired degradation of misfolded CryABR120G, we assessed the impact of CSN8 down-regulation on the protein stability of HA-tagged CryABR120G overexpressed in cultured cardiomyocytes. In NRVMs, CSN8 knockdown (CSN8KD) by small interfering RNA (siRNA) substantially increased the steady-state protein levels of CryABR120G in the soluble fraction of cardiomyocyte lysate, and the increase was even more pronounced in the insoluble fraction (Figure 5A). We also assessed the prevalence of protein aggregates in CryABR120G-overexpressed cardiomyocytes by immunostaining for HA-CryABR120G and SEC61α, the latter serving as a marker of protein aggregates.39 Immunofluorescence images showed that CSN8KD increased both the abundance and the size of protein aggregates in cardiomyocytes (Figure 5B). To dynamically assess the degradation of CryABR120G, we further performed a cycloheximide chase experiment. We found that CSN8KD substantially prolonged the half-life of CryABR120G (Figure 5C and 5D). By contrast, CSN8KD did not discernibly increase the steady state protein levels of conventional GFP and endogenous or overexpressed wild type CryAB (Online Figure I) nor did it elongate the half-life of GFP and CryAB (Online Figures II and III). Taken together, these results demonstrate that CSN8 depletion impairs degradation of CryABR120G, a bona fide cytosolic misfolded protein, and promotes protein aggregation in cardiomyocytes.
Figure 5. CSN8 knockdown stabilizes CryABR120G in cardiomyocytes.
NRVMs were infected with adenoviruses expressing HA-CryABR120G (Ad-HA- CryABR120G) or β-Gal as indicated. The cells were also transfected with siRNAs against either luciferase (siLuci) or CSN8 (siCSN8). At 72 hours after the siRNA transfection, the cells were harvested for the analyses (A, B) or treated with cycloheximide (CHX, 100 µmol/L) for the indicated times (C). (A) Representative western blot images of indicated proteins in the Triton X-100 soluble and insoluble fraction of cell lysate. GAPDH and α-actinin were probed as loading controls. (B) Immunofluorescent images showing increased protein aggregates in CSN8 knockdown cells. HA-tag (green) and Sec61α(red) were stained for CryABR120G and aggresomes, respectively. Scale bar=50 µm. (C, D) Cycloheximide (CHX) chase assay for HA-CryABR120G. HA-CryABR120G protein levels at the indicated time points were measured using western blot analyses for HA-tag. A representative image (C) and a summary of the relative levels of HA-CryABR120G (D) are shown; *p<0.05 vs. the siLuci+CryABR120G group, n=3 repeats; t-test.
CSN8/CSN and CRLs control CryABR120G ubiquitination and degradation
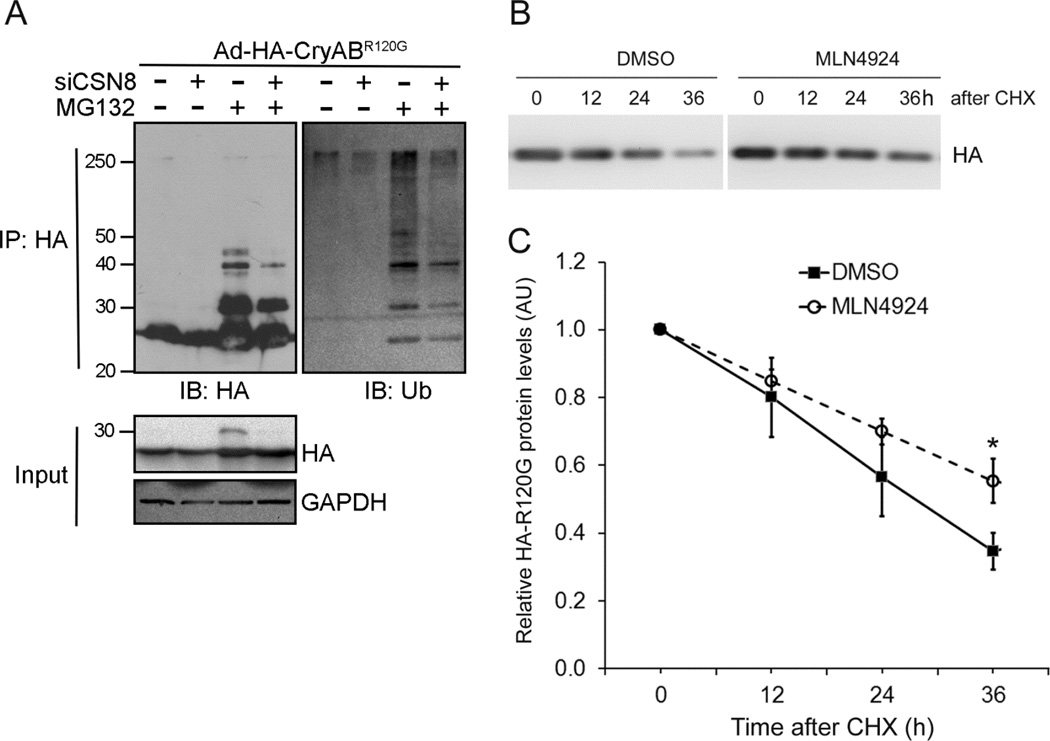
