Abstract
Human T-cells are activated by both peptide and non-peptide antigens produced by Mycobacterium tuberculosis (M.tb). T-cells recognize cell wall lipids bound to CD1 molecules, but effector functions of CD1-reactive T-cells have not been systematically assessed in M.tb-infected humans. It is also not known how these features correlate with T-cell responses to secreted protein antigens. We developed a flow cytometric assay to profile CD1-restricted T cells ex-vivo and assessed T-cell responses to five cell wall lipid antigens in a cross-sectional study of 19 M.tb-infected and 22 M.tb-uninfected South African adolescents. We analyzed six T cell functions using a recently developed computational approach for flow cytometry data in high-dimensions. We compared these data with T-cell responses to five protein antigens in the same cohort. We show that CD1b-restricted T-cells producing anti-mycobacterial cytokines IFN-γ and TNF-α are detectable ex-vivo in CD4+, CD8+, and CD4-CD8- T-cell subsets. Glucose monomycolate was immunodominant among lipid antigens tested, and polyfunctional CD4 T-cells specific for this lipid simultaneously expressed CD40L, IFN-γ, IL-2, and TNF-α. Lipid-reactive CD4+ T-cells were detectable at frequencies of 0.001–0.01%, and this did not differ by M.tb infection status. Finally, CD4 T-cell responses to lipids were poorly correlated with CD4 T-cell responses to proteins (Spearman’s rank correlation −0.01; p=0.95). These results highlight the functional diversity of CD1-restricted T-cells circulating in peripheral blood as well as the complementary nature of T-cell responses to mycobacterial lipids and proteins. Our approach enables further population-based studies of lipid-specific T-cell responses during natural infection and vaccination.
Introduction
Mycobacterium tuberculosis (M.tb) is a pathogen of global importance that infects more than one billion people and causes more than one million deaths annually(1). Several lines of evidence in human studies and animal challenge models underscore the importance of T cells in controlling M.tb infection(2–5). T cells recognize both peptide and non-peptide antigens produced by mycobacteria, so the potential catalog of antigens mediating protective immunity extends beyond the proteome of M.tb (6–8). Non-peptide antigens are fundamentally different from peptide antigens in their chemical structure, sub-cellular location within the pathogen, and pathways by which they are processed and presented to T cells. Thus, one hypothesis is that the adaptive immune system evolved the ability to recognize non-peptide antigens in order to diversify the T-cell response to infection.
T cells recognize mycobacterial cell wall lipids bound to CD1 proteins, which are homologous to MHC Class I but are functionally non-polymorphic(9). The human CD1 locus codes for four proteins (CD1a, CD1b, CD1c, CD1d) that are expressed at the cell surface and are capable of presenting lipid antigens to T cells. At least eight cell wall lipids have been identified as CD1 antigens for human T cells. Five of these lipids are presented by CD1b: mycolic acid (MA), glucose monomycolate (GMM), glycerol monomycolate (GroMM), diacylated sulfoglycolipids (Ac2SGL), and phosphatidyl-myo-inositol mannosides (PIMs)(6, 10–13). Information regarding the frequency and phenotype of CD1b-restricted T cells targeting mycobacterial glycolipids is limited. CD1b tetramers were used to estimate the frequency of GMM-specific T cells to be 0.01% in four subjects with positive purified protein derivative (PPD) skin tests(14). T cell clones that recognize GMM using a semi-invariant T cell receptor express CD4(15). However, other CD1b-reactive T cells with lower affinity for GMM express CD8, or are CD4-CD8- (10, 15, 16). A small cross-sectional study revealed that proliferative T-cell responses to GMM were present in both PPD-positive and PPD-negative study subjects(17). By contrast, lymphocytes from PPD-positive but not PPD-negative subjects produced IFN-γ in response to mycobacterial sulfoglycolipid and glycerol monomycolate(11, 12). More recently, it was demonstrated that mycolic acid induced IFN-γ production from lymphocytes in patients with tuberculosis but not healthy donors, and that these responses decreased while receiving anti-tuberculous therapy(18). Collectively, these studies provide clear evidence that populations of CD1b-restricted T cells targeting mycobacterial glycolipids are expanded in vivo, but no study has systematically assessed the phenotype of these T cells ex vivo or examined how lipid-specific T-cell responses compared with T-cell responses to protein antigens with regard to magnitude or timing.
