Abstract
Human integrase interactor 1 (INI1/SMARCB1/SNF5) is a chromatin-remodeling molecule that binds to HIV-1 integrase and enhances proviral DNA integration. INI1 is also known as a tumor suppressor gene and has been found to be mutated in several aggressive tumors such as rhabdoid and lymphoid tumors. To study the function of simian INI1, we screened and cloned simian INI1 cDNA from B lymphoma cells of rhesus monkeys using RT-PCR. Sequence analysis showed 23 single nucleotide differences compared to the human ortholog, which, however, did not result in amino acid changes, and the amino acid sequence is therefore 100 % conserved between human and simian INI1. Two alternatively spliced isoforms, INI1a and INI1b, were also found in simian INI1. These two isoforms did not show any functional difference in HIV-1 proviral DNA integration and nuclear localization, suggesting that the specificity of simian INI1 would not be a factor preventing HIV-1 infection of a simian host. Nevertheless, INI1b is expressed only in established cancer cell lines such as Jurkat and COS-7 cells, and not in primary cells, suggesting that INIlb could be an indicator of cell transformation.
Keywords: HIV-1; Simian INI1/SMARCB1/SNF5; Integrase, integration of provirus
Introduction
Integrase interactor 1 (INI1), also called human SNF5/SMARCB1, was discovered by yeast two-hybrid screening as a binding protein of HIV-1 integrase (INT), a critical viral enzyme that is absolutely required for integration of the reverse transcribed, double stranded viral DNA into the host chromosome [4]. INI1 binds not only to HIV-1 INT but also to DNA, and this interaction dramatically enhances in vitro DNA strand transfer by stimulating HIV-1 INT activity [2, 4] and integration of proviral DNA into the host chromosome by releasing target DNA strands from their tightly packed chromatin structure [2, 4]. INI1 is also known to be one of the core subunits of the SWI/SNF chromatin remodeling complex, which is required for regulation of mating type switching, sucrose-dependent growth, and transcription in yeast [5]. Mutations such as biallelic deletions or missense, nonsense, splicing and frameshift mutations in INI1, have been implicated in pediatric cancers such as rhabdoid tumors and other cancers of the soft tissue [9]. Although the molecular mechanisms by which mutations in INI1 lead to cancer are not fully understood, previous studies in mice have suggested that impairment of INI1-mediated transcriptional regulation of the cell cycle regulatory protein cyclin D1 could be involved [10, 11]. Taken together, these reports indicate that INI1 is critical not only for the HIV-1 life cycle but also for tumorigenesis.
Our recent studies have revealed that the Nef protein of simian immunodeficiency virus PBj1.9 enhanced proviral DNA integration into the host chromosome in association with INT and human INI1 [8]. To verify that simian INI1 also has the same function in proviral DNA integration, we sequenced simian INI1 cDNA and compared its function with that of its human counterpart. Like human INI1 (GenBank: AJ011738.1), simian INI1 also has a distinct isotype – INI1b, with a 9-amino-acid deletion between amino acids 69 and 77 of INI1a – which is generated by alternative splicing. We also compared the functions of simian INI1a and INI1b in terms of integration, nuclear localization, and differential expression in primary and cancer cells. Our data indicate that simian INI1a and INI1b are functionally equivalent to the human counterparts in proviral DNA integration. However, INI1b was detected only in transformed cells, and not primary cells, suggesting that INI1b could be employed as a prognostic and diagnostic marker of tumorigenesis.
Materials and methods
Cells and media
Simian B cell lymphoma cell line LCL8664 (ATCC CRL-1805) was purchased to screen for simian INI1 mRNA. Human peripheral blood mononuclear cells (hPBMC) were isolated from the blood of normal volunteers using Ficoll-Hypaque gradient centrifugation and stimulated with 500 ng of phytohemagglutinin (PHA) per ml as described [7]. The Jurkat T cell line (ATCC TIB-152) and COS-7 cell line (ATCC CRL-1651) were purchased, and interleukin-2 (IL-2)-dependent simian T cells (mm155.90) were received from Dr. Jae U. Jung (University of Southern California) as a gift. The LCL8664, hPBMC, and Jurkat cells were maintained in RPMI1940 with 10 % FBS, and COS-7 cells were maintained in DMEM with 10 % FBS. IL-2-dependent T cells were maintained in RPMI1940 with 10 % FBS and 50 ng of IL-2 per ml.
