Abstract
Glioblastoma (GBM) is a deadly and debilitating brain tumor with an abysmal prognosis. The standard therapy for GBM is surgery followed by radiation and chemotherapy with temozolomide (TMZ). Treatment of GBMs remains a challenge, largely due to the fast degradation of TMZ, inability to deliver an effective dose of TMZ to tumors, and lack of target specificity which may cause systemic toxicity. Here, we present a simple method to synthesize a nanoparticle-based carrier that can protect TMZ from rapid degradation in physiological solutions and can specifically deliver them to GBM cells through the mediation of a tumor targeting peptide chlorotoxin (CTX). Our nanoparticle, namely NP-TMZ-CTX, had a hydrodynamic size of less than 100 nm, exhibited sustained stability in cell culture media for up to two weeks, and could accommodate stable drug loading. TMZ bound to nanoparticles showed much higher stability at physiological pH, with a half-life 7-fold greater than free TMZ. NP-TMZ-CTX was able to target GBM cells and achieved 2–6-fold higher uptake and 50–90% reduction of IC50 at 72 h post-treatment as compared to non-targeted NP-TMZ. NP-TMZ-CTX showed great promise in its ability to deliver a high therapeutic dose of TMZ to GBM cells, and could serve as a template for targeted delivery of other therapeutics.
Keywords: Glioblastoma, Drug Delivery, Therapeutics, Chlorotoxin, MGMT, Nanomedicine
INTRODUCTION
Glioblastoma (GBM) is a deadly and debilitating brain tumor with 14,000 new cases diagnosed annually in the United States. Despite considerable efforts in patient care, the prognosis of GBM has remained dismal with a median survival of 12–14 months and a five-year survival rate of less than 3%.1–2 Standard treatment of GBM starts with surgery followed by concurrent radio- and chemo-therapy. The widespread use of more effective, targeted treatments in the clinic has been hindered by: (1) the heterogeneous cytogenetic nature of GBM that limits the use of pathway-specific targeted agents; (2) the diffuse infiltration of invasive GBM cells that precludes complete resection of cancerous tissue and thus recurrence after surgical resection is nearly inevitable, and (3) the presence of a blood-brain barrier (BBB) that renders the majority of chemotherapeutics and targeted agents ineffective.2–5 Most recently the anti-angiogenic antibody, bevacizumab (Avastin®), which facilitated improved outcomes in recurrent GBM,6 did not improve overall survival in newly diagnosed patients.7 Therefore, there is an urgent need to develop novel therapeutic strategies to improve the survival and quality of life of GBM patients.
Temozolomide (TMZ) is the standard-of-care chemotherapy for GBM as first line treatment with radiotherapy or as a single agent for maintenance therapy.4 As a DNA-alkylating agent that methylates guanine and adenine bases of DNA, TMZ treatment of GBM cells leads to DNA double strand breaks, cell cycle arrest, and eventual cell death.8 However, TMZ suffers from several drawbacks. TMZ attacks DNA in an indiscriminate manner and has been shown to cause damage to hematopoietic stem cells in patients9 resulting in dose-limiting hematological toxicity. Furthermore, TMZ is poorly soluble in physiological conditions and is subject to rapid hydrolysis that further limits its anti-tumor efficacy. Various strategies have been used to make TMZ target specific by conjugating it to targeting moieties and to improve the aqueous stability of TMZ by incorporation into a macromolecule delivery vehicle.10–11 However, these strategies did not address the necessity of glioblastoma-specific targeting or the problem of target antigen expression on normal cells.10 These problems create a strong incentive to design a nanoparticle-based TMZ delivery system that is specific for glioblastoma and improves the solubility and stability of TMZ.
Previously, we demonstrated that nanoparticles (NPs) with chitosan-based coatings could be used for imaging and as effective delivery vehicles for transport of gene therapeutic agents to the brain.12–13 Chitosan (Chi) is a cationic polysaccharide widely used in tissue engineering and drug/gene delivery applications. Chitosan contains abundant amine groups that can be utilized for covalent attachment of drugs, imaging agents, and targeting moieties. A number of targeting molecules have been evaluated to target GBM including chlorotoxin (CTX),12–20 anti-epidermal growth factor receptor (EGFR) antibody,21 and anti-transferrin receptor antibody.10, 22 Among these targeting ligands, CTX has several advantages. It has the ability to specifically recognize a variety of tumors with neuroectodermal origin including GBM, while showing little affinity towards normal brain.18 After binding to GBM cells, NPs functionalized with CTX are readily internalized,13 a favorable attribute for delivering drugs to their intracellular site of action. CTX has also been shown to inhibit GBM cell invasion, providing an additional therapeutic benefit.19 Most importantly, the attachment of CTX to NPs coated with chitosan enables them to bypass the BBB,12 CTX provides an ideal platform for further development of chitosan-based systems as drug delivery vehicles for GBM.
Here we present a targeted, chitosan-based NP TMZ delivery system (NP-TMZ-CTX) that can improve stability of TMZ in physiological conditions and the efficacy of TMZ against GBM. NP-TMZ-CTX was assembled via biotin-neutravidin interaction without the need for additional purification. The size and stability of NP-TMZ-CTX was characterized using dynamic light scattering (DLS). The targeting ability and therapeutic efficacy for human GBM cells were assessed through flow cytometry and the CellTiter-Glo viability assay, respectively. Specific targeting was quantified with a microplate reader and intracellular distribution was visualized by deconvolution fluorescence microscopy. The ability of NP-TMZ-CTX to cross BBB was demonstrated in vivo and examined via immunohistochemistry (IHC) and confocal fluorescent microscopy.
EXPERIMENTAL SECTION
Materials
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise noted. Chitosan oligosaccharide (MW 3,500) was purchased from Acmey Industrial Co. (Shanghai, China). PEG was grafted onto chitosan by reductive alkylation with methoxy-PEG-aldehyde (MW 2,000) following our previously established method.23 Temozolomide (TMZ) was purchased from TCI America (Portland, OR). Temozolomide acid (TMZA) was synthesized from TMZ following a previously established method.24 Biotin-PEG-valeric acid succinimidyl ester (Bt-PEG-SVA, MW 5,000) was purchased from Laysan Bio (Arab, AL). DMEM media, antibiotic-antimycotic (AA), Fetal bovine serum (FBS), trysin/EDTA solution, Alexa Fluor 647 N-hydroxysuccinimidyl ester (AF647-NHS), wheat germ agglutinin Alexa Fluor 488 conjugates (WGA-AF488), Prolong Gold with DAPI (4′,6-diamidino-2-phenylindole dihydrochloride), and methanol-free formaldehyde (10%) were purchased from Life Technologies (Carlsbad, CA). The CellTiter-Glo Luminescent Cell Viability Assay Kit was purchased from Promega (Madison, WI). Poly-L-lysine-treated glass coverslips were purchased from Becton Dickinson (Franklin Lakes, NJ). Traut’s reagent, SMPEG24, Zeba spin columns (40k MWCO) were purchased from Thermo Fisher (Rockford, IL). Rabbit anti-mouse CD31 antibody and goat anti-rabbit Fc antibody FITC conjugates were purchased from Abcam (Cambridge, MA). CTX (recombinant) was produced from Escherichia Coli according to an established protocol.25
Synthesis of Chitosan-TMZ Conjugates
To synthesize chitosan-TMZ-Bt conjugates, the molar ratio of precursors was fixed at 1:7:40 (chitosan:Bt-PEG-VAS:TMZ). To make chitosan-biotin conjugates (Chi-Bt), 1000 mg chitosan and 1867.2 mg Bt-PEG-VAS were placed in a 20 mL vial. Triethylamine (219 μL) dissolved in 5 mL DMSO was then added to the vial, and the vial was sealed and purged with nitrogen. The reaction was allowed to proceed in a shaker at room temperature for 24 h. The crude reaction product was precipitated from DMSO solution using ether, vacuum-dried, and dissolved in 5 mL of 0.1 N HCl. This 0.1N HCl solution was then dialyzed against deionized water overnight to remove any unreacted precursors. The solution was then lyophilized to obtain intermediate Chi-Bt in form of a yellow solid. To synthesize Chi-TMZ-Bt, TMZA (50 mg) was first activated using thionyl chloride (500 μL) and DMF (31 μL).24 Chi-Bt (200 mg) was dissolved in 4 mL DMSO and the resultant mixture was added to freshly prepared, activated TMZA in a 100 mL three-neck flask. Pyridine (219 μL) was added to the flask as catalyst, and the reaction was allowed to proceed for 24 h to generate Chi-TMZ-Bt. The crude reaction product was precipitated from the DMSO solution using ether, vacuum-dried, and dissolved in 5 mL of 1 N HCl. The 1 N HCl solution of Chi-TMZ-Bt was dialyzed against 1 mM HCl for 6 h to remove unreacted precursors. The solution was then lyophilized to obtain the final product Chi-TMZ-Bt as a yellow solid.