Ex vivo analysis of CD1b-restricted T cells has been hampered by the lack of specific surface markers and challenges inherent to identifying and cloning cells in-vitro. Further, ex-vivo analysis typically requires the generation of autologous dendritic cells (DCs) to facilitate antigen presentation, and this approach carries inherent difficulties. First, the antigen-presenting molecule on DCs is difficult to define because DCs simultaneously express CD1a, CD1b, CD1c, and CD1d. Second, trace amounts of contaminating peptides in the lipid preparations would be efficiently presented to MHC-restricted T cells, confounding interpretations of whether responses are due to lipids or peptides. Third, and most importantly, generating DCs is a time and reagent intensive process that renders the use of cryopreserved peripheral blood mononuclear cells (PBMC) virtually impossible. Thus, a major barrier in the field is the lack of an activation-based assay that would enable large human cohort studies of lipid-specific T cells.
To enable large scale ex vivo studies of CD1b-restricted T cells in human populations, we took advantage of an assay using K562 cells, which are a human myelogenous leukemic cell line that expresses very low levels of MHC Class I and MHC Class II and does not express CD1. Thus, these cells do not efficiently elicit allogeneic T-cell responses(19). When stably transfected with single isoforms of human CD1 proteins, these cells are capable of lipid antigen presentation to T cell clones and T cell lines derived after long-term and short term in-vitro culture, respectively(14, 19). We modified this assay to enable the detection of 64 cytokine-defined CD1b-restricted CD4+, CD8+, and CD4-CD8- T cells and conducted a cross-sectional study of ex vivo T-cell responses to five mycobacterial glycolipids in South African adolescents. We integrated this assay with a novel analytic platform, providing a comprehensive study of human T cells targeting pathogen-derived lipids. Our findings reveal a previously unappreciated functional diversity among circulating CD1-restricted T cells that are not redundant with T-cell responses to proteins. These results support the premise that the CD1 antigen presentation system evolved to complement functions performed by the MHC locus, ultimately leading to a diverse T-cell response to mycobacterial infection.
Materials and Methods
Clinical Cohort
As recently detailed, 6363 adolescents were enrolled into a study that aimed to determine the incidence and prevalence of tuberculosis infection and disease(20). Twelve to 18 year old adolescents were enrolled at eleven high schools in the Worcester region of the Western Cape of South Africa. Subjects were screened for the presence of latent tuberculosis by a tuberculin skin test and QuantiFERON-TB GOLD In-Tube (Cellestis Inc.) at study entry. Subjects were all healthy without signs or symptoms of active tuberculosis. After matching for age and gender and ensuring sufficient PBMC for the planned studies, we selected 19 M.tb-infected and 22 M.tb-uninfected subjects for intensive immunologic study (Table 1). PBMC from 34 (83%) of 41 subjects underwent both lipid and peptide T cell profiling, while the remaining seven were used for one or the other assay. Data from one M.tb-uninfected subject was dropped prior to analysis because of documented technical problems with the data.
Table 1. Demographic and Clinical Description of Study Cohort.
Study subjects were stratified based on the result of QuantiFERON-TB GOLD InTube (QFT-GIT) testing according to the manufacturer recommended cutoff of 0.35 IU/mL for a positive test. For each parameter, values are reported as either N (%) or median (IQR).
| QuantiFERON-TB GOLD InTube | ||
|---|---|---|
|
| ||
| Negative | Positive | |
|
| ||
| N | 19 | 22 |
|
| ||
| Female (%) | 8 (42%) | 11 (50%) |
|
| ||
| BMI | 20.4 (19.1–23.2) | 18.4 (16.7–22.4) |
|
| ||
| Ethnicity | ||
| Black (%) | 4 (21%) | 10 (46%) |
| White (%) | 13 (68%) | 10 (46%) |
| Mixed (%) | 2 (11%) | 2 (9%) |
|
| ||
| TST (mm) | 0 | 15 (12.8–15.10) |
Ethics
The study was approved by the IRB of the University of Washington and the University of Cape Town. Written informed consent was obtained from the parents of the adolescents and assent was obtained from the adolescents for the parent study.
