Summary
A collection of tagged deletion mutant strains was created in Streptococcus mutans UA159 to facilitate investigation of the aciduric capability of this oral pathogen. Gene-specific barcoded deletions were attempted in 1432 open reading frames (representing 73% of the genome), and resulted in the isolation of 1112 strains (56% coverage) carrying deletions in distinct non-essential genes. Since S. mutans virulence is predicated upon the ability of the organism to survive an acidic pH environment, form biofilms on tooth surfaces, and out-compete other oral microflora, we assayed individual mutant strains for the relative fitness of the deletion strain, compared to the parent strain, under acidic and oxidative stress conditions, as well as for their ability to form biofilms in glucose- or sucrose-containing medium. Our studies revealed a total of 51 deletion strains with defects in both aciduricity and biofilm formation. We have also identified 49 strains whose gene deletion confers sensitivity to oxidative damage and deficiencies in biofilm formation. We demonstrate the ability to examine competitive fitness of mutant organisms using the barcode tags incorporated into each deletion strain to examine the representation of a particular strain in a population. Co-cultures of deletion strains were grown either in vitro in a chemostat to steady-state values of pH 7 and 5 or in vivo in an animal model for oral infection. Taken together, this data represents a mechanism for assessing the virulence capacity of this pathogenic microorganism and a resource for identifying future targets for drug intervention to promote healthy oral microflora.
Keywords: genomics, pathogenesis, physiology
Introduction
Streptococcus mutans is found in the vast majority of humans living in industrialized nations in which refined sugar consumption is high, with a correspondingly high prevalence of dental caries (Nyvad, et al. 2013; Takahashi & Nyvad 2008; Takahashi & Nyvad 2011). S. mutans uses sugars in the human diet to produce extracellular glucans that act to irreversibly bind the organism to tooth surfaces. S. mutans also ferments sugars to produce organic acids, which accumulate at the tooth surface, resulting in a localized low pH environment, dissolution of enamel, and, eventually, the initiation of dental caries. Metagenomic sequencing projects have readily identified the organism in dental plaque samples from humans, and demonstrated that its relative abundance appears to increase with increasing severity of the disease, concomitant with decreased species diversity (Gross, et al. 2012; Nyvad, et al. 2013).
Streptococcus mutans has evolved specific mechanisms to cope with the acidic and oxidative stress it faces in the oral cavity (Lemos, et al. 2013). While the vast majority of genetic studies involving stress-adaptive pathways in S. mutans have involved the explorations of one to several genes in a given study, it is clear from experiments involving genome-wide transcriptional reporting that the loss of a single gene frequently affects mRNA levels from hundreds of additional genes (Hughes, et al. 2000). Comparison of the relative contribution of each gene to the overall fitness, or virulence potential, of S. mutans would profit from the availability of a collection of strains carrying unique mutations in the genetic complement of the organism.
The elucidation of the genomic sequence for Streptococcus mutans (Ajdic, et al. 2002) provided the information necessary to attempt precise genetic deletion of single open-reading frames of S. mutans on a genome-wide scale. Such a collection of strains could greatly facilitate analysis of the role of each gene in the organism’s competition with other members of the oral flora, as well as the contribution of the gene product to colonization and persistence (Aas, et al. 2008; Becker, et al. 2002; Costalonga & Herzberg 2014; Gross, et al. 2012; Kanasi, et al. 2010; Liu, et al. 2012). The genomic type strain of S. mutans, UA159 (Murchison, et al. 1986), has been extensively utilized in studies of the physiological, genetic, and virulence attributes of the organism. S. mutans UA159 has a malleable genetic system facilitating its transformation with either linear or circular DNA (Murchison, et al. 1986). In the constellation of modern microbiological studies, the number of bacterial mutant collections, or libraries, is still relatively small, and in general, has favored the use of transposon mutant libraries that offer a rapid means by which to construct mutant strains. However, the transposons may exert effects on genes downstream of the insertion site and may fail to reveal particular mutations due to preferred transposition sites (Bose, et al. 2013; Fey, et al. 2013; Ge, et al. 2008; Klein, et al. 2012; Valentino, et al. 2014). The use of DNA-barcoded deletion libraries, pioneered in yeast (Giaever, et al. 2002; Pierce, et al. 2007; Smith, et al. 2010), requires considerably more effort, but offers the advantages of defined mutations and the ability to control deletions in places such as over-lapping reading frames or the in-frame deletion of genes located in polycistronic messages.
Here, we report the construction of a DNA barcoded genomic collection of 1112 strains deleted for non-essential genes and the identification of 320 putative essential genes, together constituting 73% of the S. mutans UA159 genome. The remaining 27% of the genome was refractory to our approach for constructing PCR primers to attempt gene deletion. We have tested the surviving strains for their ability to survive in specific in vitro tests, as predictors of their ability to survive in vivo models. These assessments were chosen to examine the effect of the loss of each gene on the ability of the organism to withstand stresses encountered in the oral cavity: acid, oxidative damage (here, by chemical agents, particularly H2O2), and the presence or absence of atmospheric levels of oxygen. Further, we have tested two different subsets of deletion mutants in mixed-strain experiments to determine their abilities to compete with one another in vitro, and also to maintain themselves in vivo, in the oral cavity of rats, as a model of human oral infection. The collection is a powerful tool to study the ecology of S. mutans and to direct systems approaches to define key mechanisms involved in the pathogenesis of this organism. Our working hypothesis is that potential targets for the therapeutic intervention of dental disease might be more quickly identified by the elucidation of the set of essential genes, and genes that contribute to the overall fitness of S. mutans.
Methods
Bacterial strains
Streptococcus mutans UA159 (Murchison, et al. 1986), the genomic type strain (Ajdic, et al. 2002), was used in this study. All of the deletion strains described were derived from UA159. Deletion of each open reading frame (ORF) was performed essentially as previously detailed (Lau, et al. 2002; Sheng, et al. 2010) and the reading frame was replaced with an erythromycin resistance marker (ErmR) flanked by common, as well as unique, barcode sequences, selected from the barcodes used to create the yeast deletion library (detailed in Fig. 1) (Giaever, et al. 2002). Strains were maintained on Brain Heart Infusion (BHI) agar medium (BD/Difco, Franklin Lakes, NJ) supplemented with erythromycin (Erm) at 5 µg/mL. BHI medium was used to prepare overnight cultures for frozen stocks of the deletion strains.
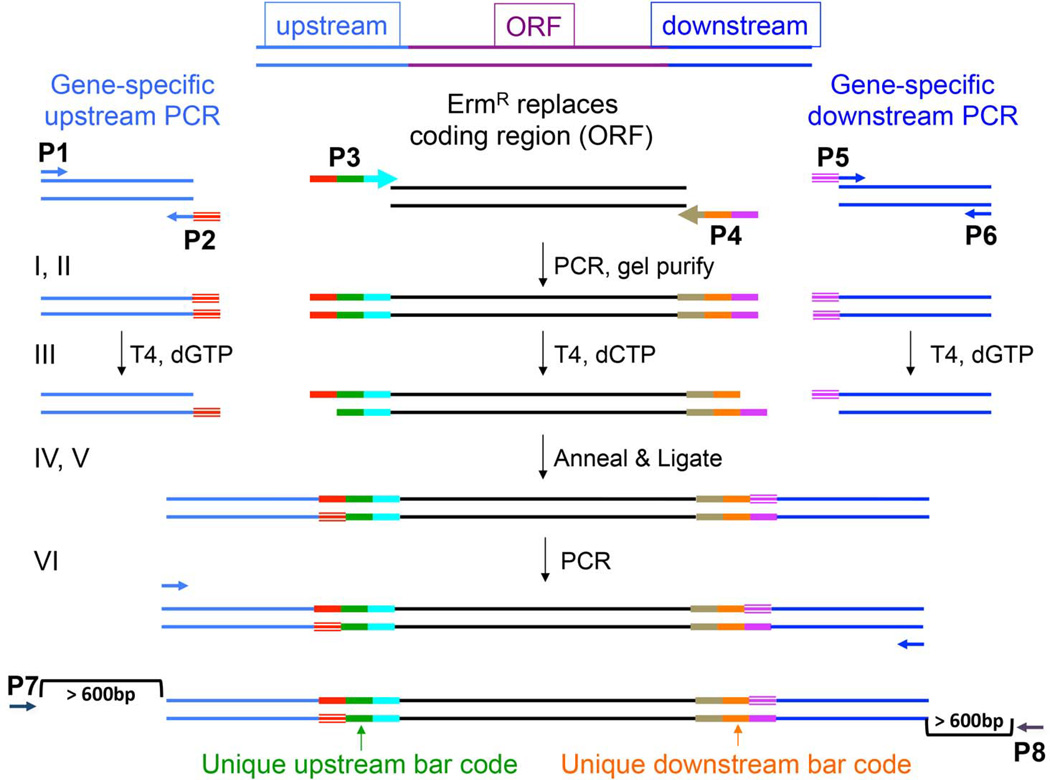
Fig. 1. Scheme for generation of deletion constructs by ligation-independent cloning (LIC).
Panel A. Oligonucleotide primers P1, P2, P3, P4, P5, and P6 were designed according to specifications outlined in Methods. The method involves 6 sequential steps, depicted in the figure by I – VI. (I) The designed primers are utilized in 3 separate PCR reactions. (II) PCR products are gel purified. (III) The amplicons from the sequences upstream and downstream of the target gene are treated with T4 DNA Polymerase in the presence of dGTP. The amplicon arising from the amplification of the erythromycin resistance cassette (ErmR) is treated with T4 DNA Polymerase in the presence of dCTP. This enzymatic treatment allows the digestion of the overhanging ends of the double-stranded DNA fragment to a G residue on the 5’ end of the ErmR cassette and to a C residue on the 3’ end. The result is a single-stranded overhang at the 5’ and 3’ ends homologous to the ends created at the 5’ and 3’ ends of the ErmR. (IV) The 3 double-stranded DNA fragments are allowed to anneal their complementary overhangs. (V) The annealed DNA is treated with T4 DNA Ligase to close the gaps. (VI) Primer pair P1/P6 was used to amplify across the linear double-stranded DNA fragment. This final product was called the deletion construct and used in subsequent transformation into Streptococcus mutans. The lowermost diagram depicts the approx. location of the P7 and P8 primers used to screen the integrated deletion construct in positive transformant colonies.
Primer design
The primer design algorithm was based on Primer3 software (Rozen & Skaletsky 2000). Primers were, in general, designed to have a Tm= 62–68°C, no predicted secondary structure, an inability to form primer-dimers, and no primers could have a 10 base pair homology in the S. mutans genome. Oligonucleotide primers were designated P1 through P6, as shown in Fig. 1. The P1, P2, P5, and P6 primers were constructed to have a predicted annealing temperature of 62–64°C and did not contain an ATG. The P3 and P4 primers were constructed to have a predicted annealing temperature of 62–68°C, and without an ATG.
The P3 and P4 primers were designed to amplify the erythromycin-resistance (ErmR) cassette (Aoki, et al. 1986) used in the study, and to add unique 20 bp upstream and downstream barcodes. Each barcode was flanked by 18 bp conserved DNA sequences that were common to all of the constructs (UpUp (red), UpDn (blue), DnUp (brown), DnDn (pink) in Fig. 1).
The P1/P2 primer pair functioned to amplify the region upstream of the ORF to be deleted, and via P2, added DNA sequence homologous to the 5’ end of P3 (red striped region) (Fig. 1). The P5/P6 primer pair amplified the region downstream of the gene to be deleted, where the P5 primer contained DNA sequence homologous to P4 (purple striped region). The sequences flanking the target genes were approx. 400 bp in length, depending on the local sequence and the ability of the primer-calling algorithm to design oligonucleotides within our parameters.
In addition to the 6 primers synthesized for assembly of the deletion construct, 2 additional primers, P7 and P8, were created to screen the resulting transformants and verify proper integration of the ErmR cassette (Fig. 1). P7 and P8 were designed to bind to sequences outside the region amplified by the P1/P2 and P5/P6 primer pairs, designated the recombination zone (400bp both upstream and downstream of the ORF to be deleted, respectively). These screening primers were designed to work with primers inside the ErmR cassette, Erm Left (Erm L) and Erm Right (Erm R) (Table S2), and result in amplicons that would possess a Tm of approx. 50oC. In addition, the primers were designed such that they were 16–21bp long, containing no secondary structure, and preferably not forming a self-duplex.
Assembly of deletion constructs
The linear, double-stranded DNA fragment used to replace each open reading frame (ORF) was constructed via a six-step process, illustrated in Fig. 1 and described below. Amplicons arising from the three PCR reactions were purified and treated with T4 DNA polymerase in the presence of dGTP for the P1/P2 product; in the presence of dCTP for the P3/P4 product; and in the presence of dGTP for the P5/P6 product. Thus, a G (coding strand) was placed at the 5’ end of the unique barcode and a C (non-coding strand) was placed at the 3’ end of the unique barcode. Treatment of the PCR products with T4 DNA polymerase would result in a PCR product with 18–20 bp single-strand overhangs at the 5’ and 3’ ends that would be homologous, respectively, to the upstream and downstream fragments that were similarly prepared.