Cloning and sequencing of simian INI1a and INI1b
Total RNA was purified from the simian B cell lymphoma cell line according to the manufacturer's protocol (Life Technologies, NY), and an RT-PCR reaction was performed with the purified RNA to generate complementary DNA (cDNA) of INI1a and INI1b, using the INI1-specific primers 5′-ATGATGATGATGGCGCTGAGCAAG-3′ and 5′-TTACCAGGCCGGGGCCGTGTTGG CAA-3′. To detect alternative splicing in INI1, the internal primers 5′-GTATGTTC CGAGGTTCTCTGTAC-3′ and 5′-CTG ATGGACACAGCCTTGTACT-3′ were employed. The PCR products were then cloned into TA cloning vector pCR2.1 (Invitrogen, Carlsbad, CA) and sequenced, and the simian INI1a and INI1b sequences were compared and aligned with that of human INI1, using NIH Blast2, ClustalW, and Jellyfish software (Biowire.com, Mountain View, CA).
Protein purification and integration assay
Full-length simian INI1a and INI1b genes were cloned into pGEX-5X (Sigma-Aldrich, St. Louis, MO), and INI1a- and INI1b-GST fusion proteins were purified from bacterial culture of the strain Rosetta transformed with each plasmid, using glutathione Sepharose beads (Amersham Pharmacia Biotech, Piscataway, NJ). His-tagged HIV-1 INT was purified using Ni-NTA agarose beads (QIAGEN, Valencia, CA) from overnight culture of BL21 cells transformed with pINSD.His.Sol, obtained from Dr. Robert Craigie through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. The in vitro DNA strand transfer assay was then performed, using purified INT and INI1 as described previously [4, 8]. Briefly, the HIV-1 INT substrate U5.5 (5′-GGATCCGGAAAA TCTCTAGCA) was labeled with 50 μCi of [γ-32P]ATP (NEN) by incubating with 10 U of T4 polynucleotide kinase (New England Biolabs, Beverly, MA). After kinase inactivation at 70 °C, U5.5 was annealed with U5.4 (ACTGCTAGAGATTTTCCGGATCC), and the annealed, double-stranded DNA was purified using a NucTrap probe purification column (Stratagene, La Jolla, CA). Different amounts of the proteins GST-INI1a, GST-INI1b, and