Abstract
falafel (flfl) encodes a Drosophila homolog of human SMEK whose in vivo functions remain elusive. In this study, we performed gain-of-function and loss-of-function analysis in Drosophila and identified flfl as a negative regulator of JNK pathway-mediated cell death. While ectopic expression of flfl suppresses TNF-triggered JNK-dependent cell death, loss of flfl promotes JNK activation and cell death in the developing eye and wing. These data report for the first time an essential physiological function of flfl in maintaining tissue homeostasis and organ development. As the JNK signaling pathway has been evolutionary conserved from fly to human, a similar role of PP4R3 in JNK-mediated physiological process is speculated.
1. Introduction
falafel (flfl) is a Drosophila protein phosphatase 4 (PP4) regulatory subunit 3 (PP4R3) [1], which specifically mediates Miranda (Mira) localization and determinants cell fate during both interphase and mitosis [2]. flfl binds to CENP-C with its EVH1 domain [3] that is crucial for PP4 catalytic activity to centromeres at chromosomes during mitosis. Previous study proposed that PP4 functions through the modular activity of its component subunits [3]. Although in vitro studies have reported that PP4 is involved in a variety of molecular and cellular processes including regulation of c-Jun N-terminal kinase (JNK) pathway [4], NF-kB pathway [5], hematopoietic progenitor kinase 1 [6], apoptosis [7], and cell division [8], flfl's in vivo functions remain poorly understood. The human homolog of flfl is SMEK, which recruits PP4c to promote neuronal differentiation by dephosphorylating Par3 [9]. However, other in vivo functions of SMEK remain largely elusive.
The JNK pathway is evolutionary conserved from Drosophila to mammal [10]. As its genome has low redundancy, Drosophila has been used as an excellent genetic model to study tumor necrosis factor- (TNF-) induced cell death in development. In Drosophila, the TNF ortholog Eiger (Egr) triggers cell death through its receptor Grindelwald (Grnd) [11], the E2 ubiquitin conjugating enzyme complex Bendless/dUev1a [12, 13], the E3 ubiquitin ligase dTRAF2 [14], the TAK1-associated binding protein 2 Tab2 [15], and the dTAK1-Hep-Bsk (Drosophila homologs of JNKKK-JNKK-JNK) kinase cascade [16, 17]. In developing eyes, ectopically expressing Egr by GMR-Gal4 (GMR > Egr hereafter) induces JNK-dependent cell death and produces small eyes in adult [16, 17].
To identify additional factors that regulate Egr-triggered JNK-mediated cell death, we performed a genetic screen for dominant modifiers of the GMR > Egr small eye phenotype. From the screen, we found that expression of flfl suppresses Egr-triggered cell death. On the other hand, knocking down flfl induced JNK activation and JNK pathway-dependent cell death, suggesting a physiological function of flfl in animal development. To our knowledge, this is the first report that flfl negatively regulate TNF-JNK signaling-induced cell death in vivo.
2. Materials and Methods
2.1. Drosophila Strains
All stocks were raised on standard Drosophila media, and crosses were performed at 25°C. UAS-flfl-IR (V103793) was obtained from Vienna Drosophila Research Center, UAS-flfl-IR (31690), flfl EY03585, UAS-GFP-IR, and ap-Gal4 were obtained from Bloomington Stock Center, and UAS-bsk-IR (5680R-2) was from Fly Stocks of National Institute of Genetics (NIG). puc E69 [18], GMR-Gal4, en-Gal4, pnr-Gal4, UAS-GFP [19, 20], UAS-Egr [16], UAS-Egrw [6], UAS-Hep, and UAS-BskDN [21] were previously described.
2.2. AO Staining
Eye discs from 3rd instar larvae were dissected in 1% PBS buffer. AO staining procedure was based on previous assay [22]. Florescent image of eye discs labeled with AO was collected with Olympus Microscope BX51. 10 discs of each genotype were collected for statistics analysis.
2.3. Light Image
3-day-old flies of each genotypes were collected and immediately frozen at −80°C. For the image, flies were mounted on 1% agarose plates. Light images of eye and thorax were documented with OLYMPUS stereo microscope SZX16.
2.4. X-Gal Staining
X-Gal staining was performed as previously described with minor modification [23, 24]. Wing imaginal discs from 3rd instar larvae were dissected in 1% PBS buffer and fixed with 1% glutaraldehyde for 15 minutes at room temperature and incubated with β-galactosidase at 37°C for 24 hours.