The fragments were then allowed to anneal to complementary overhangs and precipitated. The annealed product was reconstituted and a portion was ligated. Finally, primer pair P1/P6 was used to PCR amplify across the ligated, double-stranded DNA. Purified PCR products were analyzed by gel electrophoresis and quantitated by ethidium bromide staining.
Deletion mutant construction
DNA deletion constructs, as detailed above, were transformed into S. mutans UA159 as previously described (Perry & Kuramitsu 1981). Positive transformants were selected on BHI agar medium containing erythromycin (Erm) at 5 µg/mL. Colonies arising after 2 days were picked to fresh agar plates and then at least 2 isolates were chosen for each construct transformation to be analyzed by colony PCR.
A second attempt was made to delete ORFs that were not successfully deleted in the first attempt. If no colonies were obtained following the 2-day incubation, the transformation was performed again, with the addition of 1 µg/mL competence stimulating peptide (CSP) (Li, et al. 2002). If this subsequent transformation with CSP failed to result in ErmR colonies, deletion of that particular ORF was categorized as a potential lethal mutation and coded red in our database. Of the 1432 ORFs attempted, this occurred in 301 cases (21%).
For some ORFs, the deletion construct could not be produced in sufficient abundance for transformation or was not verified by PCR prior to transformation. Therefore, these ORFs (5%) were characterized as having technical issues and coded purple in our database.
Screening and verification of deletion mutant strains of S. mutans
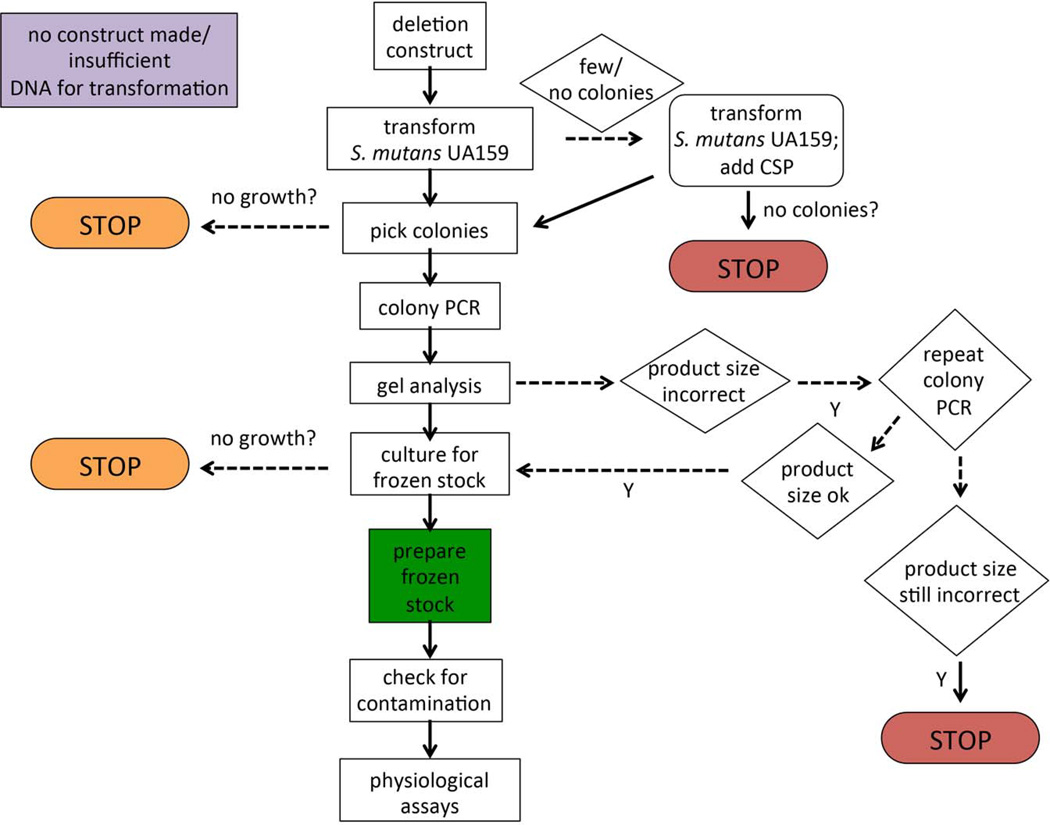
The procedure we employed to verify the deletion constructs in the collection is depicted in the flowchart in Fig. 2. Colony PCR was used to screen ErmR colonies arising from the transformation of S. mutans UA159 with each of the deletion constructs. The P7 and P8 primers (described above) were used in conjunction with Erm Left and Erm Right primers, respectively, to verify that the DNA construct containing the ErmR cassette, in place of the coding region for the ORF, had recombined into the proper site. Colony PCR was performed essentially as previously described (Buckley, et al. 2014; Santiago, et al. 2012). Briefly, a colony resulting from the transformation of S. mutans was boiled for 10 minutes in dH2O. Primers (P7/Erm Left, “left” reactions, or P8/Erm Right, “right” reactions; final concentration 0.5µM) and 1 µL from the lysed colony mixture were added to Platinum PCR SuperMix 96 tubes (Life Technologies, Carlsbad, CA) and PCR amplification was performed using the following conditions: 94°C 1 min; 55°C 1 min.; 72°C 1 min. for a total of 35 cycles. The PCR products were analyzed by gel electrophoresis. If both the “left” and “right” reactions yielded products of the expected size, the isolate was grown up for frozen stock preparation. If the products obtained were the incorrect size, colony PCR was repeated a second time to test additional isolates. A second failure to obtain amplicons of the appropriate size constituted a failure and the transformation step was repeated as detailed above to include CSP. If colonies isolated from this subsequent transformation also failed to yield the proper PCR fragments, the strain was categorized as a potential lethal mutation.
Fig. 2. Decision flow chart for deletion mutant construction.
Linear, double-stranded DNA fragments were created as described in Fig. 1. If the construct was not produced in sufficient abundance for transformation or there was a technical problem with the preparation of the DNA, these open reading frames (ORFs) are coded purple in Table S1. After transformation, if there were no colonies, the transformation was repeated with the addition of competence stimulating peptide (CSP). If no colonies arose from this attempt, then the deletion was presumed to be lethal and coded red in Table S1. If after transformation, the picked colonies did not grow on the BHI agar medium supplemented with Erm, then the gene was coded orange in Table S1 for presumed nutritional requirement. Likewise, if the resulting transformants did not grow in liquid culture for the preparation of a frozen stock, then the gene was also coded orange in Table S1. Finally, during PCR verification of the transformants, if the resulting amplicons were incorrect in size or did not appear after 2 attempts, then the gene was coded red in Table S1 to indicate a putative lethal mutation.
In addition to colony PCR validation of the strains containing deletions, we also performed colony blot analysis for some of the ORFs (25 deletion strains were analyzed in this manner) (Quivey, et al. 1991), or sequenced PCR-amplified DNA containing the deletion region via Sanger sequencing. Strains of S. mutans that had been verified for the proper integration of the deletion construct, 77% of the attempted ORFs, were stored frozen. In approx. 1% of the verified deletions, cultures for frozen stock preparation did not grow. These were coded orange in the database to indicate a potential nutritional deficiency.
Physiological characterization of deletion strains
Each of the deletion strains of S. mutans was categorized with respect to several growth characteristics. Specifically, strains were grown in BHI medium + 44mM glucose, either aerobically or micro-aerobically (with a mineral oil overlay), in the following conditions: (i) unbuffered medium, pH 7.4; (ii) medium titrated to pH 5.4 with HCl; (iii) medium titrated to pH 5.6 with HCl; (iv) medium containing 0.5 mM H2O2; and (v) medium containing 5 µM 8-hydroxyquinoline (8-HQ). Conditions chosen for testing the relative fitness of the deletion strains were based on results from empirically-derived pilot studies for this study. Growth rate and yield were assessed using a Bioscreen C automated growth reading system (Growth Curves USA, Piscataway, NJ) essentially as described elsewhere (Buckley, et al. 2014; Derr, et al. 2012; Kajfasz, et al. 2010). Growth studies for each deletion strain in each growth condition were repeated twice with 3 replicates each time.
The deletion strains were also characterized with respect to their ability to form model biofilms on a solid, plastic surface using a modification of an assay previously described (Ahn, et al. 2005). Deletion strains, and the parent strain, UA159, were grown in a microtiter dish in 0.5× tryptone-yeast extract (TY) medium with the addition of either 56 mM glucose (TYG) or 29 mM sucrose (TYS). Quantitation of biofilm formation was assessed by measuring the optical density (OD575) of wells stained with crystal violet using a BenchMark Plus microplate reader (BioRad, Hercules, CA). The ability of the parent strain, UA159, to form biofilms was normalized to a value of 1.0.
Finally, the endpoint, minimum glycolytic pH for each of the deletion strains was determined using a modification of a previously established method (Belli & Marquis 1991). The value obtained was normalized to the minimum glycolytic pH of the parent strain.
For each of the physiological characterizations performed above, the parent strain, S. mutans UA159, was grown and assessed each time a group of the mutant strains was analyzed. Values for relative doubling time, final yield (indicated by optical density of the culture), and biofilm (BF) formation were compared to the parent strain, UA159, grown under the same conditions.
Establishing the relative fitness score for each deletion strain of S. mutans
A scoring system was devised to give each of the deletion strains a phenotypic rating based on the ability of the strain to grow under the various conditions described above, as compared to the parent strain, UA159 (Table S1, Columns J-S). A rating of 0 signifies that there was no growth under the specified condition (red cell). A score of 1 indicates poor growth where the yield of the culture is <50% of control and/or the growth rate is 76–550% of control and/or a lag time greater than control of 4 hours (orange cell). A score of 2 indicates impaired growth where the yield and/or growth rate are 26–50% that of control and/or lag time is between 2.1 to 3.9 hr of control (yellow-orange cell). A rating of 3 signifies reduced growth, as compared to the parent strain, with a yield and/or growth rate 51–75% that of control and/or lag time was between 1.1 to 2 hr of control (yellow cell). A score of 4 indicates normal growth, i.e., similar to parent strain where the yield was 76–110% of control and/or the doubling time was −25 to 25% of control and/or lag time was between −1 to 1 hr of control (green cell). In three instances, deletion of the gene resulted in enhanced growth, as compared to UA159, which was given a score of 5 (blue cell). Yields were greater than 110% of control and/or doubling time was >25% lower and/or a lag time < −1 hr of control. For all 3 cases, this occurred when the cultures were grown aerobically in the presence of 8-HQ. The average of the scores determined for each growth condition was calculated and each deletion strain was assigned a mean growth score (Table S1, Column U). Cells colored grey indicate the inability to obtain a deletion mutant and cells colored white indicate that the construct was not attempted due to inability of our primer-calling algorithm to design primers for the deletion construct
The relative ability of the deletion strains to form single-strain model biofilms in medium containing glucose or sucrose, described above, was scored on a different scale, based on the magnitude and significance (p-value determined by Student’s t-test) of the difference between biofilm formation by the parent strain, UA159 (control), and the specific deletion mutant strain. No statistical difference between control and deletion strain earned a score of 4 (green). For significant variations from control (p-value < 0.05), the score was assigned as follows: relative biofilm formation 0–10% versus control = 0 (red); relative biofilm formation 11–25% versus control = 1 (orange); relative biofilm formation 26–50% versus control = 2 (yellow-orange); relative biofilm formation 51–75% versus control = 3 (yellow); and relative biofilm formation greater than 110% of control = 5 (blue).
