Abstract
Fibroblast growth factor 2 (FGF2), which is one of the 22 members of the FGF family, functions as an extracellular molecule involved in canonical receptor tyrosine kinase signaling. It has been implicated in angiogenesis and the development of the central nervous system. Here, we reveal that cytosolic LMW FGF2 (18 kD), not its secreted form, plays an unexpected role in the innate immune response. Cytosolic LMW FGF2 directly associated with inactivated RIG-I under physiological conditions, which enhanced RIG-I protein stability, thereby maintaining basal RIG-I levels. However, during RIG-I activation induced by viral RNA, cytosolic FGF2 bound to the caspase recruitment domains (CARDs) of activated RIG-I, which blocked RIG-I-MAVS complex formation. LMW FGF2 deficiency increased type I interferon production, whereas the overexpression of LMW FGF2 exerted the opposite effect. Cytosolic LMW FGF2 functions as a negative regulator in RIG-I-mediated antiviral signaling. This work provides insight into the role of FGF2 in innate immune response.
Introduction
Growth factors are natural proteins that are capable of stimulating cell growth, proliferation and differentiation. However, few have been reported to participate in immune regulation to date. Transforming growth factor β (TGF-β) plays a role in the regulation of immune responses, especially in the T cell response (1, 2). Vascular endothelial growth factor (VEGF) is believed to inhibit T cell development and contribute to tumor-induced immune suppression (3). Recently, nerve growth factor β (NGF-β) was shown to participate in Staphylococcus aureus infection by stimulating proinflammatory cytokine production in macrophages and promoting neutrophil recruitment (4).
Fibroblast growth factor 2 (FGF2 or basic FGF) is a pleiotropic signaling molecule known to be involved in multiple biological processes, including angiogenesis, embryonic development and wound healing (5–7). FGF2 activities are known to be mediated through binding to the high-affinity transmembrane tyrosine kinase receptors (FGFRs) that promote the activation of FGFR signaling pathways. However, whether FGF2 plays a role in immune responses remains unknown.
Viral infection can be rapidly detected by pattern recognition receptors (PRRs), which subsequently induce the innate immune response. RIG-I-like receptors (RLRs), which comprise RIG-I, MDA5 and LGP2, serve as cytoplasmic RNA sensors that detect viral RNA in the cytosol of most cell types (8–10). RIG-I is a key sensor of paramyxoviruses, influenza virus, hepatitis C virus (HCV) and Japanese encephalitis virus (11). Viral RNA binding to the RIG-I C-terminal regulatory domain results in a conformational change that in turn enables RIG-I binding to the signal adaptor MAVS (also called VISA, IPS-1, or CARDIF) through their N-terminal caspase recruitment domains (CARDs) (12–15). Such binding eventually leads to the production of type I interferon and inflammatory cytokines.
Here, we determined that cytosolic LMW FGF2 plays a dual functional role in RIG-I-mediated signaling through a potent interaction with RIG-I. On the one hand, LMW FGF2 stabilizes RIG-I and prevents proteasome-mediated RIG-I degradation. On the other hand, cytosolic LMW FGF2 serves as a negative regulator of RIG-I-mediated signaling pathway to keep it in an autoinhibitory state; after viral infection, LMW FGF2 is crucial for limiting the type I interferon production. Our results confirm that cytosolic FGF2 instead of secreted FGF2 functions to maintain the innate immune homeostasis of antiviral immunity.
Materials and Methods
Mice
Pathogen-free 10-week-old Fgf2lmw−/− mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA) and caged in a pathogen-free facility with air filtration. Mouse genotype was identified by Pyrosequencing. The primer sequence was as follows: forward, 5′-GCAGGAGCGGAGAAAGTTAGA -3′, and reverse, 5′-TTGCAGTAGAGCCGCTTGG -3′. The mutant sequence GCA encodes an alanine in place of the methionine ATG which is the translational start site of FGF2 18kDa low molecular weight isoform (LMW). Consequently, the FGF2 LMW is not expressed in Fgf2lmw−/− mice, whereas the FGF2 high molecular weight isoforms (HMW) are still expressed. The mice were fully backcrossed up to twelve generations onto the C57BL/6 genetic background. Fgf2lmw−/− were identified as knockout genotype (KO), and Fgf2lmw+/+ littermates were used as wild-type control (WT). All procedures including animal studies were conducted following the National Guidelines for Care of Laboratory Animals (2006-398) and performed in accordance with institutional regulations after protocol review and approval by the Institutional Animal Care and Use Committee (IACUC) of Academy of Military Medical Sciences (project no. 2012-005).
Cells, viruses and reagents
Mouse embryonic fibroblasts (MEFs) were isolated from embryonic day 14 pregnant mice, and their use was restricted to within three passages to avoid replicative senescence. Peritoneal macrophages were obtained from the mouse peritoneal cavity four days after thioglycollate (Sigma) injection. A549, 293T, HeLa, MEF and macrophage cells were cultured in Dulbecco’s Modified Eagle’s medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS) and penicillin-streptomycin (100 units/ml). Sendai virus (SeV, Cantell strain) and Influenza A virus (IAV) A/Puerto Rico/8/1934 (PR8) were propagated in 10-day-old specific-pathogen-free (SPF) chicken embryos. Other reagents used were as follows: polyinosinic-polycytidylic acid (polyIC) with low molecular weight (Invivogen); Lipofectamine 2000 (Invitrogen); recombinant human FGF2 (154aa) and IFN-β proteins (Peprotech); cycloheximide (CHX, Sigma).
Plasmids
DNA fragments corresponding to the coding sequence of ATG-initiated low molecular weight isoform of human and mouse FGF2 genes were obtained from A549 and mouse lung cDNA, respectively, by PCR amplification and subcloned into plasmid pWPXL between restriction sites PacI and EcoRI. The primer sequences used were as follows: LMW hFGF2 forward, 5′-GCCTTAATTAAGCCATGGCAGCCGGGAGC-3′ and reverse, 5′-CCGGAATTCTCAGCTCTTAGCAGA -3′; LMW mFGF2 forward, 5′-GCTTAATTAACCATGGCTGCCAGCGGCATCAC -3′ and reverse, 5′-CCGGAATTCCCAGTGTCTCAGTGACAGTGTC -3′. The LMW FGF2 Δ46 mutant was constructed by depleting LMW FGF2 C-terminal 46 amino acid from residues 109 to 155. The LMW FGF2 Δ46 DNA was amplified using the reverse primer 5′-CCGGAATTCTCAAGATTCCAATCG -3′. V5-tagged human HMW FGF2 mutant in which LMW FGF2 initiation codon ATG was substituted with GCG was synthesized by Invitrogen (Shanghai, China) and cloned into the pWPXL vector. The Flag-RIG-I, Flag-RIG-I 2CARD, Flag-RIG-I Δ2CARD and pCAGGS-PR8-NS1 (16, 17) plasmids were generously provided by Michaela U. Gack (Harvard Medical School, Boston, USA). Flag-MAVS 2CARD (18), HA-RIG-I and Flag-MDA5 (19) were kindly provided by Qinmiao Sun (Chinese Academy of Sciences, Beijing, China) and Danying Chen (Peking University, Beijing, China), respectively. The pGL3-IFN-β-Luc, pGL3-NF-κB-Luc and pISRE-Luc (20) plasmids were generously provided by Jianwei Wang (Chinese Academy of Medical Sciences, Beijing, China). In addition, the Flag-Ub WT, 48only, K48R, 63only, and K63R mutants (21) were kindly provided by Hong-Bing Shu (Wuhan University, Wuhan, China).