2.5. Data Analysis
Invasive breast carcinoma stroma versus normal data was obtained from Oncomine database (https://www.oncomine.org/).
3. Results and Discussion
3.1. flfl Suppresses Egr-Induced Cell Death in Eye Development
As previous study showed, ectopic expression of Egr under the control of GMR-Gal4 induced a small eye phenotype [17]. This phenotype is mostly suppressed by coexpressing a dominant negative allele of Bsk (BskDN) encoding the Drosophila JNK ortholog [21], which indicates Egr-induced cell death is mainly mediated by JNK signaling [25]. To identify additional components of the Egr-JNK pathway or factors interacting with the pathway, we performed a genetic screen for dominant modifiers of the GMR > Egr small eye phenotype and identified Nopo, Ben, Wnd, and Wg signaling as essential regulator of Egr-JNK pathway induced cell death [21, 26].
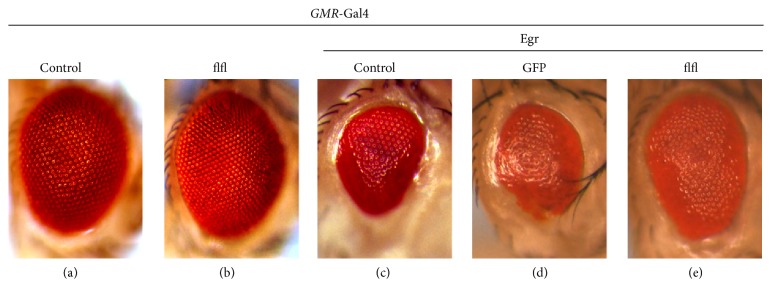
From the screen, we also found that the GMR > Egr small eye phenotype (Figure 1(c)) was significantly suppressed by flfl EY03585 (Figure 1(e)), a P-element inserted in the first intron of flfl. This P-element carries the UAS sequence located about 1 kb upstream of the coding region and is able to drive the expression of flfl by the GMR-Gal4 driver. However, expression of flfl by itself had no effect on the eye size (Figure 1(b)), compared to the GMR-Gal4 control (Figure 1(a)). As a negative control, coexpressing GFP did not suppress GMR > Egr-triggered small eye phenotype (Figure 1(d)). Thus, the data indicate that flfl is able to suppress Egr-induced cell death in the eye.
Figure 1.
flfl suppress Egr-induced cell death in Drosophila eye. Light micrographs of Drosophila eyes are shown. Compared with the GMR-Gal4 control (a), GMR > Egr triggered cell death and produced a small eye phenotype (c), which was suppressed by expressing flfl (e) but not GFP (d). Expression of flfl produced no noticeable phenotype (b). Genotypes: GMR-Gal4/+ (a); GMR-Gal4/flfl EY03585 (b); UAS-Egr/+; GMR-Gal4/+ (c); UAS-Egr/UAS-GFP; GMR-Gal4/+ (d); UAS-Egr/+; GMR-Gal4/flfl EY03585 (e).
3.2. Loss of flfl Enhances Egr-Induced Cell Death in Eye Development
As flfl gain of function suppressed Egr-induced cell death, we wonder whether loss of flfl could enhance Egr-triggered cell death. To this end, we knocked down flfl in the eye by expressing flfl RNAi with GMR-Gal4 and observed a rough eye phenotype (Figure 2(d)), compared to the control (Figure 2(a)). Consistent with previous reports, expression of a weaker UAS-Egr allele (UAS-Egrw) driven by GMR-Gal4 resulted in a rough eye phenotype (Figure 2(b)). This phenotype is severely enhanced by knocking down flfl as there was almost no eye tissue left (Figure 2(e)). As a negative control, expressing a RNAi sequence specifically targeting green fluorescent protein (GFP) has no effect on GMR > Egrw-triggered rough eye phenotype (Figure 2(c)). These results show that flfl loss of function rigorously enhances Egr-triggered eye phenotype.
Figure 2.