Competition experiments using pools of deletion strains
(i) Survival of selected strains in chemostat culture
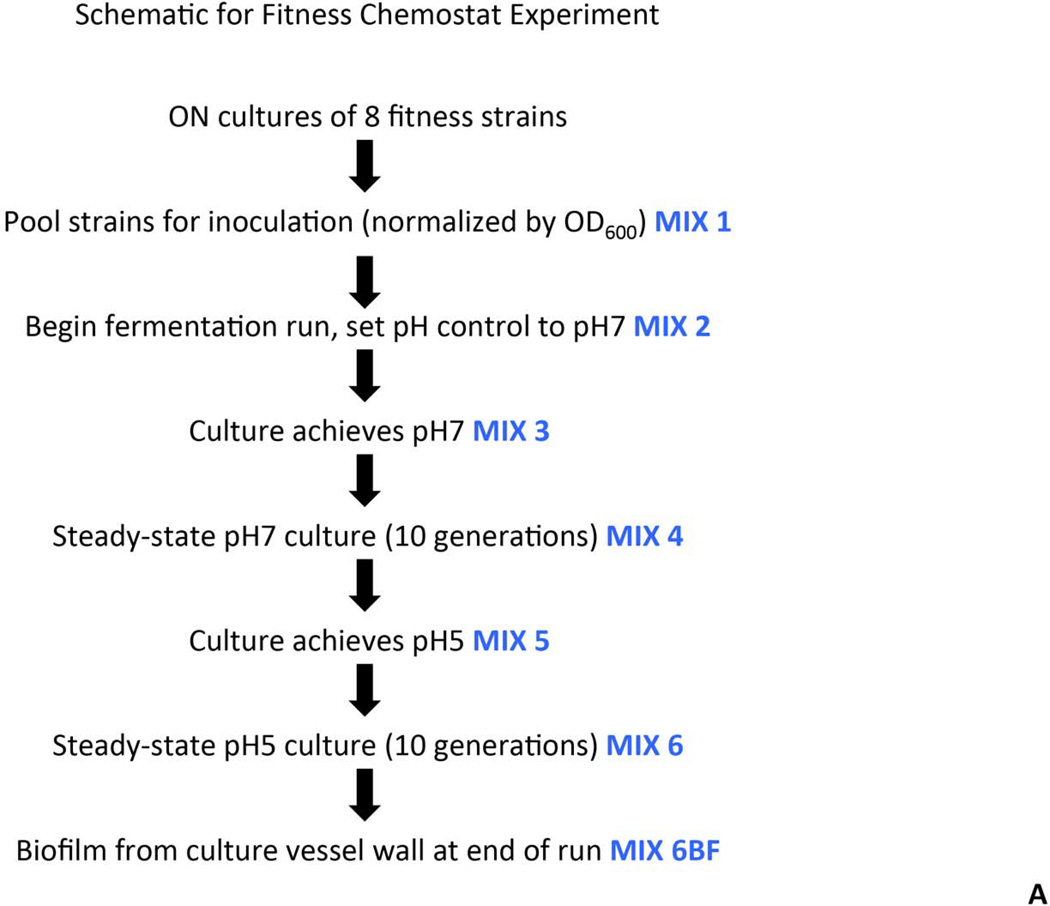
Selected strains carrying deletions in genes involved in fatty acid biosynthesis and cell wall assembly were used to examine relative fitness in steady-state culture. The strains used were MX804 (ErmR knock-in strain), MU0022 (∆plsX; SMU.26), MU0023 (∆acpP; SMU.27), MU0898 (∆cls; SMU.988), MU1020 (∆nox; SMU.1117), MU1591 (∆fabT; SMU.1745c), MU1592 (∆fabM; SMU.1746c), and MU1593 (∆pgmB; SMU.1747c). Strains were grown overnight in batch culture in TY medium containing 1% glucose. Cell density was measured at OD600 and cultures were normalized such that 25 mL would be equivalent to an OD600 = 1. Equal volumes (25 mL) of each normalized culture were used to inoculate the vessel of a BioFlo 2000 fermentor (New Brunswick Scientific, Edison, NJ). The mixed culture was grown in TY medium with limiting glucose to steady-state (10 generations) at pH values of 7 and 5 at a dilution rate of 0.144 hr−1, as previously described (Fozo, et al. 2004). Samples were removed from the mixed culture at various time points (as diagrammed in Fig. 3A), pelleted, and stored frozen.
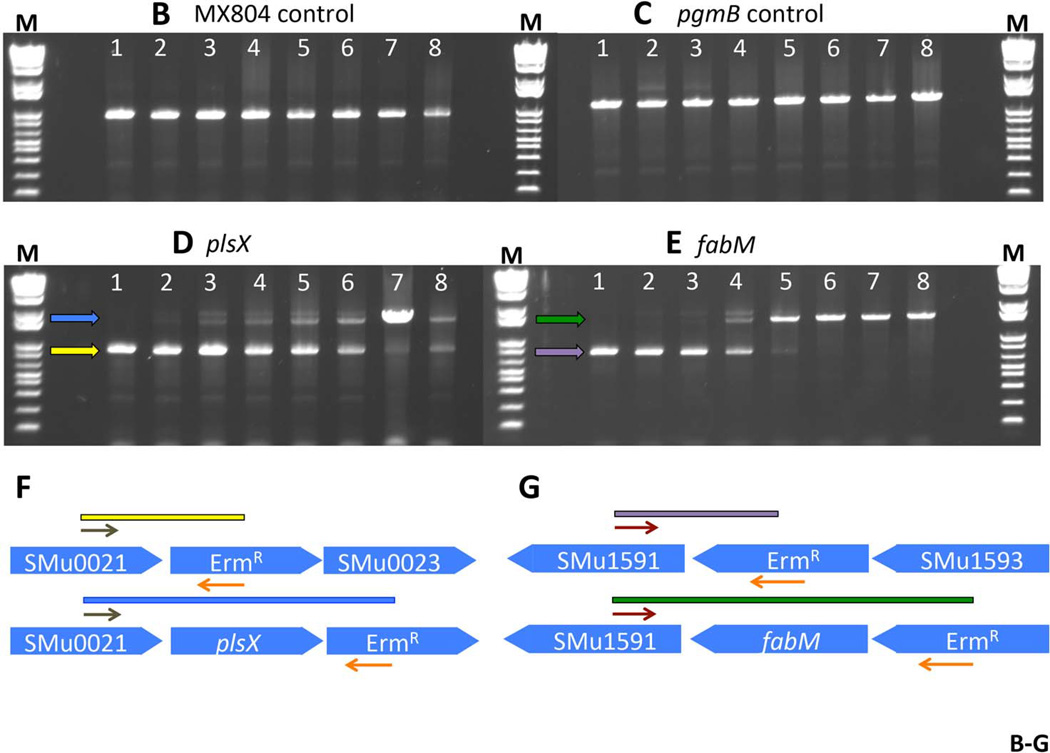
Fig. 3. Fitness testing of a subset of deletion mutant strains in chemostat culture.
Panel A. Experimental design and sampling for the chemostat culture. Eight deletion strains were individually cultured, normalized, and used to inoculate a fermenter vessel. This co-culture was sampled and designated Mix 1. The culture was allowed to grow in batch culture before turning on the pH control. At the point that the pH control was activated, and set to pH 7, another sample was taken (Mix 2). Once the culture had reached pH 7, a sample was removed and designated Mix 3. A sample from the steady-state pH 7 culture was called Mix 4. Glucose was added to the culture vessel to reduce the pH to 5, and another sample was removed (Mix 5). A sample from the steady-state pH 5 culture was called Mix 6. Finally, a sample of the biofilm that formed on the wall of the fermenter vessel was removed and called Mix 6BF. Panel B. PCR amplification of DNA representing MX804 (ErmR knock-in strain) in the samples derived from the chemostat culture. DNA prepared from the samples recovered at each step described in Panel A (above) was used as a template in a reaction with primer pair MX0804P7UPFG and Erm L to determine the relative amount of the MX804 strain at each time point. Lane order is as follows: lane 1: Genomic DNA control reaction; lane 2: sample from Mix 1; lane 3: sample from Mix 2; lane 4: sample from Mix 3; lane 5: sample from Mix 4; lane 6: sample from Mix 5; lane 7: sample from Mix 6; lane 8: sample from Mix 6 BF. Panel C. PCR amplification of DNA representing the MU1593 deletion strain (∆pgmB) (Buckley, et al. 2014) in the samples derived from the chemostat culture. DNA prepared from the samples recovered at each step described in Panel A (above) was used as a template in a reaction with primer pair MU1593P7UPFG and Erm L to determine the relative amount of the MU1593 strain at each time point. Lane order is as detailed in Panel B above. Panel D. PCR amplification of DNA representing the MU0022 deletion strain (∆plsX) in the samples derived from the chemostat culture. DNA prepared from the samples recovered at each step described in Panel A (above) was used as a template in a reaction with primer pair MU0022P7UPFG and Erm L to determine the relative amount of the MU0022 strain at each time point. Lane order is as detailed in Panel B above. Panel E. PCR amplification of DNA representing the MU1592 deletion strain (∆fabM) in the samples derived from the chemostat culture. DNA prepared from the samples recovered at each step described in Panel A (above) was used as a template in a reaction with primer pair MU1592P7UPFG and Erm L to determine the relative amount of the MU1592 strain at each time point. Lane order is as detailed in Panel B above. Panel F. Graphical representation of the origin of the bands seen in the gel photo in Panel D. The brown arrows represent the MU0022P7UPFG screening oligonucleotide and the orange arrows represent the Erm L oligonucleotide. If MU0022 DNA is present in the pooled sample, then the yellow bar and yellow arrow in the gel photograph indicate the size and migration of that specific amplicon. However, if MU0022 DNA is no longer present in the pooled sample, then we would expect to obtain the amplicon represented by the blue bar (and the blue arrow on the gel photograph), indicating DNA derived from the presence of MU0023 would arise. Panel G. Graphical representation of the origin of the bands seen in the gel photo in Panel E. The red arrows represent the MU1592P7UPFG screening oligonucleotide and the orange arrows represent the Erm L oligonucleotide. If MU1592 DNA is present in the pooled sample, then the purple bar and purple arrow in the gel photograph indicate the size and migration of that specific amplicon. However, if MU1592 DNA is no longer present in the pooled sample, then we would expect that the amplicon represented by the green bar (and the green arrow on the gel photograph), indicating DNA derived from MU1593 would arise.
DNA was prepared from samples taken from the continuous culture of the mixed pool using the Promega Wizard Genomic DNA Purification Kit (Promega, Madison, WI) and used in PCR with primers listed in Table S2 for each of the deletion mutant strains.
(ii) Survival of selected strains in the rat model of oral infection
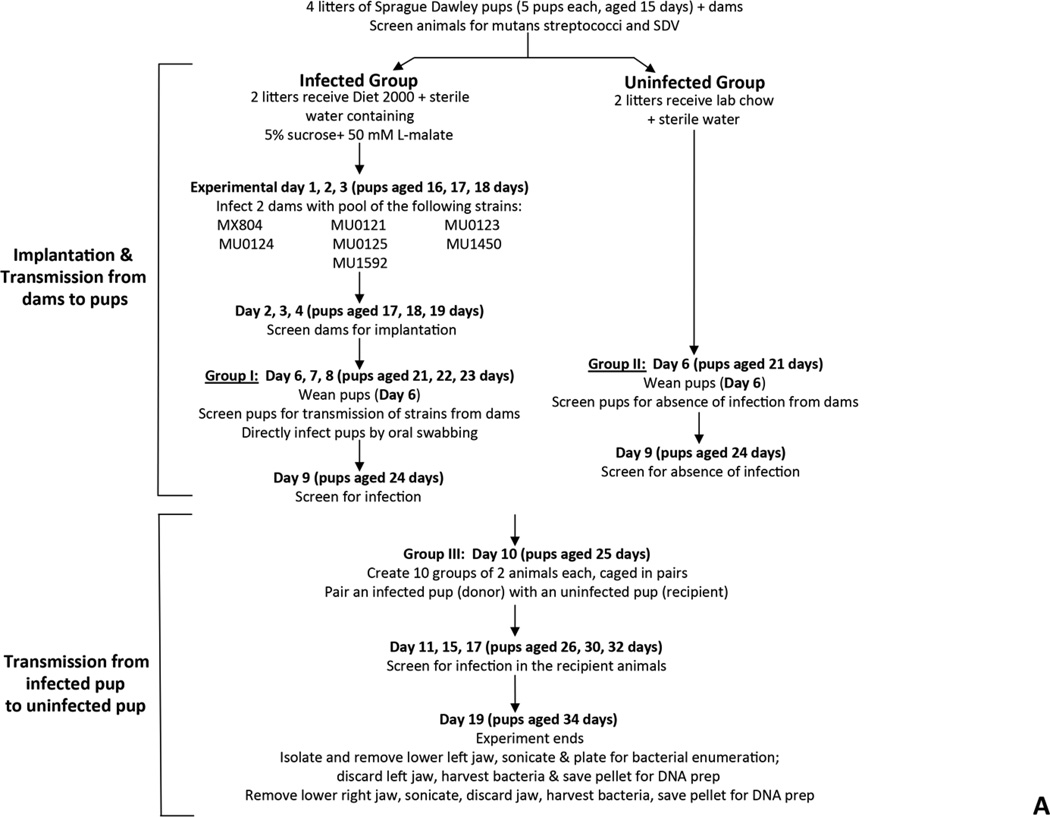
Two approaches were used to examine transmissibility, between rats, of tagged strains of S. mutans carrying deletions in genes of the malolactate fermentation pathway. A pilot study was employed to determine the ability of the strains to implant and be transmitted from an infected pup to an uninfected cagemate. The methodology used was identical to that used in a previous study from our group (Fozo, et al. 2007). The following S. mutans deletion strains created in this study were utilized: MX804 (ErmR knock-in strain), MU0121 (∆mleR; SMU.135), MU0123 (∆mleS; SMU.137), MU0124 (∆mleP; SMU.138), MU0125 (∆oxdC; SMU.139), MU1592 (∆fabM; SMU.1746c), and MU1450 (∆cah; SMU.1595) (Sheng, et al. 2010). The overall scheme for the experiment is detailed in Fig. 4A.