Quantitative real-time PCR
Total RNA was obtained from cultured cells with TRIzol reagent (Invitrogen). cDNA was generated by reverse transcription with commercial PrimeScript RT Master Mix (Takara). The primer pairs IFN-α, IFN-β, FGF2, RIG-I, IL-6, TNF-α and GAPDH (Table 1) were designed using Primer Premier software 5.0 (Premier Biosoft International, Palo Alto, CA, USA) and synthesized by Invitrogen. Quantitative real-time PCR was performed in triplicate wells of a 96-well reaction plate on an ABI 7500 PCR System (Applied Biosystems). The quantification data were analyzed with ABI 7500 SDS software v.1.3.
Table 1.
Primer pairs used for real-time PCR.
| Forward1 | Reverse | |
|---|---|---|
| Human | ||
| IFN-α | GTGAGGAAATACTTCCAAAGA | TCTCATGATTTCTGCTCTGACA |
| IFN-β | AGCTGAAGCAGTTCCAGAAG | AGTCTCATTCCAGCCAGTGC |
| FGF2 | GTGTGTGCTAACCGTTACCT | GCTCTTAGCAGACATTGGAAG |
| RIG-I | TTGCACAGTGCAATCTTGTCATCC | GTCTGTATATGCAGAATCTTTTCCC |
| GAPDH | GGTGGTCTCCTCTGACTTCAACA | GTTGCTGTAGCCAAATTCGTTGT |
| Mouse | ||
| IFN-α | TCTGATGCAGCAGGTGGG | AGGGCTCTCCAGACTTCTGCTCTG |
| IFN-β | GCACTGGGTGGAATGAGACT | AGTGGAGAGCAGTTGAGGACA |
| FGF2 | AGCGACCCACACGTCAAACTAC | CAGCCGTCCATCTTCCTTCATA |
| RIG-I | TTGCTGAGTGCAATCTCGTC | GTATGCGGTGAACCGTCTTT |
| IL-6 | TGACAACCACGGCCTTCCCTAC | CTCATTTCCACGATTTCCCAGA |
| TNF-α | CCACCACGCTCTTCTGTCTACTG | GTGGGCTACAGGCTTGTCACTC |
| GAPDH | ACAGCCGCATCTTCTTGTGCAGTG | GGCCTTGACTGTGCCGTTGAATTT |
Primer pairs of IFN-α, IFN-β, FGF2, RIG-I, IL-6, TNF-α and GAPDH were designed using Primer 5.0 and presented in Table 1.
ELISA for interferon production
IFN-β production measurements of primary MEF and macrophage culture supernatants were performed using a commercially available IFN-β ELISA kit, mouse (Invitrogen) according to the manufacturer’s instructions. The assay plate was analyzed using a Thermo Scientific Multiskan FC Microplate Photometer. The detection limit of IFN-β concentration was 15.6 pg/ml.
siRNA knockdown and lentivirus-mediated overexpression of FGF2
The FGF2-specfic siRNA#1, 5′-GCACUGAAACGAACUGGGCAGUAU-3′, and siRNA#2, 5′-GGAGUGUGUGCUAACCGUUTT-3′, were synthesized by RiboBio (Guangzhou, China). For FGF2 silencing, A549 cells were transfected with 20 nM siRNA using Lipofectamine RNAiMax (Invitrogen). After a 48-h transfection, FGF2 repression efficiency was detected by real-time PCR, followed by SeV infection experiment. For lentivirus-mediated FGF2 overexpression, 293T cells were transfected with pWPXL-mFGF2LMW or pWPXL-GFP, and with plasmids psPAX2 and pMD2.G. After a 48-h transfection, recombinant lentivirus was obtained. MEF−/− were incubated with lenti-mFGF2LMW or lenti-GFP as a control for 48 h. Then, the cells were harvested to determine FGF2 expression efficiency by real-time PCR, followed by polyIC simulation or SeV infection experiments.
Dual luciferase reporter assay
293T cells were transfected in 24-well plates with the expression plasmids LMW FGF2, HMW FGF2, or LMW FGF2 Δ46 (50 ng) and with the reporter plasmid Luc-IFN-β, Luc-NF-κB, or Luc-IRSE (50 ng) along with pRL-TK1 plasmid (20 ng) for Renilla luciferase expression using Lipofectamine 2000. After a 24-h transfection, cells were treated with polyIC (2 μg/ml) or infected with SeV (50 HA U/ml) for another 24 h. Subsequently, the cells were lysed for luciferase activities measurement using Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase activities were normalized to Renilla values. Densitometric analysis relative to values of unstimulated samples was expressed as fold change. For experiments without polyIC or SeV simulation, the reporter plasmid and pRL-TK1 were co-transfected with full-length RIG-I, RIG-I 2CARD, MDA5, or MAVS. After a 48-h transfection, cell lysates were analyzed in the dual luciferase assay.
Immunoprecipitation and immunoblotting assay
For the immunoprecipitation assays, 293T cells were collected after 48 h and then lysed in lysis buffer (25 mM Tris·HCl pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 5% glycerol) supplemented with protease inhibitor cocktail (Thermo Scientific) on ice for 30 min with periodic mixing. The whole-cell lysates (WCL) were centrifuged and then incubated with an anti-Flag (Sigma) or an anti-Rig-I (Millipore) antibody overnight at 4°C, followed by further incubation with protein A/G beads (Sigma) for 2 h at 4°C. Precipitates were washed with lysis buffer three times, and then boiled in 5× Protein SDS-PAGE Loading Buffer for 5 min. For subsequent immunoblotting assays, the proteins were resolved on an SDS-PAGE gel, and transferred onto 0.2-μm NC membrane (Whatman). The membranes were blocked by 5% (w/v) non-fat dry milk (GE Healthcare) for 1 h at room temperature and then incubated overnight at 4°C with the indicated antibodies. The primary antibodies used were as followed: anti-Flag and anti-HA (1:5000; Sigma); anti-RIG-I, anti-MDA5, anti-phospho-IRF3, and anti-IRF3 (1:1000; Cell Signaling Technology); anti-FGF2 (1:1000; Millipore); anti-V5 (1:5000; Invitrogen); anti-ubiquitin, Lys63 specific (1:1000; Millipore); anti-NS1 (1:200; Santa Cruz Biotechnology); and anti-β-actin (1:5000; Sigma). After three washes with TBST (TBS containing 0.1% Triton X-100), incubation of goat anti-rabbit or anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibody (1:5000; Cell Signaling Technology) followed for 1 h at room temperature. The final detection of protein was performed using the Signal Boost Immunoreaction Enhancer Kit (Millipore).
Confocal immunofluorescence microscopy
After a 24-h transfection, HeLa cells were fixed with 4% paraformaldehyde at room temperature for 15 min, permeabilized with PBS containing 10% FBS, 3% BSA, and 0.5% Triton X-100 for 15 min, and then incubated with diluted primary antibody overnight at 4°C. After two washes with PBS, the cells were incubated with secondary antibodies conjugated to Alexa Fluor 488 and Alexa Fluor 594 (Invitrogen) at room temperature for 1 h. Cell nuclei were stained with DAPI (Roche) for 10 min followed by three PBS washes. Images were captured on Fluoview 1000 laser scanning confocal microscope (Olympus) at a magnification of 100× and analyzed by FV10-ASW (Ver 3.0) software (Olympus).
Statistical analyses
Statistical analysis of the results was performed in GraphPad Prism software 4.0 (GraphPad Software, CA). The data are shown as the mean ± s.e.m. Statistical significance was assessed by unpaired Student’s t-test. * P<0.05; ** P<0.01; *** P<0.001.