Loss of flfl enhances Egr-triggered cell death in the developing eye. Light micrographs of Drosophila eyes ((a)–(e)) or acridine orange staining of eye discs from 3rd instar larvae ((f)–(j)) are shown. Compared with the GMR-Gal4 control ((a) and (f)), GMR > Egrw induced rough eye phenotype in adulthood (b) and cell death in larval eye disc (g) was not affected by expressing GFP RNAi ((c) and (h)) but was strongly enhanced by expressing flfl RNAi ((e) and (j)). Knocking down flfl alone caused a rough eye phenotype (d) and mild cell death in eye discs (i). Genotypes: GMR-Gal4/+ ((a) and (f)); UAS-Egrw/+; GMR-Gal4/+ ((b) and (g)); UAS-Egrw/UAS-GFP-IR; GMR-Gal4/+ ((c) and (h)); UAS-flfl-IR/+; GMR-Gal4/+ ((d) and (i)); UAS-Egrw/UAS-flfl-IR; GMR-Gal4/+ ((e) and (j)).
It was previously reported that ectopic Egr-induced eye phenotype is caused by cell death [16]. To examine cell death in vivo, we performed acridine orange (AO) staining that specifically labels dying cell. As reported previously [12], ectopic expression of a weak UAS-Egr transgene (UAS-Egrw) driven by GMR-Gal4 induced mild cell death in eye discs posterior to the morphogenetic furrow (MF), as revealed by AO staining (Figure 2(g)). Egr-triggered cell death was rigorously enhanced by expressing flfl RNAi (Figure 2(j)) but remained unaffected by expressing GFP RNAi (Figure 2(h)). Consistent with its rough eye phenotype, knocking down flfl provoked weak cell death (Figure 2(i)). These data suggest that loss of flfl enhances Egr-induced cell death in eye development.
3.3. Loss of flfl Enhances JNK-Mediated Cell Death in Thorax Development
To investigate whether flfl suppresses JNK-mediated cell death in other tissues, we activated JNK signaling in the notum with pannier-Gal4 (pnr-Gal4). Expression of Hep, the Drosophila homolog of JNK, driven by pnr-Gal4 induced cell death and produced a small scutellum in adult fly (Figure 3(d)) [21]. Knocking down flfl by pnr-Gal4 slightly decreased scutellum size (Figure 3(c)) and dramatically enhanced Hep-induced cell death by producing a no scutellum phenotype as well as a split thorax in adult flies (Figure 3(f)). As a negative control, expression of a GFP RNAi did not produce any effect on scutellum size (Figures 3(b) and 3(e)). Together, the results indicated that flfl negatively regulates JNK-mediated cell death in thorax development.
Figure 3.
Loss of flfl enhances JNK-mediated cell death in thorax. Light images of Drosophila adult thoraxes are shown. Compared with the wild type (a) and pnr > GFP-IR control (b), expression of Hep induced a small scutellum (d), which was dramatically enhanced by the expression of flfl RNAi (f), while expression of flfl RNAi slightly decreased scutellum size (c). Dashed rectangle indicates the scutellum. Genotypes: pnr-Gal4/+ (a); UAS-GFP-IR/+; pnr-Gal4/+ (b); UAS-flfl-IR/+; pnr-Gal4/+ (c); UAS-Hep/+; pnr-Gal4/+ (d); UAS-Hep/UAS-GFP-IR; pnr-Gal4/+ (e); UAS-Hep/UAS-flfl-IR; pnr-Gal4/+ (f).
During Drosophila imaginal discs development, slow-proliferating cells are eliminated by a process called “cell competition” [27], which regulates tissue's homeostasis and organs' fitness and final cell number. JNK pathway was shown to play a crucial role in cell competition by eliciting cell death in “loser cells” [28, 29]. Since our data suggest that flfl impedes JNK-mediated cell death in a nontissue specific manner, flfl is likely a negative regulator of JNK-dependent cell competition and tissue homeostasis.
3.4. Loss of flfl Induces JNK Pathway Activation and Cell Death in Wing Development
To investigate the physiological functions of flfl in wing development, we specifically knocked down flfl in the posterior compartment of wing discs by engrailed-Gal4 (en-Gal4) and checked cell death with AO staining. We found that loss of flfl triggered extensive cell death in the posterior compartment of wing discs (Figure 4(c)), compared with the en-Gal4 control (Figure 4(a)) and en > GFP-IR (Figure 4(b)). These results suggest that flfl is physiologically required for cell survival in Drosophila wing development.
Figure 4.