CSN is known to control the stability of a number of native proteins by regulating their respective CRLs’ activity.27 We next tested if CSN could regulate the ubiquitination of a bona fide misfolded protein. Immunoprecipitation of HA-CryABR120G followed by western blot analyses showed that proteasome inhibition by MG132 largely increased high-molecular-weight species of CryABR120G. Probing the immunoprecipitates with anti-Ub antibodies identified these high-molecular-weight species as ubiquitinated forms of CryABR120G. Furthermore, CSN8KD significantly reduced the ubiquitinated forms of HA-CryABR120G in both absence and presence of proteasome inhibition (Figure 6A). These data indicate that the proteasome is responsible for degradation of ubiquitinated CryABR120G and that CSN8/CSN is required for CryABR120G ubiquitination. Since CSN per se does not ubiquitinate any proteins but rather it regulates the activity of CRLs, an important family of Ub ligases,26 these in vivo and in vitro findings led us to hypothesize that CRLs are responsible for the ubiquitination of CryABR120G. To examine this hypothesis, we assessed the effect of CRLs inactivation via inhibiting their neddylation using MLN4924, a specific NAE inhibitor.24 We found that NAE inhibition significantly elongated the half-life of CryABR120G expressed in cultured NRVMs (Figure 6B and 6C). These results demonstrate that the activation of CRLs is required for the degradation of CryABR120G, suggesting that CSN controls CryABR120G ubiquitination via its regulation on CRLs.
Figure 6. Inhibition of CRLs impairs CryABR120G ubiquitination and degradation in cardiomyocytes.
(A) CSN8 knockdown impairs CryABR120G ubiquitination. NVRMs were treated as described in Figure 5. Seventy-two hours after siRNA transfection, MG132 (5 µmol/L) treatment was initiated and lasted for 6 hours before the cells were harvested. Representative images of western blot analyses (IB) of the indicated proteins in immunoprecipitated (IP) HA-CryABR120G are shown. (B, C) MLN4024 stabilizes CryABR120G in cultured NRVMs. MLN4924 (1µmol/L) or vehicle control (DMSO) treatment was initiated at 48 hours after Ad-HA-CryABR120G infection in cultured NRVMs. Cycloheximide (CHX, 50µmol/L) was added to the culture media at 30min after initiating MLN4924 treatment. Cells were harvested at the indicated time points for extraction of total proteins. Representative images (B) and pooled densitometry data (C) of western blot analyses of HA-CryABR120G are shown. *p<0.05 vs. the DMSO group, n=3 repeats; t-test.
CSN8 hypomorphism blunts autophagic responses in cardiomyocytes under stress
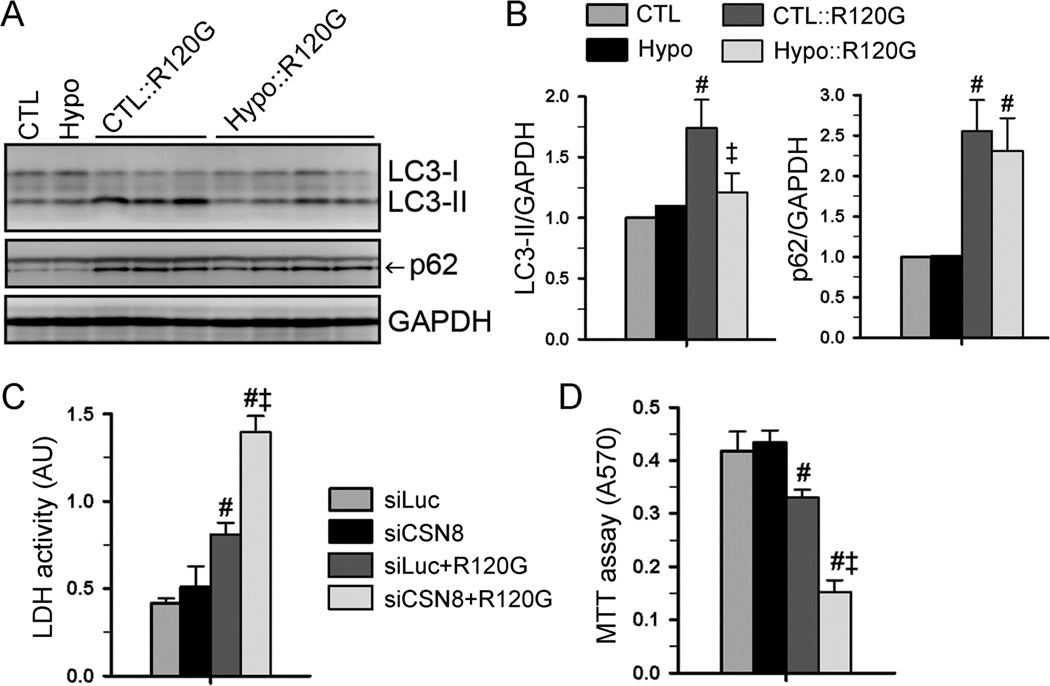
We have previously demonstrated that Cops8 loss-of-function impairs autophagosome-lysosome fusion, thereby accumulating Ub conjugates, LC3-II (a marker of autophagosomes), and p62/SQSTM1 (a substrate of autophagy) in the heart.29, 41 Hence, we examined these parameters in CSN8hypo hearts but we found none of them were discernibly altered at baseline (Online Figure IV), suggesting that autophagic activity is not perturbed in CSN8hypo mouse hearts under basal condition. Myocardial LC3-II, p62, and autophagic activity are known to increase in DRC mice.33, 39 When coupled with the CryABR120G-based DRC, CSN8hypo significantly suppressed the increase of LC3-II, but not that of p62, in the heart (Figure 