Synthesis of NA-AF647-CTX and Formation of NPs
To synthesize NA-AF647-CTX conjugates, stock neutravidin (NA) solutions were prepared in deionized water at 25 mg/mL. The NA solution was then diluted in borate buffered saline with EDTA (25 mM sodium tetraborate, 150 mM sodium chloride, 5 mM EDTA, pH 8.3), to make a final NA concentration of 10 mg/mL. AF647-NHS (100 μL at 10 mg/mL in DMSO) was added to 900 μL NA solution and the reaction was allowed to proceed for 3 h at room temperature. NA-AF647 was purified by Zeba spin columns (40k MWCO) equilibrated with 50 mM sodium phosphate (pH 7.4). At the same time, 1 mg of CTX in 100 μL 50 mM sodium phosphate (pH 7.4) was reacted with 2.15 μL of Traut’s reagent solution (10 mg/mL in 50 mM sodium phosphate, pH 7.4) for 1 h. To synthesize NA-AF647-CTX, 1.96 μL of SMPEG24 (250 mM in DMSO) was added to 1 mg of NA-AF647 solution, and the reaction was allowed to proceed in the dark for 30 min. The resulting NA-AF647-PEG24Mal was purified by Zeba spin columns (40k MWCO) equilibrated with 0.9% saline.
The AF647 loading of NA-AF647-CTX was determined by UV-vis absorbance at 649 nm (AF647 absorbance maximum) and 280 nm (protein absorbance). The amount of CTX grafted onto NA was determined by SDS-PAGE of the NA-AF647-CTX reaction mixture. Subtracting the concentration of free CTX remaining in the solution from the concentration of CTX at the beginning of reaction provided the amount of immobilized CTX.
To synthesize NP-TMZ-CTX, the Chi-TMZ-Bt solution was reconstituted in pH 4.0 saline and the pH of NA-CTX solution was adjusted to 4.0 with 1N HCl. These two solutions were combined at a NA:Bt molar ratio of 1:1. The final product was obtained after a 30-min incubation. Synthesis of NP-TMZ-NA followed the same method as NP-TMZ-CTX.
Characterizations of NPs and Controlled Drug Release
The sizes and zeta potentials of NPs were measured by dynamic light scattering (DLS) using a DTS Zetasizer Nano (Malvern Instruments, Worcestershire, UK). NP concentrations were 100 μg/mL for all measurements. NP stability was determined in PBS and DMEM containing 2.5% FBS and 1% AA.
Transmission electron microscopy (TEM) imaging was done by an FEI Tecnai G2 F20 200 kV scanning transmission electron microscope (FEI Company, Hillsboro, OR). Images of NPs were captured by a liquid nitrogen-cooled charge-coupled device (CCD) camera. NP samples in water were deposited on carbon-coated copper grid (Ted Pella Inc., Redding, CA) and negatively stained by uranyl acetate.
Drug loading was determined by measuring the absorbance of polymer or dissolved NP solutions using a UV-vis spectrometer (Agilent Technologies, Santa Clara, CA). The wavelength of UV absorption for TMZ was 328 nm.
For the measurement of drug release, 1 mL of NP samples (equivalent TMZ concentration: 1 mg/mL) were dispersed in PBS buffer at pH 7.4, 6.4, 5.5 or 4.5, and incubated at 37°C. Aliquots (100 μL) were removed from the bulk solution at 0.5 h, 1 h, 2 h, 3 h, 6 h, 9 h, 12 h and 24 h. Absorbance was obtained using the UV-vis spectrometer at 328 nm for active TMZ (the degradation product of TMZ, MTIC (5-(3-methyltriazen-1-yl)imidazole-4-carboxamide), does not absorb at 328 nm), and corrected with the intrinsic absorbance of chitosan at 328 nm. The percent release of TMZ (R%) was calculated using the following equation:
where ANP-TMZ is the absorbance of TMZ in bulk solution, AChi is the absorbance of chitosan copolymer without TMZ at same concentration, and ETMZ is extinction coefficient of TMZ.
Dose-Response Experiment
The human GBM cell line U-118 MG was obtained from American Type Culture Collection (ATCC, Manassas, VA). SF767 and GBM6 were gifts from Brain Tumor Research Laboratory, UCSF 26 and Sarkaria laboratory, Mayo Clinic,27 respectively. GBM cells were maintained in DMEM containing 2.5% FBS and 1% AA at 37°C in 95%/5% humidified air/CO2.
All GBM cells were plated at 5,000 cells per well in 96-well plates immediately prior to treatment. Cells were then exposed to free TMZ, NP-TMZ or NP-TMZ-CTX at 0, 20, 200, 2,000, 20,000, and 200,000 ng/mL TMZ in 200 μL fully supplemented DMEM. Cell viability was determined 72 h after initial exposure using the CellTiter-Glo viability assay following the manufacturer’s protocol. IC50 values were calculated from dose-response curves generated with a polynomial dose-response four-parameter approximation using the GraphPad Prism software package (GraphPad Software Inc., La Jolla, CA).
Cell Uptake Experiment and Fluorescence Imaging
For cell uptake quantification experiments, all cells were plated at 100,000 cells per well in 24-well plates the night before treatment. Cells were then treated with 5 μM, 25 μM, or 250 μM NP-TMZ-NA or NP-TMZ-CTX in 1 mL fully supplemented DMEM for 12 h. Cells were washed three times with PBS and then solubilized with 500 μL of 2.4 M HCl. The fluorescence intensity of AF647 was measured with a SpectraMax M5 microplate reader (Molecular Devices, Union City, CA), using excitation and emission wavelengths of 647 nm and 670 nm, respectively. The cell number per well was assessed using a Coomassie Blue protein quantification assay and calculated based on a previously prepared standard curve.