Loss of flfl induces JNK pathway activation and cell death in wing development. Drosophila 3rd instar wing discs with AO ((a)–(c)) and X-Gal staining ((d)–(f)) are shown. Knocking down flfl in the posterior compartment of wing discs by en-Gal4 induced extensively cell death (c) and puc-LacZ expression (f), while expressing a GFP RNAi failed to do so ((b) and (d)). en-Gal4 ((a) and (d)) served as controls. Dashed line indicates the anterior-posterior boundary of wing discs ((c) and (f)). Anterior boundary is to the left in all panels. Genotypes: en-Gal4/+ (a); en-Gal4/UAS-GFP-IR (b); en-Gal4/UAS-flfl-IR (c); en-Gal4/+; puc E69/+ (d); en-Gal4/+; puc E69/UAS-GFP-IR (e); en-Gal4/+; puc E69/UAS-flfl-IR (f). SMEK1 (g) and DUSP1 (h) relative expression level in invasive breast carcinoma stroma compared to normal tissue in Finak Breast dataset are shown. Reporter: A_24_P36961 and A_23_P110712 are probes used in the study to detect SMEK1 and DUSP1, respectively. Breast stands for normal samples. The number in the parenthesis represents the total number of samples.
To examine whether JNK signaling plays a role in loss of flfl induced cell death, we checked the expression of puc, a transcriptional target of JNK pathway [30]. puc E69 is a puc mutant allele with a LacZ bearing P-element inserted into the puc locus and serves as a puc-LacZ reporter [31] whose expression could be easily visualized by X-Gal staining. We found that knocking down flfl in the posterior compartment of wing discs resulted in upregulated puc-LacZ expression (Figure 4(f)), compared with the en-Gal4 control (Figure 4(d)) and en > GFP-IR (Figure 4(e)), suggesting that loss of flfl promotes JNK pathway activation.
The JNK pathway is evolutionary conserved from fly to human. Compared with the compact Drosophila genome, there are three homologs of flfl, SMEK1, SMEK2, and SMEK3P, and dozens of Puc homologs named dual specificity phosphatase (DUSP) in human. Previous study has reported that JNK signaling is essential for cell migration and tumor invasion [32]. Based on the above data, we speculate that SMEK is downregulated and DUSP is upregulated in metastatic tumor. Consistent with the hypothesis, we found from the Oncomine database (https://www.oncomine.org/) that SMEK1 expression is indeed downregulated whereas DUSP1 is upregulated in invasive breast carcinoma stroma compared to normal tissue (Figures 4(g) and 4(h)) [33]. These data imply that the role of flfl in modulating JNK pathway is likely conserved by SMEK1 from Drosophila to human.
Although our data mining and previous study found that JNK activity is elevated in several cancer cell lines, its role in tumor development is context-dependent [8]. JNK pathway was implicated as both procancer and anticancer signaling in cancer development for its regulation on cell proliferation and cell death, respectively [6]. In certain mouse models of cancer, JNK deficiency enhances tumor formation and metastasis [20, 34]. In Drosophila, clones with ectopic oncogene Src expression induce no-autonomous tumor growth [35], while Src expression also induces cell death through JNK pathway [22]. Cells in Src clone could escape from cell death if JNK pathway is blocked [35]. Intriguingly, another important oncogene Ras can also switch JNK pathway from anti- to protumor signaling [6]. Thus, upon the presence of different regulating factor(s), JNK pathway modulates cell death, tumor genesis, and progression in a cell context-dependent manner.
3.5. Loss of flfl Induced Cell Death Is JNK Pathway-Dependent
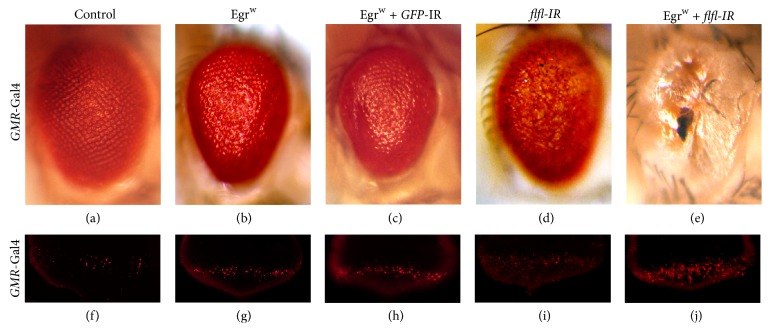
Knocking down flfl by GMR-Gal4 induced cell death in eye discs (Figure 2(i)) and produced a rough eye phenotype in adults (Figure 2(d)). These results were confirmed by another independent line of flfl RNAi (Figures 5(b) and 5(b′)). To understand whether loss of flfl induced cell death is JNK pathway dependent, we blocked JNK signaling by expressing a bsk RNAi or a dominant negative allele of Bsk (BskDN). We found that loss of flfl triggered rough eye phenotype (Figure 5(b)) and increased cell death in eye discs (Figure 5(b′)) were significantly suppressed by compromised JNK activity (Figures 5(c)–5(e)). As a control, GFP RNAi and loss of Bsk signaling produced no evident phenotype in adult eyes (Figures 5(f)–5(h)). These results indicate that depletion of flfl induced cell death is JNK pathway-dependent.