Antigens
Mycolic acid (MA), glucose monomycolate (GMM), glycerol monomycolate (GroMM), and phosphatidyl-myo-inositol mannosides (PIM) were purified from mycobacterial cell wall extracts as previously described(6, 10, 12, 21). SL37 was synthesized as an analog of a purified diacylated sulfoglycolipid antigen(22). We generated peptide pools of 15 amino acids overlapping by 12 for five immunodominant proteins secreted by M. tuberculosis (Ag85A, Ag85B, CFP-10, ESAT-6, and TB10.4). For the analysis of CMV-specific T-cell responses, cryopreserved PBMC from healthy donors known to be high-responders to CMV were stimulated with a peptide pool targeting pp65 consisting of 138 peptides 15 amino acids in length overlapping by 11 peptides.
Cellular Assays
K562 cells stably transfected with empty vector (K562-EV) or CD1b (K562-CD1b)(19) were maintained in RPMI 1640 (GIBCO) supplemented with 10% fetal calf serum and G418 (Sigma) at a concentration of 200 mcg/ml and periodically assessed for CD1b expression by flow cytometry.
On Day 0, cryopreserved PBMCs were thawed, counted, and assessed for viability assessed by Guava easyCyte (EMD Millipore). PBMCs were rested in RPMI + 10% fetal calf serum overnight and reassessed again by Guava. After resting, if there were not enough cells to test five antigens or if viability was less than 70%, the sample was discarded.
Lipids were evaporated to dryness from chloroform-based solvents under a sterile nitrogen stream and then sonicated into media. This lipid suspension was added to 50,000 K562-EV or K562-CD1b cells at final concentrations that were previously determined to be greater than those yielding maximal T-cell responses: 5 μg/ml for MA and GMM and 10 μg/ml for GroMM, PIM, and SL37(6, 10–12, 21). K562 cells were incubated for 18 hours at 37°C, 5% CO2 to facilitate lipid loading. The following day, rested PBMCs were split and added to the K562 cells without washing at a final density of 1 million per well. Thus, each subject’s PBMCs were tested against ten conditions. The cell mixture was allowed to incubate for six hours in the presence of anti-CD28/Ly29d antibodies (BD Biosciences), Brefeldin A at a final concentration of 10 μg/ml (Sigma), and GolgiStop containing Monensin (BD) after which EDTA, at a final concentration of 2mM, was added to disaggregate cells. Plates were stored at 4°C until the following day when they were stained and acquired by FACS in duplicate. A total of 35 study subjects were analyzed in nine batches in which the number of samples from M.tb-infected and M.tb-uninfected subjects were matched.
For peptide stimulations, PBMCs were thawed and rested as above. The following day, PBMC were distributed at a final density of 1 million cells per well and peptide pools dissolved in DMSO were added in single wells at a final concentration of 1 μg/ml per peptide. A negative control containing only DMSO was also plated. Cells were allowed to incubate with peptide for six hours in the presence of anti-CD28/Ly29d, Brefeldin A, and GolgiStop as above. A total of 40 study subjects were analyzed in five batches in which the number of M.tb-infected and M.tb-uninfected samples were matched.
The T-cell clone LDN5 was maintained in culture through a combination of rapid expansion and antigen stimulation, as previously described(10). Polyclonal T cells were purified from total PBMCs obtained from a healthy donor by removing cells expressing CD14, CD16, CD19, CD36, CD56, CD123, and Glycophorin A using Pan T cell Isolation Kit II (Miltenyi)
Flow Cytometry
We have previously published an optimized and validated 12-color intracellular cytokine staining assay to quantify antigen-specific T cells after vaccination(23, 24). This panel was modified to include anti-IL-17a and anti-IL-22 instead of anti-MIP-1β and anti-CD107a (Supp Table 1). Briefly, cells were first stained with Avid Live/Dead (Life Technologies) viability dye and anti-CD14. After washing, the cells were permeabilized with FACS Perm II (BD) and stained for the remaining markers (CD3, CD4, CD8, CD154, IFN-γ, TNF-α, IL-2, IL-4, IL-17a, and IL-22). Fully stained cells were washed and resuspended in 1% paraformaldehyde. Data were acquired on LSR II (BD) equipped with a high-throughput sampler and configured with violet (405nm, 50mW), blue (488nm, 20mW), green (561nm, 50mW), and red (641nm, 150mW) lasers. Raw data were manually gated in FlowJo (TreeStar Inc.). The raw FCS files and manual gates were imported into the R environment using the OpenCyto framework for data visualization and downstream analysis(25). For some of the assays with lipid antigens, we noted tandem dye breakdown within APC-Cy7 CD4 leading to spurious signal within the APC IL-4 gate, so this cytokine was not included in downstream analysis. Even though this artifact was not observed with peptide antigens, we removed IL-4 signal from peptide data to maintain consistency.