For flow cytometry experiments, all cells were plated at 100,000 cells per well in 6-well plates the night before treatment. Cells were then treated with 25 μM NP-TMZ-CTX or NP-TMZ-NA in 1 mL fully supplemented DMEM for 4 h. Cells were washed three times with PBS containing 2.5% FBS and detached with 1 mL trypsin/EDTA solution. Cells were suspended in 1 mL PBS containing 2.5% FBS. Analysis of at least 10,000 cells for each sample was performed on a BD LSR II flow cytometer (Becton Dickinson). NP fluorescence was detected in the CY5 channel and data was analyzed using the FlowJo software package (Tree Star Inc., Ashland, OR).
For fluorescence imaging experiments, all cells were plated at 100,000 cells per well on poly-L-lysine-coated glass coverslips in 24-well plates the night before treatment. Cells were then treated with 100 μM NP-TMZ or NP-TMZ-CTX in 1 mL fully supplemented DMEM for 12 h. Cells were washed three times with PBS and fixed in 4% formaldehyde for 30 min. Cell membranes were stained with WGA-AF488 for 15 min. The cells were then mounted on microscope slides using Prolong Gold anti-fade solution containing DAPI for cell nuclei staining. NP-TMZ-CTX uptake was analyzed using fluorescence microscopy on a DeltaVision Elite (GE Healthcare, Piscataway, NJ) deconvolution imaging system with Olympus IX71 inverted microscope with a 60× oil-immersion lens. Fluorophores were excited by a solid-state light engine with seven excitation bands at 390, 440, 475, 510, 545, 580, and 640 nm. Emissions from DAPI, WGA-488, and AF647 were detected by a standard filter set (DAPI, FITC, TRITC, Cy5).
In Vivo BBB Permeability Study
All mice were housed at the University of Washington in a pathogen-free environment under protocols approved by the Institutional Animal Care and Use Committee (IACUC). C57BL6 wild type mice (Charles River, Wilmington, MA) were injected through the tail vein with 100 μl of 2 mg/ml nanoparticles. Each animal was euthanized at 0.5 h and 2 h post-injection and brain tissues were dissected. Tissues were then embedded in OCT and kept frozen at −80°C until needed. The frozen tissues were sliced in 12 μm thick sections and mounted onto glass slides. Slides were then washed with TBS with 0.025% Triton X-100 and blocked in 10% normal serum with 1% BSA in tris(hydroxymethyl)aminomethane-buffered saline (TBS) for 2 h. Slides were then rinsed and stained with rabbit anti-mouse CD31 primary antibody and goat anti-rabbit Fc secondary antibody-FITC conjugate according to instructions provided by Abcam. Cover slips were then mounted on microscope slides using Prolong Gold antifade solution containing DAPI for cell nuclei staining. Images were acquired on an LSM 780 NLO confocal fluorescence microscope (Carl Zeiss Inc., Peabody, MA) equipped with a 63 x oil immersion lens and appropriate filters.
Statistical Analysis
All experiments were run in triplicate unless stated otherwise. Acquired data were expressed as the means ± standard deviation. Statistical significance was determined using Student’s t-test. Values of * P < 0.05, ** P < 0.01, and *** P < 0.001 were considered significant.
RESULTS
Synthesis of Polymers, Protein Conjugates, and NPs
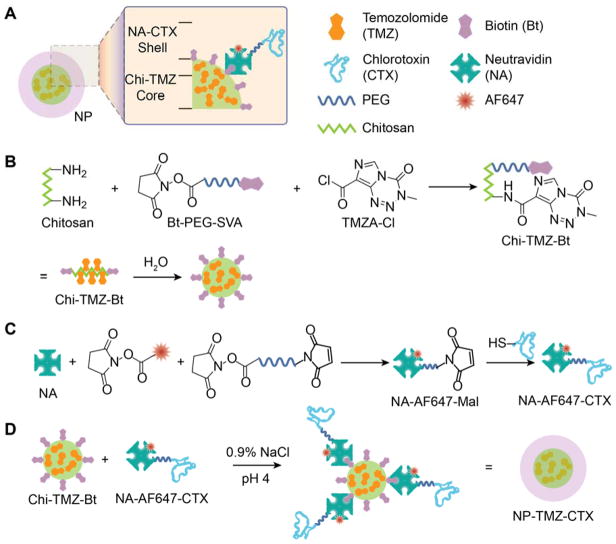
The structure and synthesis of NP-TMZ-CTX is shown in Scheme 1. NP-TMZ-CTX is comprised of a chitosan core loaded with TMZ and biotin (Chi-TMZ-Bt) and a shell containing CTX targeting ligands attached to AF647-conjugated neutravidin (NA-AF647-CTX, Scheme 1A). Chi-TMZ-Bt was synthesized through nucleophilic reactions of amine groups on chitosan-g-PEG with biotin-PEG-valeric acid succinimidyl ester and TMZA chloride (Scheme 1B). NA-AF647-CTX was synthesized through linking NA with thiolated CTX using a short, heterobifunctional PEG chain (Scheme 1C). The final product, NP-TMZ-CTX, was synthesized by simply combining Chi-TMZ-Bt and NA-AF647-CTX at a 1:1 molar ratio of Bt to NA (Scheme 1D) and did not require subsequent purification.
Scheme 1.
Schematic of NP-TMZ-CTX synthesis. A) The structure of NP-TMZ-CTX. B) Synthesis procedure for Chi-TMZ-Bt. C) Synthesis procedure for NA-AF647-CTX. D) Formation of NP-TMZ-CTX by linking Chi-TMZ-Bt and NA-AF647-Bt via Bt-NA interactions.
NP Properties and Controlled Drug Release
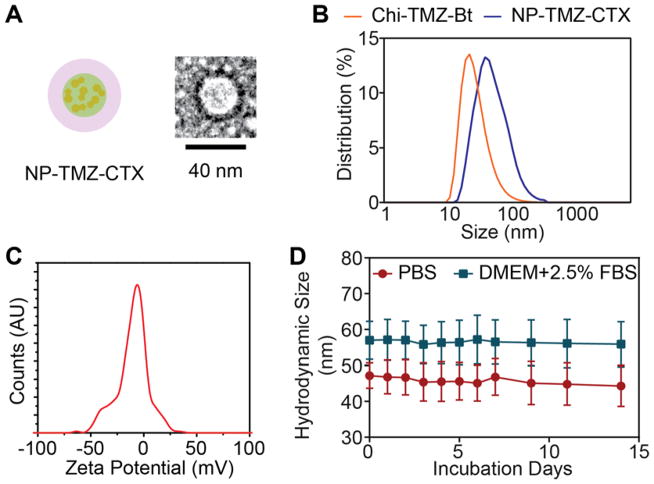
The intermediates Chi-TMZ-Bt and NA-AF647-CTX were characterized by UV-vis to quantify the ratio of their components. The TMZ loading of Chi-TMZ-Bt was 5.4 ± 0.6 wt% (Table 1) as determined by UV-vis absorbance at 328 nm of active TMZ (degradation product MTIC does not absorb at this wavelength). The size and zeta potential of Chi-TMZ-Bt, as characterized by DLS, were 38 ± 7 nm and 3.9 ± 5.3 mV, respectively (Table 1 and Fig. 1B and C). There were 1.1 AF647 fluorophore and 2.1 CTX molecules per NA-AF647-CTX molecule, respectively.