Figure 5.
flfl loss-of-function induced cell death was suppressed by compromised JNK activity. Light micrographs of Drosophila eyes ((a)–(d) and (f)–(h)) or acridine orange staining of eye discs from 3rd instar larvae ((a′)–(d′) and (f′)–(h′)) are shown. Compared with the control (a), knocking down flfl induced cell death in eye discs (b′) and a rough eye phenotype in adult (b), which were significantly suppressed by knocking down bsk ((c′) and (c)) or coexpressing a dominant negative form of Bsk ((d′) and (d)). Expressions of GFP-IR, bsk-IR, or BskDN were included as controls ((f)–(h′)). (e) is the statistical analysis of acridine orange positive cells in the posterior part of eye discs from the indicated panels. Column shows mean + SEM and significance was tested by unpaired Student t-test; ∗∗∗ P ≤ 0.001; ∗∗ P ≤ 0.01.
4. Conclusions
In this study we have identified flfl as a negative regulator of TNF-trigger JNK-mediated cell death in Drosophila. While ectopic expression of flfl impedes JNK signaling-induced cell death, loss of flfl induces JNK pathway activation and cell death in Drosophila eye and wing discs and produced morphological defects in the adult eye. These data suggest an important physiological function of flfl in maintaining tissue homeostasis in Drosophila organ development. flfl's ability to inhibit JNK signaling is likely retained by its human homolog SMEK1. Consistently, while activated JNK pathway promotes dermal fibroblasts cell migration in wound healing [36], ectopic expression of SMEK1 significantly decreased the migration ability of carcinoma cells [37]. In addition, we found from Oncomine database that SMEK1 is downregulated whereas JNK signaling target gene DUSP1 is upregulated in human invasive carcinoma [33].
Acknowledgments
The authors thank the Bloomington Drosophila Stock Center, Vienna Drosophila Research Center, and National Institute of Genetics (NIG-FLY) for fly stocks and members of the Xue Laboratory for comments and discussion. This work is supported by the National Basic Research Program of China (973 Program) (2011CB943903), National Natural Science Foundation of China (31171413 and 31371490), the Specialized Research Fund for the Doctoral Program of Higher Education of China (20120072110023), and Shanghai Committee of Science and Technology (09DZ2260100 and 14JC1406000).
Conflict of Interests
The authors declare no conflict of interests.