GST were added to the integration reaction mixture consisting of purified HIV-1 INT, DNA substrate, and integration buffer (20 mM HEPES, pH 7.5, 10 mM MnCl2, 10 mM dithiothreitol, 0.05 % NP-40). After a 1-h incubation, an equal volume of loading buffer (98 % deionized formamide, 10 mM EDTA, 0.05 % bromophenol blue, 0.05 % xylene cyanol) was added to stop the reaction, and the samples were analyzed by 20 % polyacrylamide urea gel electrophoresis and autoradiography.
Fluorescence microscopy analysis
To analyze the subcellular distribution of the INI1 proteins, INI1a and INI1b genes were placed in-frame in front of the green fluorescent protein (gfp) gene of pEGFP-N3 (Clontech, Palo Alto, CA). The INI1-gfp fusion plasmids were then introduced into 293T cells by transfection, and the subcellular localization of these fusion proteins was determined by fluorescence microscopy.
Results and discussion
Sequence analysis of simian INI1
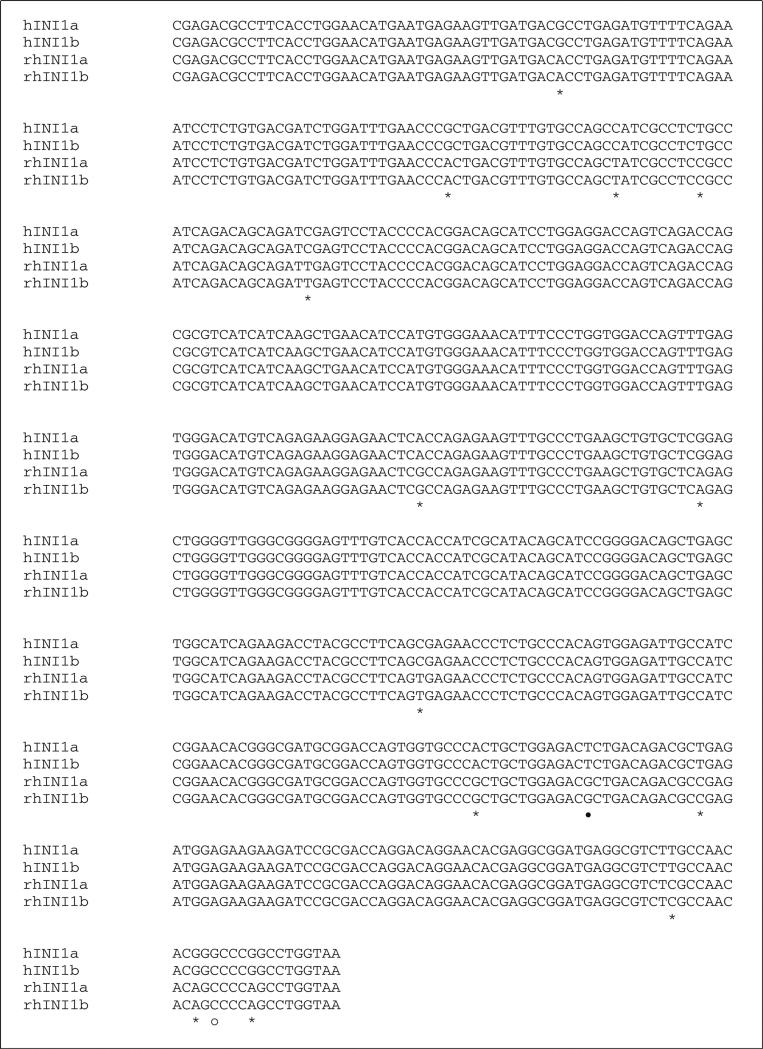
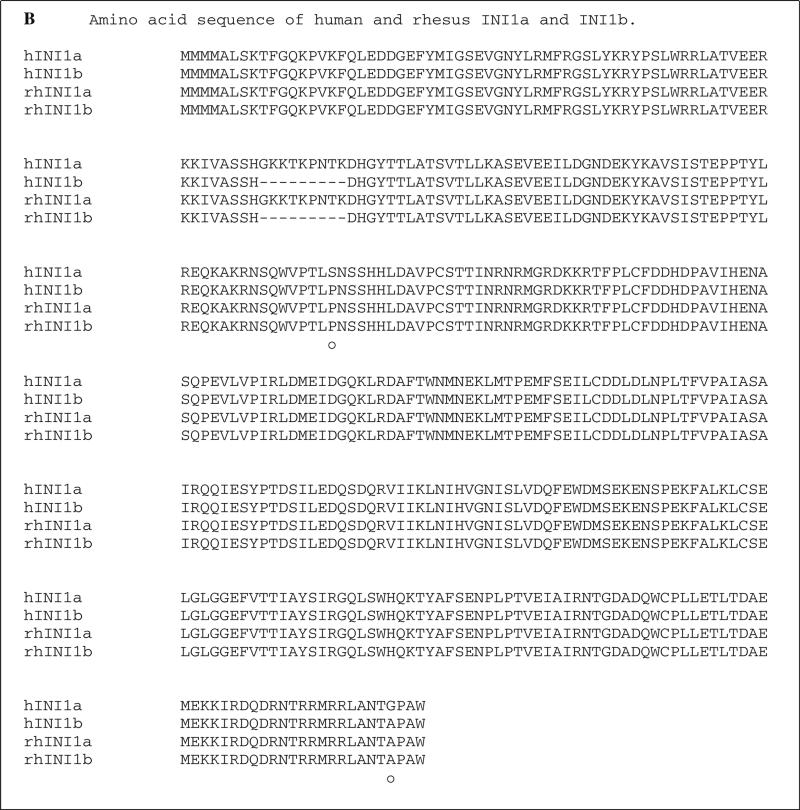
Simian INI1 cDNA was generated from the simian B cell lymphoma cell line LCL8664 by RT-PCR, and the size of the amplified product of simian INI1 was identical to the human ortholog (data not shown). Nine individual clones of simian INI1 cDNA were recovered using the PCR cloning vector pCR2.1. Sequence analysis showed that 23 bases differed from those in the human ortholog, and these were mostly located in the middle and C-terminal regions (Fig. 1A). However, these 23 single nucleotide differences did not result in any amino acid changes, as shown in Fig. 1B. Like human INI1b, simian INI1 has an alternatively spliced isoform, simian INI1b, with a 9-amino-acid deletion. The sequence of murine INI1b has already been reported and is 100 % identical to the human orthologs of INI1b [1]. These similarities indicate that INI1 is well conserved across species and might retain the same functions in vivo despite the different host specificities of the viruses.
Fig. 1.