7A and 7B), suggesting that CSN8hypo may impair cardiac autophagy under a stress condition. To examine this postulate further, we performed cell culture experiments. CSN8KD in cultured NRVMs did not reduce LC3-II flux or p62 flux at baseline but did so during simulated starvation (Online Figure V). Similar evidence was obtained in mouse embryonic fibroblasts (MEFs) in which autophagic flux was monitored using the tandem fluorescence protein-fused LC3 (tf-LC3) as a reporter.42 During autophagic activation triggered by simulated starvation, autophagosome-lysosome fusion impairment was detected in CSN8hypo MEFs (Online Figure VI). Moreover, CSN8KD significantly decreased LC3-II flux in NRVMs overexpressing CryABR120G (Online Figure VII). Taken together, these results indicate that impairment of autophagic flux by CSN8hypo becomes discernible only when the demand for autophagy is elevated. This also implicates that UPS impairment in Csn8hypo mouse hearts, which is discernible at baseline, is not secondary to potential ALP impairment.
Figure 7. CSN8 deficiency mitigates autophagic responses in mouse hearts and augments proteotoxicity in cardiomyocytes.
(A, B) Western blot analyses for myocardial LC3 and p62. CSN8 hypomorphism was cross-bred into CryABR120G (R120G) mice. Ventricular myocardial total protein extracts from 8-week-old mice were used. Representative images (A) and pooled densitometry data (B) are shown. n=4~6 mice/group. #p<0.01 vs. CTL; ‡p<0.05 vs. CTL::R120G. (C, D) CSN8 knock-down augments CryABR120G-induced cardiomyocyte injury. NRVMs were treated as described in Figure 5. The assays were performed 72 hours after siRNA transfection. LDH activities in the cultured medium (C) were measured to assess cell injury. Cell viability was assessed by the MTT assay (D). #p<0.01 vs. siLuci; ‡p<0.01 vs. siLuci+R120G; n=4~6. ANOVA followed by the Scheffé test.
CSN8 deficiency sensitizes cardiomyocytes to proteotoxic stress
Protein misfolding and aberrant aggregation are associated with cytotoxicity and heart failure in DRC.31, 32, 39 Given the critical role of CSN8/CSN in the removal of misfolded protein CryABR120G, we then tested if CSN8/CSN protects cardiomyocytes from proteotoxic stress-induced cytotoxicity. The proteotoxic stress was imposed to cultured cardiomyocytes by overexpression of CryABR120G. Cell death and cell viability were respectively assessed with the lactate dehydrogenase (LDH) leakage assay and the MTT assay. CSN8KD did not affect the survival of cardiomyocytes in basal condition. However, expression of CryABR120G significantly increased cytotoxicity, as evidenced by a significant increase of LDH activity in media and a decrease of cell viability. CSN8 depletion further aggravated the CryABR120G–induced cytotoxicity, as revealed by a ~70% increase of LDH release and a ~54% decrease of MTT reading compared to CryABR120G-expressed cardiomyocytes (Figure 7C and 7D). These data indicate that CSN8-deficient cardiomyocytes are more susceptible to proteotoxicity.