Statistical Analysis
Cell counts were analyzed using COMPASS (COMbinatorial Polyfunctionality Analysis of Antigen-Specific T-cell Subsets) (26). COMPASS is a novel computational approach developed for high-dimensional flow cytometry data analysis that can be used to detect changes across all observable functional T-cell subsets, without the need to limit our analysis to very specific subsets based on expected biological significance. In COMPASS, responses are quantified using posterior probabilities that summarize for each subject and subset the evidence that the corresponding response is antigen-specific by comparing the proportion of cytokine positive cells in the antigen sample to the corresponding proportion in the control sample. Notably, COMPASS only reports the probability of detecting T-cell responses with a particular functional profile rather than the frequency, which can be separately calculated. For a given subject, COMPASS can also be used to compute a functionality score that summarize the entire functionality profile into a single number. Here, we have applied COMPASS to each of the five lipids in three (CD4+, CD8+, CD4-CD8-) T cell subsets and five peptide pools in the CD4+ T cell subset, leading to 20 analyses. Each one of the analyses was unbiased and considered all of the 64 possible cytokine functions (defined as Boolean combination). Magnitudes of T-cell responses were calculated independent of COMPASS as the maximum of zero or the proportion of gated events in the stimulated condition minus the proportion of gated events in the unstimulated condition.
Results
A flow cytometric assay facilitates ex-vivo analysis of CD1-restricted T cells
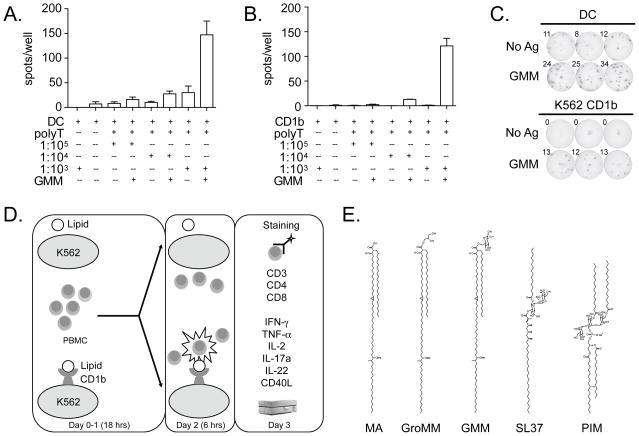
To determine if lipid-specific T cells present at low frequency could be detected with sufficient sensitivity, we tested the ability of both K562 cells and DCs to activate LDN5, a T cell clone specific for GMM, at limiting concentrations. Using IFN-γ ELISPOT, we found that DCs and K562-CD1b cells were able to detect activated LDN5 T cells at a precursor frequency as low as 1:10,000 (Fig.1A and Fig.1B). Notably, background responses seen in the presence of DCs were lower or absent when K562 cells were used as APCs (Fig.1C and data not shown). As a negative control, we then mixed K562 cells with PBMC from healthy individuals known to have high frequency responses to the immunodominant CMV protein pp65. Even at an effector to target ratio of 1:5, there was no effect on responding CD4 or CD8 T cells (data not shown). Thus, the mere presence of K562 cells does not lead to non-specific secondary stimulation of T cells. We modified this assay to enable the analysis of cryopreserved PBMC by multi-parameter flow cytometry (Fig.1D and Supp Fig.1). We did not find systematic evidence of NK cell activation as measured by IFN-γ production by CD3- cells, nor did we find evidence of secondary stimulation of T cells by mock-transfected K562 cells after six hours (data not shown). These data validate a sensitive and specific method of using CD1-transfected K562 cells to facilitate the detection of lipid-specific T cells within a polyclonal mixture of T cells.
Figure 1. K562 cell transfectants provide a sensitive and specific means to detect CD1b-restricted T cells.