Table 1.
Physicochemical properties of Chi-TMZ-Bt, NP-TMZ-NA, and NP-TMZ-CTX.
| Name | TMZ loading (wt %)a | Loading in (%) of TMZ addeda | Hydrodynamic diameter in PBS (nm)b | Hydrodynamic diameter in DMEM, 2.5% FBS (nm)b | Zeta potential at pH 7.4 (mV)c |
|---|---|---|---|---|---|
| Chi-TMZ-Bt | 5.4 ± 0.6 | 24.2 ± 2.1 | 38 ± 7 | 40 ± 5 | 3.9 ± 5.3 |
| NP-TMZ-NA | 5.0 ± 0.5 | N/A | 45 ± 8 | 53 ± 9 | − 2.9 ± 3.7 |
| NP-TMZ- CTX | 4.9 ± 0.5 | N/A | 47 ± 8 | 57 ± 18 | − 1.8 ± 4.3 |
Mean ± standard deviation;
Size ± PDI width;
Zeta potential ± zeta deviation
Figure 1.
Physicochemical properties: A) Schematic representation of NP-TMZ-CTX and TEM image of a single NP. B) Hydrodynamic size distributions for NP-TMZ-Bt (orange) and NP-TMZ-CTX (blue) in PBS. C) Zeta potential distribution of as synthesized NP-TMZ-CTX. D) The hydrodynamic size of NP-TMZ-CTX in PBS (orange) and DMEM with 2.5% FBS (blue) over a time period of 14 days. Error bars represent the standard error of means.
NP-TMZ-CTX was characterized by TEM (Fig. 1A) as roughly spherical shape. The surface of NP was enhanced by uranyl acetate staining as dark fringe. The size of NP under TEM was around ~35 nm. NP-TMZ-CTX and NP-TMZ-NA were also characterized by UV-vis for TMZ loading and DLS for hydrodynamic size and zeta potential. Both NP-TMZ-CTX and NP-TMZ-NA contained similar amounts of TMZ (4.9 ± 0.5 wt% vs 5.0 ± 0.5 wt%, respectively, Table 1). The difference between TEM size and hydrodynamic size (a few nanometers) is caused by hydration layers associated with NPs. Additionally, the hydrodynamic sizes in PBS and zeta potentials at pH 7.4 of NP-TMZ-CTX and NP-TMZ-NA were similar (Table 1 and Fig. 1A and 1B). Importantly, NP-TMZ-CTX was highly stable in PBS and cell culture media, showing no change in size over 14 days (Fig 1C).
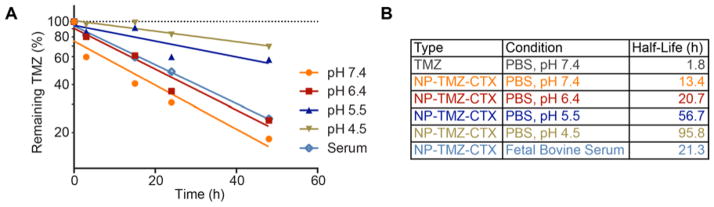
The aqueous stability of TMZ in NP-TMZ-CTX was tested at pH 7.4, 6.4, 5.5 and 4.5, as well as in serum. The results (Fig. 2A) showed that the stability of TMZ, as measured by the half-life of remaining active TMZ, was significantly increased when bound to the NP (Fig. 2B). For comparison, the half-life of free TMZ at pH 7.4 is only 1.8 h,10 while the half-life of NP-TMZ-CTX is 13.4 h. At pH 4.5, TMZ bound to the NP had a marked half-life of 95.8 h. In serum, the half-life of NP-bound TMZ was 21.3 h.
Figure 2.
TMZ degradation profile under various conditions. A) Remaining active TMZ vs. time plot of NP-TMZ-CTX incubated at pH 7.4, 6.4, 5.5 and 4.5 in PBS, as well as in fetal bovine serum. B) Calculated half-lives of TMZ and NP-TMZ-CTX at various conditions. The half-life of free TMZ was reported previously 10.
Dose-Response Results
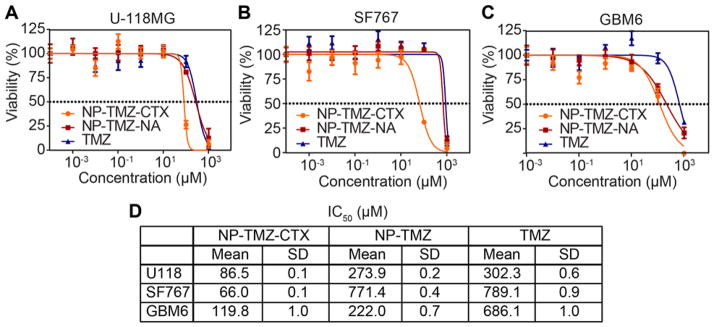
To determine the targeted therapeutic efficacy of NP-TMZ-CTX, human GBM cells were treated with various doses of NP-TMZ-CTZ, NP-TMZ-NA, and free TMZ for 72 hrs and cell viability was determined using the CellTiter Glo assay. The dose-response curves for NP-TMZ-CTX, non-targeting control NP-TMZ-NA, and free TMZ are shown in Fig. 3A. NP-TMZ-CTX achieved over 50% cell killing (IC50) at the lowest TMZ dose indicating NP-TMZ-CTX was most effective against all three cell lines. The IC50 values for NP-TMZ-CTX in U-118 MG, SF767, and GBM6 cells were 86.5 μM, 66.5 μM, and 119.8 μM, respectively. The IC50 values of NP-TMZ-NA and free TMZ were in the range of 220 to 800 μM for all three cell lines. NP-TMZ-CTX achieved at least a 50% reduction in IC50 as compared to NP-TMZ-NA and free TMZ.
Figure 3.
Therapeutic efficacy of NP-TMZ-CTX, NP-TMZ, and free TMZ as determined by CellTiter-Glo. Dose-response curves in A) U-118MG, B) SF767, and C) GBM6 cell lines. D) IC50 values for NP-TMZ-CTX, NP-TMZ, and free TMZ in the three cell lines.
Cellular Uptake of NP-TMZ-CTX and NP-TMZ-NA
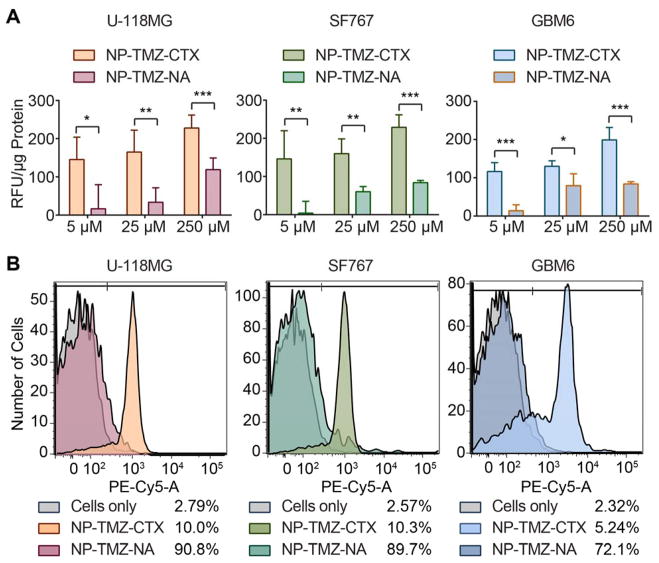
U-118 MG, SF767, and GBM6 cells were incubated with NP-TMZ-CTX or NP-TMZ-NA as a control at TMZ concentrations of 5 μM, 25 μM, and 250 μM. The targeting ability of NP-TMZ-CTX was assessed by fluorescence. A ~2-fold increase in fluorescence intensity was observed in cells treated with NP-TMZ-CTX as compared to NP-TMZ-NA for all three cell types (Fig. 4A). The relative fluorescence values (RFU/μg protein) of NP-TMZ-CTX ranged from 120–150 at 5 μM TMZ to 200–230 at 250 μM TMZ, demonstrating a dose-dependent uptake. NP-TMZ-NA showed much lower uptake in these cells, ranging from 30–60 RFU/μg protein at 5 μM TMZ to 85–120 at 250 μM TMZ.