DISCUSSION
Aberrant protein aggregation is best exemplified in DRC but it is also implicated in the heart of humans with CHF of common causes,6, 7 suggesting that cardiac proteotoxicity is pathogenic in a large subset of cardiac disease. Indeed, improving UPS performance was recently shown to protect against not only DRC but also myocardial ischemia/reperfusion injury.14 Hence, a better understanding of the clearance of misfolded proteins may provide new strategies to treat heart disease or prevent cardiotoxicity of the treatment of non-cardiac disease. Here we have shown that CSN8hypo impairs the ubiquitination and degradation of a surrogate misfolded protein (GFPdgn) as well as a bona fide misfolded protein CryABR120G, leading to accumulation of protein aggregates and exacerbation of DRC in mice. Our data demonstrate that CSN8/CSN is essential to the ubiquitination and degradation of misfolded proteins via UPS and ALP and that CRLs participate in the degradation of cytosolic misfolded proteins.
CSN8 is an indispensable subunit of the CSN holoenzyme for cullin deneddylation;23 hence CSN8 deficiency is expected to compromise CRLs catalytic dynamics, thereby affecting ubiquitination efficiency of a large family of proteins.24, 43 Somewhat surprisingly, CSN8hypo mice do not display abnormal phenotypes for at least the first 6 months of age. Nevertheless, CSN8hypo exacerbates cardiac proteinopathy and reduced ubiquitination emerges as a primary defect caused by CSN8hypo during proteotoxic stress. This is supported by (1) myocardial UPS performance in CSN8hypo mice was decreased without altering proteasomal activities; (2) Ub conjugates were markedly decreased in both protein aggregates and total myocardial protein extracts from CryABR120G::CSN8hypo mice; and (3) CSN8KD reduced ubiquitinated forms of CryABR120G in cultured NRVMs.
Ubiquitination not only is essential to proteasomal degradation but also can indirectly promote ALP-mediated degradation.19, 20 Based on the prevalent model, when escaped form proteasomal degradation, misfolded proteins form aggregates and their Ub chains may bind p62/SQSTM1 which, in turn, recruits LC3-II-positive phagophores to trigger autophagosomal engulfment and degradation of the aggregates (Online Figure VIII). Our results show that the CSN8hypo-derived defect in ubiquitination reduces primarily UPS performance but does not affect ALP activity at baseline; however, CSN8hypo discernibly limits autophagic activity under a stress condition (e.g., nutrient deprivation, misfolded protein overexpression) that normally upregulates autophagy. This expands our prior findings from cardiac-specific CSN8 knockout mice that CSN8/CSN is essential to both UPS and ALP.29, 41 This also explains why CSN8hypo significantly decreased LC3-II protein levels in CryABR120G tg mouse hearts. Via binding ubiquitinated proteins, p62 facilitates aggresome formation and is often enriched in aberrant aggregates,39 whereas p62 is stabilized by ALP impairment.44 It is likely that p62 stabilizing factors (e.g., reduced autophagic flux) and destabilizing factors (e.g., reduction of Ub chains in the aggregates) counter each other, resulting in unaltered p62 protein levels in CSN8hypo::CryABR120G hearts, compared with CTL::CryABR120G hearts.
In cardiomyocytes, the degradation of overexpressed misfolded proteins such as CryABR120G depends on both UPS and ALP.13, 14, 19, 20 CSN8hypo reduces baseline myocardial UPS performance and limits stress-induced ALP activity in cardiomyocytes. Hence, we submit that reduced UPS- and ALP-mediated degradation of CryABR120G contribute to the exacerbation of protein aggregation and disease progression in DRC mouse hearts by CSN8hypo.
The specificity of ubiquitination is determined principally by Ub ligases. In mammals, HRD1, Parkin, and CHIP (C-terminus of Hsp70-interacting protein) have been shown to serve as the Ub ligases of the endoplasmic reticulum (ER) associated degradation (ERAD), responsible for ubiquitination of misfolded proteins retro-translocated from the ER to the cytosolic side.22 However, the identities of Ub ligases responsible for the degradation of cytosolic misfolded proteins are virtually unknown although CHIP is implicated in cytosolic PQC.22 Notably, none of these PQC ligases reported so far belongs to CRLs. Here we present multiple lines of strong evidence to support an important role of CRLs in the ubiquitination and degradation of cytosolic misfolded proteins. First, the degradation of a surrogate misfolded protein (GFPdgn) by the UPS is impaired in CSN8hypo mouse hearts with unaltered proteasome peptidase activities; second, reduction of CSN8/CSN and its deneddylation function decreased the misfolded proteins-induced protein ubiquitination in the heart of intact animals; third, disruption of CSN-mediated deneddylation activities by CSN8KD inhibited the ubiquitination and increased aberrant aggregation of a bona fide misfolded protein in cultured cardiomyocytes; and lastly, inhibition of CRLs by a NAE-specific inhibitor MLN4924 significantly slowed down the degradation of a bona fide misfolded protein in cardiomyocytes. It is unlikely that a single E3 ligase can account for the ubiquitination of all misfolded proteins, given the multitude of conformations that misfolded proteins can assume. There are 7 cullin proteins in the cullin family, each assembled with multiple substrate-recognizing adaptors to regulate the ubiquitination of the substrates.43 Therefore, the diversity and flexibility of CRLs seem well suited to accommodate the multitude of conformations that misfolded proteins may assume. It will be important to identify specific cullins and adaptors responsible for ubiquitination of misfolded proteins.