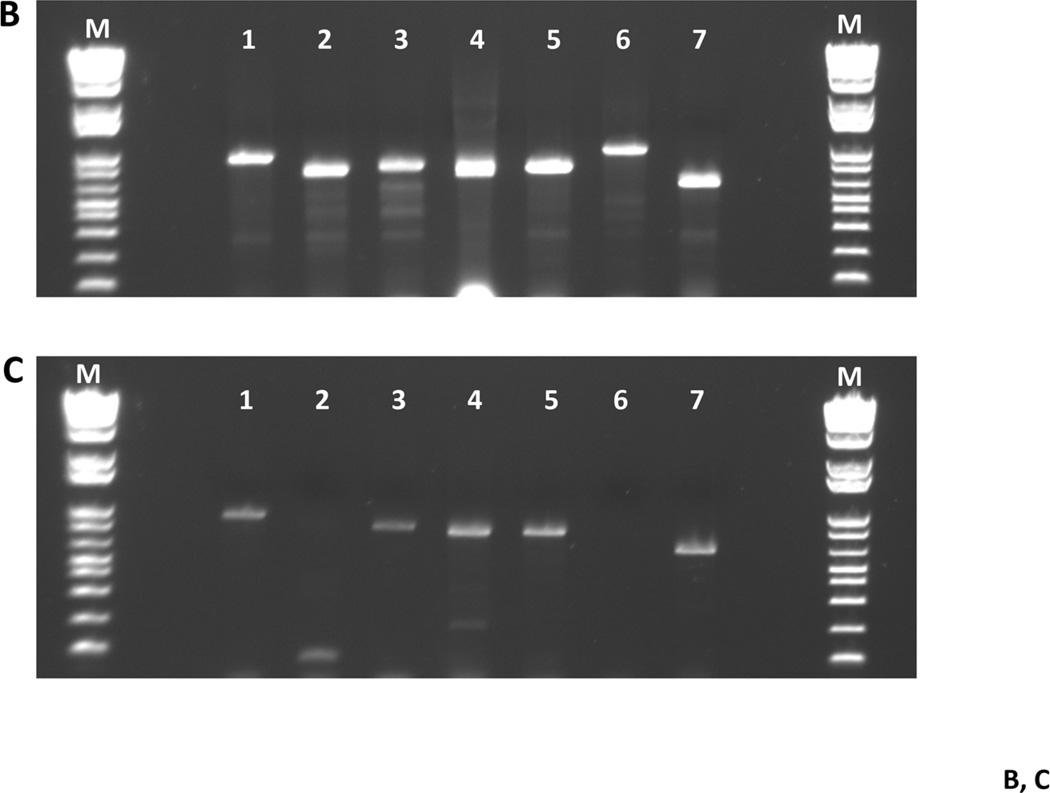
Fig. 4. Fitness testing of a subset of deletion mutant strains in a rat model for oral infection.
Panel A. Transmission experiment outline for 7 deletion mutant strains. SDV = sialoacroadenitis virus. Panel B. Gel photograph showing the presence of each of the deletion strains in the pre-infection pool used to inoculate rat pup jaws (from Day 8; see Panel A). Lane contents are as follows: lane 1: MX804-specific primers (MX0804P7UPFG/ Erm L); lane 2: MU0121-specific primers (MU0121P7UPFG/ Erm L); lane 3: MU0123-specific primers (MU0123P7UPFG/ Erm L); lane 4: MU0124-specific primers (MU0124P7UPFG/ Erm L); lane 5: MU0125-specific primers (MU0125P7UPFG/ Erm L); lane 6: MU1592-specific primers (MU1592P8DNRH/ Erm L); lane 7: MU1450-specific primers (MU1450P7UPFG/ Erm L). Panel C. Representative gel photograph showing amplified DNAs isolated from strains surviving after 19 days in the rat model. The left hemi-jaw from rat #13 (in Group II uninfected pups) was sonicated and DNA prepared for use as a template in a PCR reaction with the gene-specific primers for each of the 7 deletion strains utilized in the experiment. Lane order and primer pairs are as detailed in Panel B (above).
Strains used in the experiment were grown overnight in BHI medium. The overnight cultures were diluted 1:40 (or 1:20 for MU1592) into a total volume of 20 mL fresh BHI medium the following morning and allowed to grow to mid-logarithmic phase (OD600 ~ 0.6–0.8). Aliquots of 1 mL cells were harvested and pellets were saved as “pre-infection pools” from each of the 3 days of infection (Day 1, 2, and 3 for infection of the dams; Day 6, 7, 8 for infection of the pups). Another aliquot of the pooled culture was serially diluted and plated on BHI agar medium to enumerate input bacteria.
Two dams were infected with an actively growing, mid-logarithmic pooled culture by oral swabbing on experimental days 1, 2, 3 (pups were aged 16, 17, 18 days). Two additional dams were not infected. Infection in the dams was confirmed by streaking oral swabs on MSE agar on days 2, 3 and 4 of the experiment (pups aged 17, 18, 19 days). Infected dams were given Diet-2000 and drinking water sweetened with 5% (w/v) sucrose containing 50 mM L-malate, ad libitum; the uninfected dams were fed lab chow and water ad libitum.
On day 6 of the experiment, pups (aged 21 days) associated with the infected dams (Group I) were weaned, then screened for infection from dams by oral swabbing and plating on MSE agar medium. The pups fed by uninfected dams (Group II) were also weaned and screened for absence of infection by plating oral swabs on MSE agar medium. The pups from Group I were then infected with the actively growing pooled culture of 7 strains over 3 consecutive days (experimental Day 6, 7, and 8). Infection was confirmed in the Group I pups by Day 10. The dams were sacrificed and the lower left jaws were transferred to a saline solution and sonicated to free bacteria. The jaws were removed and the sonicate solution centrifuged with resulting pellets stored at −20°C for use in DNA purification.
On experimental day 10, infected pups from Group I (aged 25 days), referred to as donors, were paired with uninfected pups, and referred to as recipients, from Group II. The new group, Group III, consisted of 10 pairs of donor and recipient animals. All animals were given Diet-2000 containing 5% sucrose water supplemented with 50 mM L-malate, ad libitum. The previously uninfected pups, or recipients, were screened for S. mutans infection by oral swabbing on day 11, 15, 17 (pups aged 26, 30, 32 days), respectively. Animals were sacrificed by CO2 asphyxiation on Day 21 (animals aged 36 days).
The lower jaw was removed and separated into 2 hemi-jaws. The left lower hemi-jaw was placed into 5 mL sterile saline and sonicated three times for 10 seconds each. Jaw sonicates were then serially diluted and plated onto BHI agar medium containing Erm (5µg/mL) to determine streptococcal colony forming units (CFUs), as established in previous studies. The remaining sonicate for each of the 20 rat pups was centrifuged and resulting pellets were stored at −20°C until used for DNA purification. The right lower hemi-jaw was also placed in 5 mL sterile saline and sonicated. The sonicates were passed through a 0.8µm syringe filter to clarify the particulate matter, centrifuged, and the resulting pellets were stored at −20°C as a back up, if needed.
DNA was isolated from frozen samples using the Promega Wizard Genomic DNA Purification Kit (Promega, Madison, WI) and used in PCR with primers listed in Table S2 to ascertain presence or absence of a given deletion mutant in the mixed population.
Results and Discussion
Construction of the collection
The methodology that we employed to create deletion strains, shown in Fig. 1 and detailed in METHODS, is generally similar to the approach used to create the Saccharomyces cerevisiae deletion library (Giaever, et al. 2002) and to create single genetic deletion mutations in S. mutans (Lau, et al. 2002). Precise gene deletions in which ErmR replaces the coding sequence were targeted to particular chromosomal locations by homologous recombination between sequences that flank the coding region and sequences that flank the ErmR gene. Using PCR to amplify the regions upstream and downstream of each gene also provided the opportunity to create two unique DNA tags for each deletion, one upstream of the coding region of a given gene and the other downstream, that could be used as primers for assaying the relative abundance of each deletion strain in mixed populations (Fig. 1). Furthermore, the constructs also contained upstream and downstream common sequences that could be utilized to PCR amplify the S. mutans-specific DNA sequences from a co-culture.
One challenge in creating the collection in S. mutans was the requirement for relatively long flanking sequences to correctly target the ErmR gene to specific locations. This was solved by independently amplifying ~400 base pairs of the upstream and downstream sequences, then using ligation independent (LIC) cloning methods to link the three fragments, followed by ligation and PCR amplification. Using this protocol, we generated 1432 constructs of the expected size at sufficient DNA concentrations to transform S. mutans. We were unable to generate constructs for 530 genes.
The methodology and decision-making processes used to create the deletion strain collection are illustrated in the flow chart in Fig. 2. From this approach, 1112 strains were recovered that were capable of growth in suspension culture; 301 genes were identified as putative essential genes; and 19 genes exhibited difficulties in the recovery of a stable strain; i.e., no growth. These 19 strains that we were unable to cultivate in liquid culture could represent a nutritional requirement that was not met in our high-throughput approach.
The distribution of essential and non-essential genes is shown in the heat map in Fig. S1. It can be immediately seen that the putative essential genes are non-randomly distributed throughout the chromosome. As can also be seen in Fig. S1, the gaps in deletion coverage from the lack of deletion cassettes to begin the process are distributed around the chromosome. Therefore, we presume that inability to produce a deletion cassette for a particular ORF is due to the A-T rich nature of the organism and not a consequence of the methodology. Current availability of economical synthetic gene constructs would preclude this obstacle.
Each deletion was designed to be non-polar, although we did not test downstream effects of specific deletions on all genes. However, for example, the deletion of fabT (SMU.1745c) did not affect transcription of fabH (SMU.1744c), which it overlaps by 1 bp (Faustoferri, et al. 2014). Further, we have verified in separate experiments that the phenotype that resulted from the deletion of a particular ORF was attributable to loss of that gene product via genetic complementation of the ORF either in an ectopic locus in the genome (Buckley, et al. 2014; Derr, et al. 2012; Faustoferri, et al. 2014; Santiago, et al. 2012; Santiago, et al. 2013) or in situ (MacGilvray, et al. 2012).
In vitro characterization of the collection of genome-wide deletion mutant strains of S. mutans
The ability of S. mutans to respond to stress in its environment represents a key element of the organism’s virulence potential. Thus, our primary interest was to assay the ability of each deletion strain to grow in conditions thought to affect virulence. We measured growth rates in the following conditions: in acidic medium, in medium containing compounds known to inhibit growth of the organism, and under reduced oxygen tensions. The deletion strains were grown, individually, to assess doubling times vis-à-vis the parent strain grown under the same conditions. The strains were then assigned overall scores, using these data, to indicate the relative fitness of each strain, compared to the parent, UA159, which earned a score of 4 (see further details in METHODS). We also created an ErmR cassette knock-in in a non-coding region upstream of gtfA (SMU.881), designated MX804, to use as a bar-coded “parent” strain in mixed culture experiments. This strain displayed survival capacity similar to S. mutans UA159 in all conditions tested for the single deletion strain assays (data not shown).
The summary statistics deriving from in vitro growth experiments for 1059 surviving strains are shown in Table S1. These strains contain deletions in putative non-essential genes that were tested for their ability to grow in suspension culture under specific conditions. Of these, the majority (84%) (861) grew as well as the parent strain in rich medium. The remaining 16% of the strains (168) were statistically different from wild-type in terms of growth rates and yields in rich medium, with 40 strains improving to the level of the parent strain when provided a reduced oxygen environment.
Growth of the strains under oil-overlay, to create a microaerobic environment, resulted in improved growth for some strains under the conditions tested, but in general, did not result in dramatic movement of strains from one phenotypic rating level to another (Table S1, compare columns J-K, L-M, N-O, P-Q, R-S). Reduced oxygen tensions did appear to make a difference in growth of strains in medium containing the metal chelator 8-hydroxyquinoline (8-HQ, Table S1, columns R-S). The most dramatic effect of reducing oxygen tensions in 8-HQ-containing cultures was in elevating 212 strains to a phenotypic rating comparable to the parent strain (a rating of 4), suggesting that chelation of metals represented a strongly oxidative environment for S. mutans, and that reduction of oxygen levels alleviated, to some extent, that stress.
