Abstract
Curli are functional amyloid fibers assembled by many Gram-negative bacteria as part of an extracellular matrix that encapsulates the bacteria within a biofilm. A multi-component secretion system ensures the safe transport of the aggregation-prone curli subunits across the periplasm and outer membrane, and coordinates subunit self-assembly into surface-attached fibers. To avoid the buildup of potentially toxic intracellular protein aggregates, the timing and location of the interactions of the different curli proteins are of paramount importance. Here we review the structural and molecular biology of curli biogenesis, with a focus on the recent breakthroughs in our understanding of subunit chaperoning and secretion. The mechanistic insight in the curli assembly pathway will provide tools for new biotechnological applications, and inform the design of targeted inhibitors of amyloid polymerization and biofilm formation.
Keywords: functional amyloid, biofilm matrix, protein secretion, peptide diffusion channel, amyloid chaperone
Biofilm formation
To aid colonization and persistence in host or environmental niches, many bacteria coalesce to form encapsulated communities embedded within a complex hydrated matrix of proteins, nucleic acids, and polysaccharides. These communities, known as biofilms, protect the bacteria within from physical and chemical stresses, such as oxidative damage and desiccation [1,2]. Build-up of these biofilms on household, industrial, and medical equipment results in a potential reservoir of infectious agents recalcitrant to traditional cleaning techniques. Further, biofilm formation by pathogenic bacterial strains within hosts drastically reduces their susceptibility to host immune responses and therapeutic antimicrobial agents and constitutes a major health concern. [3–10].
Biofilm formation is comprised of a series of related steps. Following the reversible cell-surface and/or cell-cell adherence of planktonic microoganisms, filamentous structures known as pili mediate a robust surface attachment and inclusion in an extracellular matrix [11]. In Escherichia coli and Salmonella enterica biofilms, aggregative fibers known as curli (sometimes referred to by their now-obsolete name, tafi) constitute the major proteinaceous component of this extracellular matrix [12]. These matrices promote the formation of floating biofilms (pellicles) at the air-liquid interface of static liquid cultures and can mediate the adhesion of solid cultures to biotic and abiotic surfaces, such as animal and plant tissue, stainless steel, and glass [13–20].
Curli fibers are produced by a dedicated secretion pathway known as the nucleation-precipitation mechanism, or the type VIII secretion system [21,22]. In E. coli, seven curli-specific genes (csg) make up the structural components and assembly apparatus of the curli fibers and are encoded by two divergently transcribed operons (csgBAC and csgDEFG), respectively [21] (Figure 1A). Two recent papers in the field by Goyal et al. (2014) and Evans et al. (2015) have greatly advanced our understanding of the structural components of curli transport and secretion [23,24]. In this work, we review the structural and mechanistic aspects of curli fiber structure and assembly, and the applicability of these exciting findings to the study of bacterial biofilms and human pathogenic amyloids.
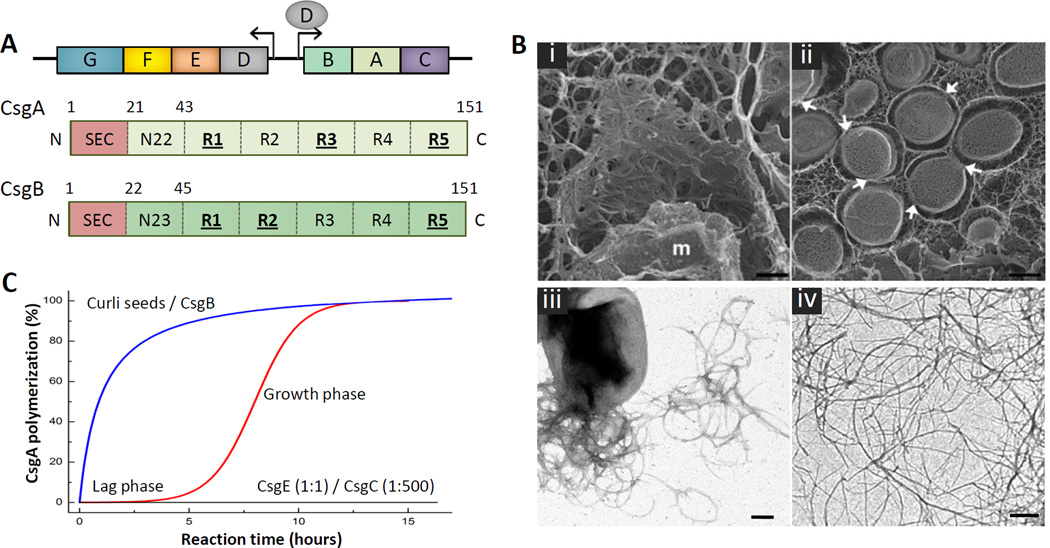
Figure 1. Curli composition and structure.
(A) Organization of the csgBAC and csgDEFG curli operons and architecture of the curli subunits CsgA (light green) and CsgB (dark green). The N-terminal signal sequence (SEC; red) is cleaved after export into the periplasm. The mature subunits contain an N-terminal curli-specific targeting sequence (N22 or N23 in CsgA and CsgB, respectively) that is followed by a pseudo-repeat region (R1 to R5) that forms the amyloidogenic core of the curli subunits (green). Repeats that efficiently self-polymerize in vitro are underscored. (B) Electron microscopy of curli fibers. (i, ii) Freeze-fracture EM of E. coli biofilms shows bacterial cells are encased in a matrix supported by interwoven curli. Bacteria appear to come into contact with the matrix only at discrete locations (white arrows); (m: fractioned bacterial membrane); scale bars 500 nm. (reproduced from [12]). (iii, iv) Transmission EM of individual E. coli cells producing curli fibers (iii), and curli-like fibers grown in vitro from purified CsgA (iv); scale bars: 200 nm. (C) Representation of typical in vitro CsgA polymerization profiles under different conditions. The addition of preformed fibers or the CsgB nucleator removes the lag phase preceding exponential fiber growth (blue curve). In the presence of CsgE (1:1 ratio) or CsgC (1:500 ratio), no CsgA polymerization is observed (black curve) [24].
Curli fibers
Upon visualization by electron microscopy, E. coli-associated curli usually appear as a tangled mass of linear, surface-associated fibers of 4–6 nm width and several microns length (Figure 1B). Within biofilms, curli fibers form an interwoven mesh that supports the extracellular matrix and encapsulate individual cells (Figure 1B). Solid-state NMR experiments have demonstrated that curli fibers make up roughly 85% of the matrix material in curli-associated biofilms [12]. In these matrices, curli frequently appear detached from the embedded cells. It is unclear if this observation represents a regulated process necessary for efficient biofilm matrix formation, or is an artefact of cell desiccation during sample preparation procedures.
Curli are non-covalent heteropolymeric filaments of CsgA and CsgB subunits, present at ratios of approximately 20:1 CsgA:CsgB in in vivo wild-type fibers [25]. Curli belong to a class of stable, ordered protein aggregates known as amyloids [21,26]. Although commonly associated with pathological protein misfolding in human diseases [27–30], a significant body of research now suggests that amyloids are also intentionally produced by a variety of organisms to fulfil important physiological functions, such as regulation of hydrophobicity during fungal reproduction or transcriptional regulation [31–33]. Amyloid filaments are characterized structurally by their ‘cross-β spine’ architecture, in which repeating β-strand units are oriented perpendicular to the fiber axis [34–36]. This repetitive, tightly-ordered packing of β-strands confers high stability and physical robustness to the filaments. Indeed, curli are resistant to proteolytic degradation or dissolution by sodium dodecyl sulphate (SDS), and instead must be subjected to harsh treatment with formic acid or hexafluoroisopropanol (HFIP) in order to depolymerize fibers into individual subunits. The extracellular, in vivo fiber formation can be macroscopically monitored by the staining of bacterial cultures with Congo red, a dye that undergoes a spectral shift upon binding β-rich polymers such as curli and other amyloid species. Though a convenient research tool, precaution is needed due to the nonspecific binding of Congo red to other polymers including biofilm matrix components such as cellulose.