Figure 4.
Quantification of cellular uptake of NP-TMZ-CTX and NP-TMZ-NA. A) Quantification of NP uptake by microplate reader based on fluorescence intensity. B) Flow cytometry analysis of U-118 MG, SF767, and GBM6 cells treated with NPs. The percentages of positive cells are provided on the right side of legend. N.S. indicates not significant, * indicates P < 0.05, ** indicates P < 0.01, *** indicates P < 0.001, compared to NP-TMZ-NA as determined by Student’s t-test.
The target specificity of NP-TMZ-CTX was further confirmed by flow cytometry. The proportion of cells that contained NP-TMZ-CTX based on Cy5 fluorescence was 90.8%, 89.7% and 72.1% for U-118 MG, SF767, and GBM6, respectively (Fig. 4B). For cells treated with NP-TMZ-NA, the proportion of positive cells remained below 10.5%.
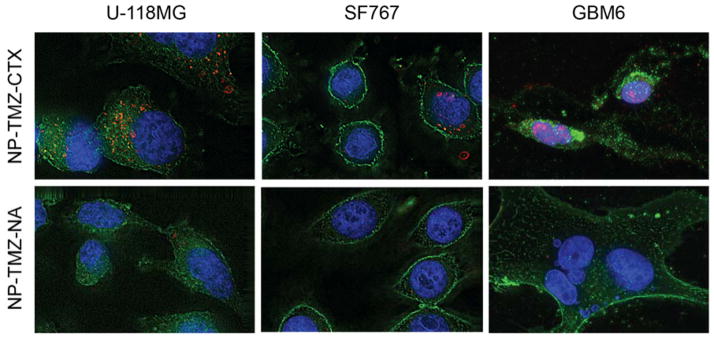
The distributions of NP-TMZ-CTX and NP-TMZ-NA within the cells were examined by deconvolution fluorescence imaging. NP-TMZ-CTX displayed a strong signal in all three cell lines whereas little NP-TMZ-NA could be visualized. For U-118 MG and SF767 cells, NPs appeared in the perinuclear region, while for GBM6 cells, the NP signal overlapped with DAPI signal, indicating their accumulation in the nuclei.
In Vivo BBB Permeability Study
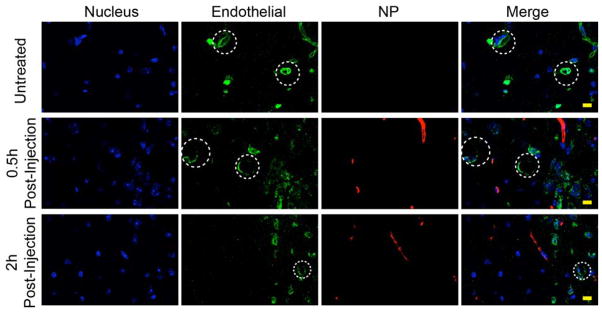
To evaluate BBB permeability of NP-TMZ-CTX, analysis was performed on brain sections of mice 0.5 and 2 hours post injections of NPs intravascularly (Fig. 6). Brain tissue from a mouse receiving no injection showed no signal (red) from NPs (top panel). Images obtained from the mouse 0.5 h after NP administration showed distribution of NPs throughout the entire image area (not shown) and near blood vessels (middle panel). The extravasation of NPs from blood vessels was more apparent on images obtained from the mouse 2 h after NP administration. There was a significant amount of NPs located either in the region distal from blood vessels or the avascular region of the brain (bottom panel).
Figure 6.
In vivo BBB permeability study: Confocal fluorescent microscopy images of mouse brain tissue sections with one of three treatment conditions: Untreated (top panel), 0.5 h after injected with NP-TMZ-CTX (middle panel), 2 h after injected with NP-TMZ-CTX (bottom panel). Cell nuclei are shown in blue, CD31 (endothelial cells) in green, and NP-TMZ-CTX in red. Some blood vessels are highlighted in dashed circles. The scale bar corresponds to 10 μm.
DISCUSSION
Synthesis of a target-specific TMZ delivery platform requires rapid and compatible chemical methods to attach targeting molecules to TMZ-delivery vehicles. The most commonly used methods for targeting agent conjugation utilizes nucleophilic reaction under neutral or slightly basic conditions, which is detrimental to the stability of TMZ. Therefore, novel chemical methods are needed to synthesize targeted TMZ-delivery vehicles in order to preserve the activity of TMZ. Here we utilized the strong interaction between biotin (Bt) and neutravidin (NA) to attach targeting molecules to our TMZ delivery vehicles. NA is a deglycosylated version of avidin with a higher isoelectric point (pI) with minimal nonspecific interactions.28 In vivo applications of NA have been reported with no noticeable safety issues.29–30 Bt-NA binding is the strongest among all noncovalent interactions (Kd = 10−15 M) and also possesses fast reaction kinetics. Additionally, the Bt-NA interaction is stoichiometric and stable across a broad pH range31 as well as at physiological conditions, making it an optimal linker chemistry for targeting agent attachment to TMZ-bound NPs.
The synthesis of our NP (NP-TMZ-CTX) is a modular process where therapeutic cores (Chi-TMZ-Bt) and targeting moieties (NA-AF647-CTX) are synthesized independently. The end product is easily formed in an “on-demand” fashion by mixing the two components at a predetermined ratio to achieve high drug loading. This self-assembly has proven to be effective as many of the targeted nanomedicines in clinical trials utilize the “on-demand” strategy.32–33 Our NPs do not require subsequent purification and the end product is monodisperse with a small hydrodynamic size (Fig. 1A and 2B). No excessive crosslinking was observed between the four biotin-binding sites on each NA molecule and three biotin sites on each chitosan chain. The size of NP-TMZ-CTX remained around 67.2 nm, even in biological fluid, suggesting it should be able to avoid elimination by the reticuloendothelial system.34 Furthermore, NP-TMZ-CTX had a near neutral zeta potential that further reduces non-specific binding to serum proteins, extracellular matrix, and healthy cells.