Mutants were identified with impaired growth in two or more conditions relevant to virulence
With the establishment of the growth rates for the strains in rich medium, we began our assessment of the fitness of the individual strains in conditions known to be involved with the virulence potential of the organism: aciduricity (Fozo, et al. 2007), ability to form an anaerobic environment (Marquis 1995), and biofilm formation (Bowen & Koo 2011). We examined the ability of the deletion strains to grow in medium titrated to an acidic pH value or in the presence of hydrogen peroxide or 8-HQ. We also determined the ability of deletion strains to produce model biofilms in mono-organism culture, in the presence of glucose or sucrose. The majority of the deletion strains produced quantities of biofilm comparable to the parent strain, in cultures growing in the presence of glucose or sucrose. However, we identified 101 deletion strains that were defective in biofilm formation in the presence of glucose and 64 strains defective in the presence of sucrose.
One of the valuable assets of the deletion collection was that we are able to identify mutants with defects in two or more growth characteristics. In this case, we were interested in mutations that cause defects in multiple aspects of virulence. We found 51 strains (Table 1A) that were both acid-sensitive (a phenotypic rating of ≤2) and biofilm-defective (a biofilm formation rating of ≤2) in the presence of sucrose. We found 49 strains were both defective in oxidative-stress resistance (a phenotypic rating of ≤2) and biofilm-defective (a biofilm formation rating of ≤2) (Table 1B). These two subsets represent mutants of S. mutans carrying deletions in genes that are likely involved in the pathogenicity of the organism. Furthermore, there are 39 genes whose deletion caused a defect in all 3 of the conditions; of these, only 6 have established functions, 24 have been annotated and have been assigned a putative function, and 9 are completely unknown or even hypothetical, representing potential targets for future investigations into their role in virulence (Table 1C).
Table 1.
| A. Deletion strains exhibiting poor acid survival and deficiencies in biofilm formation | ||||
|---|---|---|---|---|
| Oralgen ID | Locus | Gene Name(s) | Common Name | Functional Class |
| 0022 | SMU.26 | plsX ylpD | putative fatty acid/phospholipid synthesis protein | Fatty acid and phospholipid metabolism: Biosynthesis |
| 0072 | SMU.80 | hrcA | transcriptional regulator repressor (HrcA) of class I | Regulatory functions: DNA interactions |
| 0085 | SMU.94c | tpn | hypothetical protein putative transposase fragment | Mobile and extrachromosomal element functions: Transposon functions |
| 0137 | SMU.154 | rpsO | 30S ribosomal protein S15 | Protein synthesis: Ribosomal proteins: synthesis and modification |
| 0153 | SMU.170 | rpsl | 30S ribosomal protein S9 | Protein synthesis: Ribosomal proteins: synthesis and modification |
| 0223 | SMU.246 | rgpG | putative glycosyl transferase N-acetylglucosaminyltransferase), RgpG | Cell envelope: Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides |
| 0275 | SMU.304 | cdd | putative deaminase | Unknown function: Enzymes of unknown specificity |
| 0315 | SMU.350 | hypothetical protein | Hypothetical proteins | |
| 0317 | SMU.352 | rpe | putative ribulose-phosphate-3-epimerase | Energy metabolism: Pantose phosphate pathway |
| 0318 | SMU.353 | conserved hypothetical protein | Biosynthesis of cofactors, prosthetic groups, and carriers: Thiamine | |
| 0431 | SMU.475 | conserved hypothetical protein | Unknown function: General | |
| 0434 | SMU.480 | priA | primosomal replication factor Y (primosomal protein N) | DNA metabolism: DNA replication, recombination, and repair |
| 0437 | SMU.483 | pphA ppL | putative phosphoprotein phosphatase (pppL protein) | Regulatory functions: Protein interactions |
| 0500 | SMU.550 | divlB ftsQ | putative cell division protein FtsQ (DivlB) | Cellular processes: Cell division |
| 0510 | SMU.561c | mutT | putative hydrolase (MutT family) | DNA metabolism: DNA replication, recombination, and repair |
| 0521 | SMU.572 | folD | putative tetrahydrofolate dehydrogenase/cyclohydrolase | Biosynthesis of cofactors, prosthetic groups, and carriers: Folic acid |
| 0545 | SMU.598 | recR | putative recombination protein RecM | DNA metabolism: DNA replication, recombination, and repair |
| 0571 | SMU.629 | sod sodA | putative manganese-type superoxide dismutase, Fe/Mn-SOD | Cellular processes: Detoxification |
| 0645 | SMU.706c | conserved hypothetical protein | Transport and binding proteins: Other | |
| 0666 | SMU.731 | putative ABC transporter, ATP-binding protein | Transport and binding proteins: Unknown substrate | |
| 0672 | SMU.739c | hypothetical protein | Hypothetical proteins: Conserved | |
| 0698 | SMU.770c | hitA | putative manganese transporter | Transport and binding proteins: Cations and iron carrying compounds |
| 0705 | SMU.777 | aroD | putative 3-dehydroquinate dehydratase | Amino acid biosynthesis: Aromatic amino acid family |
| 0706 | SMU.778 | aroE | putative shikimate 5-dehydrogenase | Amino acid biosynthesis: Aromatic amino acid family |
| 0708 | SMU.780 | aroC | putative chorismate aynthase | Amino acid biosynthesis: Aromatic amino acid family |
| 0740 | SMU.816 | arcT | putative aminotransferase | Amino acid biosynthesis: Aspartate family |
| 0749 | SMU.825 | rgpAc | putative RgpAc glycosyltransferase | Unclassified: Role category not yet assigned |
| 0750 | SMU.826 | rgpBc | rhamnosyltransferase | Cell envelope: Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides |
| 0754 | SMU.830 | rgpFc | RgpFc protein | Unclassified: Role category not yet assigned |
| 0792 | SMU.869 | trxB | putative thioredoxin reductase | Unknown function: General |
| 0985 | SMU.1077 | pgmA | putative phosphoglucomutase | Cell envelope: Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides |
| 0988 | SMU 1080c | conserved hypothetical protein possible transposon-related protein | Hypothetical proteins: Domain | |
| 0991 | SMU 1083c | conserved hypothetical protein | Unknown function: General | |
| 1016 | SMU.1113 | srtA | putative sortase | Cell envelope: Other |
| 1035 | SMU.1133 | phoU | putative phosphate transport system regulatory protein | Regulatory functions: Other |
| 1092 | SMU.1193 | yhcF | putative transcriptional regulator | Regulatory functions: DNA interactions |
| 1166 | SMU.1276c | ezrA | putative septation ring formation regulator | Cellular processes: Cell division |
| 1209 | SMU.1324 | ftsX | putative cell-division protein FtsX | Cellular processes: Cell division |
| 1210 | SMU. 1325 | ftsE | putative ABC transporter, ATP-binding component | Cellular processes: Cell division |
| 1329 | SMU.1460 | rmlC | putative dTDP-4-keto-L-rhamnose reductase | Cell envelope. Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides |
| 1330 | SMU.1461 | rfbA rmlA | putative glucose-1-phosphate thymidyltransferase | Cell envelope: Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides |
| 1444 | SMU.15B9c | cpoA ywaF | putative hexosyltransferase | Cell envelope: Biosynthenis and degradation of surface polysaccharides and lipopolysaccharides |
| 1483 | SMU 1629c | putative cell division protein DNA segregation ATPaae | Cellular processes: Cell division | |
| 1538 | SMU.1688 | dlt4 dltD | putative extramembranal protein, DltD protein | Cell envelope: Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides |
| 1541 | SMU.1691 | dlt1 dltA | putative D-alanine-D-alanyl carrier protein ligase | Cell envelope: Biosynthesis and degradation of murein sacculus and peptidoglycan |
| 1551 | SMU.1703c | conserved hypothetical protein | Hypothetical proteins | |
| 1587 | SMU.1741 | fabD | putative malonyl-CoA) acyl-carrier-protein) transacylase | Fatty acid end phospholipid metabolism: Biosynthesis |
| 1592 | SMU.1746c | fabM phaB | putative enoyl-CoA hydratase | Fatty acid and phospholipid metabolism: Degradation |
| 1662 | SMU.1824c | codY | putative transcriptional regulator | Regulatory functions: DNA interactions |
| 1823 | SMU.2007 | rl15 rplO | 50S ribosomal protein L15 | Protein synthesis: Ribosomal proteins: synthesis and modification |
| 1886 | SMU.2078c | conserved hypothetical protein | Hypothetical proteins: Conserved | |
| B. Deletion strains exhibiting poor survival in oxidative stress conditions and in biofilm formation | ||||
|---|---|---|---|---|
| Oralgen ID | Locus | Gene Name(s) | Common Name | Functional Class |
| 0012 | SMU.14 | hgt hprT | putative hypoxanthine-guanine phosphoribosyltransferase | Purines. pyrimidines, nucleosides, and nucleotides: Salvage of nucleosides and nucleotides |
| 0022 | SMU 26 | plsX ylpD | putative fatty acid/phospholipid synthesis protein | Fatty and and phospholipid metabolism Biosynthesis |
| 0137 | SMU.154 | rpsO | 30S ribosomal protein S15 | Protein synthesis: Ribosomal proteins: synthesis and modification |
| 0153 | SMU 170 | rpsl | 30S ribosomal proton S9 | Protein synthesis: Ribosomal proteins: synthesis and modification |
| 0223 | SMU.246 | rgpG | putative glycosyl transferase N-acetylglucosaminyltransferase), RgpG | Cell envelope Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides |
| 0275 | SMU 304 | cdd | putative deaminease | Unknown function: Enzymes of unknown specificity |
| 0317 | SMU.352 | rpe | putative ribulose-phosphate-3-epimerase | Energy metabolism Pentose phosphate pathway |
| 0318 | SMU.353 | conserved hypothetical protein | Biosynthesis of cofactors, prosthetic groups, and carriers: Thiamine | |
| 0378 | SMU 417 | conserved hypothetical protein | Unknown function General | |
| 0431 | SMU.475 | conserved hypothetical protein | Unknown function: General | |
| 0434 | SMU.480 | priA | primosomal replication factor Y (primosomal protein N) | DNA metabolism: DNA replication, recombination, and repair |
| 0437 | SMU.483 | pphA ppL | putative phosphoprotein phosphatase (pppL protein) | Regulatory functions. Protein interactions |
| 0491 | SMU 540 | dpr | peroxide resistance protein Dpr | Transport and binding proteins: Cations and iron carrying compounds |
| 0500 | SMU.550 | divlB ftsQ | putative cell division protein FtsQ (DivlB) | Cellular processes: Cell division |
| 0521 | SMU.572 | folD | putative tetrahydrofolate dehydrogenase/cyclohydrolase | Biosynthesis of cofactors, prosthetic groups, and carriers: Folic acid |
| 0555 | SMU.609 | bsp | putative 40K cell wall protein precursor | Unknown function: General |
| 0571 | SMU 629 | sod sodA | putative manganese type superoxide dismutase Fe/Mn-SOD | Cellular processes: Detoxification |
| 0645 | SMU.706c | conserved hypothetical protein | Transport and binding proteins: Other | |
| 0672 | SMU 739c | hypothetical protein | Hypothetical proteins: Conserved | |
| 0685 | SMU.754 | hprK ptsK | HPr(serine) kinase/phosphatase | Regulatory functions: Protein interactions |
| 0705 | SMU 777 | aroD | putative 3-dehydroquinate dehydratase | Amino acid biosynthesis: Aromatic amino and family |