Results
Cytosolic LMW FGF2 interacts with inactive RIG-I
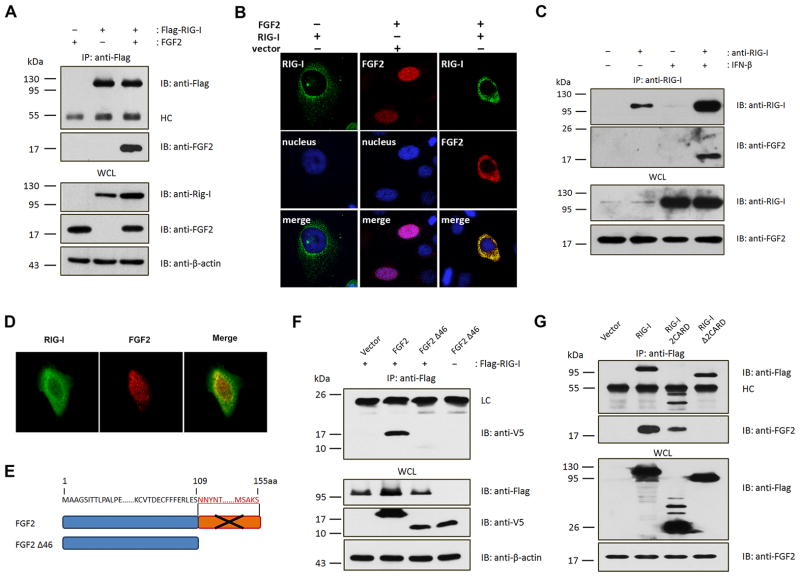
At the beginning of delving into the role of FGF2 in innate immune responses, first of all we speculated that FGF2 participated in RIG-I-mediated signaling, then we investigated whether or not there was a possibility of an potential interaction between FGF2 and RIG-I. Immunoprecipitation assays revealed an potent interaction of full-length RIG-I with LMW FGF2 (Fig. 1A). Additionally, we examined whether HMW FGF2 interacted with RIG-I. To avoid any potential impact of LMW FGF2, we constructed an HMW FGF2 (22 kDa)-only plasmid by replacing the LMW FGF2 initiation codon ATG with GCG, by which HMW FGF2 but not LMW FGF2 is expressed (Fig. S1A and B). The results showed that there was undetectable interaction between HMW FGF2 and RIG-I (Fig. S1C). Confocal microscopy showed that the expression of LMW FGF2 alone resulted in nuclear localization with a minor cytoplasmic component, whereas the co-expression of RIG-I and LMW FGF2 led to a shift in FGF2 cytoplasmic localization and extensive co-localization of LMW FGF2 and RIG-I (Fig. 1B). Furthermore, the endogenous interaction between RIG-I and LMW FGF2 rather than HMW FGF2 was confirmed (Fig. 1C). Confocal microscopy also showed an intracellular co-localization of endogenous FGF2 and RIG-I (Fig. 1D). Crystallography studies on the three-dimensional structure of human LMW FGF2 have revealed that it is composed entirely of a β-sheet structure that comprises three copies of a four-stranded β-meander motif, which correspond to residues 18–59, 60–100, and 101–143; the third repeat of FGF2 is believed to be a very important functional unit (22). Our studies showed that LMW FGF2 Δ46 in which the C-terminal 46 aa residues were deleted barely interacted with RIG-I (Fig. 1E and F). In addition, RIG-I consists of an N-terminal 2CARD domain as signal activation unit, a central DExH/C helicase domain and a C-terminal domain for viral RNA recognition (23). LMW FGF2 specifically interacted with full-length RIG-I and RIG-I 2CARD domain, but that RIG-I Δ2CARD was incapable of binding to LMW FGF2 (Fig. 1G). These results together indicate that the RIG-I 2CRAD and FGF2 C-terminal region primarily contributes to the interaction of RIG-I with LMW FGF2. Besides that, we verified that LMW FGF2 could also interacted with MDA5 (Fig. S1D), but whether LMW FGF2 had an specific effect on RIG-I-mediated signaling was necessary to be verified.
Figure 1. Cytosolic LMW FGF2 interacts with RIG-I.
(A) 293T cells were transfected with Flag-RIG-I (500 ng) and LMW FGF2 (500 ng) or vector control (500 ng) in cell culture dish. Whole cell lysates (WCL) were subjected to immunoprecipitation (IP) using anti-Flag antibodies. WCLs and immunoprecipitates of 293T cells were separated by 12% SDS-PAGE and analyzed by immunoblotting (IB) using antibodies against Flag, FGF2, RIG-I and β-actin.
(B) HeLa cells were transfected with Flag-RIG-I and/or LMW FGF2. Twenty-four hours after transfection, HeLa cells were immunostained with anti-Flag (green) and anti-FGF2 (red) antibodies and DAPI for nuclei staining (blue). Original magnification, 100×.
(C) The interaction between endogenous RIG-I and FGF2 was assessed in A549 cells. The cells were treated with or without IFN-β (10 ng/ml). WCLs of A549 cells were immunoprecipitated with an anti-RIG-I antibody or rabbit IgG as control, followed by IB with anti-RIG-I and anti-FGF2 antibodies.
(D) HeLa cells were immunostained with anti-RIG-I (green) and anti-FGF2 (red) antibodies. Original magnification, 100×.
(E) V5-tagged LMW FGF2 Δ46 mutant was constructed by depletion of C-terminal 46 amino acid corresponding to residues 109–155.
(F) 293T cells were transfected with Flag-RIG-I and V5-LMW FGF2 or V5-LMW FGF2 Δ46. WCLs of 293T cells were immunoprecipitated with an anti-Flag antibody, followed by IB with anti-V5, anti-Flag, and anti-β-actin antibodies.
(G) 293T cells were transfected with LMW FGF2 and Flag-full-length RIG-I, RIG-I 2CARD or RIG-I Δ2CARD, as indicated. WCLs of 293T cells were immunoprecipitated with anti-Flag antibodies, followed by IB with anti-Flag and anti-FGF2 antibodies.