The fast degradation kinetics of TMZ in physiological conditions would limit its therapeutic potential and hinder its use in standard NP delivery vehicles. Therefore, achieving slower TMZ degradation kinetics is crucial for NP-TMZ-CTX to succeed as a viable drug delivery system. In aqueous solutions, TMZ molecules are converted to 5-(3-methyltriazen-1-yl)imidazole-4-carboxamide (MTIC) and then rapidly to 4-amino-5-imidazole-carboxamide (AIC) that release the methyldiazonium ion responsible for DNA methylation.10 The conversion of TMZ to MTIC is pH-dependent, with increasing reaction rates at higher pH values. Premature methyldiazonium ion release in the circulatory system could cause off-target DNA damage on hematopoietic stems cells, resulting in neutropenia and other unwanted toxicity, as well as reducing the amount of TMZ available to reach target GBM cells. NP-TMZ-CTX improved the half-life of TMZ at pH 7.4 by almost seven-fold (Fig. 2), so it could potentially reduce methyldiazonium ion release in circulation. The improvement of half-life was probably caused by the encapsulation of TMZ within the NP core preventing water from reaching TMZ. Furthermore, the increased stability of TMZ when bound to the NP at an acidic pH mimicking the tumor microenvironment is highly important to NP trafficking into the cell. The 20 h half-life of NP-bound TMZ at pH 6.5 would ensure active TMZ could be delivered to the intracellular site of action of target cells prior to conversion to MTIC.
The targeting ability of NP delivery vehicles is critical to ensure uptake of NPs by tumor cells and distribution throughout the tumor. CTX is a targeting ligand that binds specifically to glioma and other malignant cells.18 Conjugating CTX to NP surfaces has shown to help the intratumoral diffusion of these NPs35. By combining a greater half-life of TMZ and specific targeting of CTX, the overall administrated dose of TMZ could be reduced. Therefore, it could potentially reduce its bone marrow toxicity and other off-target effects. We examined the uptake of NP-TMZ-CTX as compared to NP-TMZ-NA in various human GBM cells including U-118 MG, SF767, and GBM6. U-118 MG is a commonly used GBM cell line that is sensitive to TMZ.36 SF767 is a commonly used therapy-resistant cell line because of its resistance to radiation and TMZ therapies.26 GBM6 is a primary GBM culture that shows high resistance to TMZ.27 CTX targeting drastically increased the accumulation of NPs in GBM cells by 2–6-fold (Fig. 4A–B) as compared to the NP-TMZ-NA, resulting in more TMZ available for DNA methylation. Furthermore, fluorescence microscopy revealed strong signals from NP-TMZ-CTX in all three cell lines, whereas minimal signals from NP-TMZ-NA could be detected. These results indicated that CTX promoted specific targeting and subsequent uptake of NPs into GBM cells.
NP-TMZ-CTX showed at least 50–90% reduction of the IC50 when compared with NP-TMZ and free TMZ indicating that the improved uptake observed with NP-TMZ-CTX correlated with higher cell killing. The improvement of IC50 on SF767 and GBM6 cell lines (Fig. 3B) are significant since the therapy-resistant subpopulations of GBM cells are difficult to eradicate and are responsible for recurrence of tumor.37 It is intriguing that NP-TMZ-CTX was able to reduce the IC50 of TMZ resistant cells as resistance in SF767 and GBM6 is mediated by high expression of O6-methylguanine-DNA-methyltransferase (MGMT) that removes the cytotoxic methylated DNA. Therefore, the vast majority of research in overcoming TMZ resistance has focused on inhibiting MGMT activity.38–39 However, our data suggest that NP-mediated delivery of TMZ can also improve the effects of TMZ on these resistant cells, perhaps because of significantly larger doses of active TMZ reaching the intracellular site of action. These large doses of TMZ are thought to deplete intracellular MGMT. Indeed, this was the rationale behind clinical trials studying how dose-dense schedules of TMZ therapy improve patient outcome.40 Although the trial did not show extended survival with dose-escalation, it is likely that sufficient concentrations could not be delivered to the tumor because of toxicity concerns.41 NP-TMZ-CTX might offer a solution by improving target specific uptake and minimizing dose limiting off-target effects.
We confirmed the ability of NP-TMZ-CTX to escape the neural vasculature by performing BBB permeability study in wild-type mice injected with NP-TMZ-CTX intravascularly. Since wild-type mice have intact BBB, the accumulation of NPs could not be attributed to BBB disruption, as in case of advanced stage glioblastoma. At 0.5 h after NP administration, NPs were near blood vessels (Fig. 6, middle panel). This indicates NPs started the process of passing through the BBB. At 2 h after NP administration, NPs appeared in regions distant from blood vessels (Fig. 6, bottom panel). This result suggests that the NP could traverse through avascular region of the brain for relatively long distance. The BBB permeability of the NP might be attributed to the small size of NP-TMZ-CTX (~50 nm) and multiple BBB permeation molecules attached on NPs including TMZ39, 42 and CTX via receptor-mediated mechanism,39 and chitosan via absorptive-mediated mechanism.43
CONCLUSIONS
We have introduced a novel synthetic method to develop a targeted TMZ delivery system that could protect TMZ from fast degradation at physiological conditions and improve the therapeutic efficacy of TMZ against GBM. The NP contains a biotin-functionalized chitosan core that is covalently conjugated with TMZ that were linked to GBM-specific, surface-bound CTX ligands with fast, stable, and stoichiometric biotin-neutravidin interactions. NP-TMZ-CTX showed higher uptake in GBM cells and was more effective in cell killing than non-targeted NPs and free drug. We further demonstrated that NP-TMZ-CTX can cross the BBB and deliver TMZ into avascular region of brain. NP-TMZ-CTX offers hope to deliver effective doses of TMZ specifically to GBM cells while minimizing dose-limiting toxicity on other healthy tissues. This NP-based delivery system can be broadly applied to various applications as it can be tailored to incorporate various therapeutics that are otherwise unstable or insoluble in physiological conditions. Additionally, the targeting ligand attachment method could be easily modified to other bioorthogonal linkage chemistries such as trans-cyclooctyne-tetrazine reaction pairs.
Figure 5.
Deconvolved fluorescence images of U-118 MG, SF767, and GBM6 cells treated with NP-TMZ-CTX (top row) or NP-TMZ-NA (bottom row). Cell nuclei were stained with DAPI (blue), cell membranes were stained with WGA-AF488 (green), and AF647 signal from NP were shown as red.
Acknowledgments
The work is supported by grants R01CA161953, R01EB006043, R21EB014572 and R01CA134213 from the NIH/NCI, and UW-CGF 65-6330 from the University of Washington. C. F. acknowledges support from an NSF/NCI IGERT Fellowship and F. M. K., Z. R. S and Q. M. acknowledge support from an NIH Ruth L. Kirschstein T32 Fellowship (T32CA138312). F. M. K. also acknowledges support from an American Brain Tumor Association Fellowship in Honor of Susan Kramer. K. W. acknowledges support from the University of Washington College of Engineering Dean’s Fellowship. We acknowledge lab assistance from Chris Dayringer and Zhixing Wang. We thank FHCRC Shard Resources Scientific Imaging Core for their assistance on deconvolution microscopy, confocal microscopy and image analysis. We thank the Sarkaria lab at the Mayo clinic for providing the GBM6 cells.
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Conflict of Interest Disclosure
The authors declare no competing financial interest.