Curli-like fibers can also be formed in vitro, starting from the purified major structural subunit CsgA [37,38]. This process can be monitored by the fluorescence increase from the Thioflavin T (ThT) dye as it binds the forming amyloid structure (Figure 1C). Following an extended lag time that is associated with the formation of a metastable oligomeric nucleus, in vitro fiber formation occurs spontaneously and follows an exponential growth phase. Prior to nucleation, concentrated CsgA samples frequently form amorphous aggregates that remain competent for incorporation into curli-fibers. [37,38]. The lag time preceding fibrillation can be eliminated by the addition of pre-formed curli fragments or purified CsgB protein, which accelerates fiber formation in a process known as heteronucleation [38–40]. This suggests that curli polymerization is a templated process, as is also seen for other amyloid structures. Deletion of csgB in vivo results in CsgA secretion away from the cell in an SDS-soluble, unpolymerized form [39]. However, when csgB− cells are placed in the vicinity of cells encoding csgB but lacking csgA, these soluble CsgA subunits can polymerize on the surface of CsgB(+) cells in a process known as interbacterial complementation [21].
The exact structure of curli fibers has not yet been elucidated with molecular resolution. X-ray fiber diffraction, solid state NMR and electron microscopy data on CsgA and CsgB fibers grown in vitro point to a cross-β architecture that is most consistent with stacked β-helical subunits rather than stacked parallel in-register β-sheet structures as often seen in other amyloids [41,42]. However, it should be noted that in vitro fibers are frequently more variable in width than their wild-type counterparts, and often show 3–4 nm protofilaments branching off from thicker filament bundles (Figure 1B). These observations suggest that in vivo assembled curli may not be fully structurally isomorphous with in vitro grown curli-like fibers [41]. This is consistent with the observation that the presence or absence of periplasmic, non-structural accessory proteins such as CsgE or CsgC in vivo can alter curli morphology [21,43].
The master biofilm regulator CsgD
CsgD is the master transcriptional regulator of curli-associated biofilm assembly, acting as a positive regulator of csgBAC operon to promote the transcription of the curli structural components CsgA and CsgB [44,45]. The csgD gene is itself a member of the csgDEFG operon [44]. CsgD also transcriptionally regulates adrA, a posttranscriptional regulator of cellulose synthesis that controls the synthesis of cyclic diguanylate [46,47]. By controlling both curli and cellulose production, CsgD is able to modulate the levels of both major structural components of biofilms [47–50]. The C-terminal part of CsgD is homologuous to the conserved helix-turn-helix-DNA-binding motif of transcriptional regulators belonging to the LuxR family [44]. CsgD activity is itself highly regulated by a variety of regulatory systems tasked with mediating diverse cellular responses to a variety of cellular stressors. Included among these are the EnvZ/OmpR and CpxA/CpxR two-component systems, in which a sensing kinase prosphorylates a general transcription factor in response to changes in the cellular environment [14,51,52]. One such kinase, EnvZ, phosphorylates OmpR in response to changes in the osmolarity of the extracellular space, which subsequently induces CsgD expression, stimulates porin production, and affects many other downstream tagets [49,53–55]. Similarly, CpxA responds to outer membrane stress resulting from an alkaline pH and overabundance of secreted proteins, by phosphorylating CpxR, which in turn stimulates CsgD expression, amongst other roles [49,56,57]. Additionally, many other proteins have been implicated for their roles in the regulation of curli expression and biofilm formation, including those that regulate cellular responses to temperature and pH [58–60].
The major curli subunit CsgA
CsgA is the major structural subunit of the curli fiber, forming homopolymers of several hundred units long both in vivo and in vitro. The amino acid sequence of CsgA contains three well-described domains; a signal peptide, an N-terminal 22-amino-acid targeting sequence (N22), and the C-terminal amyloid core domain [61] (Figure 1A). Following export into the periplasm through the SecYEG apparatus [21], the signal peptide is proteolytically cleaved, resulting in a 13.1 kDa mature periplasmic CsgA subunit. CsgA subunits then reach the cell surface in a process dependent on the outer membrane (OM) curli secretion pore CsgG [62], and are nucleated by CsgB to assemble extracellular fibers. The N22 sequence is not part of the core amyloid fold, and remains susceptible to proteinase K degradation in the curli fibers [21,63]. When fused to heterologous polypeptides, the N22 sequence is able to direct their secretion to the cell surface, indicating a role for substrate targeting through the curli secretion machinery [62].
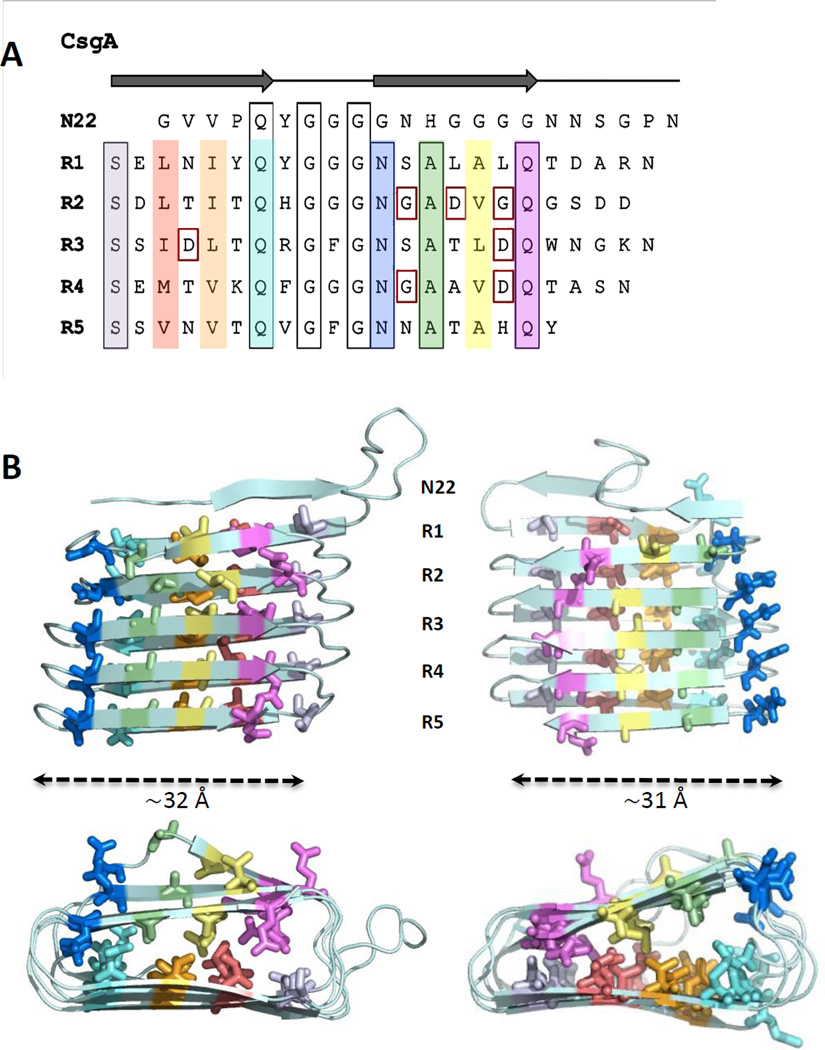
The CsgA amyloid core consists of five repeating units, each with ~30% amino acid identity to one another. These units, designated R1–R5, each contain a common Ser-X5-Gln-X-Gly-X-Gly-Asn-X-Ala-X3-Gln motif that is highly conserved in known homologs from Gammaproteobacteria [38,63] (Figure 2A). Synthetic peptides corresponding to R1 and R5 can efficiently assemble into amyloid-like fibers [38]. Peptides corresponding to the R3 sequence form only short fibrous aggregates, while R2 and R4 peptides cannot form any fibers [38]. No experimental structures are currently available for the CsgA amyloid core. Based on the estimated mass-per-length of individual curli fibers of 1.5 kDa/Å, the lower limit for the length of an individual subunit along the fibrils axis was estimated to be ~9 Å (ie. the width of two stacked β-strands) [41]. More detailed structural information of the CsgA fibril has been provided by solid state NMR experiments. Interresidue (or nearest neighbour) distances between 13C-labelled carbonyls was experimentally found to be ~7 Å separately for all Val, Leu, and Phe residues [41]. These distances were found to be in good overall agreement with computational models based on studies by Tian et al., who used the covariation of amino acids in CsgA homologues to extract predicted amino acid contacts [64]. Simulations designed to find compatible low-energy conformations in CsgA consistently resulted in a predicted β-helical structure, the handedness of which remains unknown [64] (Figure 2B). Further experimental work will be needed to validate these models.