In conclusion, here we demonstrate that upregulation of CSN8/CSN is adaptive in DRC hearts and CSN8hypo exacerbates cardiac proteinopathy; the exacerbation is associated with augmented accumulation of protein aggregates, increased NEDD8 conjugates, and reduced levels of total Ub conjugates in the heart. Cardiomyocyte culture experiments further show that both CSN8 deficiency and CRLs inhibition suppress the ubiquitination and degradation of CryABR120G, resulting in accumulation of protein aggregates and exacerbation of CryABR120G cytotoxicity. Hence, we have obtained compelling evidence that CSN8/CSN is essential to the ubiquitination and clearance of misfolded cytosolic proteins and protects against proteotoxicity and that CRLs participate in the degradation of cytosolic misfolded proteins.
Increased production and impaired removal of misfolded proteins in the heart due to genetic mutations or acquired causes are highly conceivable and, in some cases, well-demonstrated in cardiac remodeling and heart failure.1, 2, 6 However, presently there is no treatment specifically aiming at enhancing degradation of misfolded proteins in the heart. Meanwhile, the firs-in-class NAE inhibitor (MLN4924), which inhibits neddylation and activation of CRLs, is in clinical trials for treating cancers.45 Increased NEDD8 conjugates in end-stage failing human hearts were recently reported.46 The present study identifies that CSN8/CSN and CRLs contribute to degradation of cytosolic misfolded proteins. This represents one major step closer to identification of specific Ub ligases for targeted degradation of toxic misfolded proteins; on the other hand, this also cautions that NAE inhibition may potentially exert cardiotoxicity, just like proteasome inhibitors.16, 17
Supplementary Material
Novelty and Significance.
What Is Known?
The COP9 signalosome holocomplex (CSN) consisting of 8 unique proteins (CSN1 through CSN8), which via cullin deneddylation, regulates cullin-RING ligases (CRLs), the largest family of ubiquitin ligases.
Cardiac ablation of Cops8 impairs myocardial protein degradation by the ubiquitin-proteasome system (UPS) and autophagic-lysosomal pathway (ALP), resulting in cardiac hypertrophy, heart failure, and premature death in mice.
Neither CSN nor CRLs are known to control the ubiquitination of cytosolic misfolded proteins.
What New Information Does This Article Contribute?
Myocardial CSN8/CSN is increased in a mouse model of proteinopathy whereas down-regulation of CSN8/CSN impairs the ubiquitination and degradation of a bona fide misfolded cytosolic protein in cardiomyocytes.
CRLs participate in ubiquitination and degradation of a bona fide misfolded cytosolic protein.
CSN8/CSN protects against cardiac proteinopathy.
UPS- and ALP- mediated protein degradation constitutes the last line of defense in protein quality control (PQC) which acts to minimize the level and the toxicity of misfolded proteins in the cell. Increases of misfolded proteins in cardiomyocytes are an inevitable consequence and cause of increased cardiac stress, whereas PQC inadequacy has been implicated in the progression to heart failure from a large subset of heart diseases. However, no specific treatment aimed at improving cardiac PQC is currently available. The present study reveals that (1) myocardial CSN8 is upregulated in a classical mouse model of proteinopathy and, when introduced into this mouse, CSN8 hypomorphism hastens disease progression; (2) the degradation of a bona fide misfolded protein in cardiomyocytes is suppressed by CSN8 knockdown or the inhibition of CRLs using a NEDD8 activating enzyme (NAE) inhibitor which is in clinical trial to treat cancers; and (3) CSN8 increases ubiquitination of misfolded proteins, thereby promoting their degradation by the proteasome and ALP. These findings suggest that both CSN8/CSN and CRLs may represent key targets for improving cardiac PQC and caution that NAE inhibition may yield cardiac toxicity.
Acknowledgments
SOURCES OF FUNDING
This work was in part supported by NIH grants R01HL085629 and R01HL072166 (to X.W.) and R01HL124248 (to H.S.), and American Heart Association grants 0740025N (to X.W.) and 11SDG6960011 (to H.S.).