Intracellular LMW FGF2 specifically antagonizes RIG-I-mediated antiviral signaling
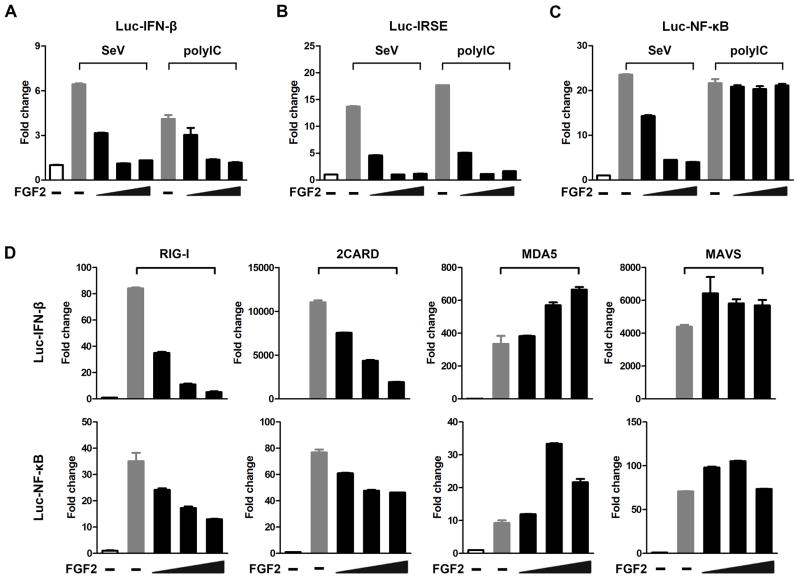
RIG-I recognizes RNA from a broad range of viruses, such as SeV and influenza A virus (IAV), and also viral dsRNA analog polyIC. Upon RNA binding, RIG-I 2CARD is released from the helicase domain and forms a filament, which subsequently promotes downstream signaling activation. To determine how LMW FGF2 functions in antiviral signaling, we assessed the effect of LMW FGF2 on IFN-β-, NF-κB-, and IFN-sensitive response element (IRSE) promoter activity in 293T cells in response to SeV and polyIC. The protein level of FGF2 in 293T cells is undetectable by immunoblotting (Fig. 1A and F); therefore, it is an ideal cellular model for these investigations. The study showed that FGF2 expression markedly decreased both IFN-β (Fig. 2A) and IRSE (Fig. 2B) promoter activity in 293T with polyIC and SeV stimulation in a dose-dependent manner. However, NF-κB promoter activation (Fig. 2C) as induced by SeV infection, but not polyIC stimulation, was decreased by FGF2 expression in a dose-dependent manner. We speculated that there might be an opposite effect of LMW FGF2 on some unknown signaling in which NF-κB would be subjected to activation in response to polyIC, and that thereby ultimately resulted in unchanged luc NF-kB activity. These results indicate that FGF2 might exert a suppressive effect on certain RNA virus-induced antiviral responses. To date, which step of antiviral signaling is intervened by FGF2 has remained unclear. RIG-I is known to mediate the recognition of SeV and short dsRNAs and the activation of downstream adaptor MAVS. Accordingly, to explore this more in detail, we examined the IFN-β and IRSE promoter activity in 293T cells that were transfected with full-length RIG-I, RIG-I 2CARD, MDA5, or MAVS. The study revealed that FGF2 expression notably inactivated the IFN-β and NF-κB promoter activity induced by full-length RIG-I and RIG-I 2CARD in a dose-dependent manner (Fig. 2D), whereas FGF2 activated MDA5-mediated IFN-β and NF-κB promoter activity in a dose-dependent manner. Moreover, FGF2 had little effect on IFN-β and NF-κB promoter activity induced by MAVS. These results suggest that FGF2 negatively regulates RIG-I- but not MDA5-mediated antiviral responses and that the inhibitory effect occurs upstream of MAVS. Additionally, we investigated the effect of HMW FGF2 on IFN-β and NF-κB promoter activity. The result showed that HMW FGF2 had little impact on IFN-β promoter activity but that it activated NF-κB promoter activity in a dose-dependent manner (Fig. S2A and B), which was completely different from the effect of LMW FGF2. These results indicate that LMW FGF2 but not HMW FGF2 specifically suppresses RIG-I-mediated antiviral signaling. Moreover, extracellular secretory FGF2 is known to bind to high-affinity receptor (FGFR) accompanied by HSPG and then activate downstream signaling. However, whether extracellular FGF2 contributes to antiviral signaling has not been confirmed. Thus, we further confirmed that IFN-β and NF-κB promoter activity could not be inactivated by recombinant human LMW FGF2 protein (rFGF2) treatment (Fig. S2C and D), which was totally different from intracellular LMW FGF2 exogenous expression. Our results collectively suggest that intracellular LMW FGF2 specifically antagonizes RIG-I-mediated antiviral signaling. In addition, the expression of LMW FGF2 Δ46 led to the complete loss of inhibitory effect on the IFN-β promoter activity stimulated by SeV or RIG-I 2CARD (Fig. S2E and F). The results indicate that FGF2 C-terminal 46 aa residues possess an essential structure to sustain the functional integrity of interaction between FGF2 and RIG-I, which is crucial for suppressing RIG-I-mediated antiviral signaling.
Figure 2. Intracellular LMW FGF2 specifically antagonizes RIG-I-mediated antiviral signaling.
(A–C) 293T cells were transfected with plasmids encoding luc-IFN-β, luc-IRSE or luc-NF-κB (50 ng) with varying amounts of a plasmid encoding LMW FGF2 (20, 50, and 100 ng). Twenty-four hours after transfection, cells were treated with polyIC or infected with SeV (50 HA unit/ml). Cells were harvested after 48 h of treatment. The IFN-β, IRSE and NF-κB promoter activity in the lysates was then measured using a dual luciferase assay.
(D) 293T cells were transfected with plasmids expressing full-length RIG-I (or RIG-I 2CARD, MDA5 or MAVS), and luc-IFN-β (or luc-NF-κB), and various amounts of LMW FGF2 (20, 50, and 100 ng). Forty-eight hours after transfection, cells lysates were prepared for luciferase activity detection.
The binding of LMW FGF2 to RIG-I can be impaired by polyIC, SeV, IAV, and IAV nonstructural protein 1 (NS1)
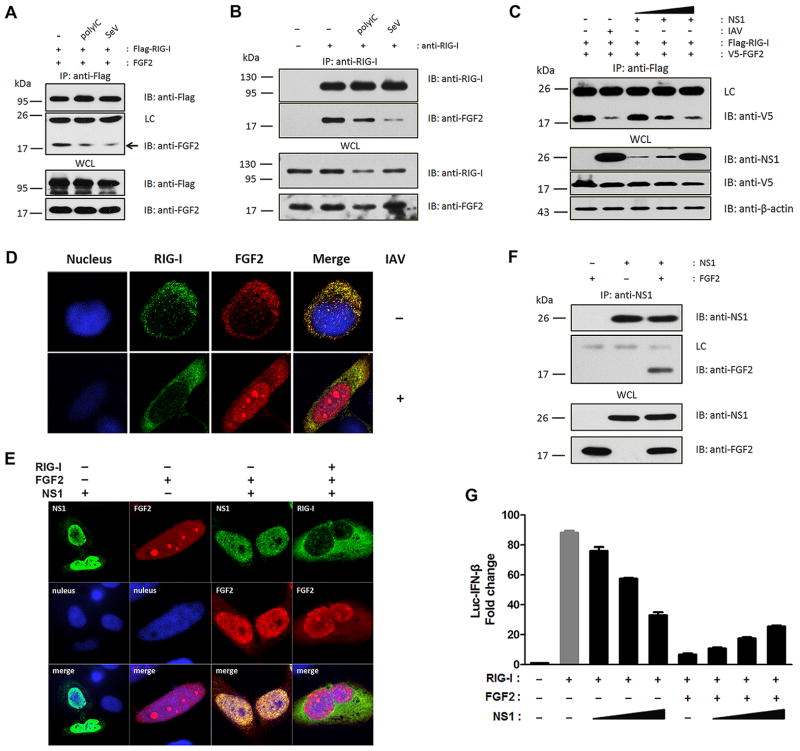
The interaction between RNA-free inactivate RIG-I and LMW FGF2 has been identified previously. However, it is still unclear whether FGF2 remains bound to RIG-I after RNA recognition or is released from activated RIG-I. To investigate these possibilities, we first examined the binding of activated RIG-I with LMW FGF2 induced by polyIC and SeV. The overexpression and endogenous immunoprecipitation assays showed that the interaction between RIG-I and LMW FGF2 was impaired in response to polyIC and SeV (Fig. 3A and B), which suggested that cytosolic LMW FGF2 may also interact with activated RIG-I after viral RNA conjugation, but their binding was partially inhibited. Furthermore, we confirmed a weaker interaction between LMW FGF2 and RIG-I upon IAV infection (Fig. 3C). These results indicate that cytosolic LMW FGF2 is a potent binding partner of inactive RIG-I when cells are in a resting state, while after RIG-I activation the binding was impaired to some extent. Given that IAV appeared to be more efficient at blocking the interaction of LMW FGF2 and RIG-I than SeV and polyIC, we speculated that certain IAV proteins may contribute to the inhibitory effect on LMW FGF2 and RIG-I binding besides viral RNA. IAV NS1 is known to suppress RIG-I activation by blocking TRIM25-mediated antiviral interferon response (17). Thus, we investigated the effect of IAV NS1 expression on the interaction of LMW FGF2 with RIG-I. Interestingly, NS1 expression markedly diminished their interaction in a dose-dependent manner (Fig. 3C). In addition, confocal microscopy revealed that IAV infection resulted in the primary nuclear localization of LMW FGF2 with a minor cytoplasmic component and that less LMW FGF2 co-localized with RIG-I (Fig. 3D). Moreover, NS1 expression had a similar effect on FGF2 localization as IAV infection (Fig. 3E). IAV NS1 expression alone mainly located in the nucleus, as shown previously (17), whereas NS1 co-expression with LMW FGF2 led to a striking nuclear co-localization between NS1 and LMW FGF2, which resulted in FGF2 retention in the nucleus and thereby prevented LMW FGF2 and RIG-I co-localization in cytoplasm. To further explore the NS1 inhibition mechanism, we investigated the potential interaction between NS1 and LMW FGF2. Immunoprecipitation assays showed that NS1 potently interacted with LMW FGF2 (Fig. 3F). Moreover, consistent with the effect of NS1 expression on LMW FGF2 and RIG-I interaction, LMW FGF2-inhibited IFN-β promoter activity increased on NS1 expression in a dose-dependent manner, while remained at a low level compared to RIG-I only induced IFN-β promoter activity (Fig. 3G). These results indicate that IAV NS1 inhibits LMW FGF2 function on Rig-I-mediated signaling probably by interacting with LMW FGF2 and thereby blocking FGF2 export from the cell nucleus to the cytoplasm.