Figure 2. Architecture and 3D model of curli subunit CsgA.
(A) CsgA amino acid sequence aligned to highlight the conserved motifs (black boxes) in N22 and the pseudo-repeat structure in R1 to R5. Residues forming the predicted inner core of the β-helix in the modeled CsgA structures are differentiated in color. Indicated in red squares are gatekeeper residues that reduce aggregation propensity of individual repeats [85]. (B) Structural models of CsgA. Lowest-energy conformations of theoretical CsgA models predicted based on amino acid covariation analysis [64]. The MD simulations and covariance restraints were compatible with both left-handed (left) and right-handed (right) β-helical structures. For both, the width of the β-helical core is around 31 Å. The ’rectangular’ hydrophobic cores are primarily formed by side chains of Ala, Ile, Leu, Met and Val, and colored as in (A). Future experimental characterization is required to validate whether either model is representative of the CsgA structure in the curli fibers.
The nucleator protein CsgB
Similar to CsgA, CsgB’s amino acid sequence contains a signal peptide, an N22 targeting sequence, and a C-terminal amyloid core domain (Figure 1A). Interestingly, unlike its counterpart in CsgA, the N22 sequence on CsgB is not strictly required for biogenesis of wild-type fibers [65]. Like CsgA, the core amyloidogenic domain of CsgB is comprised of five repeating units [25,39]. While R1–R4 contain a Ala-X3-Gln-X-Gly-X2-Asn-X-Ala-X3-Gln motif similar to that in CsgA, R5 is markedly different, instead containing four positively charged amino acids (Lys133, Arg140, Arg147, and Arg151) absent in other repeating units. Deletion of this R5 unit results in a truncated CsgB that no longer associates with the surface of the cell [65], but can still seed CsgA polymerization in vitro [39].
CsgB is also capable of self-assembly into ordered amyloid fibrils [40,65]. However, CsgB homopolymers assemble with a much shorter lag time [40]. In vitro, synthetic CsgB peptides corresponding to R1, R2, and R4 are capable of self-assembly into ordered amyloid fibers, while peptides corresponding to R3 and R5 do not [65] (Figure 1A). In CsgA, deletion of a single repeat does not abrogate in vitro fiber formation, although a CsgA-ΔR5 mutant forms fibers much more slowly than its wild-type counterparts. Conversely, in CsgB, deletion of either R4 or R5 in vivo results in loss of curli assembly on the cell surface, with both CsgA and CsgB being secreted into the surrounding agar [65]. The ability of CsgB to nucleate cell-surface associated curli requires the presence of CsgF, an extracellular accessory protein that is bound to the CsgG secretion pore [66]. In its absence, CsgB is secreted away from the cell, as is CsgA. Thus, it is possible that R4 or R5 are responsible for the mediation of a hitherto undiscovered interaction between CsgB and CsgF.
The amyloid inhibitor CsgC
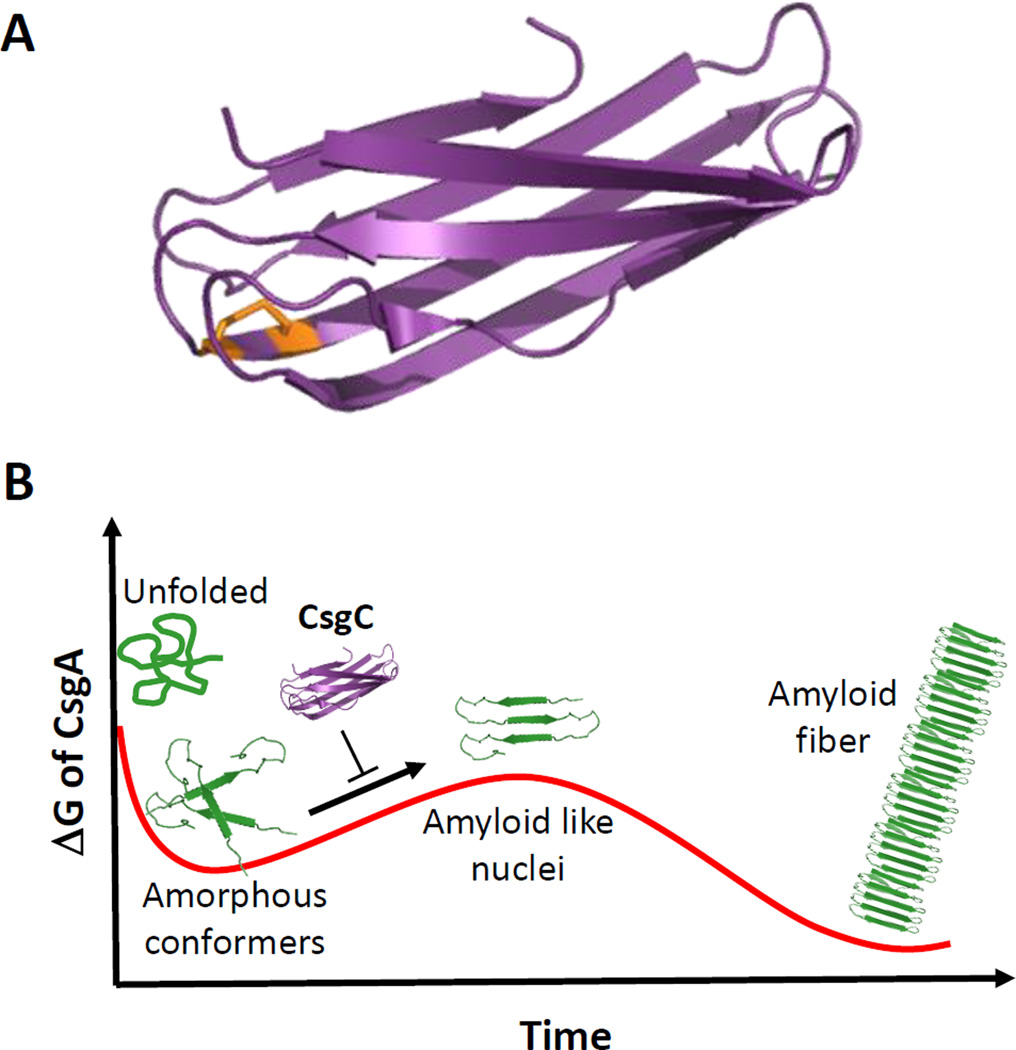
Until recently, the role of the third gene in the csgBAC operon, csgC, has been the subject of intense debate. In the absence of csgC, fibers formed in vivo show an increase in diameter and a complete lack of CsgB incorporation [43]. A recent paper by Evans et al. has provided a breakthrough in our understanding of CsgC function [24]. Based upon the decade-old observation that deletion of the CsgG secretion channel does not result in the cellular toxicity expected from intracellular amyloid aggregation, it was hypothesized that periplasmic factors must exist to prevent premature amyloid formation. To verify the presence of one or more such factors, periplasmic extracts from E. coli cells were added to purified CsgA, which failed to polymerize normally. When the csgC gene was deleted from these cells and the periplasmic extracts tested for amyloid inhibition, it was found that CsgA self-polymerized into fibers [24]. In vitro, purified CsgC is able to completely inhibit CsgA polymerization at substoichiometric molar ratios as low as 1:500. CD and NMR studies indicate CsgA is being held in a β-deficient, SDS-soluble state in presence of CsgC. Thus, it would appear that CsgC acts as a periplasmic chaperone, preventing CsgA nucleation events and ensuring periplasmic pools of CsgA remains in a secretion- and degradation-competent form (Figure 3).
Figure 3. Structure and function of the curli chaperone CsgC.
(A) Cartoon representation of the X-ray structure of oxidized CsgC (PDB 2Y2Y; [67]). The disulfide bond connecting C29 to C31 is shown in orange. (B) Schematic representation of proposed role of CsgC in CsgA chaperoning, showing a predicted energy landscape of CsgA amyloidogenesis where CsgA forms dynamic, amorphous aggregates before assembling into amyloid-like, prefibrillar oligomers. CsgC is believed to avoid periplasmic CsgA amyloidosis by transiently interacting with a prefibrillar CsgA and inhibiting its progression as a template for CsgA fibrillation. Figure adapted from Evans et al. [24].