(A) In-vitro derived DCs (DC) or (B) K562 cells stably transfected with CD1b (CD1b) were co-incubated with purified polyclonal T cells from a healthy donor (polyT) spiked with titrating quantities of clone LDN5 at ratios ranging from 1:1000 to 1:100,000 and GMM prior to IFN-γ ELISPOT. (C) Shown are triplicate measurements when DCs or K562+CD1b was used to detect LDN5 at a precursor frequency of 1:10,000 among polyclonal T cells. (D) Schematic illustrating lipid-specific T cell profiling assay. Briefly, cryopreserved PBMC are thawed, washed, and rested overnight. At the same time, K562 cells (stably transfected with empty vector or CD1b) are loaded with one of the five glycolipid antigens. The following day, PBMC are harvested, counted, and added to wells with lipid loaded K562 cells for a six-hour stimulation. On the final day, cells are stained and acquired on a flow cytometer. (E) Representative structures of five mycobacterial glycolipid antigens known to be presented by CD1b to T cells: mycolic acid (MA), glycerol monomycolate (GroMM), glucose monomycolate (GMM), diacylated sulfotrehalose (SL37), and phosphatidyl-myo-inositol mannosides (PIM).
CD1b-restricted T-cell responses in CD4+, CD8+, and CD4-CD8- T cell subsets
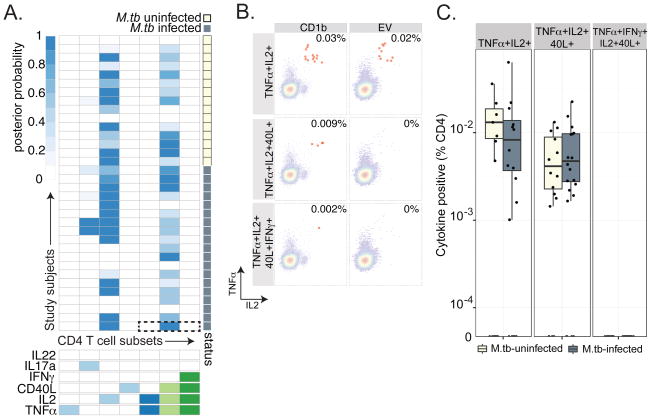
We used this assay to perform a cross-sectional analysis of T-cell responses to five mycobacterial glycolipid antigens presented by CD1b (Fig.1E) in South African adolescents with known M.tb infection status. Our goal was to assess the frequency and cytokine profile of T cells recognizing each lipid antigen, the effect of M.tb infection, and the correlation with T-cell responses to mycobacterial proteins. We employed a computational framework we recently developed called COMPASS (COMbinatorial Polyfunctionality Analysis of Antigen-Specific T-cell Subsets) to thoroughly characterize the functional profiles of CD1b-restricted CD4+ T cells (26). We found a high probability of detecting CD1b-restricted responses among CD4 T cells in only seven of 64 possible functional profiles (Fig.2A). Some of these responses were unique to a single lipid antigen and some were shared among many lipids. For example, CD4 T cells expressing CD40L and producing TNF-α and IL-2 were detected in response to MA and GMM, but T cells that additionally produced IFN-γ were only detected in response to GMM (Fig.2B, 2C and Fig.3A, Fig.3B). Because CD40L expression is transiently induced upon activation of T cells by antigen, we can infer that these responses were due to lipid-antigen specific stimulation, a finding further supported by their absence in the presence of PIM and GroMM antigens(27). These data are consistent with the cytokine-producing profiles of GMM and MA-specific T-cell clones derived after long term in vitro culture and so confirm that the phenotype exists in the ex-vivo state(15, 16). Notably, the magnitude of T-cell responses containing CD40L was not significantly different between M.tb infected and M.tb uninfected adolescents (Fig.2D and Fig.3C). We also detected CD4 T cells expressing CD40L but no cytokines in the presence of SL37, suggesting that T cells specific to this lipid might have functional properties other than the ones we chose to study (Fig.2A). Finally, we noted a high probability of detecting CD4 T cells producing only IL-2 in most individuals and in the presence of most lipids (Fig.2A, Supp Fig.2A and 2B). These results suggested that T cells restricted by CD1b with unknown antigen specificity might be present at low frequency in all subjects. Indeed, follow up experiments confirmed that CD4 T cells producing only IL-2 were detectable in the presence of CD1b without the addition of a foreign lipid antigen (Supp Fig.2C). Taken together, these data reveal the range of functional profiles exhibited by CD1b-restricted CD4 T cells, some of which are specific to mycobacterial lipid antigens.
Figure 2. COMPASS analysis reveals lipid-antigen specific CD4 T-cell responses.