References
- 1.Ostrom QT, Gittleman H, Liao P, Rouse C, Chen Y, Dowling J, Wolinsky Y, Kruchko C, Barnholtz-Sloan J. Cbtrus Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2007–2011. Neuro Oncol. 2014;16(Suppl 4):iv1–63. doi: 10.1093/neuonc/nou223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wen PY, Kesari S. Malignant Gliomas in Adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 3.Huse JT, Holland E, DeAngelis LM. Glioblastoma: Molecular Analysis and Clinical Implications. Annual Rev Med. 2013;64:59–70. doi: 10.1146/annurev-med-100711-143028. [DOI] [PubMed] [Google Scholar]
- 4.Omuro A. Glioblastoma and Other Malignant Gliomas. JAMA. 2013;310:1842. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 5.Parsons DW, Jones S, Zhang X, Lin JCH, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Hartigan J, Smith DR, Strausberg RL, Marie SKN, Shinjo SMO, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chamberlain MC. Bevacizumab for the Treatment of Recurrent Glioblastoma. Clin Med Insights Oncol. 2011;5:117–29. doi: 10.4137/CMO.S7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, Vogelbaum MA, Colman H, Chakravarti A, Pugh S, Won M, Jeraj R, Brown PD, Jaeckle KA, Schiff D, Stieber VW, Brachman DG, Werner-Wasik M, Tremont-Lukats IW, Sulman EP, Aldape KD, Curran WJ, Jr, Mehta MP. A Randomized Trial of Bevacizumab for Newly Diagnosed Glioblastoma. N Engl J Med. 2014;370:699–708. doi: 10.1056/NEJMoa1308573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Stevens MF, Bradshaw TD. Temozolomide: Mechanisms of Action, Repair and Resistance. Curr Mol Pharmacol. 2012;5:102–14. doi: 10.2174/1874467211205010102. [DOI] [PubMed] [Google Scholar]
- 9.Quinn JA, Jiang SX, Reardon DA, Desjardins A, Vredenburgh JJ, Rich JN, Gururangan S, Friedman AH, Bigner DD, Sampson JH, McLendon RE, Herndon JE, 2nd, Walker A, Friedman HS. Phase Ii Trial of Temozolomide Plus O6-Benzylguanine in Adults with Recurrent, Temozolomide-Resistant Malignant Glioma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2009;27:1262–7. doi: 10.1200/JCO.2008.18.8417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patil R, Portilla-Arias J, Ding H, Inoue S, Konda B, Hu J, Wawrowsky KA, Shin PK, Black KL, Holler E, Ljubimova JY. Temozolomide Delivery to Tumor Cells by a Multifunctional Nano Vehicle Based on Poly(β-L-Malic Acid) Pharm Res. 2010;27:2317–2329. doi: 10.1007/s11095-010-0091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braun K, Wiessler M, Ehemann V, Pipkorn R, Spring H, Debus J, Didinger B, Koch M, Muller G, Waldeck W. Treatment of Glioblastoma Multiforme Cells with Temozolomide-Bioshuttle Ligated by the Inverse Diels-Alder Ligation Chemistry. Drug Des Dev Ther. 2009;2:289–301. doi: 10.2147/dddt.s3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veiseh O, Sun C, Fang C, Bhattarai N, Gunn J, Kievit F, Du K, Pullar B, Lee D, Ellenbogen RG, Olson J, Zhang M. Specific Targeting of Brain Tumors with an Optical/Magnetic Resonance Imaging Nanoprobe across the Blood-Brain Barrier. Cancer Res. 2009;69:6200–7. doi: 10.1158/0008-5472.CAN-09-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kievit FM, Veiseh O, Fang C, Bhattarai N, Lee D, Ellenbogen RG, Zhang M. Chlorotoxin Labeled Magnetic Nanovectors for Targeted Gene Delivery to Glioma. ACS Nano. 2010;4:4587–4594. doi: 10.1021/nn1008512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graf N, Mokhtari TE, Papayannopoulos IA, Lippard SJ. Platinum(Iv)-Chlorotoxin (Ctx) Conjugates for Targeting Cancer Cells. J Inorg Biochem. 2012;110:58–63. doi: 10.1016/j.jinorgbio.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang R, Ke W, Han L, Li J, Liu S, Jiang C. Targeted Delivery of Chlorotoxin-Modified DNA-Loaded Nanoparticles to Glioma Via Intravenous Administration. Biomaterials. 2011;32:2399–2406. doi: 10.1016/j.biomaterials.2010.11.079. [DOI] [PubMed] [Google Scholar]
- 16.Sun C, Fang C, Stephen Z, Veiseh O, Hansen S, Lee D, Ellenbogen RG, Olson J, Zhang M. Tumor-Targeted Drug Delivery and Mri Contrast Enhancement by Chlorotoxin-Conjugated Iron Oxide Nanoparticles. Nanomedicine. 2008;3:495–505. doi: 10.2217/17435889.3.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun C, Veiseh O, Gunn J, Fang C, Hansen S, Lee D, Sze R, Ellenbogen RG, Olson J, Zhang M. In Vivo Mri Detection of Gliomas by Chlorotoxin-Conjugated Superparamagnetic Nanoprobes. Small. 2008;4:372–379. doi: 10.1002/smll.200700784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veiseh M, Gabikian P, Bahrami SB, Veiseh O, Zhang M, Hackman RC, Ravanpay AC, Stroud MR, Kusuma Y, Hansen SJ, Kwok D, Munoz NM, Sze RW, Grady WM, Greenberg NM, Ellenbogen RG, Olson JM. Tumor Paint: A Chlorotoxin:Cy5.5 Bioconjugate for Intraoperative Visualization of Cancer Foci. Cancer Res. 2007;67:6882–6888. doi: 10.1158/0008-5472.CAN-06-3948. [DOI] [PubMed] [Google Scholar]
- 19.Veiseh O, Gunn JW, Kievit FM, Sun C, Fang C, Lee JSH, Zhang M. Inhibition of Tumor-Cell Invasion with Chlorotoxin-Bound Superparamagnetic Nanoparticles. Small. 2008;5:256–264. doi: 10.1002/smll.200800646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veiseh O, Sun C, Gunn J, Kohler N, Gabikian P, Lee D, Bhattarai N, Ellenbogen R, Sze R, Hallahan A, Olson J, Zhang M. Optical and Mri Multifunctional Nanoprobe for Targeting Gliomas. Nano Let. 2005;5:1003–1008. doi: 10.1021/nl0502569. [DOI] [PubMed] [Google Scholar]
- 21.Mortensen JH, Jeppesen M, Pilgaard L, Agger R, Duroux M, Zachar V, Moos T. Targeted Antiepidermal Growth Factor Receptor (Cetuximab) Immunoliposomes Enhance Cellular Uptake in Vitro and Exhibit Increased Accumulation in an Intracranial Model of Glioblastoma Multiforme. J Drug Deliv. 2013;2013:209205. doi: 10.1155/2013/209205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoon DJ, Kwan BH, Chao FC, Nicolaides TP, Phillips JJ, Lam GY, Mason AB, Weiss WA, Kamei DT. Intratumoral Therapy of Glioblastoma Multiforme Using Genetically Engineered Transferrin for Drug Delivery. Cancer Res. 2010;70:4520–7. doi: 10.1158/0008-5472.CAN-09-4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhattarai N, Matsen FA, Zhang M. Peg-Grafted Chitosan as an Injectable Thermoreversible Hydrogel. Macromol Biosci. 2005;5:107–11. doi: 10.1002/mabi.200400140. [DOI] [PubMed] [Google Scholar]