CsgC’s inhibitory activity is not specific for E. coli CsgA. E. coli CsgC is capable of inhibiting CsgA produced from S. enterica and Citrobacter coseri, which share 75% sequence identity with their E. coli counterpart, at similar molar ratios [24]. Despite sharing only 30% sequence identity with CsgA, CsgB oligomerization can also be inhibited, albeit at higher molar ratios of 1:10 (CsgC:CsgBR1–R4) [24]. Finally, CsgC can also inhibit the Parkinson’s disease-associated protein α-synuclein, at ratios of 1:10 [24]. Sequence analysis of these proteins indicates that a conserved Q-X-G-X1,2-N-X5-Q motif found on all repeating units of CsgA and CsgB and in the solubilizing domain of α-synuclein is likely responsible for CsgC-mediated amyloid inhibition. Amyloids lacking this sequence, such as amyloid-β, are not inhibited by any concentration of CsgC. The molecular mechanism by which CsgC maintains CsgA in this ‘molten oligomeric’ state is still unknown. No stable interaction between CsgC and CsgA has been identified using native gel shift assays [24], suggesting that CsgC may not function as a traditional chaperone. Instead, CsgC may transiently interact with a soluble, oligomeric pool of CsgA to prevent seeding and ordered polymerization. Alternatively, CsgC may induce a structural change within a subpopulation of CsgA, which then biases the entire population away from amyloidogenesis. The structure of CsgC, solved in 2011, demonstrates that CsgC is a soluble, β-rich protein containing an immunoglobulin-like fold and a conserved CxC motif similar to that seen in the redox-active DsbD protein [67] (Figure 3A). Interestingly, all E. coli strains expressing CsgC also contain a free cysteine in the CsgG pore, while species lacking CsgC often have this residue mutated, hinting at a possible role for a CsgG-CsgC interaction in regulation of curli secretion [67].
The secretion channel CsgG
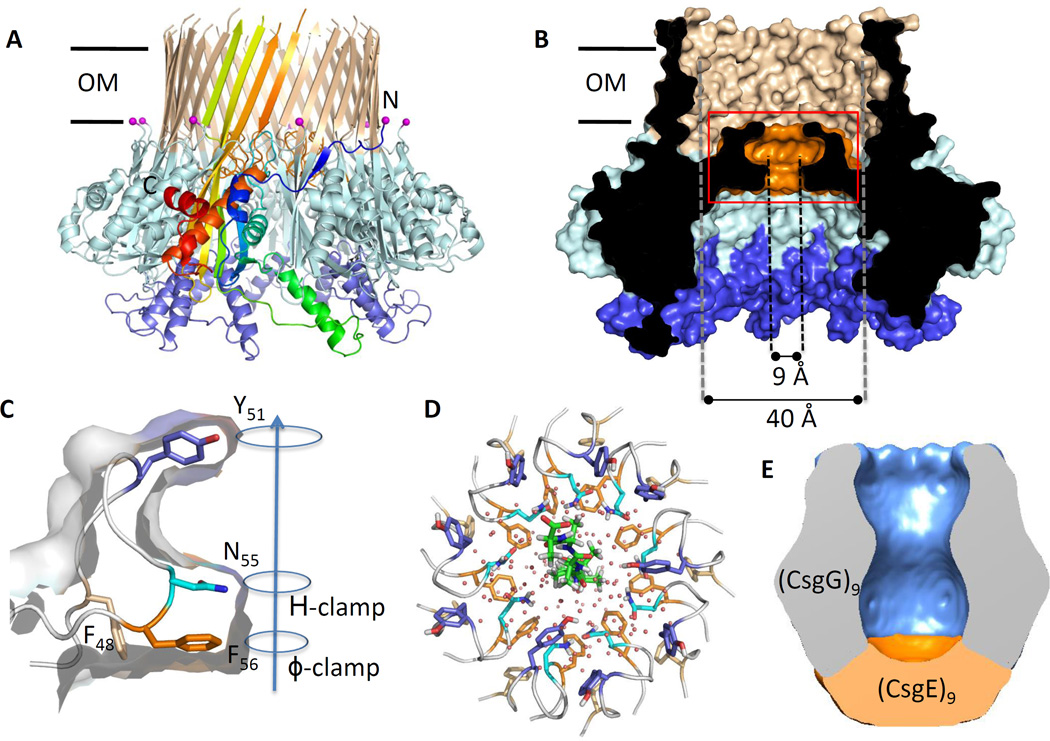
Secretion of CsgA, CsgB and CsgF across the OM requires CsgG, a 262-residue pore-forming lipoprotein [23,62,66,68]. CsgG crystal structures, recently reported by Goyal et al. and Cao et al., have provided insight on its role within the secretion machinery. CsgG forms a nonameric transport complex of 120 Å in diameter and 85 Å in height, that traverses the OM through a 36-stranded β-barrel of 40 Å inner diameter [23,69] (Figure 4A, B). A periplasmic domain of the channel, separated from the transmembrane β-barrel by an iris-like diaphragm, forms a large solvent-accessible cavity of 24,000 Å3. This diaphragm is formed by a conserved 12-residue ‘constriction loop’ (CL) found in each subunit (Figure 4B, C), that together form a funnel-shaped secretion conduit with solvent-excluded diameter of 9 Å (Figure 4B) [23,69]. These minimum pore dimensions are in agreement with those calculated from single channel currents measured from reconstituted CsgG, and would be compatible with the accommodation of one to two (e.g. a looped structure) extended polypeptide segments during substrate passage [23] (Figure 4D). The luminal lining of the constriction is composed of three stacked concentric rings formed by the side chains of residues Y51, N55 and F56 [23,69] (Figure 4C). Strikingly, a structurally equivalent concentric Phe-ring, termed the ϕ-clamp, is seen at the channel entrance of the Anthrax toxin, where it is proposed to facilitate peptide recruitment and progression in the translocation channel ([70,71];Box 1). In CsgG, sequence conservation and directed mutagenesis indicate the stacked configuration of a ϕ-clamp followed by a H-bond donor/acceptor in the channel constriction are required for productive CsgA secretion [23]. The structural data and single-channel recordings infer CsgG forms an ungated peptide diffusion channel [23]. Indeed, elevated concentrations of in vivo CsgG facilitate non-selective diffusive leakage of periplasmic polypeptides. At physiological concentrations CsgG-mediated secretion is specific for CsgA and CsgB and requires the periplasmic factor CsgE [72]. A fundamental question underlying CsgG-mediated polypeptide transport concerns the energy source driving the process. Unlike the chaperone/usher pilus secretion pathway [73], csgB and csgF knockout mutants demonstrate the secretion process is not intrinsically coupled to the polymerization process [21]. Rather, the prevailing data point to a passive, diffusion-based transport mechanism of individual CsgA subunits (Box 1) [23].
Figure 4. The structure of curli secretion channel CsgG.
(A, B) Side and cross-sectional view of CsgG nonamers in ribbon and surface representation, respectively. (PDB: 4UV3; [23]) (A) A single protomer is colored from N- to C-terminus in a gradient from blue to red, respectively. The N-terminal lipidation sites are marked by magenta spheres. (B) Helix 2, the core domain and transmembrane (TM) β-hairpins are shown in blue, light blue and tan, respectively. Helix 2 forms a docking side for a CsgE nonamer (panel D) Abbreviations: OM, outer membrane. (C) Close-up view of the constriction loop of a single CsgG protomer. The channel constriction (boxed region in panel B) is formed of stacked rings F56, N55 and Y51; arrow indicates direction of CsgA transport. H-clamp: conserved H-bond donor/acceptor. (D) Top view of the CsgG channel constriction, with a modelled polyalanine chain residing in the channel, viewed form the extracellular side. (E) 3D cryo-EM reconstruction of the CsgG:CsgE complex. The model shows a nonameric particle comprised of CsgG (blue) and an additional density assigned as a CsgE nonamer (orange), encapsulating a pre-constriction chamber of approximately 24.000 Å3. Figures adapted from Goyal et al. [23].
Box 1. Anthrax toxin, a model peptide diffusion channel.
Channel proteins utlilize a diffusion-based transport mechanism, facilitating substrate passage without the need for a conformational cycle and associated powerstroke from an external energy source (ie. ATP hydrolysis, co-transport or an electrochemical gradient). For channels carrying large, extended polymers such as polypeptides, the substrate threads the pore in smaller diffusive steps that require a rectifying mechanism to avoid trapping the chain in a futile equilibrium of forward and reverse steps [84].