| 0706 | SMU.778 | aroE | putative shikimate 5-dehydrogenase | Amino acid biosynthesis: Aromatic amino and family |
| 0708 | SMU.780 | aroC | putative chorismate synthase | Amino acid biosynthesis: Aromatic amino and family |
| 0749 | SMU.825 | rgpAc | putative RgpAc glycosyltransferase | Unclassified: Roe category not yet assigned |
| 0750 | SMU.826 | rgpBc | rhamnosyltransferase | Cell envelope: Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides |
| 0753 | SMU.829 | putative glycosyltransferase | cell envelope: Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides | |
| 0754 | SMU.830 | rgpFc | RgpFc protein | Unclassified: Role category not yet assigned |
| 0792 | SMU.889 | trxB | putative thioredoxin reductase | Unknown function: General |
| 0985 | SMU.1077 | pgmA | putative phosphoglucomutase | Cell envelope: Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides |
| 0988 | SMU.1080c | conserved hypothetical protein possible transposon-related protein | Hypothetical proteins: Domain | |
| 0991 | SMU.1083c | conserved hypothetical protein | Unknown function: General | |
| 1035 | SMU.1133 | phoU | putative phosphate transport system regulatory protein | Regulatory functions: Other |
| 1092 | SMU.1193 | yhcF | putative transcriptional regulator | Regulatory functions: DNA interactions |
| 1166 | SMU.1276c | ezrA | putative septation ring formation regulator | Cellular processes: Cell division |
| 1209 | SMU.1324 | ftsX | putative cell-division protein FtsX | Cellular processes: Cell division |
| 1210 | SMU.1325 | ftsE | putative ABC transporter ATP-binding component | Cellular processes: Cell division |
| 1329 | SMU.1460 | rmlC | putative dTDP-4-keto-L-rhamnose reductase | Cell envelope: Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides |
| 1330 | SMU.1461 | rfbA rmlA | putative glucose-1-phosphate thymidyltransferase | Cell envelope Biosynthesis and degradation of surface polysaccharides and lipoyolysaccharides |
| 1444 | SMU.1589c | cpoA ywaF | putative hexosyltransferase | Cell envelope: Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides |
| 1468 | SMG.1614 | fpg mutM | putative formamidopyrimidine-DNA glycosylase | DNA metabolism: DNA replication, recombination, and repair |
| 1483 | SMU.1629c | putative cell division DNA segregation ATPase | Cellular processes: Cell division | |
| 1551 | SMU.1703c | conserved hypothetical protein | Hypothetical proteins | |
| 1587 | SMU.1741 | fabD | putative malonyl-COA (acyl-carrier-protein) transacylase | Fatty acid and phospholipid metabolism: Biosynthesis |
| 1592 | SMU.1746c | fabM phaB | putative enoyl-CoA hydratase | Fatty acid and phospholipid metabolism: Degradation |
| 1769 | SMU.1947 | nusG | putative transcription antitermination factor | Transcription: Transcription factors |
| 1771 | SMU.1949 | pbp2a | putative membrane carboxypeptidase, penicillin-binding protein 2a | Cell envelope: Biosynthesis and degradation of murein sacculus and peptidoglycan |
| 1812 | SMU.1995c | adcR zitR | putative transcriptional regulator | Regulatory functions. Other |
| 1823 | SMU.2007 | rl15 rplO | 50S ribosomal protein L15 | Protein synthesis: Ribosomal proteins: synthesis and modification |
| 1086 | SMU.2078c | conserved hypothetical protein | Hypothetical: Conserved | |
| C. Deletion strains exhibiting poor survival in acid, peroxide and poor biofilm formation | ||||
|---|---|---|---|---|
| ID | Locus | Gene Name(s) | Description | Functional Class |
| 0022 | SMU.26 | plsX ylpD | putative fatty acid/phospholipid synthesis protein | Fatty acid and phospholipid metabolism: Biosynthesis |
| 0137 | SMU.154 | rpsO | 30S ribosomal protein S15 | Protein synthesis: Ribosomal proteins: synthesis and modification |
| 0153 | SMU.170 | rpsl | 30S ribosomal protein S9 | Protein synthesis: Ribosomal proteins: synthesis and modification |
| 0223 | SMU.246 | rgpG | putative glycosyl transferase N-acetylglucosaminyltransferase), RgpG | Cell envelope: Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides |
| 0275 | SMU 304 | cdd | putative deaminase | Unknown function: Enzymes of unknown specificity |
| 0317 | SMU.352 | rpe | putative ribulose-phosphate-3-epimerase | Energy metabolism: Pentose phosphate pathway |
| 0318 | SMU.353 | conserved hypothetical protein | Biosynthesis of cofactors, prosthetic groups, and carriers: Thiamine | |
| 0431 | SMU.475 | conserved hypothetical protein | Unknown function: General | |
| 0434 | SMU.480 | priA | primosomal replication factor Y (primosomal protein N) | DNA metabolism: DNA replication, recombination, and repair |
| 0437 | SMU.483 | pphA pppL | putative phosphoprotein phosphatase (pppL protein) | Regulatory functions. Protein interactions |
| 0500 | SMU.550 | diviB ftsQ | putative cell division protein FtsQ (DivlB) | Cellular processes Cell division |
| 0521 | SMU 572 | folD | putative tetrahydrofolate dehydrogenase/cyclohydrolase | Biosynthesis of cofactors, prosthetic groups, and carriers: Folic acid |
| 0571 | SMU 629 | sod sodA | putative manganese-type superoxide dismutase, Fe/Mn SOD | Cellular processes Detoxification |
| 0645 | SMU.706c | conserved hypothetical protein | Transport and binding proteins: Other | |
| 0672 | SMU.739c | hypothetical protein | Hypothetical proteins: Conserved | |
| 0705 | SMU.777 | aroD | putative 3-dehydroquinate dehydratase | Amino acid biosynthesis Aromatic amino acid family |
| 0706 | SMU.778 | aroE | putative shikimate 5-dehydrogenase | Amino acid biosynthesis: Aromatic amino acid family |
| 0708 | SMU.780 | aroC | putative chorismate synthase | Amino acid biosynthesis: Aromatic amino acid family |
| 0749 | SMU.825 | rgpAc | putative RgpAc glycosyltransferase | Unclassified: Role category not yet assigned |
| 0750 | SMU.826 | rgpBc | rhamnosyltransferase | Cell envelope: Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides |
| 0754 | SMU 830 | rgpFc | RgpFc protein | Unclassified: Role category not yet assigned |
| 0792 | SMU.869 | trxB | putative thioredoxin reductase | Unknown function: General |
| 0985 | SMU.1077 | pgmA | putative phosphoglucomutase | Cell envelope: Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides |
| 0988 | SMU.1080c | conserved hypothetical protein possible transposon-related protein | Hypothetical proteins: Domain | |
| 0991 | SMU.1083c | conserved hypothetical protein | Unknown function General | |
| 1035 | SMU.1133 | phoU | putative phosphate transport system regalatory protein | Regulatory functions: Other |
| 1092 | SMU.1193 | yhcF | putative transcriptional regulator | Regulatory functions: DNA interactions |
| 1166 | SMU.1276c | ezrA | putative septation ring formation regulator | Cellular processes: Cell division |
| 1209 | SMU.1324 | ftsX | putative cell-division protein FtsX | Cellular processes: Cell division |
| 1210 | SMU 1325 | ftsE | putative ABC transporter, ATP-binding component | Cellular processes: Cell division |
| 1329 | SMU.1460 | rmlC | putative dTDP-4-keto-L-rhamnose reductase | Cell envelope: Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides |
| 1330 | SMU.1461 | rfbA rmlA | putative glucose 1-phosphate thymidyltransferase | Cell envelope: Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides |
| 1444 | SMU.1589c | cpoA ywaF | putative hexosytransterase | Cell envelope: Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides |
| 1483 | SMU.1629c | putative cell division protein DNA segregation ATPase | Cellular processes: Cal division | |
| 1551 | SMU 1703c | conserved hypothetical protein | Hypothetical proteins | |
| 1587 | SMU.1741 | fabD | putative malonyl-CoA (acyl-carrier-protein) transacylase | Fatty acid and phospholipid metabolism: Biosynthesis |
| 1592 | SMU.1746c | fabM phaB | putative enoyl-CoA hydratase | Fatty acid and phospholipid metabolism: Biosynthesis |
| 1823 | SMU 2007 | rl15 rplO | 50S ribosomal protein L15 | Protein synthesis: Ribosomal proteins: synthesis and modification |
| 1886 | SMU.2078c | conserved hypothetical protein | Hypothetical proteins: Conserved | |
51 deletion mutant strains of S. mutans that displayed relative fitness score values of 2 or less for both aerobic growth at pH 5.4 and biofilm formation in 0.5× tryptone-yeast extract (TY) medium with 29 mM sucrose (TYS).
49 deletion mutant strains of S. mutans that displayed relative fitness score values of 2 or less for aerobic growth in medium containing 0.5 mM H2O2 or medium containing 5 µM 8-hydroxyquinoline (8-HQ) and also for biofilm formation in 0.5× tryptone-yeast extract (TY) medium with 29 mM sucrose (TYS).
49 deletion mutant strains of S. mutans that displayed relative fitness score values of 2 or less for aerobic growth in either medium titrated to pH 5.4, medium containing 0.5 mM H2O2, or medium containing 5 µM 8-hydroxyquinoline (8-HQ) and also for biofilm formation in 0.5× tryptone-yeast extract (TY) medium with 29 mM sucrose (TYS).
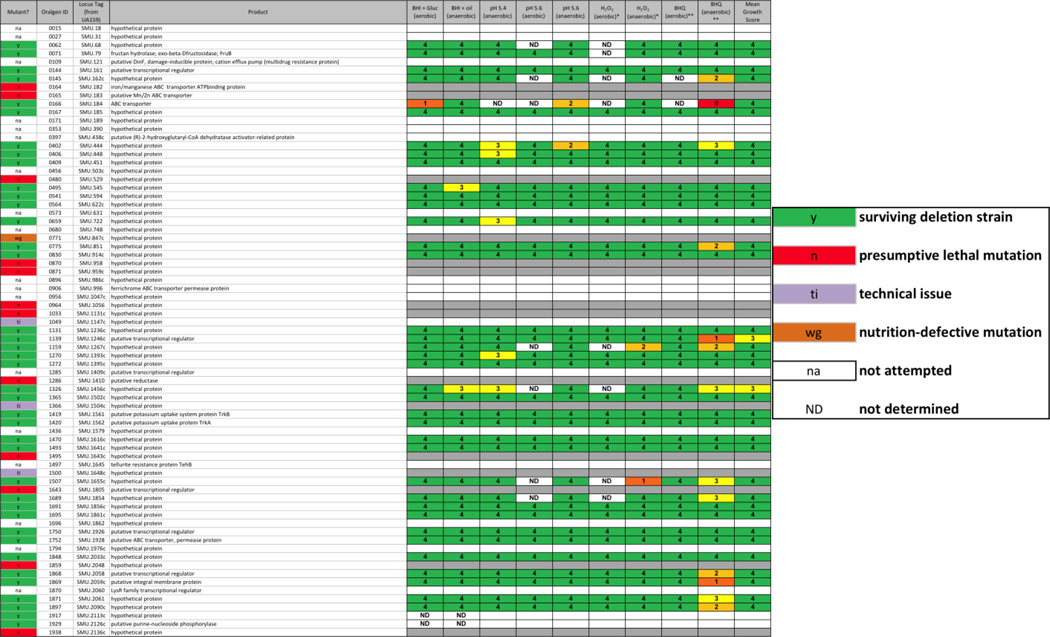
Recently, Cornejo, et al. published a pan-genome sequence for mutans streptococci based on sequences from 57 strains (Cornejo, et al. 2013). The core genome contained 1490 genes common to all strains and 73 conserved hypothetical genes, most of unknown function, though some with an assigned putative function. Examination of these genes in the present database shows that we attempted to delete 55 of the 73 genes, resulting in the isolation of 39 surviving strains, 4 strains that could not be sub-cultured following transformation, and 12 genes that could not be deleted (Table 2). Of the surviving strains, few of them exhibited strong phenotypic differences from the parent strain, though the greatest number of altered phenotypes were seen when the strains were grown in the presence of 8-hydroxyquinoline and oxygen (13 strains), the majority of which reverted to parental rates of growth in the micro-aerobic condition (11 strains), suggesting a role for metals and/or oxygen by these gene products. These results provide evidence toward the utility of the deletion collection for the identification and characterization of hypothetical proteins that would otherwise be missed.
TABLE 2. Unique core gene from S. mutans identified in the deletion strain collection.
In a study by Cornejo, et al., 73 genes were identified from a core S. mutans genome that had been annotated as encoding conserved hypothetical proteins (Cornejo, et al. 2013). Table 2 contains these 73 genes and the results from our deletion mutant study. In the far left column, red (n) cells indicate a putative lethal mutation, green (y) cells indicate successful mutation, purple (ti) cells indicate a technical issue that precluded the making of a deletion strain, orange (wg) cells indicate that resulting transformants were unable to grow in liquid medium for the preparation of a frozen stock, and white (na) cells indicate that the construct was not attempted due to inability of our primer calling algorithm to design primers for the deletion construct. Scoring for each physiological screen was performed as described in METHODS, on a scale from 0 – 5; ND = not determined.