Nonstandard Abbreviations and Acronyms
- ALP
the autophagic-lysosomal pathway
- CHX
cycloheximide
- CRLs
cullin-based RING ligases
- CryAB
αB-crystallin
- CSN
the COP9 signalosome
- CSN8hypo
CSN8 hypomorphism
- DRC
desmin-related cardiomyopathy
- GFP
green fluorescent protein
- GFPdgn
GFP modified by carboxyl fusion of degron CL1
- HF
heart failure
- LDH
lactate dehydrogenase
- LV
left ventricle
- MTT
3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide
- NEDD8
neural precursor cell expressed, developmentally down-regulated 8
- NAE
NEDD8 activating enzyme
- NEO
neomycin phosphotransferase II gene
- PQC
protein quality control
- siRNA
small interfering RNA
- Ub
ubiquitin
- UPS
the ubiquitin proteasome system
Footnotes
DISCLOSURE
The authors declare there is no conflict of interest to disclose.
REFERENCES
- 1.Wang X, Pattison JS, Su H. Posttranslational modification and quality control. Circ Res. 2013;112:367–381. doi: 10.1161/CIRCRESAHA.112.268706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willis MS, Patterson C. Proteotoxicity and cardiac dysfunction--alzheimer's disease of the heart? N Engl J Med. 2013;368:455–464. doi: 10.1056/NEJMra1106180. [DOI] [PubMed] [Google Scholar]
- 3.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 4.Chen Q, Liu JB, Horak KM, Zheng H, Kumarapeli AR, Li J, Li F, Gerdes AM, Wawrousek EF, Wang X. Intrasarcoplasmic amyloidosis impairs proteolytic function of proteasomes in cardiomyocytes by compromising substrate uptake. Circ Res. 2005;97:1018–1026. doi: 10.1161/01.RES.0000189262.92896.0b. [DOI] [PubMed] [Google Scholar]
- 5.Liu J, Tang M, Mestril R, Wang X. Aberrant protein aggregation is essential for a mutant desmin to impair the proteolytic function of the ubiquitin-proteasome system in cardiomyocytes. J Mol Cell Cardiol. 2006;40:451–454. doi: 10.1016/j.yjmcc.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 6.Sanbe A, Osinska H, Saffitz JE, Glabe CG, Kayed R, Maloyan A, Robbins J. Desmin-related cardiomyopathy in transgenic mice: A cardiac amyloidosis. Proc Natl Acad Sci U S A. 2004;101:10132–10136. doi: 10.1073/pnas.0401900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gianni D, Li A, Tesco G, McKay KM, Moore J, Raygor K, Rota M, Gwathmey JK, Dec GW, Aretz T, Leri A, Semigran MJ, Anversa P, Macgillivray TE, Tanzi RE, del Monte F. Protein aggregates and novel presenilin gene variants in idiopathic dilated cardiomyopathy. Circulation. 2010;121:1216–1226. doi: 10.1161/CIRCULATIONAHA.109.879510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ranek MJ, Zheng H, Huang W, Kumarapeli AR, Li J, Liu J, Wang X. Genetically induced moderate inhibition of 20s proteasomes in cardiomyocytes facilitates heart failure in mice during systolic overload. J Mol Cell Cardiol. 2015;85:273–281. doi: 10.1016/j.yjmcc.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian Z, Zheng H, Li J, Li Y, Su H, Wang X. Genetically induced moderate inhibition of the proteasome in cardiomyocytes exacerbates myocardial ischemia-reperfusion injury in mice. Circ Res. 2012;111:532–542. doi: 10.1161/CIRCRESAHA.112.270983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taneike M, Yamaguchi O, Nakai A, Hikoso S, Takeda T, Mizote I, Oka T, Tamai T, Oyabu J, Murakawa T, Nishida K, Shimizu T, Hori M, Komuro I, Takuji Shirasawa TS, Mizushima N, Otsu K. Inhibition of autophagy in the heart induces age-related cardiomyopathy. Autophagy. 2010;6:600–606. doi: 10.4161/auto.6.5.11947. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi M, Imanaka-Yoshida K, Yoshida T, Wood M, Fearns C, Tatake RJ, Lee JD. A crucial role of mitochondrial hsp40 in preventing dilated cardiomyopathy. Nat Med. 2006;12:128–132. doi: 10.1038/nm1327. [DOI] [PubMed] [Google Scholar]