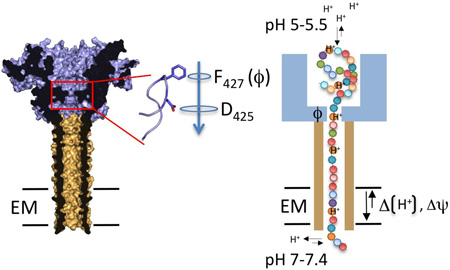
A model peptide diffusion channel is the anthrax protective antigen (PA; Figure I), the pore-forming component of the anthrax toxin complex that also includes edema factor (EF) and lethal factor (LF) [74]. After endocytosis of the toxin by the host cell, PA forms the channel for EF and LF delivery to the host cytosol. Threading of EF and LF (or synthetic peptides) through the PA channel requires an electric potential (ΔΨ) and/or a proton gradient [70,71]. The substrate capture and the processivity of its translocation through the channel critically depend on the presence of a concentric ring of phenylalanines (dubbed ϕ-clamp) at the pore entrance, and a cation selective channel [70]. Together these elements form a Brownian ratchet where diffusive steps of the translocating polypeptide inside the channel, are forward rectified towards the cytosol as a result of the polypeptide protonation and deprotonation at the endosomal (pH 5–5.5) and cytosolic (pH 7–7.4) side of the channel, respectively. The ϕ-clamp and cation-selective channel form a barrier to back-slipping of the translocating chain after it deprotonates at the cytosolic exit of the channel [74].
Figure I. Structure and transport model for the anthrax toxin. Left, cross-section of the solvent-accessible surface of the anthrax toxin protective antigen (PA; PDB: 3C9J, [71]). PA forms a heptameric β-barrel (shown in tan) that traverses the endosomal membrane (EM) and provides the conduit for translocation of the lethal factor and edema factor (not shown) that are bound to PA’s endosomal domain (shown in blue). A concentric 7-membered ring composed of F427 (ϕ-clamp) and D425 form an inner constriction (boxed region) preceding the PA polypeptide diffusion channel. Arrow indicates direction of polypeptide transport. Right, schematic representation of the protein translocation mechanism operated by PA. Amino acids along the length of a translocating polypeptide are represented as spheres. The ϕ-clamp (labelled ϕ) aids substrate recruitment and the forward rectification of the polypeptide’s diffusion in the translocation channel, which is driven by a pH gradient over the EM.
The secretion factor CsgE
CsgE is a non-secreted accessory factor essential to curli biogenesis. When added to purified CsgA monomers at 1:1 stoichiometric ratios, mature CsgE purified as a soluble protein from the periplasm completely inhibits in vitro fiber formation (Figure 1C) [72]. This finding suggests the presence of a direct interaction between individual CsgE and CsgA subunits, rather than an interaction with early fibrillation intermediates as is hypothesized to occur during CsgC substoichiometric chaperone activity [24]. In cells lacking CsgE, a paucity of CsgA reaching the cell surface drastically reduces curli production [43,72]. At elevated levels of OM-localized CsgG, this secretion deficiency in csgE mutants can be rescued, though this is associated with a more generalized, non-specific leakage of periplasmic proteins [72]. Co-overexpression of CsgE suppresses this non-specific efflux and restores substrate specificity in CsgG towards CsgA, CsgB, or heterologous polypetides fused to CsgA’s N22 region [72]. A direct interaction between CsgG and CsgE has also been seen by co-immunoprecipitation [62]. Thus, these data argue that CsgE serves as a secretion adaptor that interacts with curli subunits during their periplasmic transit, and helps target them to the CsgG channel (see below, Figure 4E).
Goyal et al. confirmed the existence of a dynamic CsgG:CsgE complex that is present in a 9:9 stoichiometry [23]. Size-exclusion chromatography and cryo-electron microscopy show purified CsgE is present in an equilibrium of a low molecular weight species (likely monomers) and a concave, cup-like nonameric complex. When bound to CsgG, this CsgE nonamer serves as a capping adaptor that closes off the periplasmic vestibule of the CsgG secretion channel, creating a pre-constriction chamber of ~24.000 Å3 [23] (Figure 4E). Single channel recordings show the binding of the CsgE nonamer to the CsgG channel results in the complete silencing of ion conductance and provides a molecular explanation to CsgE’s gating of non-specific influx and efflux seen in vivo at elevated CsgG concentrations [23]. Single channel current traces show an equilibrium of discrete binding and dissociation steps around 10nM CsgE, and also purified CsgG:CsgE complexes readily exchange with free CsgG and CsgE nonamers; both suggesting that CsgG and CsgE are not in an obligate, constitutive interaction. It is not currently known whether CsgE and CsgG form an intricate complex in vivo or whether consecutive cycles of CsgE binding and unbinding events are part of the translocation mechanism.
The assembly factor CsgF
CsgF represents a third component to the curli secretion apparatus. The protein reaches the periplasm via the SEC pathway, after which mature CsgF (12.9 KDa) is found as a surface-exposed protein in a CsgG-dependent manner [66]. In presence of CsgG, CsgF fractionates with the OM, and co-immunoprecipitation experiments show the two proteins are in direct contact [62]. Unlike CsgE, CsgF is expected to form a constitutive complex with the CsgG translocation channel. Curli assembly phenotypes of csgF deletion mutants resemble those observed for csgB mutants, where CsgA accumulates as a soluble protein in the extracellular environment. This secreted CsgA is assembly-competent and able to form curli-like fibers on a CsgB recipient cell [21]. Nenninger et al. showed that CsgB is no longer found as a surface associated protein in absence of CsgF and is then no longer able to exert its role as a curli nucleator [66]. Thus, available data demonstrate CsgF is non-essential for productive subunit secretion, but rather suggest the protein forms a coupling factor between CsgA secretion and extracellular polymerization into curli fibers by coordinating or chaperoning the nucleating function of the CsgB subunit.
Integrated model for curli secretion and assembly
The prevailing biochemical, genetic and structural information can be integrated into a mechanistic model for CsgA secretion and curli assembly (Figure 5, Key Figure). Though the tentative model is compatible with available data and allows for the iterative secretion of polypeptide chains across the non-energized OM, several questions are unanswered and/or await experimental validation (Outstanding Questions Box). The proposed pathway and accompanying discussion should therefore be seen as a working model only.
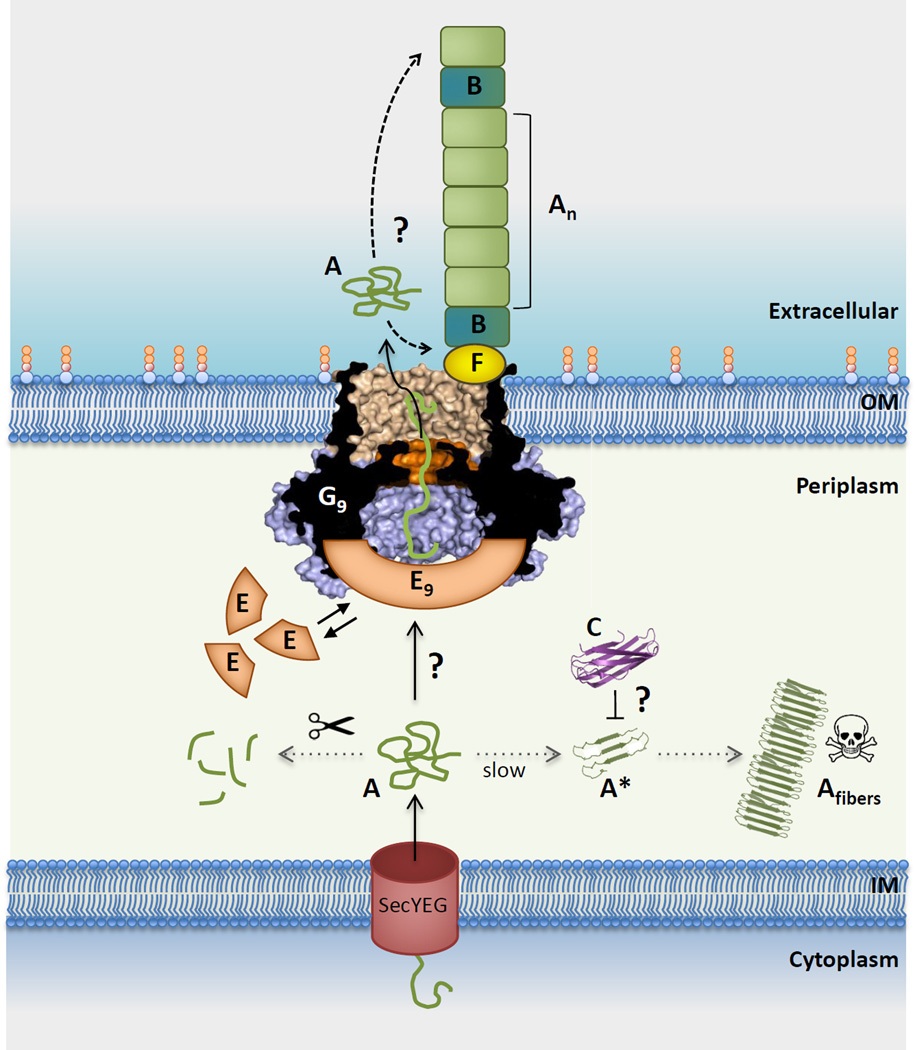
Figure 5. Integrated model for curli subunit secretion.