In vitro growth and testing of deletion mutant strains
The primary purpose of creating the S. mutans deletion collection was to investigate which of the deletions would indicate presumptive “essential” genes, and which of the surviving “non-essential” genes would be affected in the ability to grow in competition with other deletion strains. We conducted a pilot experiment to evaluate the ability of specific deletion strains to survive growth in a culture consisting of a mixed population of deletion strains under fixed-pH chemostat conditions (Fozo & Quivey 2004b; Quivey, et al. 1995). We chose the following strains: MX804 (ErmR knock-in strain), MU0022 (∆plsX; SMU.26), MU0023 (∆acpP; SMU.27), MU0898 (∆cls; SMU.988), MU1020 (∆nox; SMU.1117), MU1591 (∆fabT; SMU.1745c), MU1592 (∆fabM; SMU.1746c), and MU1593 (∆pgmB; SMU.1747c) (Buckley, et al. 2014; Derr, et al. 2012; Faustoferri, et al. 2014; MacGilvray, et al. 2012). This subset of strains was chosen to examine the effects of deletion of particular genes involved in cell membrane homeostasis. The experimental plan for the chemostat run is detailed in Fig. 3. Samples were removed from the chemostat culture at various times during the run to ascertain which strains were still able to thrive under the test conditions: steady-state pH values of 7 and 5 in the presence of other strains carrying deletions in gene products involved in cell wall synthesis (∆plsX, ∆acpP, ∆cls, ∆fabT, ∆fabM), or in transport or breakdown of metabolic intermediates (∆nox, ∆pgmB). PCR performed with primers specific to the ErmR cassette contained in the control strain, MX804, yielded an amplicon in all samples removed at every time point (Fig. 3B). Similarly, gene-replacement-specific primers for pgmB (SMU.1747c) were able to amplify a product at all time points, representing the presence of the MU1593 strain (Fig. 3C). However, in the case of plsX, the amplicon decreased in predominance as the culture pH was reduced to 5. In fact, as the plsX-ErmR cassette became less abundant, another band appeared on the gel representing the ErmR cassette in the adjacent ORF, acpP (Fig. 3D, F). This finding was mirrored in the results obtained with the fabM-ErmR cassette: as the culture pH decreased, the strain lacking fabM was lost in the mixed culture (Fig. 3E, G). We have previously demonstrated that loss of fabM results in severe acid-sensitivity (Fozo & Quivey 2004a); therefore, the outcome here was anticipated.
Results from this experiment demonstrate the potential uses for the deletion collection in mixed culture experiments to examine the relative fitness of individual deletion strains; for example, the fluctuating environmental conditions mimicking the oral cavity.
In vivo growth and testing of deletion mutant strains
Examination of the pathophysiology of S. mutans should, by necessity, include determination of the fitness of a particular strain in the rat model of oral infection. Consequently, we conducted a pilot study to test infection and transmission of a subset of deletion strains, particularly those associated with the malolactic acid pathway, in the rat model (Lemme, et al. 2010; Sheng, et al. 2010; Sheng & Marquis 2007). The malolactate pathway is controlled by MleR (SMU.135) (Sheng, et al. 2010), and consists of a malate permease (mleP; SMU.138) and the malolactic synthase (mleS; SMU.137), which splits malate into carbon dioxide and lactic acid. The resulting carbon dioxide is available to carbonic anhydrase (cah; SMU.1595) for conversion to bicarbonate, which would buffer the cytoplasm. The malolactate fermentation system has been shown to be an important aspect of acid tolerance in S. mutans and, indeed, it has been demonstrated that the enzymatic activity is induced in low pH growth conditions (Sheng & Marquis 2007). The experimental design is detailed in Fig. 4A and is essentially similar to published work from our laboratory (Fozo, et al. 2007). Here, we were interested in examining whether the presence of other deletion strains in the inoculum used to infect rats would have an effect on the ability of competing members of the pool to infect or to be transmitted to non-infected cage mates. Presence or absence of a particular deletion strain at the completion of the experimental period was assayed by PCR using gene-deletion-specific primers for the individual strains in the inoculum pool, similar to the method used above for the mixed chemostat culture experiment. As shown in Fig. 4B, all the strains were identified in the inoculum pool at the start of the experiment; however, after 19 days in vivo on rat jaws, 2 members of the pool were no longer detected by PCR amplification (Fig. 4C), the fabM (SMU.1746c; MU1592) and mleR (SMU.135; MU0121) strains. These data indicate the further utility of the collection: to investigate differences in the competitive fitness, in vivo, of specific members of the deletion collection.
Conclusions
Fitness profiling of genomic deletion collections has provided a wealth of information on everything from responses to chemicals or drugs (Lopez, et al. 2008; Nichols, et al. 2011; Rooney, et al. 2008; Xu, et al. 2007), to genes involved in particular processes (Valentino, et al. 2014; Korte, et al. 2014) (Muller-Herbst, et al. 2014), to interactions between gene networks (Gagarinova, et al. 2012; Kuzmin, et al. 2014; Wang, et al. 2013). We report here the application of these principles to S. mutans, an organism with a critical role in dental caries. We have constructed barcoded deletions of 1112 genes in S. mutans and demonstrated that these can be used to assess defects in growth in conditions relevant to virulence. The value of the mutant collection is immediately obvious in the ability to identify mutations that cause defects in several conditions related to virulence. Moreover, the ability to mix mutant strains and then assess growth effects either in vitro or in the rat model, more accurately mimics the consequences of drug targeting in which only a portion of the population may be affected. Mutants that are not rescued by the presence of other wild-type strains are likely to represent the best effective drug targets.
Supplementary Material
The color-coding refers to the result of attempts to delete each gene in the S. mutans UA159 genome. Green indicates a surviving mutant strain, red indicates a putative lethal mutation (as described in Fig. 2 and in Methods), orange indicates a presumed nutritional requirement (as described in Fig. 2 and in Methods), purple indicates a technical issue with construction of the deletion construct that did not result in a deletion mutant strain, and white indicates that the gene was not attempted due to constraints in the design of primers required for the deletion construction.
The complete set of data obtained from the Streptococcus mutans deletion collection is presented, sorted by Los Alamos Oralgen ID number (http://www.oralgen.org/). Column A (Mutant?): Letter/color-coded entry to indicate ability to create a deletion strain of S. mutans UA159 in that particular ORF: red (n) cells indicate a putative lethal mutation, green (y) cells indicate successful mutation, purple (ti) cells indicate a technical issue that precluded the making of a deletion strain, orange (wg) cells indicate that resulting transformants were unable to grow in liquid medium for the preparation of a frozen stock, and white (na) cells indicate that the construct was not attempted due to inability of our primer calling algorithm to design primers for the deletion construct. Column B (Oralgen ID): corresponds to ORF designation from the Los Alamos Oral Pathogen Database for each particular gene, format SMu####. Column C (Locus): corresponds to NCBI designation for that particular ORF in reference sequence NC_004350.2. Column D (Upstream Barcode): 20 bp unique sequence incorporated upstream of the particular ORF. Column E (Downstream Barcode): 20 bp unique sequence incorporated downstream of the particular ORF. Column F (Gene Name(s)): annotated gene names corresponding to the specific locus. Column G (Common Name): descriptor for gene function as annotated by Los Alamos Oral Pathogens Database. Column H (Functional Class): predicted functional class for the specific locus. Columns J – S: relative fitness score (as detailed in Methods), on a scale from 0 – 5, ND = not determined, for each of the individual physiological screens used to test the survival of each deletion strain, as compared to the parent strain, UA159. Aerobic = growth in a Bioscreen C automated plate reader without mineral oil overlay; Anaerobic = growth in a Bioscreen C automated plate reader with a mineral oil overlay to create a micro-aerobic environment; BHI = Brain Heart Infusion medium; Gluc = 44 mM glucose added to culture medium; pH 5.4/pH 5.6 = BHI medium titrated to these pH values with HCl; H2O2 = 0.5 mM hydrogen peroxide added to BHI culture medium; 8HQ = 5 µM 8-hydroxyquinoline added to BHI culture medium. Column U (Mean Growth Score) was calculated from the relative fitness scores after each physiological assay, as compared to the parent strain, UA159. Column W and X (BF Formation Score (TYG) and BF Formation Score (TYS), respectively): relative ability of the deletion strain to form a biofilm on polystyrene microtiter plates (described in Methods), as compared to the parent strain, in 0.5× tryptone-yeast extract (TY) medium with the addition of either 56 mM glucose (TYG) or 29 mM sucrose (TYS). Column Z (Min pH): Minimum endpoint glycolytic pH value for individual deletion strains as determined by the method of Belli and Marquis (Belli and Marquis 1991). Column AA (Diff from Control (Mutant – UA159)): difference in minimum endpoint glycolytic pH between the deletion strain and the parent strain. Column AB (p-value): statistical significance of the variation in the minimum endpoint glycolytic pH value from the deletion strain vs. the parent strain, as determined by Student’s t-test.
List of primers used for polymerase chain reactions using DNA derived from samples taken during in vitro and in vivo growth and testing experiments using deletion mutant strains. Presence or absence of a particular deletion strain was assayed by PCR using gene-deletion-specific primers for the individual strains in inoculum pools used in chemostat growth experiments and in the rat model of infection for dental caries. The Erm Left (ErmL) and Erm Right (ErmR) primers were used in all phases of the study from screening the constructs to detection in the mixed pool experiments. These primers were paired with gene-deletion-specific primers as noted in the “Paired Primer” column, where “a” denotes a pairing with ErmL and “b” denotes a pairing with ErmR.
Acknowledgements
This study was supported by Public Health Service grants from the National Institute for Dental and Craniofacial Research DE017425 (R.G.Q), DE017157 (R.G.Q.), and T90 DE-021985 (B.S.). Support also came from CAPES BEX2827/07-7 and CNPq (302222/2008-1) Brazil (P.L.R.).
The authors thank Dr. Gene Watson, U. of Rochester Eastman Institute for Oral Health, for his assistance with the rat infection experiments. We also thank Dr. Eric Phizicky (U. of Rochester Dept. of Biochemistry and Biophysics) for his helpful suggestions and discussion, throughout.
Footnotes
Table S1. S. mutans fitness database table.
Table S2. Primers used in in vitro and in vivo fitness competition experiments.