Figure 3. The interaction between LMW FGF2 and RIG-I can be impaired by polyIC, SeV, IAV and NS1.
(A) 293T cells were transfected with Flag-RIG-I and LMW FGF2. Thirty hours after transfection, cells were treated with polyIC or infected with SeV (50 HA unit/ml) for another 18 h. WCLs of 293T cells were immunoprecipitated with an anti-Flag antibody, followed by IB with anti-Flag and anti-FGF2 antibodies. The arrows indicated LMW FGF2 bound with RIG-I.
(B) The interaction between endogenous RIG-I and FGF2 was assessed in A549 cells. The cells were treated with IFN-β, and with or without polyIC and SeV. WCLs of A549 cells were immunoprecipitated with an anti-RIG-I antibody or rabbit IgG as control, followed by IB with anti-RIG-I and anti-FGF2 antibodies.
(C) 293T cells were transfected with Flag-Rig-I and V5-LMW FGF2 and with or without IAV infection after 36 h of transfection, or co-transfected with various amounts of NS1 (50, 100, 500 ng). WCLs of 293T cells were immunoprecipitated with an anti-Flag antibody, followed by IB with anti-V5, anti-NS1 and anti-β-actin antibodies.
(D) HeLa cells were transfected with Flag-RIG-I and LMW FGF2. After 12 h, the cells were infected with or without IAV for another 12 h, and then immunostained with anti-Flag (green) and anti-FGF2 (red) antibodies, and DAPI for nucleus staining (blue). Original magnification, 100×.
(E) HeLa cells were transfected with NS1, LMW FGF2 alone, or NS1 together with LMW FGF2, or NS1 together with Flag-RIG-I and LMW FGF2. Twenty-four hours later, cells were stained with anti-NS1 (green), anti-FGF2 (red), and anti-Flag (green) antibodies and DAPI for nuclei staining (blue). Original magnification, 100×.
(F) 293T cells were transfected with NS1 and LMW FGF2 or vector control. WCLs of 293T cells were immunoprecipitated with an anti-NS1 antibody or mouse IgG as control, followed by IB with anti-NS1 and anti-FGF2 antibodies.
(G) 293T cells were transfected with plasmids expressing RIG-I, luc-IFN-β and various amounts of NS1 (0, 20, 50, and 100 ng), and with or without LMW FGF2 co-expression. Forty-eight hours after transfection, cells lysates were prepared for luciferase activity detection.
FGF2 suppresses the interaction of activated RIG-I with MAVS 2CARD, but not Lys 63-linked RIG-I ubiquitination
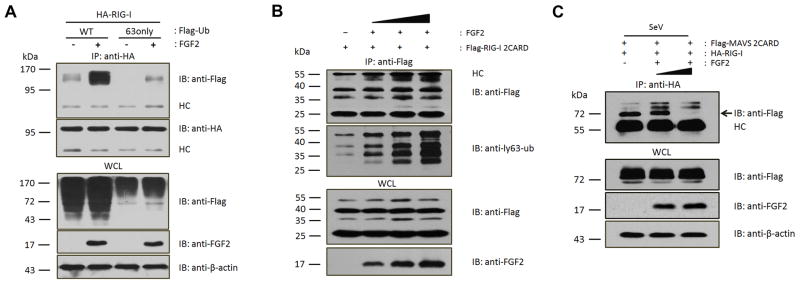
To elucidate the mechanism by which LMW FGF2 inhibits RIG-I-mediated signaling, we first investigated whether LMW FGF2 reduced Lys 63-linked ubiquitination of RIG-I in view of the fact that the Lys 63-linked ubiquitination of RIG-I is crucial for RIG-I-mediated antiviral signaling (16). However, contrary to our expectations, LMW FGF2 expression markedly elevated both total RIG-I ubiquitination and Lys 63-linked ubiquitination (Fig. 4A). Additionally, we examined the effect of LMW FGF2 on the Lys 63-linked ubiquitination of RIG-I 2CARD. The results showed that Lys 63-linked ubiquitin of RIG-I 2CARD increased upon the expression of LMW FGF2 in a dose-dependent manner (Fig. 4B). These results suggest that LMW FGF2 did not inhibit the Lys 63-linked ubiquitination of RIG-I or RIG-I 2CARD. It is known that activated RIG-I in the presence of RNA results in 2CARD oligomerization, which then interacts with the downstream signaling adaptor MAVS 2CARD and thereby activates interferon signaling (24). Accordingly, we questioned whether the binding of LMW FGF2 to RIG-I suppressed the interaction between RIG-I and MAVS 2CARD. Thus, we investigated the effect of LMW FGF2 on RIG-I/MAVS 2CARD interaction upon SeV infection. The results showed that LMW FGF2 expression notably inhibited the binding of RIG-I with MAVS 2CARD in a dose-dependent manner (Fig. 4C). Collectively, these results indicate that cytosolic LMW FGF2 suppresses antiviral signaling likely by inhibiting the interaction of activated RIG-I with downstream MAVS, but not Lys 63-linked RIG-I ubiquitination.
Figure 4. LMW FGF2 suppresses the interaction between activated RIG-I and MAVS 2CARD.
(A) 293T cells were transfected with HA-RIG-I or Flag-Ub (WT or 63only) with or without LMW FGF2. Forty-eight hours later, WCLs of 293T cells were immunoprecipitated with an anti-HA antibody, followed by IB with anti-Flag, anti-HA, anti-FGF2 and anti-β-actin antibodies.