CsgA (A) subunits enter the periplasm via the SecYEG translocon, from where they progress to the cell surface via the curli transporter CsgG (G) or are proteolytically degraded (left dotted line). Premature polymerization of CsgA in the periplasm (right dotted line) is subverted by CsgC (C), probably by the binding and neutralization of early assembly intermediates (labeled A*). CsgG forms a nonameric complex (G9) that acts as a non-specific peptide diffusion channel. A nonameric CsgE complex (E9) acts as specificity and secretion factor to the CsgG channel and forms a capping structure to a pre-constriction cavity in the CsgG complex. Recruitment and (partial) enclosure of CsgA in the pre-constriction cavity is proposed to create an entropy gradient over the channel that favors CsgA’s outward diffusion as an unfolded, soluble polypeptide. Once secreted, curli fiber formation and elongation is templated by CsgB (B), in a CsgF (F)-dependent manner. It is unknown whether fibers grow from the proximal or distal end (dashed arrows). Also, the sequence that leads to the assembly of the CsgF:CsgG:CsgE:CsgA secretion complex, or its stoichiometry (i.e. G9:E9 + n CsgA subunits) are currently unknown [23]), with three possible scenarios as follows: (1) does an encounter of CsgE and CsgA in the periplasm lead to their docking onto CsgG?; (2) or does CsgG itself recruit CsgA prior to the binding of the capping CsgE nonamer?; (3) or are CsgG and CsgE in a constitutive complex that is dynamically gated in function of CsgA binding? Further experimental and structural work is needed to resolve these mechanistic questions in the curli secretion pathway. Abbreviations: IM, inner membrane; OM, outer membrane.
Outstanding Questions Box.
What is the mode of action of CsgC? Does CsgC’s prevention of periplasmic CsgA amyloidosis result from an on pathway function by interacting with all subunits destined for secretion, or does this represent the salvage or neutralization of off-pathway subunits transitioning to polymerization nuclei? How does this work?
What is the composition, structure and order of assembly of the transient CsgF:CsgG:CsgE:CsgA secretion complex? At what point and how is the CsgA substrate recruited to the complex?
Does the secretion adaptor CsgE cycle between a periplasmic and CsgG-bound form, or does it form a constitutive complex with the CsgG channel? How is this coordinated with subunit recruitment?
How are secreted curli subunits incorporated into surface-bound curli fibers? Does fiber extension occur distal or proximal to the secretion channel?
What is the structure of the CsgG:CsgF complex and how does CsgF perform its role as assembly factor coordinating CsgG-mediated subunit secretion and CsgB-mediated curli nucleation?
Is the minor curli subunit CsgB found throughout the curli fiber or only at the base of the fiber? CsgA and CsgB are found at approximate 20:1 ratios, but the distribution of CsgB within the curli fiber is unclear.
What is the ultrastructure of native curli fibers? Curli morphology has been found to alter upon loss of non-structural components in the curli secretion machinery such as CsgE and CsgC. Are the altered curli morphologies a result of disrupted secretion and polymerization kinetics, or do CsgE and/or CsgC alter the pre-assembly conformation of secreting CsgA subunits?
Upon translocation into the periplasm via the Sec translocon, CsgA is expected to interact with the periplasmic factor CsgE, which destines the subunits to the CsgG translocation pore [72]. In case of secretion defects (eg. as seen in csgG or csgE mutants) a potentially toxic periplasmic accumulation of the pro-amylodogenic subunits is prevented by means of a rapid proteolytic degradation [68]. Additionally, the periplasmic component CsgC performs a chaperoning function that suppresses the formation or function of nucleating CsgA oligomers that would otherwise set off a mislocalized, periplasmic fibrillation of nascent subunits [24].
When CsgA reaches the CsgG:CsgF:CsgE secretion complex, the ϕ-clamp at the entrance of the CsgG channel constriction is expected to facilitate its binding into the channel lumen, in analogy to what is observed in the anthrax protective antigen (PA) [70,74] (Box1). Whether the recruitment into the channel lumen is directional or involves specific sequence elements in CsgA is presently unknown. At any given time, an ungated channel constriction as seen in the X-ray structures would be compatible with the binding of a maximum of 4 to 8 residues (ie. in an extended, α-helical or hairpin conformation) in the passing substrate [23]. Passage of the entire subunit (137 residues in E. coli CsgA) thus requires a stepwise Brownian diffusion of this interaction window along the length of the chain. In PA, such Brownian diffusion is forward rectified by means of a pH gradient (Box 1). In case of CsgG, where no such electrochemical gradients can operate due to its location at the outer membrane, the forward rectification of the substrate diffusion is proposed to depend on an entropy gradient across the channel, in a process that involves the secretion factor CsgE. In addition to its substrate targeting or recruiting function, CsgE is seen to form a capping structure that gives rise to an extensive pre-constriction chamber in the CsgG translocator, which would be compatible with at least a partial entrapment of the secretion substrate [23] (Figure 5, Key Figure). Together, such CsgE-mediated substrate targeting and enclosure at the secretion complex are proposed to raise an entropic potential from the periplasm to the extracellular milieu. In our model, we expect the initial cost of engaging CsgA to the secretion pathway to be compensated by binding energy released during assembly of the CsgG:CsgF:CsgE:CsgA secretion complex. In a scenario where periplasmic CsgA would be prevented from attaining its full conformational freedom by binding to CsgE and/or CsgG as it emerges from the Sec translocon, this would create a net positive ΔS across the cell envelope that would drive secretion of nascent CsgA. Whether the CsgE:CsgA interaction is through a periplasmic pool of CsgE, CsgA is directly recruited to a CsgG:CsgF:CsgE complex, or it is the binding of CsgA to CsgG:CsgF that recruits CsgE cannot presently be determined (Figure 5, Key Figure).
Under native conditions, secreted CsgA rapidly incorporates into growing curli at the cell surface. This process depends both on the nucleating action of CsgB, as well as on CsgF, which acts as an interface between the secretion pore, CsgB and/or the growing curli fibers. Whether subunit addition occurs at a distal or proximal side relative to the secretion pore is currently unknown. Though the accumulation of unfolded extracellular CsgA in csgF and csgB mutants demonstrates that substrate secretion is not intrinsically coupled to its polymerization on the cell surface, a model in which proximal polymerization at the exit of the secretion pore facilitates substrate diffusion under physiological conditions cannot be excluded based on available data.
Concluding remarks
Amyloidogenic proteins readily self-assemble into fibers that are thermodynamically favorable and highly resistant to chemical and physical perturbations [21,37,75–79]. These properties makes the amyloid architecture particularly fit for the fiber formation in the harsh extracellular environment encountered by bacteria. The unique secretion and assembly properties of the curli biogenesis pathway are rapidly gaining interest for biotechnological applications [80–82]. A detailed understanding of the mechanisms of curli biogenesis in bacteria will not only aid the optimization of these applications, but is anticipated to aid the design of potential inhibitors or regulators that can be used as therapeutics to prevent biofilm formation in case of pathogenic bacteria. Finally, to avoid potential cytotoxic species associated with amyloidogenesis, the curli assembly pathway appears to employ a unique nucleation safeguard in the periplasm, CsgC [24]. The molecular principles underlying such amyloid chaperoning function can be of interest with respect to future treatments of pathological amyloid deposits seen in human wasting diseases. Although their molecular mode of action remains to be determined, small compound inhibitors of CsgA polymerization have indeed been described [83].