References
- Aas JA, Griffen AL, Dardis SR, et al. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol. 2008;46:1407–1417. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SJ, Lemos JA, Burne RA. Role of HtrA in growth and competence of Streptococcus mutans UA159. J Bacteriol. 2005;187:3028–3038. doi: 10.1128/JB.187.9.3028-3038.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajdic D, McShan WM, McLaughlin RE, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki H, Shiroza T, Hayakawa M, Sato S, Kuramitsu HK. Cloning of a Streptococcus mutans glucosyltransferase gene coding for insoluble glucan synthesis. Infect Immun. 1986;53:587–594. doi: 10.1128/iai.53.3.587-594.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker MR, Paster BJ, Leys EJ, et al. Molecular analysis of bacterial species associated with childhood caries. J Clin Microbiol. 2002;40:1001–1009. doi: 10.1128/JCM.40.3.1001-1009.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belli WA, Marquis RE. Adaptation of Streptococcus mutans and Enterococcus hirae to acid stress in continuous culture. Appl Environ Microbiol. 1991;57:1134–1138. doi: 10.1128/aem.57.4.1134-1138.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose JL, Fey PD, Bayles KW. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl Environ Microbiol. 2013;79:2218–2224. doi: 10.1128/AEM.00136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen WH, Koo H. Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 2011;45:69–86. doi: 10.1159/000324598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen WH, Madison KM, Pearson SK. Influence of desalivation in rats on incidence of caries in intact cagemates. J Dent Res. 1988a;67:1316–1318. doi: 10.1177/00220345880670101401. [DOI] [PubMed] [Google Scholar]
- Bowen WH, Pearson SK, Young DA. The effect of desalivation on coronal and root surface caries in rats. J Dent Res. 1988b;67:21–23. doi: 10.1177/00220345880670010301. [DOI] [PubMed] [Google Scholar]
- Buckley AA, Faustoferri RC, Quivey RG., Jr beta-Phosphoglucomutase contributes to aciduricity in Streptococcus mutans. Microbiology. 2014;160:818–827. doi: 10.1099/mic.0.075754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo OE, Lefebure T, Bitar PD, et al. Evolutionary and population genomics of the cavity causing bacteria Streptococcus mutans. Mol Biol Evol. 2013;30:881–893. doi: 10.1093/molbev/mss278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costalonga M, Herzberg MC. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol Lett. 2014;162:22–38. doi: 10.1016/j.imlet.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derr AM, Faustoferri RC, Betzenhauser MJ, Gonzalez K, Marquis RE, Quivey RG., Jr Mutation of the NADH oxidase gene (nox) reveals an overlap of the oxygen- and acid-mediated stress responses in Streptococcus mutans. Appl Environ Microbiol. 2012;78:1215–1227. doi: 10.1128/AEM.06890-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustoferri RC, Hubbard CJ, Santiago B, Buckley AA, Seifert TB, Quivey RG. Regulation of fatty acid biosynthesis by the global regulator CcpA and the local regulator FabT in Streptococcus mutans. Mol Oral Microbiol. 2014 doi: 10.1111/omi.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey PD, Endres JL, Yajjala VK, et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio. 2013;4:e00537-00512. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozo EM, Kajfasz JK, Quivey RG., Jr Low pH-induced membrane fatty acid alterations in oral bacteria. FEMS Microbiol Lett. 2004;238:291–295. doi: 10.1016/j.femsle.2004.07.047. [DOI] [PubMed] [Google Scholar]
- Fozo EM, Quivey RG., Jr The fabM gene product of Streptococcus mutans is responsible for the synthesis of monounsaturated fatty acids and is necessary for survival at low pH. J Bacteriol. 2004a;186:4152–4158. doi: 10.1128/JB.186.13.4152-4158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozo EM, Quivey RG., Jr Shifts in the membrane fatty acid profile of Streptococcus mutans enhance survival in acidic environments. Appl Environ Microbiol. 2004b;70:929–936. doi: 10.1128/AEM.70.2.929-936.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozo EM, Scott-Anne K, Koo H, Quivey RG., Jr Role of unsaturated fatty acid biosynthesis in virulence of Streptococcus mutans. Infect Immun. 2007;75:1537–1539. doi: 10.1128/IAI.01938-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagarinova A, Babu M, Greenblatt J, Emili A. Mapping bacterial functional networks and pathways in Escherichia coli using synthetic genetic arrays. J Vis Exp. 2012 doi: 10.3791/4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Kitten T, Chen Z, Lee SP, Munro CL, Xu P. Identification of Streptococcus sanguinis genes required for biofilm formation and examination of their role in endocarditis virulence. Infect Immun. 2008;76:2551–2559. doi: 10.1128/IAI.00338-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Gross EL, Beall CJ, Kutsch SR, Firestone ND, Leys EJ, Griffen AL. Beyond Streptococcus mutans : dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS One. 2012;7:e47722. doi: 10.1371/journal.pone.0047722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Kajfasz JK, Rivera-Ramos I, Abranches J, et al. Two Spx proteins modulate stress tolerance, survival, and virulence in Streptococcus mutans. J Bacteriol. 2010;192:2546–2556. doi: 10.1128/JB.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanasi E, Dewhirst FE, Chalmers NI, et al. Clonal analysis of the microbiota of severe early childhood caries. Caries Res. 2010;44:485–497. doi: 10.1159/000320158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein BA, Tenorio EL, Lazinski DW, Camilli A, Duncan MJ, Hu LT. Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC Genomics. 2012;13:578. doi: 10.1186/1471-2164-13-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte HL, Fels SR, Christensen GA, et al. Genetic basis for nitrate resistance in Desulfovibrio strains. Front Microbiol. 2014;5:153. doi: 10.3389/fmicb.2014.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin E, Sharifpoor S, Baryshnikova A, et al. Synthetic genetic array analysis for global mapping of genetic networks in yeast. Methods Mol Biol. 2014;1205:143–168. doi: 10.1007/978-1-4939-1363-3_10. [DOI] [PubMed] [Google Scholar]
- Lau PC, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods. 2002;49:193–205. doi: 10.1016/s0167-7012(01)00369-4. [DOI] [PubMed] [Google Scholar]
- Lemme A, Sztajer H, Wagner-Dobler I. Characterization of mleR a positive regulator of malolactic fermentation and part of the acid tolerance response in Streptococcus mutans. BMC Microbiol. 2010;10:58. doi: 10.1186/1471-2180-10-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos JA, Quivey RG, Jr, Koo H, Abranches J. Streptococcus mutans : a new Gram-positive paradigm? Microbiology. 2013;159:436–445. doi: 10.1099/mic.0.066134-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Tang N, Aspiras MB, et al. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J Bacteriol. 2002;184:2699–2708. doi: 10.1128/JB.184.10.2699-2708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Faller LL, Klitgord N, et al. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS One. 2012;7:e37919. doi: 10.1371/journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez A, Parsons AB, Nislow C, Giaever G, Boone C. Chemical-genetic approaches for exploring the mode of action of natural products. Prog Drug Res. 2008;66:237, 239–271. doi: 10.1007/978-3-7643-8595-8_5. [DOI] [PubMed] [Google Scholar]
- MacGilvray ME, Lapek JD, Jr, Friedman AE, Quivey RG., Jr Cardiolipin biosynthesis in Streptococcus mutans is regulated in response to external pH. Microbiology. 2012;158:2133–2143. doi: 10.1099/mic.0.057273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis RE. Oxygen metabolism, oxidative stress and acid-base physiology of dental plaque biofilms. J Ind Microbiol. 1995;15:198–207. doi: 10.1007/BF01569826. [DOI] [PubMed] [Google Scholar]
- Muller-Herbst S, Wustner S, Muhlig A, et al. Identification of genes essential for anaerobic growth of Listeria monocytogenes. Microbiology. 2014;160:752–765. doi: 10.1099/mic.0.075242-0. [DOI] [PubMed] [Google Scholar]
- Murchison HH, Barrett JF, Cardineau GA, Curtiss R., 3rd Transformation of Streptococcus mutans with chromosomal and shuttle plasmid (pYA629) DNAs. Infect Immun. 1986;54:273–282. doi: 10.1128/iai.54.2.273-282.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols RJ, Sen S, Choo YJ, et al. Phenotypic landscape of a bacterial cell. Cell. 2011;144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyvad B, Crielaard W, Mira A, Takahashi N, Beighton D. Dental caries from a molecular microbiological perspective. Caries Res. 2013;47:89–102. doi: 10.1159/000345367. [DOI] [PubMed] [Google Scholar]
- O'Connell AC, Van Wuyckhuyse BC, Pearson SK, Bowen WH. The effect of propranolol on salivary gland function and dental caries development in young and aged rats. Arch Oral Biol. 1993;38:853–861. doi: 10.1016/0003-9969(93)90094-3. [DOI] [PubMed] [Google Scholar]
- Perry D, Kuramitsu HK. Genetic transformation of Streptococcus mutans. Infect Immun. 1981;32:1295–1297. doi: 10.1128/iai.32.3.1295-1297.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce SE, Davis RW, Nislow C, Giaever G. Genome-wide analysis of barcoded Saccharomyces cerevisiae gene-deletion mutants in pooled cultures. Nat Protoc. 2007;2:2958–2974. doi: 10.1038/nprot.2007.427. [DOI] [PubMed] [Google Scholar]
- Quivey RG, Jr, Faustoferri RC, Belli WA, Flores JS. Polymerase chain reaction amplification, cloning, sequence determination and homologies of streptococcal ATPase-encoding DNAs. Gene. 1991;97:63–68. doi: 10.1016/0378-1119(91)90010-9. [DOI] [PubMed] [Google Scholar]
- Quivey RG, Jr, Faustoferri RC, Clancy KA, Marquis RE. Acid adaptation in Streptococcus mutans UA159 alleviates sensitization to environmental stress due to RecA deficiency. FEMS Microbiol Lett. 1995;126:257–261. doi: 10.1111/j.1574-6968.1995.tb07427.x. [DOI] [PubMed] [Google Scholar]
- Rooney JP, Patil A, Zappala MR, Conklin DS, Cunningham RP, Begley TJ. A molecular bar-coded DNA repair resource for pooled toxicogenomic screens. DNA Repair (Amst) 2008;7:1855–1868. doi: 10.1016/j.dnarep.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Santiago B, MacGilvray M, Faustoferri RC, Quivey RG., Jr The branched-chain amino acid aminotransferase encoded by ilvE is involved in acid tolerance in Streptococcus mutans. J Bacteriol. 2012;194:2010–2019. doi: 10.1128/JB.06737-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago B, Marek M, Faustoferri RC, Quivey RG., Jr The Streptococcus mutans aminotransferase encoded by ilvE is regulated by CodY and CcpA. J Bacteriol. 2013;195:3552–3562. doi: 10.1128/JB.00394-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng J, Baldeck JD, Nguyen PT, Quivey RG, Jr, Marquis RE. Alkali production associated with malolactic fermentation by oral streptococci and protection against acid, oxidative, or starvation damage. Can J Microbiol. 2010;56:539–547. doi: 10.1139/w10-039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng J, Marquis RE. Malolactic fermentation by Streptococcus mutans. FEMS Microbiol Lett. 2007;272:196–201. doi: 10.1111/j.1574-6968.2007.00744.x. [DOI] [PubMed] [Google Scholar]
- Smith AM, Heisler LE, St Onge RP, et al. Highly-multiplexed barcode sequencing: an efficient method for parallel analysis of pooled samples. Nucleic Acids Res. 2010;38:e142. doi: 10.1093/nar/gkq368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Nyvad B. Caries ecology revisited: microbial dynamics and the caries process. Caries Res. 2008;42:409–418. doi: 10.1159/000159604. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Nyvad B. The role of bacteria in the caries process: ecological perspectives. J Dent Res. 2011;90:294–303. doi: 10.1177/0022034510379602. [DOI] [PubMed] [Google Scholar]
- Valentino MD, Foulston L, Sadaka A, et al. Genes contributing to Staphylococcus aureus fitness in abscess- and infection-related ecologies. MBio. 2014;5:e01729-01714. doi: 10.1128/mBio.01729-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang X, Arkin AP, Samoilov MS. Inference of gene regulatory networks from genome-wide knockout fitness data. Bioinformatics. 2013;29:338–346. doi: 10.1093/bioinformatics/bts634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Jiang B, Ketela T, et al. Genome-wide fitness test and mechanism-of-action studies of inhibitory compounds in Candida albicans. PLoS Pathog. 2007;3:e92. doi: 10.1371/journal.ppat.0030092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The color-coding refers to the result of attempts to delete each gene in the S. mutans UA159 genome. Green indicates a surviving mutant strain, red indicates a putative lethal mutation (as described in Fig. 2 and in Methods), orange indicates a presumed nutritional requirement (as described in Fig. 2 and in Methods), purple indicates a technical issue with construction of the deletion construct that did not result in a deletion mutant strain, and white indicates that the gene was not attempted due to constraints in the design of primers required for the deletion construction.
The complete set of data obtained from the Streptococcus mutans deletion collection is presented, sorted by Los Alamos Oralgen ID number (http://www.oralgen.org/). Column A (Mutant?): Letter/color-coded entry to indicate ability to create a deletion strain of S. mutans UA159 in that particular ORF: red (n) cells indicate a putative lethal mutation, green (y) cells indicate successful mutation, purple (ti) cells indicate a technical issue that precluded the making of a deletion strain, orange (wg) cells indicate that resulting transformants were unable to grow in liquid medium for the preparation of a frozen stock, and white (na) cells indicate that the construct was not attempted due to inability of our primer calling algorithm to design primers for the deletion construct. Column B (Oralgen ID): corresponds to ORF designation from the Los Alamos Oral Pathogen Database for each particular gene, format SMu####. Column C (Locus): corresponds to NCBI designation for that particular ORF in reference sequence NC_004350.2. Column D (Upstream Barcode): 20 bp unique sequence incorporated upstream of the particular ORF. Column E (Downstream Barcode): 20 bp unique sequence incorporated downstream of the particular ORF. Column F (Gene Name(s)): annotated gene names corresponding to the specific locus. Column G (Common Name): descriptor for gene function as annotated by Los Alamos Oral Pathogens Database. Column H (Functional Class): predicted functional class for the specific locus. Columns J – S: relative fitness score (as detailed in Methods), on a scale from 0 – 5, ND = not determined, for each of the individual physiological screens used to test the survival of each deletion strain, as compared to the parent strain, UA159. Aerobic = growth in a Bioscreen C automated plate reader without mineral oil overlay; Anaerobic = growth in a Bioscreen C automated plate reader with a mineral oil overlay to create a micro-aerobic environment; BHI = Brain Heart Infusion medium; Gluc = 44 mM glucose added to culture medium; pH 5.4/pH 5.6 = BHI medium titrated to these pH values with HCl; H2O2 = 0.5 mM hydrogen peroxide added to BHI culture medium; 8HQ = 5 µM 8-hydroxyquinoline added to BHI culture medium. Column U (Mean Growth Score) was calculated from the relative fitness scores after each physiological assay, as compared to the parent strain, UA159. Column W and X (BF Formation Score (TYG) and BF Formation Score (TYS), respectively): relative ability of the deletion strain to form a biofilm on polystyrene microtiter plates (described in Methods), as compared to the parent strain, in 0.5× tryptone-yeast extract (TY) medium with the addition of either 56 mM glucose (TYG) or 29 mM sucrose (TYS). Column Z (Min pH): Minimum endpoint glycolytic pH value for individual deletion strains as determined by the method of Belli and Marquis (Belli and Marquis 1991). Column AA (Diff from Control (Mutant – UA159)): difference in minimum endpoint glycolytic pH between the deletion strain and the parent strain. Column AB (p-value): statistical significance of the variation in the minimum endpoint glycolytic pH value from the deletion strain vs. the parent strain, as determined by Student’s t-test.
List of primers used for polymerase chain reactions using DNA derived from samples taken during in vitro and in vivo growth and testing experiments using deletion mutant strains. Presence or absence of a particular deletion strain was assayed by PCR using gene-deletion-specific primers for the individual strains in inoculum pools used in chemostat growth experiments and in the rat model of infection for dental caries. The Erm Left (ErmL) and Erm Right (ErmR) primers were used in all phases of the study from screening the constructs to detection in the mixed pool experiments. These primers were paired with gene-deletion-specific primers as noted in the “Paired Primer” column, where “a” denotes a pairing with ErmL and “b” denotes a pairing with ErmR.