References
- 1.Gingras A.-C., Caballero M., Zarske M., et al. A novel, evolutionarily conserved protein phosphatase complex involved in cisplatin sensitivity. Molecular & Cellular Proteomics. 2005;4(11):1725–1740. doi: 10.1074/mcp.m500231-mcp200. [DOI] [PubMed] [Google Scholar]
- 2.Sousa-Nunes R., Chia W., Somers W. G. Protein Phosphatase 4 mediates localization of the Miranda complex during Drosophila neuroblast asymmetric divisions. Genes and Development. 2009;23(3):359–372. doi: 10.1101/gad.1723609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lipinszki Z., Lefevre S., Savoian M. S., Singleton M. R., Glover D. M., Przewloka M. R. Centromeric binding and activity of protein phosphatase 4. Nature Communications. 2015;6, article 5894 doi: 10.1038/ncomms6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou G., Mihindukulasuriya K. A., MacCorkle-Chosnek R. A., et al. Protein phosphatase 4 is involved in tumor necrosis factor-α-induced activation of c-Jun N-terminal kinase. The Journal of Biological Chemistry. 2002;277(8):6391–6398. doi: 10.1074/jbc.m107014200. [DOI] [PubMed] [Google Scholar]
- 5.Chen L., Dong W., Zou T., et al. Protein phosphatase 4 negatively regulates LPS cascade by inhibiting ubiquitination of TRAF6. FEBS Letters. 2008;582(19):2843–2849. doi: 10.1016/j.febslet.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 6.Enomoto M., Kizawa D., Ohsawa S., Igaki T. JNK signaling is converted from anti- to pro-tumor pathway by Ras-mediated switch of Warts activity. Developmental Biology. 2015;403(2):162–171. doi: 10.1016/j.ydbio.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Ohsawa S., Sugimura K., Takino K., Xu T., Miyawaki A., Igaki T. Elimination of oncogenic neighbors by JNK-mediated engulfment in Drosophila . Developmental Cell. 2011;20(3):315–328. doi: 10.1016/j.devcel.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Weston C. R., Davis R. J. The JNK signal transduction pathway. Current Opinion in Cell Biology. 2007;19(2):142–149. doi: 10.1016/j.ceb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Lyu J., Yu X., He L., et al. The protein phosphatase activity of PTEN is essential for regulating neural stem cell differentiation. Molecular Brain. 2015;8(1, article 26) doi: 10.1186/s13041-015-0114-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su Y.-C., Treisman J. E., Skolnik E. Y. The Drosophila Ste20-related kinase misshapen is required for embryonic dorsal closure and acts through a JNK MAPK module on an evolutionarily conserved signaling pathway. Genes and Development. 1998;12(15):2371–2380. doi: 10.1101/gad.12.15.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartz D. R., Kardia S. L. R., Shedden K. A., et al. Gene expression in ovarian cancer reflects both morphology and biological behavior, distinguishing clear cell from other poor-prognosis ovarian carcinomas. Cancer Research. 2002;62(16):4722–4729. [PubMed] [Google Scholar]
- 12.Ma X., Yang L., Yang Y., Li M., Li W., Xue L. dUev1a modulates TNF-JNK mediated tumor progression and cell death in Drosophila . Developmental Biology. 2013;380(2):211–221. doi: 10.1016/j.ydbio.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 13.Ma X., Li W., Yu H., et al. Bendless modulates JNK-mediated cell death and migration in Drosophila . Cell Death and Differentiation. 2014;21(3):407–415. doi: 10.1038/cdd.2013.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue L., Igaki T., Kuranaga E., Kanda H., Miura M., Xu T. Tumor suppressor CYLD regulates JNK-induced cell death in Drosophila . Developmental Cell. 2007;13(3):446–454. doi: 10.1016/j.devcel.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 15.Geuking P., Narasimamurthy R., Basler K. A genetic screen targeting the tumor necrosis factor/eiger signaling pathway: identification of drosophila TAB2 as a functionally conserved component. Genetics. 2005;171(4):1683–1694. doi: 10.1534/genetics.105.045534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Igaki T., Kanda H., Yamamoto-Goto Y., et al. Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. The EMBO Journal. 2002;21(12):3009–3018. doi: 10.1093/emboj/cdf306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moreno E., Yan M., Basler K. Evolution of TNF signaling mechanisms: JNK-dependent apoptosis triggered by Eiger, the Drosophila homolog of the TNF superfamily. Current Biology. 2002;12(14):1263–1268. doi: 10.1016/s0960-9822(02)00954-5. [DOI] [PubMed] [Google Scholar]