Sequence alignment of human and simian INI1. A. DNA sequence alignment. Sequences of hINI1a and hINI1b were retrieved from GenBank (accession numbers U04847.1 and AB017523.1, respectively. “—” indicates an in-frame deletion in the nucleotide sequence of INI1b produced by alternative splicing. Twenty-three single nucleotide differences were identified by alignment, and of those, 21 (91 %) were transitions (*) and two (9 %) were transversions (•). differences in the nucleotide sequences of human INI1a and INI1b are indicated by “○”. B. Amino acid sequence alignment. All of the substitutions are silent mutations. However, two substitution mutations identified between human INI1a and INI1b resulted in amino acid changes, as indicated by “•”. “—” indicates the amino acid deletion in both human and simian INI1b due to the in-frame deletion of nucleotide sequences
Functional analysis of simian INI1a and INI1b
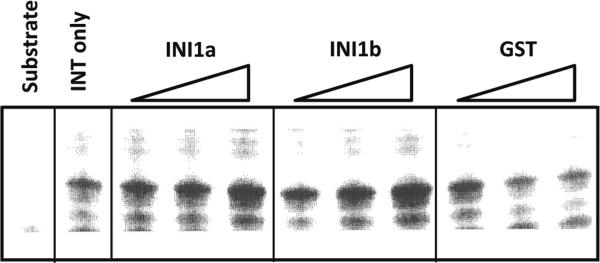
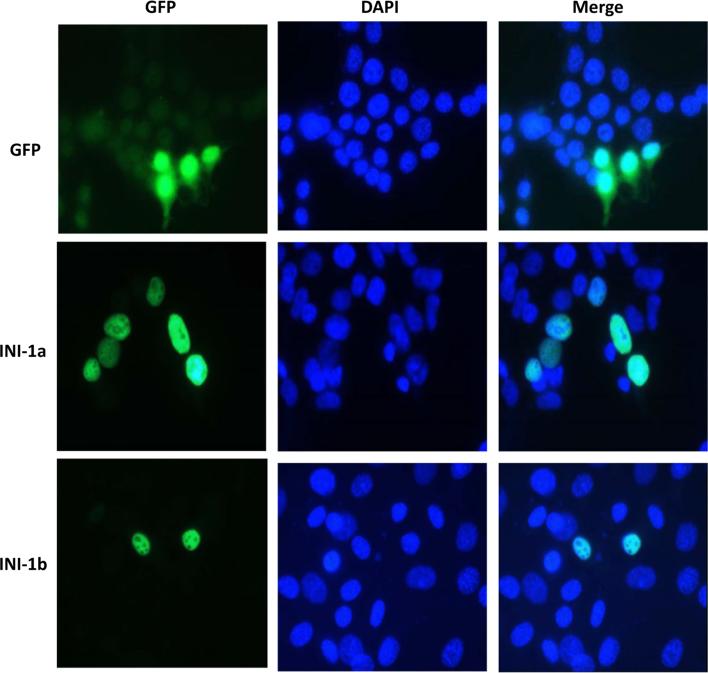
INI1a is known to enhance the activity of HIV-1 INT [4]. However, a functional comparison between INI1a and INI1b has not been carried out. To determine the functional difference in DNA incorporation between INI1a and INI1b, we performed in vitro integration assays with different concentrations of simian INI1a and INI1b using purified HIV-1 INT and radiolabeled oligonucleotides as described [4, 8]. Our data showed that simian INI1b increases HIV-1 INT activity to the same degree as INI1a does (Fig. 2), suggesting that deletion of nine amino acids (GKKTKPNTK) by the alternative splicing of INI1 does not affect INI1 function in proviral integration. We next examined whether subcellular localization of INI1 was affected by deletion of the nine amino acids, since four of them are basic amino acids, lysine (K), and the basic-rich domain is known to play an important role in the nuclear localization of the proteins [3, 6]. Our data showed that the nuclear localization of simian INI1b was indistinguishable from that of INI1a (Fig. 3), indicating that the deleted region in INI1b is not essential for nuclear localization of INI1b.
Fig. 2.
Effect of INI1a and INI1b on INT activity. An in vitro DNA strand transfer assay was performed with increasing concentrations of simian INI1a and INI1b (100, 300, and 600 ng) in the presence of HIV-1 INT. GST was used as a control at the same concentration. The reaction samples were analyzed by 20 % polyacrylamide urea gel electrophoresis and autoradiography
Fig. 3.