(B) 293T cells were transfected with Flag-RIG-I 2CARD and various amounts of LMW FGF2 (100, 200, and 500 ng). Forty-eight hours later, WCLs of 293T cells were immunoprecipitated with an anti-Flag antibody, followed by IB assay with anti-Flag, anti-ly63-Ub, and anti-FGF2 antibodies.
(C) 293T cells were transfected with Flag-MAVS 2CARD and HA-RIG-I and various amounts of LMW FGF2 (0, 200, and 500 ng). Thirty hours later, 293T cells were infected with SeV (50 HA unit/ml) for another 18 h and then immunoprecipitated with an anti-HA antibody, followed by IB with anti-Flag, anti-FGF2 and anti-β-actin antibodies.
LMW FGF2 enhances RIG-I stability by inhibition of Lys 48-linked ubiquitination-mediated RIG-I degradation
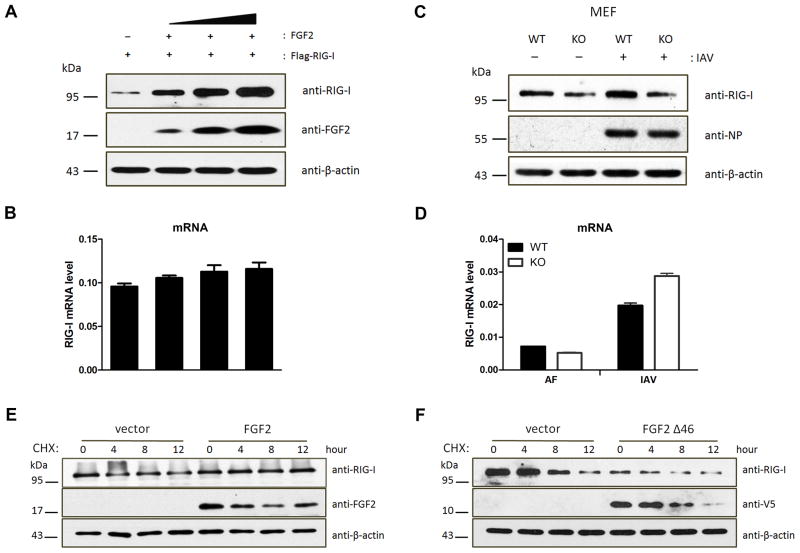
On the other hand, the RIG-I protein levels could be dramatically elevated by the exogenous expression of LMW FGF2 in a dose-dependent manner (Fig. 5A), but RIG-I mRNA levels were hardly altered (Fig. 5B). Moreover, we investigated the effect of endogenous LMW FGF2 deficiency on RIG-I expression in mouse embryonic fibroblast (MEF). Notably, three FGF2 isoforms with molecular masses of 18, 21, and 22 kDa in wild-type mice (WT) are expressed by alternative translation (25); however, in Fgf2lmw−/− mice (KO) that we used here, the LMW FGF2 rather than HMW FGF2 was not expressed (Fig. S3A). RIG-I protein levels in KO MEF were lower than those in WT MEF with IAV infection or not and that IAV infection notably increased RIG-I expression in WT MEF but not KO MEF (Fig. 5C). However, there was no consistent effect on the RIG-I mRNA levels (Fig. 5D). Additionally, RIG-I protein, but not mRNA, was downregulated by siRNA-mediated FGF2 knockdown in A549 cells with IAV infection or not (Fig. S3B–D). These results suggest that LMW FGF2 deficiency or knockdown decreases RIG-I, likely by post-translational regulation. Considering the potent interaction between LMW FGF2 and RIG-I, we questioned whether LMW FGF2 affected RIG-I protein stability. Thus, we investigated the stability of RIG-I by using a cycloheximide chase assay. The results showed that RIG-I levels in control cells declined starting at 4 h after treatment with cycloheximide (Fig. 5E); however, in LMW FGF2-expressing cells, the RIG-I levels changed minimally over time. Furthermore, we identified that the positive effect on RIG-I protein stability could not be sustained by LMW FGF2 Δ46 expression (Fig. 5F). These results indicate that the interaction of FGF2 with RIG-I may play an important role in maintaining RIG-I protein stability. To further confirm whether that is RIG-I specific, we detected the effect of LMW FGF2 on MDA5 stability, the study showed that LMW FGF2 could not increase MDA5 protein levels (Fig. S3E), suggesting that the effect of LMW FGF2 on protein stability was RIG-I specific. Finally, we explored whether FGF2 participated in the process of Lys 48-linked ubiquitination-mediated RIG-I degradation (Fig. S3F). The results showed that FGF2 expression elevated RIG-I in cells with plasmids expressing Lys 48-only ubiquitin, Lys 63-only ubiquitin and the Lys 63 (K–R) ubiquitin mutant. By contrast, FGF2 had no effect on RIG-I protein level in cells that exhibited Lys 48 (K–R) ubiquitin mutant expression. The results suggest that FGF2 may prevent Lys 48-linked ubiquitination-mediated RIG-I degradation.
Figure 5. LMW FGF2 increases RIG-I stability.
(A, B) 293T cells were transfected with Flag-RIG-I and various amounts of LMW FGF2 (20, 50, and 100 ng/μl). Forty-eight hours after transfection, (A) WCLs of 293T cells were subjected to an IB assay using anti-RIG-I, anti-FGF2, and anti-β-actin antibodies; (B) The RIG-I mRNA levels were assessed by real time-PCR.
(C, D) Primary MEFs isolated from Fgf2lmw+/+ (WT) or Fgf2lmw−/− (KO) mice were infected with IAV or allantoic fluid (AF). (C) Cell lysates were harvested at 12 hpi and then subjected to IB assay using anti-RIG-I, anti-influenza A virus nuclear protein (NP) and anti-β-actin antibodies; (D) The RIG-I mRNA levels were assessed by real time-PCR.
(E, F) 293T cells were transfected with plasmid encoding LMW FGF2 (E), LMW FGF2 Δ46 (F) or empty plasmid as control. Forty-eight hours later, 293T cells were treated with 100 μM cycloheximide (CHX). The cells were harvested at the indicated times and then subjected to an IB assay using anti-RIG-I, anti-FGF2 or anti-V5 and anti-β-actin antibodies.