Trends Box.
CsgC acts as a chaperone to prevent or neutralize premature, periplasmic amyloidosis of curli subunits. The protein does so at low substoichiometric ratios and in absence of ATP.
The CsgG lipoprotein forms a composit transmembrane β-barrel. Transition from a soluble prepore to the outer membrane-bound pore conformation entails nonamerization and extension of two adjacent β-hairpins per subunit.
The CsgG nonamer forms a constitutive peptide diffusion channel that cooperates with CsgE to expel curli subunits in an entropy-driven process.
CsgE forms a periplasmic, nonameric secretion adaptor that binds and caps a preconstriction chamber in the curli transporter CsgG.
CsgF acts as a CsgG-bound curli assembly factor that coordinates the function of the curli nucleator CsgB with CsgG’s secretion of the major curli subunit CsgA.
Acknowledgements
HR acknowledges financial support by VIB project grant PRJ9 and ERC grant 649082 BAS-SBBT. SH acknowledges financial support by National Institutes of Health RO1 grants AI099099 and AI048689.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hall-Stoodley L, Stoodley P. Evolving concepts in biofilm infections. Cell. Microbiol. 2009;11:1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 2.Branda SS, et al. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Anderson GG, O’Toole GA. Innate and induced resistance mechanisms of bacterial biofilms. Curr. Top. Microbiol. Immunol. 2008;322:85–105. doi: 10.1007/978-3-540-75418-3_5. [DOI] [PubMed] [Google Scholar]
- 4.Høiby N, et al. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents. 2010;35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert P, et al. The physiology and collective recalcitrance of microbial biofilm communities. Adv. Microb. Physiol. 2002;46:202–256. [PubMed] [Google Scholar]
- 6.Otter JA, et al. Surface-attached cells, biofilms and biocide susceptibility: implications for hospital cleaning and disinfection. J. Hosp. Infect. 2015;89:16–27. doi: 10.1016/j.jhin.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Fux CA, et al. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 8.Costerton JW, et al. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 9.Van Houdt R, Michiels CW. Biofilm formation and the food industry, a focus on the bacterial outer surface. J. Appl. Microbiol. 2010;109:1117–1131. doi: 10.1111/j.1365-2672.2010.04756.x. [DOI] [PubMed] [Google Scholar]
- 10.Hall-Stoodley L, et al. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 11.Beloin C, et al. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 2004;51:659–674. doi: 10.1046/j.1365-2958.2003.03865.x. [DOI] [PubMed] [Google Scholar]
- 12.Hung C, et al. Escherichia coli biofilms have an organized and complex extracellular matrix structure. mBio. 2013;4 doi: 10.1128/mBio.00645-13. e00645–00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olsen A, et al. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 14.Vidal O, et al. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 1998;180:2442–2449. doi: 10.1128/jb.180.9.2442-2449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryu JH, et al. Attachment and biofilm formation on stainless steel by Escherichia coli O157:H7 as affected by curli production. Lett. Appl. Microbiol. 2004;39:359–362. doi: 10.1111/j.1472-765X.2004.01591.x. [DOI] [PubMed] [Google Scholar]
- 16.Barak JD, et al. Salmonella enterica virulence genes are required for bacterial attachment to plant tissue. Appl. Environ. Microbiol. 2005;71:5685–5691. doi: 10.1128/AEM.71.10.5685-5691.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goulter-Thorsen RM, et al. CsgA production by Escherichia coli O157:H7 alters attachment to abiotic surfaces in some growth environments. Appl. Environ. Microbiol. 2011;77:7339–7344. doi: 10.1128/AEM.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cookson AL, et al. The role of type 1 and curli fimbriae of Shiga toxin-producing Escherichia coli in adherence to abiotic surfaces. Int. J. Med. Microbiol. 2002;292:195–205. doi: 10.1078/1438-4221-00203. [DOI] [PubMed] [Google Scholar]
- 19.Uhlich GA, et al. Analyses of the red-dry-rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl. Environ. Microbiol. 2006;72:2564–2572. doi: 10.1128/AEM.72.4.2564-2572.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhlich GA, et al. Escherichia coli serotype O157:H7 retention on solid surfaces and peroxide resistance is enhanced by dual-strain biofilm formation. Foodborne Pathog. Dis. 2010;7:935–943. doi: 10.1089/fpd.2009.0503. [DOI] [PubMed] [Google Scholar]
- 21.Chapman MR, et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desvaux M, et al. Secretion and subcellular localizations of bacterial proteins: a semantic awareness issue. Trends Microbiol. 2009;17:139–145. doi: 10.1016/j.tim.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Goyal P, et al. Structural and mechanistic insights into the bacterial amyloid secretion channel CsgG. Nature. 2014;516:250–253. doi: 10.1038/nature13768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans ML, et al. The bacterial curli system possesses a potent and selective inhibitor of amyloid formation. Mol. Cell. 2015;57:445–55. doi: 10.1016/j.molcel.2014.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White AP, et al. Structure and characterization of AgfB from Salmonella enteritidis thin aggregative fimbriae. J. Mol. Biol. 2001;311:735–749. doi: 10.1006/jmbi.2001.4876. [DOI] [PubMed] [Google Scholar]
- 26.Zhou Y, et al. Bacterial amyloids. Methods Mol. Biol. 2012;849:303–320. doi: 10.1007/978-1-61779-551-0_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irvine GB, et al. Protein aggregation in the brain: the molecular basis for Alzheimer’s and Parkinson’s diseases. Mol. Med. 2008;14:451–464. doi: 10.2119/2007-00100.Irvine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breydo L, et al. A-synuclein misfolding and Parkinson’s disease. Biochim. Biophys. Acta. 2012;1822:261–285. doi: 10.1016/j.bbadis.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Zuccato C, et al. Molecular Mechanisms and Potential Therapeutical Targets in Huntington’s Disease. Physiol. Rev. 2010;90:905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 30.Hardy J, Selkoe DJ. The Amyloid Hypothesis of Alzheimer’s Disease: Progress and Problems on the Road to Therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 31.Blanco LP, et al. Diversity, biogenesis and function of microbial amyloids. Trends Microbiol. 2012;20:66–73. doi: 10.1016/j.tim.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fowler DM, et al. Functional amyloid - from bacteria to humans. Trends Biochem. Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 33.Badtke MP, et al. Functional amyloids signal their arrival. Sci. Signal. 2009;2:pe43. doi: 10.1126/scisignal.280pe43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sunde M, et al. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 35.Nelson R, et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sunde M, Blake C. The structure of amyloid fibrils by electron microscopy and X-ray diffraction. Adv. Protein Chem. 1997;50:123–159. doi: 10.1016/s0065-3233(08)60320-4. [DOI] [PubMed] [Google Scholar]
- 37.Dueholm MS, et al. Fibrillation of the major curli subunit CsgA under a wide range of conditions implies a robust design of aggregation. Biochemistry. 2011;50:8281–8290. doi: 10.1021/bi200967c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, et al. In vitro polymerization of a functional Escherichia coli amyloid protein. J. Biol. Chem. 2007;282:3713–3719. doi: 10.1074/jbc.M609228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hammer ND, et al. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12494–12499. doi: 10.1073/pnas.0703310104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shu Q, et al. The E. coli CsgB nucleator of curli assembles to β-sheet oligomers that alter the CsgA fibrillization mechanism. Proc. Natl. Acad. Sci. U. S. A. 2012;109:6502–6507. doi: 10.1073/pnas.1204161109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shewmaker F, et al. The functional curli amyloid is not based on in-register parallel beta-sheet structure. J. Biol. Chem. 2009;284:25065–25076. doi: 10.1074/jbc.M109.007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toyama BH, Weissman JS. Amyloid structure: conformational diversity and consequences. Annu. Rev. Biochem. 2011;80:557–585. doi: 10.1146/annurev-biochem-090908-120656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibson DL, et al. AgfC and AgfE facilitate extracellular thin aggregative fimbriae synthesis in Salmonella enteritidis. Microbiology. 2007;153:1131–1140. doi: 10.1099/mic.0.2006/000935-0. [DOI] [PubMed] [Google Scholar]