(A) Stacked COMPASS heatmaps displaying CD4 T-cell responses to five mycobacterial glycolipids in M.tb-infected and M.tb-uninfected adolescents. In the heatmap, columns represent cytokine subsets in which responses were generally higher in the presence of K562 CD1b when compared to K562 alone, and these are displayed in order of increasing functionality from left to right. For example, the first column represents T cells that produce TNF-α but none of the other functions. Rows represent study subjects, which are stratified by a diagnosis of latent tuberculosis as defined by QuantiFERON-TB GOLD testing. The depth of shading represents the posterior probability of detecting responses to a given lipid within a given individual. The dotted box outlines the heatmap for GMM. Lipids are abbreviated as SL37 (diacylated sulfoglycolipid), GMM (glucose monomycolate), GroMM (glycerol monomycolate), PIM (phosphatidyl-myo-inositol mannoside), and mycolic acid (MA). (B) Expanded view of COMPASS analysis of GMM-specific T-cell responses. The dotted box outlines polyfunctional T-cell responses in an M.tb-infected subject. (C) Expanded view of polyfunctional T-cell responses in an M.tb-infected subject. Representative FACS plots in which CD4 T cells expressing two (TNFα+IL2+), three (TNFα+IL2+CD40L+), or four (TNFα+IL2+CD40L+IFN-γ+) functions in the presence of GMM and K562 cells transfected with CD1B or empty vector (EV) are highlighted in red. A significant difference in T cell frequencies between conditions is picked up by COMPASS even if the frequency is rare. However, high background decreases the probability of detecting a true response (D) The magnitude of CD4 T cells expressing two, three, and four functions are shown for GMM stratified by M.tb infection status. There was no statistically significant difference between the magnitude of CD4 T-cell responses and M.tb infection (TNFα+IL2+ p=0.40, TNFα+IL2+CD40L+ p=0.32, and TNFα+IL2+CD40L+IFN-α+ p=0.89 by Wilcoxon rank sums test).
Figure 3. CD4 T-cell responses to mycolic acid.
(A) COMPASS analysis reveals T-cell responses are largely limited to two T cell subsets: IL-2 single producing cells and cells expressing IL-2, TNF-a, and CD40L. The dotted box outlines polyfunctional T-cell responses in an M.tb-infected subject that does not predict 2 or 4-function T cell subsets (B) Expanded view of polyfunctional T-cell responses in an M.tb-infected subject. Representative FACS plots in which CD4 T cells expressing two, three, or four functions in the presence of MA and K562 cells transfected with CD1B or empty vector (EV) are highlighted in red. High background within the TNFa+IL2+ subset decreases the probability of detecting a true response using COMPASS. (C) The magnitude of MA-specific CD4 T cells expressing two, three, and four functions are shown. There was no association with M.tb infection (p > 0.05 for TNFα+IL2+ and TNFα+IL2+CD40L+, no data for TNFα+IL2+CD40L+IFN-γ+).
We also detected CD1b-restricted CD8 and CD4-CD8- T cells, and the pattern was different from what we observed for the CD4 T cell subset. A high probability of detecting CD8 T-cell responses were limited to only five of 64 possible functional profiles and consisted of permutations of IFN-γ, IL-2, and TNF-α (Supp Fig.3A). We found CD4-CD8- T-cell responses limited to six of 64 possible functional profiles, none of which included more than two simultaneous functions (Supp Fig.3B). As with the CD4 T cell analysis, COMPASS revealed a high probability of detecting IL-2 single producing CD8 and CD4-CD8- T cells in most individuals and in the presence of most lipids. The lack of a clear association between functional profiles and lipid antigens or M.tb infection suggested that these responses might be the result of T cells recognizing CD1b expressed on the surface of K562 cells irrespective of the bound antigen. Because CD40L is not a useful marker of antigen-specific activation of CD8 and CD4-CD8- T cells, we were unable to separate antigen-specific from antigen-non-specific T-cell responses in this analysis. Nevertheless, these data reveal that CD8+ and CD4-CD8- T cells that recognize CD1b presented antigens are likely present in all subjects and could contribute to host defense by elaborating the anti-mycobacterial cytokines IFN-γ and TNF-α.