Cytosolic LMW FGF2 suppresses antiviral innate immune response
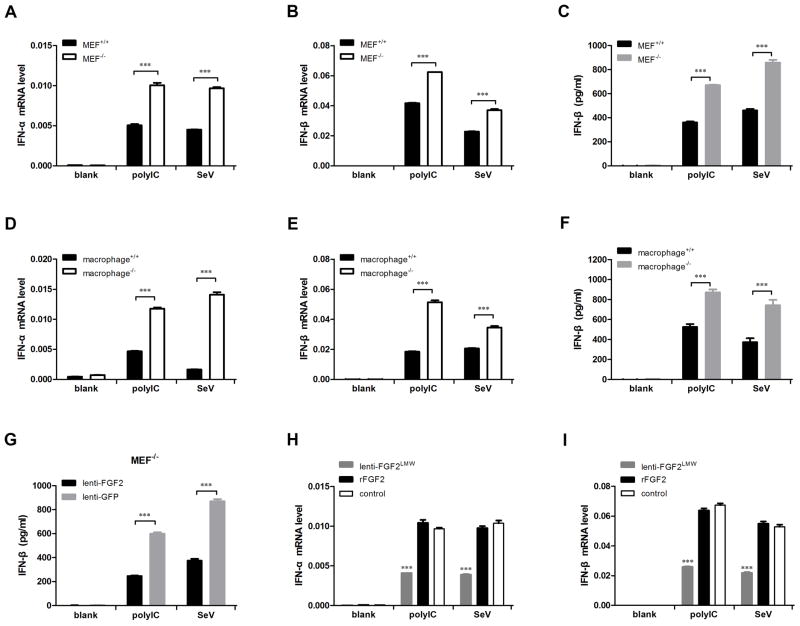
Type I interferon, including IFN-α and IFN-β, is known to play important roles in antiviral responses. Thus, we examined the production of IFN-α and IFN-β induced by SeV infection or polyIC stimulation in MEFs and macrophages, which were isolated from Fgf2lmw−/− mice and wild-type mice (referred to as MEF−/− and MEF+/+ and as macrophage−/− and macrophage+/+). Real-time PCR assays showed that both polyIC and SeV stimulated IFN-α and IFN-β expression in MEFs (Fig. 6A and B) and macrophages (Fig. 6D and E); importantly, there was substantially higher interferon expression in MEF−/− and macrophage−/− than in MEF+/+ and macrophage+/+. Furthermore, we detected extracellular IFN-β production in culture supernatants using ELISA, which confirmed that the IFN-β production from MEF−/− (Fig. 6C) and macrophage−/− (Fig. 6F) in response to polyIC and SeV was substantially higher than that observed in their wild-type counterparts. Moreover, we examined whether interferon production could be suppressed by FGF2 exogenous expression. As expected, FGF2 overexpression markedly reduced IFN-β production in MEF−/− by polyIC and SeV stimulation compared with the control groups (Fig. 6G). The effective lentivirus-mediated FGF2 overexpression on MEF−/− cells was also confirmed (Fig. S4A). In addition, we compared the effect of exogenous LMW rFGF2 protein and overexpressed-LMW FGF2 on type I interferon expression in Fgf2lmw−/− MEF cells. The results showed that only overexpressed-LMW FGF2 but not rFGF2 protein reduced type I interferon expression in response to polyIC and SeV compared to control (Fig. 6H and I), suggesting that cytosolic LMW FGF2 rather than secreted form mediated the suppressive effect on type I interferon expression. Furthermore, we examined the phosphorylation of IRF3 activation in SeV- or mock-infected Fgf2lmw−/− and Fgf2lmw+/+ MEF cells. The results showed that the phosphorylation of IRF3 was significantly activated by SeV infection with much higher levels in Fgf2lmw−/− MEF cells than in WT cells (Fig. S4B). Besides, we investigated the inflammatory cytokines IL-6 and TNF-α expression in MEF cells and macrophage in response to SeV and polyIC. The results showed that MEF−/− and macrophage−/− presented significantly higher expression levels of IL-6 and TNF-α than MEF+/+ (Fig. S4C and D) and macrophage+/+ (Fig. S4E and F). Finally, we also verified the effect of FGF2 on interferon production in A549 cells using a specific siRNA-mediated FGF2 knockdown assay. IFN-β expression was significantly increased in A549 cells by endogenous FGF2 knockdown 12 h after SeV infection (Fig. S4G). Meanwhile, LMW FGF2 protein levels were detected in SeV-infected A549 cells (Fig. S4H). These results indicate that the suppressive function of LMW FGF2 in the regulation of type I interferon is universal and is not cell type specific. Taken together, our studies indicate that cytosolic LMW FGF2 inhibits antiviral innate immune response.
Figure 6. Cytosolic LMW FGF2 suppresses type I interferon production.
(A–F) The primary MEFs (A–C) and peritoneal macrophages (D–F) isolated from Fgf2lmw+/+ or Fgf2lmw−/− mice were stimulated with or without polyIC or infected with SeV at 50 HA unit/ml. (A, B, D, and E) Twelve hours later, IFN-α and IFN-β mRNA expression levels were detected by real-time PCR assays. (C, F) After a 24-h treatment, the IFN-β production in cell culture supernatants of primary MEFs and macrophages was analyzed by ELISA.
(G) Primary Fgf2lmw−/− MEF cells (MEF−/−) were infected with recombinant lentivirus for LMW FGF2 expression, and lentivirus-GFP was used as control. MEF−/− with LMW FGF2 or GFP expression were treated with polyIC or infected with SeV. After a 24-h treatment, the IFN-β production in cell culture supernatants of MEF−/− was analyzed by ELISA.
(H, I) MEF−/− cells were infected with recombinant lentivirus for LMW FGF2 expression, or treated with recombinant human LMW FGF2 proteins (rFGF2), followed by polyIC and SeV stimulation. After a 12-h treatment, IFN-α and IFN-β mRNA expression levels were detected by real-time PCR assays.
Discussion
In our report, we revealed the unexpected connection between cytosolic LMW FGF2 and the RIG-I-mediated innate immune response. Our results show that cytosolic LMW FGF2 plays a dual functional role in regulating the RIG-I signaling pathway. On the one hand, the RIG-I protein levels are largely dependent on cytosolic LMW FGF2 expression. The direct binding of LMW FGF2 to RIG-I is crucial for maintaining RIG-I stability. On the other hand, cytosolic LMW FGF2 is a negative factor in RIG-I-mediated signaling pathway. In the absence of viral infection, FGF2 maintains the autoinhibitory state of RIG-I signaling. After infection, LMW FGF2 is important for limiting the production of RIG-I-mediated type I interferon in the innate immune response.
Several mechanisms have been shown to regulate RIG-I stability. The ISG15 conjugation system correlates with the RIG-I expression pattern (26). The ISGylation of RIG-I reduced the basal levels of RIG-I before and after viral infection and served as a negative feedback mechanism of RIG-I antiviral signaling. Another important regulatory mechanism of RIG-I stability is proteasome-dependent degradation after conjugation to ubiquitin. RNF125 functioned as an E3 ligase for RIG-I ubiquitin conjugation and regulated the cellular levels of RIG-I (27). Both the ISGylation and ubiquitin of RIG-I negatively regulate RIG-I stability. In this report, we present evidence that LMW FGF2 positively maintains RIG-I stability through a direct interaction. The interaction efficiently prevents Lys48-linked ubiquitination-mediated RIG-I degradation, thus keeping the basal level of RIG-I. The C-terminal region of FGF2 is essential for the LMW FGF2 and RIG-I interaction. Further research is needed to explore the specific amino acids that are responsible for this interaction with RIG-I.
In contrast to the strong binding of cytosolic LMW FGF2 to inactive RIG-I, the interaction of LMW FGF2 with active RIG-I is diminished. We speculate that RNA recognition-induced conformation change of RIG-I might partially impair the binding between FGF2 and RIG-I, although RIG-I 2CARD could still interact with FGF2. This result suggests that RIG-I 2CRAD is masked by the helicase domain under an auto-repressed state, and that the conformation facilitates LMW FGF2 binding to RIG-I. To obtain additional details regarding RIG-I and LMW FGF2 binding, structure studies are needed.
Numerous factors have been found to be involved in the RIG-I-MAVS signaling pathway. Most studies are based on siRNA screening or a human cDNA library screening in 293 cells. We noted that the FGF2 protein levels are undetectable in 293T cells. Therefore, the role of FGF2 in the regulation of type I interferon can easily be overlooked. FGF2 is highly expressed in epithelial cells, fibroblast cells and macrophages under normal cellular conditions. However, the other known factors involved in RIG-I-MAVS signaling occur primarily at low expression levels, but can be rapidly upregulated upon viral infection or type I interferon induction. These studies indicate the functional roles of LMW FGF2 during the resting state of cells. Upon stimulation by RIG-I signaling inducers, the interaction of FGF2 with RIG-I is weakened. Nevertheless, LMW FGF2 still presents a suppressive effect on the interaction of activated-RIG-I and the adaptor MAVS. Considering that the CRAD domain exists in a wide array of proteins, it is likely that FGF2 may interact with other CRAD-containing proteins and regulate other cell signaling. Here, we found that LMW FGF2 could also interact with MDA5, but had no similar effect on regulating protein stability or downstream signaling. In addition, FGF2-mediated cell apoptosis has been confirmed for many years (28, 29). It is possible that FGF2 associates with CARD motif-containing caspases, which results in alternative caspase activity that then regulates cell apoptosis.