Subcellular localization of simian INI1a and INI1b. Plasmids expressing simian INI1a-GFP or INI1b-GFP were introduced into 293T cells by transfection using Superfect (Clontech). Subcellular localization was determined by fluorescence microscopy, and DAPI was employed for nuclear staining
Differential expression of INI1a and INI1b
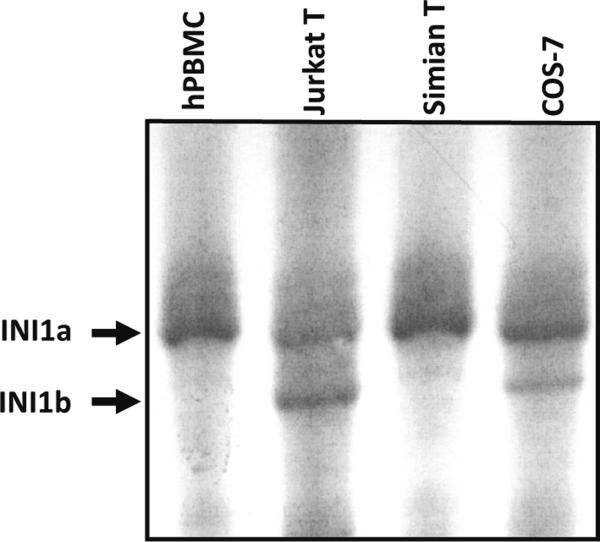
Next, we investigated the specificity of the expression of INI1a and INI1b in different tissues and cell lines. To this end, we purified mRNA from human and simian primary cells as well as from transformed cell lines and performed RT-PCR with specially designed internal primers to examine the expression of INI1a and INI1b. Electrophoresis in a 4 % polyacrylamide urea gel clearly showed the 27-base difference between INI1a (228 bp) and INI1b (201 bp) (Fig. 4). Interestingly, only established cell lines such as Jurkat, LCL8664 (data not shown), and COS-7 expressed both INI1a and INI1b, while PHA-activated hPBMC and IL-2 dependent simian primary T cells expressed only INI1a (Fig. 4). Detection of the 9-amino-acid shorter INI1b only in immortalized cells but not in primary human or simian cells suggests that the 9-amino-acid motif could be associated with transformation/immortalization of cells. If this observation is confirmed by extension of this experiment to other cancer cells and tissues, this might allow INI1 to be used as a diagnostic and prognostic marker for progression of tumors and cancers.
Fig. 4.
Expression of INI1a and INI1b. RT-PCR was performed with mRNA isolated from the indicated cells, and the cDNAs were analyzed by electrophoresis in a 4 % polyacrylamide urea gel and autoradiography. The bands of INI1a (228 bp) and INI1b (201 bp) are indicated by arrows
Acknowledgments
This work was supported by the David Geffen Research Fellowship for AIDS Research (D. P.) and NIH/NIDDK R01 DK099055 (I-W. P).
References
- 1.Bruder CE, Dumanski JP, Kedra D. The mouse ortholog of the human SMARCB1 gene encodes two splice forms. Biochem Biophys Res Commun. 1999;257:886–890. doi: 10.1006/bbrc.1999.0563. [DOI] [PubMed] [Google Scholar]
- 2.Das S, Cano J, Kalpana GV. Multimerization and DNA binding properties of INI1/hSNF5 and its functional significance. J Biol Chem. 2009;284:19903–19914. doi: 10.1074/jbc.M808141200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalderon D, Richardson WD, Markham AF, Smith AE. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- 4.Kalpana GV, Marmon S, Wang W, Crabtree GR, Goff SP. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 5.Kingston RE, Bunker CA, Imbalzano AN. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 6.Lanford RE, Butel JS. Construction and characterization of an SV40 mutant defective in nuclear transport of T antigen. Cell. 1984;37:801–813. doi: 10.1016/0092-8674(84)90415-x. [DOI] [PubMed] [Google Scholar]
- 7.Park IW, Sodroski J. Functional analysis of the vpx, vpr, and nef genes of simian immunodeficiency virus. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;8:335–344. [PubMed] [Google Scholar]
- 8.Pyeon D, Park IW. Interaction between Nef and INI1/SMARCB1 augments replicability of HIV-1 in resting human peripheral blood mononuclear cells. Arch Virol. 2015;160:727–737. doi: 10.1007/s00705-014-2315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts CW, Biegel JA. The role of SMARCB1/INI1 in development of rhabdoid tumor. Cancer Biol Ther. 2009;8:412–416. doi: 10.4161/cbt.8.5.8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsikitis M, Zhang Z, Edelman W, Zagzag D, Kalpana GV. Genetic ablation of Cyclin D1 abrogates genesis of rhabdoid tumors resulting from Ini1 loss. Proc Natl Acad Sci USA. 2005;102:12129–12134. doi: 10.1073/pnas.0505300102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang ZK, Davies KP, Allen J, Zhu L, Pestell RG, Zagzag D, Kalpana GV. Cell cycle arrest and repression of cyclin D1 transcription by INI1/hSNF5. Mol Cell Biol. 2002;22:5975–5988. doi: 10.1128/MCB.22.16.5975-5988.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]