- 44.Hammar M, et al. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- 45.Zakikhany K, et al. Unphosphorylated CsgD controls biofilm formation in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2010;77:771–786. doi: 10.1111/j.1365-2958.2010.07247.x. [DOI] [PubMed] [Google Scholar]
- 46.Simm R, et al. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 47.Römling U, et al. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 2000;36:10–23. doi: 10.1046/j.1365-2958.2000.01822.x. [DOI] [PubMed] [Google Scholar]
- 48.Brombacher E, et al. The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiol. 2003;149:2847–2857. doi: 10.1099/mic.0.26306-0. [DOI] [PubMed] [Google Scholar]
- 49.Prigent-Combaret C, et al. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J. Bacteriol. 2001;183:7213–7223. doi: 10.1128/JB.183.24.7213-7223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerstel U, Römling U. The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res. Microbiol. 2003;154:659–667. doi: 10.1016/j.resmic.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 51.Jubelin G, et al. CpxR/OmpR Interplay Regulates Curli Gene Expression in Response to Osmolarity in Escherichia coli. J. Bacteriol. 2005;187:2038–2049. doi: 10.1128/JB.187.6.2038-2049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dorel C, et al. Involvement of the Cpx signal transduction pathway of E. coli in biofilm formation. FEMS Microbiol. Lett. 1999;178:169–175. doi: 10.1111/j.1574-6968.1999.tb13774.x. [DOI] [PubMed] [Google Scholar]
- 53.Russo FD, Silhavy TJ. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J. Mol. Biol. 1991;222:567–580. doi: 10.1016/0022-2836(91)90497-t. [DOI] [PubMed] [Google Scholar]
- 54.Huang KJ, et al. Phosphorylation stimulates the cooperative DNA-binding properties of the transcription factor OmpR. Proc.Natl.Acad.Sci.U.S.A. 1997;94:2828–2832. doi: 10.1073/pnas.94.7.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mizuno T, Mizushima S. Signal transduction and gene regulation through the phosphorylation of two regulatory components: the molecular basis for the osmotic regulation of the porin genes. Mol. Microbiol. 1990;4:1077–1082. doi: 10.1111/j.1365-2958.1990.tb00681.x. [DOI] [PubMed] [Google Scholar]
- 56.Ma Q, Wood TK. OmpA influences Escherichia coli biofilm formation by repressing cellulose production through the CpxRA two-component system. Environ. Microbiol. 2009;11:2735–2746. doi: 10.1111/j.1462-2920.2009.02000.x. [DOI] [PubMed] [Google Scholar]
- 57.Hunke S, et al. Signal integration by the Cpx-envelope stress system. FEMS Microbiol. Lett. 2012;326:12–22. doi: 10.1111/j.1574-6968.2011.02436.x. [DOI] [PubMed] [Google Scholar]
- 58.Brombacher E, et al. Gene Expression Regulation by the Curli Activator CsgD Protein: Modulation of Cellulose Biosynthesis and Control of Negative Determinants for Microbial Adhesion. J. Bacteriol. 2006;188:2027–2037. doi: 10.1128/JB.188.6.2027-2037.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ishihama A. Prokaryotic genome regulation: multifactor promoters, multitarget regulators and hierarchic networks. FEMS Microbiol. Rev. 2010;34:628–645. doi: 10.1111/j.1574-6976.2010.00227.x. [DOI] [PubMed] [Google Scholar]
- 60.Liu Z, et al. CsgD regulatory network in a bacterial trait-altering biofilm formation. Emerg. Microbes Infect. 2014;3:e1. doi: 10.1038/emi.2014.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Collinson SK, et al. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J. Bacteriol. 1991;173:4773–4781. doi: 10.1128/jb.173.15.4773-4781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson LS, et al. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol. Microbiol. 2006;59:870–881. doi: 10.1111/j.1365-2958.2005.04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Collinson SK, et al. Structural predictions of AgfA, the insoluble fimbrial subunit of Salmonella thin aggregative fimbriae. J. Mol. Biol. 1999;290:741–756. doi: 10.1006/jmbi.1999.2882. [DOI] [PubMed] [Google Scholar]
- 64.Tian P, et al. Structure of a functional amyloid protein subunit computed using sequence variation. J. Am. Chem. Soc. 2015;137:22–25. doi: 10.1021/ja5093634. [DOI] [PubMed] [Google Scholar]
- 65.Hammer ND, et al. The C-terminal repeating units of CsgB direct bacterial functional amyloid nucleation. J. Mol. Biol. 2012;422:376–389. doi: 10.1016/j.jmb.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nenninger AA, et al. Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc. Natl. Acad. Sci. U. S. A. 2009;106:900–905. doi: 10.1073/pnas.0812143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taylor JD, et al. Atomic Resolution Insights into Curli Fiber Biogenesis. Structure. 2011;19:1307–1316. doi: 10.1016/j.str.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Loferer H, et al. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol. Microbiol. 1997;26:11–23. doi: 10.1046/j.1365-2958.1997.5231883.x. [DOI] [PubMed] [Google Scholar]
- 69.Cao B, et al. Structure of the nonameric bacterial amyloid secretion channel. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E5439–E5444. doi: 10.1073/pnas.1411942111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krantz BA, et al. A phenylalanine clamp catalyzes protein translocation through the anthrax toxin pore. Science. 2005;309:777–781. doi: 10.1126/science.1113380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiang J, et al. Atomic structure of anthrax protective antigen pore elucidates toxin translocation. Nature. 2015;521:545–549. doi: 10.1038/nature14247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nenninger AA, et al. CsgE is a curli secretion specificity factor that prevents amyloid fibre aggregation. Mol. Microbiol. 2011;81:486–499. doi: 10.1111/j.1365-2958.2011.07706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Phan G, et al. Crystal structure of the FimD usher bound to its cognate FimC–FimH substrate. Nature. 2011;474:49–53. doi: 10.1038/nature10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feld GK, et al. Ratcheting up protein translocation with anthrax toxin. Protein Sci. 2012;21:606–624. doi: 10.1002/pro.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dueholm MS, et al. Functional amyloid in Pseudomonas. Mol. Microbiol. 2010;77:1009–1020. doi: 10.1111/j.1365-2958.2010.07269.x. [DOI] [PubMed] [Google Scholar]
- 76.Jordal PB, et al. Widespread Abundance of Functional Bacterial Amyloid in Mycolata and Other Gram-Positive Bacteria. Appl. Environ. Microbiol. 2009;75:4101–4110. doi: 10.1128/AEM.02107-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Larsen P, et al. Amyloid adhesins are abundant in natural biofilms. Environ. Microbiol. 2007;9:3077–3090. doi: 10.1111/j.1462-2920.2007.01418.x. [DOI] [PubMed] [Google Scholar]
- 78.Romero D, et al. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schwartz K, et al. Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS Pathog. 2012;8:e1002744. doi: 10.1371/journal.ppat.1002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Van Gerven N, et al. Pili and Flagella: Biology, Structure, and Biotechnological Applications. Prog. Mol. Biol. Transl. Sci. 2011;103:21–72. doi: 10.1016/B978-0-12-415906-8.00005-4. [DOI] [PubMed] [Google Scholar]
- 81.Chen AY, et al. Synthesis and patterning of tunable multiscale materials with engineered cells. Nat. Mater. 2014;13:515–523. doi: 10.1038/nmat3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sivanathan V, Hochschild A. Generating extracellular amyloid aggregates using E. coli cells. Genes Dev. 2012;26:2659–2667. doi: 10.1101/gad.205310.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cegelski L, et al. Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat. Chem. Biol. 2009;5:913–919. doi: 10.1038/nchembio.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Van den Broeck I, et al. Insights in peptide diffusion channels from the bacterial amyloid secretor CsgG. Channels. 2015;9:65–67. doi: 10.1080/19336950.2015.1017172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang X, et al. Gatekeeper residues in the major curlin subunit modulate bacterial amyloid fiber biogenesis. Proc. Natl. Acad. Sci. U. S. A. 2010;107:163–168. doi: 10.1073/pnas.0908714107. [DOI] [PMC free article] [PubMed] [Google Scholar]