- 12.Kumarapeli AR, Su H, Huang W, Tang M, Zheng H, Horak KM, Li M, Wang X. Alpha b-crystallin suppresses pressure overload cardiac hypertrophy. Circ Res. 2008;103:1473–1482. doi: 10.1161/CIRCRESAHA.108.180117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhuiyan MS, Pattison JS, Osinska H, James J, Gulick J, McLendon PM, Hill JA, Sadoshima J, Robbins J. Enhanced autophagy ameliorates cardiac proteinopathy. J Clin Invest. 2013;123:5284–5297. doi: 10.1172/JCI70877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Horak KM, Su H, Sanbe A, Robbins J, Wang X. Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice. J Clin Invest. 2011;121:3689–3700. doi: 10.1172/JCI45709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ranek MJ, Terpstra EJ, Li J, Kass DA, Wang X. Protein kinase g positively regulates proteasome-mediated degradation of misfolded proteins. Circulation. 2013;128:365–376. doi: 10.1161/CIRCULATIONAHA.113.001971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grandin EW, Ky B, Cornell RF, Carver J, Lenihan DJ. Patterns of cardiac toxicity associated with irreversible proteasome inhibition in the treatment of multiple myeloma. J Card Fail. 2015;21:138–144. doi: 10.1016/j.cardfail.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 17.Honton B, Despas F, Dumonteil N, Rouvellat C, Roussel M, Carrie D, Galinier M, Montastruc JL, Pathak A. Bortezomib and heart failure: Case-report and review of the french pharmacovigilance database. Fundam Clin Pharmacol. 2014;28:349–352. doi: 10.1111/fcp.12039. [DOI] [PubMed] [Google Scholar]
- 18.Chen SN, Czernuszewicz G, Tan Y, Lombardi R, Jin J, Willerson JT, Marian AJ. Human molecular genetic and functional studies identify trim63, encoding muscle ring finger protein 1, as a novel gene for human hypertrophic cardiomyopathy. Circ Res. 2012;111:907–919. doi: 10.1161/CIRCRESAHA.112.270207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clague MJ, Urbe S. Ubiquitin: Same molecule, different degradation pathways. Cell. 2010;143:682–685. doi: 10.1016/j.cell.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 20.Fredrickson EK, Gardner RG. Selective destruction of abnormal proteins by ubiquitin-mediated protein quality control degradation. Semin Cell Dev Biol. 2012;23:530–537. doi: 10.1016/j.semcdb.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doroudgar S, Volkers M, Thuerauf DJ, Khan M, Mohsin S, Respress JL, Wang W, Gude NA, Muller OJ, Wehrens XH, Sussman MA, Glembotski CC. Hrd1 and er-associated protein degradation, erad, are critical elements of the adaptive er stress response in cardiac myocytes. Circ Res. 2015 doi: 10.1161/CIRCRESAHA.115.306993. 10.1161/CIRCRESAHA.1115.306993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Terpstra EJ. Ubiquitin receptors and protein quality control. J Mol Cell Cardiol. 2013;55:73–84. doi: 10.1016/j.yjmcc.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei N, Deng XW. The cop9 signalosome. Annu Rev Cell Dev Biol. 2003;19:261–286. doi: 10.1146/annurev.cellbio.19.111301.112449. [DOI] [PubMed] [Google Scholar]
- 24.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, Cullis CA, Doucette A, Garnsey JJ, Gaulin JL, Gershman RE, Lublinsky AR, McDonald A, Mizutani H, Narayanan U, Olhava EJ, Peluso S, Rezaei M, Sintchak MD, Talreja T, Thomas MP, Traore T, Vyskocil S, Weatherhead GS, Yu J, Zhang J, Dick LR, Claiborne CF, Rolfe M, Bolen JB, Langston SP. An inhibitor of nedd8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 25.Kandala S, Kim IM, Su H. Neddylation and deneddylation in cardiac biology. Am J Cardiovasc Dis. 2014;4:140–158. [PMC free article] [PubMed] [Google Scholar]
- 26.Lydeard JR, Schulman BA, Harper JW. Building and remodelling cullin-ring e3 ubiquitin ligases. EMBO Rep. 2013;14:1050–1061. doi: 10.1038/embor.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei N, Serino G, Deng XW. The cop9 signalosome: More than a protease. Trends Biochem Sci. 2008;33:592–600. doi: 10.1016/j.tibs.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 28.Su H, Li J, Menon S, Liu J, Kumarapeli AR, Wei N, Wang X. Perturbation of cullin deneddylation via conditional csn8 ablation impairs the ubiquitin-proteasome system and causes cardiomyocyte necrosis and dilated cardiomyopathy in mice. Circ Res. 2011;108:40–50. doi: 10.1161/CIRCRESAHA.110.230607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su H, Li F, Ranek MJ, Wei N, Wang X. Cop9 signalosome regulates autophagosome maturation. Circulation. 2011;124:2117–2128. doi: 10.1161/CIRCULATIONAHA.111.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLendon PM, Robbins J. Desmin-related cardiomyopathy: An unfolding story. Am J Physiol Heart Circ Physiol. 2011;301:H1220–H1228. doi: 10.1152/ajpheart.00601.