FGF2 consists of LMW and HMW isoforms produced by alternative translation. HMW isoforms are N-terminal extensions of the LMW isoform and use upstream in-frame CUG codons as alternative translational start sites (30). Interestingly, although the C-terminal region of HMW FGF2 contains the complete amino acid sequence of LMW FGF2, it does not exhibit a function similar to LMW FGF2 in antiviral signaling. We infer that LMW FGF2 may possess a unique structure distinct from HMW FGF2 for its interaction with RIG-I.
In summary, our results provide insight into the interaction between growth factor-FGF2 and the RLR-dependent immune response. The identification of cytosolic LMW FGF2 as a negative regulator in RIG-I-mediated signaling represents a new paradigm in innate immunity. It is also conceivable that other FGF family members might play similar roles in immune responses for certain pathogens. Our findings provide an important target for the modulation of interferon-induced inflammatory diseases.
Supplementary Material
Acknowledgments
We thank everyone who kindly provided plasmids, including Michaela U. Gack, Jianwei Wang, Qinmiao Sun, Danying Chen and Hong-bing Shu.
Footnotes
This work was supported by Beijing Institute of Microbiology and Epidemiology funding for overseas scholars (20110801).
References
- 1.Kehrl JH, Wakefield LM, Roberts AB, Jakowlew S, Alvarez-Mon M, Derynck R, Sporn MB, Fauci AS. Production of transforming growth factor beta by human T lymphocytes and its potential role in the regulation of T cell growth. The Journal of experimental medicine. 1986;163:1037–1050. doi: 10.1084/jem.163.5.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li MO, Wan YY, Sanjabi S, Robertson AKL, Flavell RA. Transforming growth factor-β regulation of immune responses. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 3.Ohm JE, Gabrilovich DI, Sempowski GD, Kisseleva E, Parman KS, Nadaf S, Carbone DP. VEGF inhibits T-cell development and may contribute to tumor-induced immune suppression. Blood. 2003;101:4878–4886. doi: 10.1182/blood-2002-07-1956. [DOI] [PubMed] [Google Scholar]
- 4.Hepburn L, Prajsnar TK, Klapholz C, Moreno P, Loynes CA, Ogryzko NV, Brown K, Schiebler M, Hegyi K, Antrobus R. A Spaetzle-like role for nerve growth factor β in vertebrate immunity to Staphylococcus aureus. Science. 2014;346:641–646. doi: 10.1126/science.1258705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ortega S, Ittmann M, Tsang SH, Ehrlich M, Basilico C. Neuronal defects and delayed wound healing in mice lacking fibroblast growth factor 2. Proc Natl Acad Sci U S A. 1998;95:5672–5677. doi: 10.1073/pnas.95.10.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nugent MA, Iozzo RV. Fibroblast growth factor-2. Int J Biochem Cell Biol. 2000;32:115–120. doi: 10.1016/s1357-2725(99)00123-5. [DOI] [PubMed] [Google Scholar]
- 7.Virag JA, Rolle ML, Reece J, Hardouin S, Feigl EO, Murry CE. Fibroblast growth factor-2 regulates myocardial infarct repair: effects on cell proliferation, scar contraction, and ventricular function. Am J Pathol. 2007;171:1431–1440. doi: 10.2353/ajpath.2007.070003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nature immunology. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 9.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 10.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Akira S. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. The Journal of Immunology. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 11.Rehwinkel J, e Sousa CR. RIGorous detection: exposing virus through RNA sensing. Science. 2010;327:284–286. doi: 10.1126/science.1185068. [DOI] [PubMed] [Google Scholar]
- 12.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 13.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-β signaling. Molecular cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I-and Mda5-mediated type I interferon induction. Nature immunology. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 15.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 16.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 17.Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y, Sun X, Nie X, Sun L, Tang TS, Chen D, Sun Q. COX5B regulates MAVS-mediated antiviral signaling through interaction with ATG5 and repressing ROS production. PLoS Pathog. 2012;8:e1003086. doi: 10.1371/journal.ppat.1003086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao D, Yang YK, Wang RP, Zhou X, Diao FC, Li MD, Zhai ZH, Jiang ZF, Chen DY. REUL is a novel E3 ubiquitin ligase and stimulator of retinoic-acid-inducible gene-I. PLoS One. 2009;4:e5760. doi: 10.1371/journal.pone.0005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Z, Jia X, Xue Q, Dou Z, Ma Y, Zhao Z, Jiang Z, He B, Jin Q, Wang J. TRIM14 is a mitochondrial adaptor that facilitates retinoic acid-inducible gene-I-like receptor-mediated innate immune response. Proc Natl Acad Sci U S A. 2014;111:E245–254. doi: 10.1073/pnas.1316941111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Hu MM, Wang YY, Shu HB. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem. 2012;287:28646–28655. doi: 10.1074/jbc.M112.362608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang JD, Cousens LS, Barr PJ, Sprang SR. Three-dimensional structure of human basic fibroblast growth factor, a structural homolog of interleukin 1 beta. Proc Natl Acad Sci U S A. 1991;88:3446–3450. doi: 10.1073/pnas.88.8.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peisley A, Wu B, Xu H, Chen ZJ, Hur S. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 2014;509:110–114. doi: 10.1038/nature13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peisley A, Wu B, Yao H, Walz T, Hur S. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol Cell. 2013;51:573–583. doi: 10.1016/j.molcel.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 25.Bugler B, Amalric F, Prats H. Alternative initiation of translation determines cytoplasmic or nuclear localization of basic fibroblast growth factor. Mol Cell Biol. 1991;11:573–577. doi: 10.1128/mcb.11.1.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim MJ, Hwang SY, Imaizumi T, Yoo JY. Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. Journal of virology. 2008;82:1474–1483. doi: 10.1128/JVI.01650-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arimoto K-i, Takahashi H, Hishiki T, Konishi H, Fujita T, Shimotohno K. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proceedings of the National Academy of Sciences. 2007;104:7500–7505. doi: 10.1073/pnas.0611551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sahni M, Raz R, Coffin JD, Levy D, Basilico C. STAT1 mediates the increased apoptosis and reduced chondrocyte proliferation in mice overexpressing FGF2. Development. 2001;128:2119–2129. doi: 10.1242/dev.128.11.2119. [DOI] [PubMed] [Google Scholar]
- 29.Maloof P, Wang Q, Wang H, Stein D, Denny TN, Yahalom J, Fenig E, Wieder R. Overexpression of basic fibroblast growth factor (FGF–2) downregulates Bcl–2 and promotes apoptosis in MCF–7 human breast cancer cells. Breast cancer research and treatment. 1999;56:151–165. doi: 10.1023/a:1006258510381. [DOI] [PubMed] [Google Scholar]
- 30.Liao S, Bodmer J, Pietras D, Azhar M, Doetschman T, Schultz JEJ. Biological functions of the low and high molecular weight protein isoforms of fibroblast growth factor-2 in cardiovascular development and disease. developmental dynamics. 2009;238:249–264. doi: 10.1002/dvdy.21677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.