- 24.Arrowsmith J, Jennings SA, Clark AS, Stevens MFG. Antitumor Imidazotetrazines. 41. 1 conjugation of the Antitumor Agents Mitozolomide and Temozolomide to Peptides and Lexitropsins Bearing DNA Major and Minor Groove-Binding Structural Motifs. J Med Chem. 2002;45:5458–5470. doi: 10.1021/jm020936d. [DOI] [PubMed] [Google Scholar]
- 25.Deshane J, Garner CC, Sontheimer H. Chlorotoxin Inhibits Glioma Cell Invasion Via Matrix Metalloproteinase-2. J Biol Chem. 2003;278:4135–44. doi: 10.1074/jbc.M205662200. [DOI] [PubMed] [Google Scholar]
- 26.Bobola MS, Kolstoe DD, Blank A, Silber JR. Minimally Cytotoxic Doses of Temozolomide Produce Radiosensitization in Human Glioblastoma Cells Regardless of Mgmt Expression. Mol Cancer Ther. 2010;9:1208–18. doi: 10.1158/1535-7163.MCT-10-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carlson BL, Grogan PT, Mladek AC, Schroeder MA, Kitange GJ, Decker PA, Giannini C, Wu W, Ballman KA, James CD, Sarkaria JN. Radiosensitizing Effects of Temozolomide Observed in Vivo Only in a Subset of O6-Methylguanine-DNA Methyltransferase Methylated Glioblastoma Multiforme Xenografts. Int J Radiat Oncol Biol Phys. 2009;75:212–9. doi: 10.1016/j.ijrobp.2009.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marttila AT, Laitinen OH, Airenne KJ, Kulik T, Bayer EA, Wilchek M, Kulomaa MS. Recombinant Neutralite Avidin: A Non-Glycosylated, Acidic Mutant of Chicken Avidin That Exhibits High Affinity for Biotin and Low Non-Specific Binding Properties. FEBS Letters. 2000;467:31–6. doi: 10.1016/s0014-5793(00)01119-4. [DOI] [PubMed] [Google Scholar]
- 29.Tseng CL, Wu SY, Wang WH, Peng CL, Lin FH, Lin CC, Young TH, Shieh MJ. Targeting Efficiency and Biodistribution of Biotinylated-Egf-Conjugated Gelatin Nanoparticles Administered Via Aerosol Delivery in Nude Mice with Lung Cancer. Biomaterials. 2008;29:3014–22. doi: 10.1016/j.biomaterials.2008.03.033. [DOI] [PubMed] [Google Scholar]
- 30.Hama Y, Urano Y, Koyama Y, Choyke PL, Kobayashi H. Activatable Fluorescent Molecular Imaging of Peritoneal Metastases Following Pretargeting with a Biotinylated Monoclonal Antibody. Cancer Res. 2007;67:3809–17. doi: 10.1158/0008-5472.CAN-06-3794. [DOI] [PubMed] [Google Scholar]
- 31.Nordlund HR, Hytonen VP, Laitinen OH, Uotila ST, Niskanen EA, Savolainen J, Porkka E, Kulomaa MS. Introduction of Histidine Residues into Avidin Subunit Interfaces Allows Ph-Dependent Regulation of Quaternary Structure and Biotin Binding. FEBS Letters. 2003;555:449–54. doi: 10.1016/s0014-5793(03)01302-4. [DOI] [PubMed] [Google Scholar]
- 32.Davis ME, Zuckerman JE, Choi CHJ, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Evidence of Rnai in Humans from Systemically Administered Sirna Via Targeted Nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farokhzad OC, Jon S, Khademhosseini A, Tran TN, Lavan DA, Langer R. Nanoparticle-Aptamer Bioconjugates: A New Approach for Targeting Prostate Cancer Cells. Cancer Res. 2004;64:7668–72. doi: 10.1158/0008-5472.CAN-04-2550. [DOI] [PubMed] [Google Scholar]
- 34.Petros RA, Desimone JM. Strategies in the Design of Nanoparticles for Therapeutic Applications. Nat Rev Drug Discov. 2010;9:615–627. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- 35.Fang C, Veiseh O, Kievit F, Bhattarai N, Wang F, Stephen Z, Li C, Lee D, Ellenbogen RG, Zhang M. Functionalization of Iron Oxide Magnetic Nanoparticles with Targeting Ligands: Their Physicochemical Properties and in Vivobehavior. Nanomedicine. 2010;5:1357–1369. doi: 10.2217/nnm.10.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ponten J, Macintyre EH. Long Term Culture of Normal and Neoplastic Human Glia. Acta Pathol Microbiol Scand. 1968;74:465–86. doi: 10.1111/j.1699-0463.1968.tb03502.x. [DOI] [PubMed] [Google Scholar]
- 37.Pattabiraman DR, Weinberg RA. Tackling the Cancer Stem Cells - What Challenges Do They Pose? Nat Rev Drug Discov. 2014;13:497–512. doi: 10.1038/nrd4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hegi ME, Liu L, Herman JG, Stupp R, Wick W, Weller M, Mehta MP, Gilbert MR. Correlation of O6-Methylguanine Methyltransferase (Mgmt) Promoter Methylation with Clinical Outcomes in Glioblastoma and Clinical Strategies to Modulate Mgmt Activity. J Clin Oncol. 2008;26:4189–99. doi: 10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- 39.Stephen ZR, Kievit FM, Veiseh O, Chiarelli PA, Fang C, Wang K, Hatzinger SJ, Ellenbogen RG, Silber JR, Zhang M. Redox-Responsive Magnetic Nanoparticle for Targeted Convection-Enhanced Delivery of O6-Benzylguanine to Brain Tumors. ACS Nano. 2014;8:10383–95. doi: 10.1021/nn503735w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilbert MR, Wang M, Aldape KD, Stupp R, Hegi ME, Jaeckle KA, Armstrong TS, Wefel JS, Won M, Blumenthal DT, Mahajan A, Schultz CJ, Erridge S, Baumert B, Hopkins KI, Tzuk-Shina T, Brown PD, Chakravarti A, Curran WJ, Jr, Mehta MP. Dose-Dense Temozolomide for Newly Diagnosed Glioblastoma: A Randomized Phase Iii Clinical Trial. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2013;31:4085–91. doi: 10.1200/JCO.2013.49.6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spence AM, Kiem HP, Partap S, Schuetze S, Silber JR, Peterson RA. Complications of a Temozolomide Overdose: A Case Report. J Neurooncol. 2006;80:57–61. doi: 10.1007/s11060-006-9152-y. [DOI] [PubMed] [Google Scholar]
- 42.Mrugala MM, Chamberlain MC. Mechanisms of Disease: Temozolomide and Glioblastoma--Look to the Future. Nature clinical practice Oncology. 2008;5:476–86. doi: 10.1038/ncponc1155. [DOI] [PubMed] [Google Scholar]
- 43.Agyare EK, Curran GL, Ramakrishnan M, Yu CC, Poduslo JF, Kandimalla KK. Development of a Smart Nano-Vehicle to Target Cerebrovascular Amyloid Deposits and Brain Parenchymal Plaques Observed in Alzheimer’s Disease and Cerebral Amyloid Angiopathy. Pharmaceutical research. 2008;25:2674–84. doi: 10.1007/s11095-008-9688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]