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanbe A, Osinska H, Villa C, Gulick J, Klevitsky R, Glabe CG, Kayed R, Robbins J. Reversal of amyloid-induced heart disease in desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2005;102:13592–13597. doi: 10.1073/pnas.0503324102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng H, Tang M, Zheng Q, Kumarapeli AR, Horak KM, Tian Z, Wang X. Doxycycline attenuates protein aggregation in cardiomyocytes and improves survival of a mouse model of cardiac proteinopathy. J Am Coll Cardiol. 2010;56:1418–1426. doi: 10.1016/j.jacc.2010.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tannous P, Zhu H, Johnstone JL, Shelton JM, Rajasekaran NS, Benjamin IJ, Nguyen L, Gerard RD, Levine B, Rothermel BA, Hill JA. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2008;105:9745–9750. doi: 10.1073/pnas.0706802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bova MP, Yaron O, Huang Q, Ding L, Haley DA, Stewart PL, Horwitz J. Mutation r120g in alphab-crystallin, which is linked to a desmin-related myopathy, results in an irregular structure and defective chaperone-like function. Proc Natl Acad Sci U S A. 1999;96:6137–6142. doi: 10.1073/pnas.96.11.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Osinska H, Klevitsky R, Gerdes AM, Nieman M, Lorenz J, Hewett T, Robbins J. Expression of r120g-alphab-crystallin causes aberrant desmin and alphab-crystallin aggregation and cardiomyopathy in mice. Circ Res. 2001;89:84–91. doi: 10.1161/hh1301.092688. [DOI] [PubMed] [Google Scholar]
- 36.Houser SR, Margulies KB, Murphy AM, Spinale FG, Francis GS, Prabhu SD, Rockman HA, Kass DA, Molkentin JD, Sussman MA, Koch WJ. American Heart Association Council on Basic Cardiovascular Sciences CoCC, Council on Functional G, Translational B. Animal models of heart failure: A scientific statement from the american heart association. Circ Res. 2012;111:131–150. doi: 10.1161/RES.0b013e3182582523. [DOI] [PubMed] [Google Scholar]
- 37.Menon S, Chi H, Zhang H, Deng XW, Flavell RA, Wei N. Cop9 signalosome subunit 8 is essential for peripheral t cell homeostasis and antigen receptor-induced entry into the cell cycle from quiescence. Nat Immunol. 2007;8:1236–1245. doi: 10.1038/ni1514. [DOI] [PubMed] [Google Scholar]
- 38.Kumarapeli AR, Horak KM, Glasford JW, Li J, Chen Q, Liu J, Zheng H, Wang X. A novel transgenic mouse model reveals deregulation of the ubiquitin-proteasome system in the heart by doxorubicin. FASEB J. 2005;19:2051–2053. doi: 10.1096/fj.05-3973fje. [DOI] [PubMed] [Google Scholar]
- 39.Zheng Q, Su H, Ranek MJ, Wang X. Autophagy and p62 in cardiac proteinopathy. Circ Res. 2011;109:296–308. doi: 10.1161/CIRCRESAHA.111.244707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Su H, Ranek MJ. Protein quality control and degradation in cardiomyocytes. J Mol Cell Cardiol. 2008;45:11–27. doi: 10.1016/j.yjmcc.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su H, Li J, Osinska H, Li F, Robbins J, Liu J, Wei N, Wang X. The cop9 signalosome is required for autophagy, proteasome-mediated proteolysis, and cardiomyocyte survival in adult mice. Circ Heart Fail. 2013;6:1049–1057. doi: 10.1161/CIRCHEARTFAILURE.113.000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged lc3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Martin DS. The cop9 signalosome and cullin-ring ligases in the heart. Am J Cardiovasc Dis. 2015;5:1–18. [PMC free article] [PubMed] [Google Scholar]
- 44.Tian Z, Wang C, Hu C, Tian Y, Liu J, Wang X. Autophagic-lysosomal inhibition compromises ubiquitin-proteasome system performance in a p62 dependent manner in cardiomyocytes. PLoS One. 2014;9:e100715. doi: 10.1371/journal.pone.0100715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swords RT, Erba HP, DeAngelo DJ, Bixby DL, Altman JK, Maris M, Hua Z, Blakemore SJ, Faessel H, Sedarati F, Dezube BJ, Giles FJ, Medeiros BC. Pevonedistat (mln4924), a first-in-class nedd8-activating enzyme inhibitor, in patients with acute myeloid leukaemia and myelodysplastic syndromes: A phase 1 study. Br J Haematol. 2015;169:534–543. doi: 10.1111/bjh.13323. [DOI] [PubMed] [Google Scholar]
- 46.Li J, Ma W, Li H, Hou N, Wang X, Kim IM, Li F, Su H. Nedd8 ultimate buster-1 long (nub1l) protein suppresses atypical neddylation and promotes proteasomal degradation of misfolded proteins. J Biol Chem. 2015 Aug;Oct; doi: 10.1074/jbc.M115.664375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.