- 18.Fanto M., Weber U., Strutt D. I., Mlodzik M. Nuclear signaling by Rac and Rho GTPases is required in the establishment of epithelial planar polarity in the Drosophila eye. Current Biology. 2000;10(16):979–988. doi: 10.1016/s0960-9822(00)00645-x. [DOI] [PubMed] [Google Scholar]
- 19.Motwani M., Sirotnak F. M., She Y., Commes T., Schwartz G. K. Drg1, a novel target for modulating sensitivity to CPT-11 in colon cancer cells. Cancer Research. 2002;62(14):3950–3955. [PubMed] [Google Scholar]
- 20.She Q.-B., Chen N., Bode A. M., Flavell R. A., Dong Z. Deficiency of c-Jun-NH2-terminal kinase-1 in mice enhances skin tumor development by 12-O-tetradecanoylphorbol-13-acetate. Cancer Research. 2002;62(5):1343–1348. [PubMed] [Google Scholar]
- 21.Zhang S., Chen C., Wu C., Yang Y., Li W., Xue L. The canonical Wg signaling modulates Bsk-mediated cell death in Drosophila . Cell Death and Disease. 2015;6(4) doi: 10.1038/cddis.2015.85.e1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma X., Shao Y., Zheng H., Li M., Li W., Xue L. Src42A modulates tumor invasion and cell death via Ben/dUev1a-mediated JNK activation in Drosophila . Cell Death and Disease. 2013;4(10, article e864) doi: 10.1038/cddis.2013.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xue L., Noll M. Dual role of the Pax gene paired in accessory gland development of Drosophila . Development. 2002;129(2):339–346. doi: 10.1242/dev.129.2.339. [DOI] [PubMed] [Google Scholar]
- 24.Bertram M. J., Akerkar G. A., Ard R. L., Gonzalez C., Wolfner M. F. Cell type-specific gene expression in the Drosophila melanogaster male accessory gland. Mechanisms of Development. 1992;38(1):33–40. doi: 10.1016/0925-4773(92)90036-j. [DOI] [PubMed] [Google Scholar]
- 25.Igaki T., Miura M. The Drosophila TNF ortholog Eiger: emerging physiological roles and evolution of the TNF system. Seminars in Immunology. 2014;26(3):267–274. doi: 10.1016/j.smim.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Ma X., Huang J., Yang L., Yang Y., Li W., Xue L. NOPO modulates Egr-induced JNK-independent cell death in Drosophila . Cell Research. 2012;22(2):425–431. doi: 10.1038/cr.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baillon L., Basler K. Reflections on cell competition. Seminars in Cell & Developmental Biology. 2014;32:137–144. doi: 10.1016/j.semcdb.2014.04.034. [DOI] [PubMed] [Google Scholar]
- 28.Moreno E., Basler K., Morata G. Cells compete for decapentaplegic survival factor to prevent apoptosis in Drosophila wing development. Nature. 2002;416(6882):755–759. doi: 10.1038/416755a. [DOI] [PubMed] [Google Scholar]
- 29.Moreno E., Basler K. dMyc transforms cells into super-competitors. Cell. 2004;117(1):117–129. doi: 10.1016/s0092-8674(04)00262-4. [DOI] [PubMed] [Google Scholar]
- 30.Martín-Blanco E., Gampel A., Ring J., et al. Puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila . Genes & Development. 1998;12(4):557–670. doi: 10.1101/gad.12.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agnès F., Suzanne M., Noselli S. The Drosophila JNK pathway controls the morphogenesis of imaginal discs during metamorphosis. Development. 1999;126(23):5453–5462. doi: 10.1242/dev.126.23.5453. [DOI] [PubMed] [Google Scholar]
- 32.Huang C., Jacobson K., Schaller M. D. MAP kinases and cell migration. Journal of Cell Science. 2004;117(part 20):4619–4628. doi: 10.1242/jcs.01481. [DOI] [PubMed] [Google Scholar]
- 33.Finak G., Bertos N., Pepin F., et al. Stromal gene expression predicts clinical outcome in breast cancer. Nature Medicine. 2008;14(5):518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 34.Hubner A., Mulholland D. J., Standen C. L., et al. JNK and PTEN cooperatively control the development of invasive adenocarcinoma of the prostate. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(30):12046–12051. doi: 10.1073/pnas.1209660109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Enomoto M., Igaki T. Src controls tumorigenesis via JNK-dependent regulation of the Hippo pathway in Drosophila . EMBO Reports. 2013;14(1):65–72. doi: 10.1038/embor.2012.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J.-C., Lin B.-B., Hu H.-W., et al. NGF accelerates cutaneous wound healing by promoting the migration of dermal fibroblasts via the PI3K/Akt-Rac1-JNK and ERK pathways. BioMed Research International. 2014;2014:13. doi: 10.1155/2014/547187.547187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Byun H.-J., Kim B.-R., Yoo R., Park S.-Y., Rho S. B. SMEK1 enhances gemcitabine anti-cancer activity through inhibition of phosphorylation of Akt/mTOR. Apoptosis. 2012;17(10):1095–1103. doi: 10.1007/s10495-012-0751-0. [DOI] [PubMed] [Google Scholar]