Highly polyfunctional T-cell responses against mycobacterial proteins
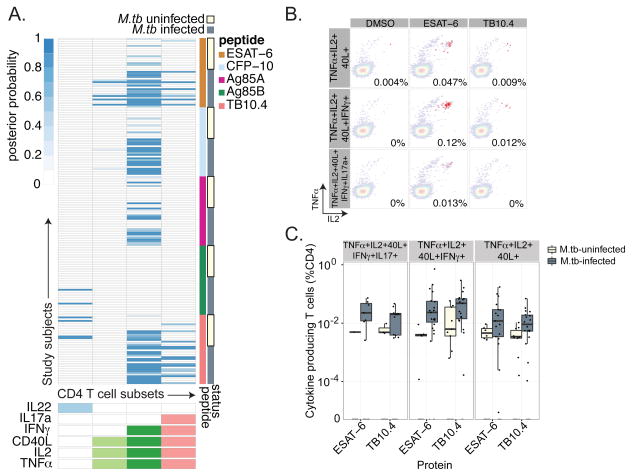
We also performed a parallel cross-sectional study of T-cell responses to the immunodominant protein antigens ESAT-6, CFP-10, TB10.4, Ag85A, and Ag85B in the same cohort of South African adolescents. We recently reported the association between a five-function T cell subset and M.tb infection using this data (26). Here, we reanalyzed this dataset with a focus on T-cell responses to individual proteins, which were exclusively detected within CD4 T cell subset (Fig.4A). We considered the possibility that the peptide pools would preferentially stimulate CD4 T cells instead of CD8 T cells based on their length, but we would not expect this to be the case based on published data with these antigens(28, 29). Further, we were readily able to detect CD8 T-cell responses against CMV using peptide pools designed and tested in the same way (data not shown). The probability of detecting CD4 T-cell responses to proteins was highest with a single highly polyfunctional T cell subset (IFNγ+IL2+TNFα+CD40L+). The five-function T-cell subset (IFNγ+IL2+TNFα+CD40L+IL17a+) was preferentially detected in the presence of ESAT-6 and TB10.4 (Fig.4A and 4B). As expected, the magnitude of highly polyfunctional T-cell responses as well as total IFN-γ production was higher among M.tb infected subjects (Fig.4C and data not shown). Thus, COMPASS analysis reveals a highly polyfunctional CD4 T-cell response that was surprisingly uniform across protein antigens.
Figure 4. CD4 T-cell responses against secreted protein antigens are highly polyfunctional.
(A) Stacked COMPASS heatmaps displaying CD4 T-cell responses to five secreted proteins in M.tb-infected and M.tb-uninfected adolescents. In the heatmap, columns represent cytokine subsets and rows represent study subjects, as in Fig 2. The depth of shading represents the posterior probability of detecting responses to a given protein within a given individual. (B) Representative FACS plots showing CD4 T cells expressing three (TNFα+IL2+CD40L+), four (TNFα+IL2+CD40L+IFN-γ+), or five (TNFα+IL2+CD40L+IFN-γ+IL17a+) functions in the presence of DMSO or peptide pools against ESAT-6 or TB10.4. (C) The magnitude of CD4 T cells expressing three, four, and five functions are shown for ESAT-6 and TB10.4 stratified by M.tb infection status. There was a statistically significant increase in the magnitude of CD4 T-cell responses among M.tb infected subjects for both ESAT-6 and TB10.4 (TNFα+IL2+CD40L p<0.01, TNFα+IL2+CD40L+IFN-γ+ p<0.01, and TNFα+IL2+CD40L+IFN-γ+IL17a+ p<0.05 by Wilcoxon rank sums test).
Quantitative comparisons of T-cell responses to mycobacterial lipids and proteins
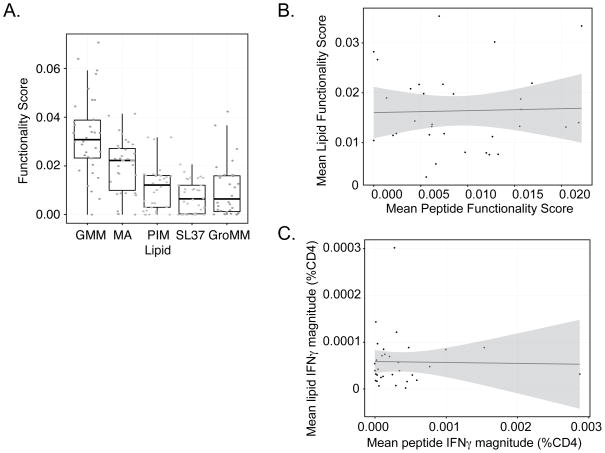
Finally, we sought to compare CD4 T-cell responses among lipids and between lipids and peptides. To account for the functional diversity of the observed T-cell responses, we used the functionality score generated by COMPASS, which summarizes the posterior probabilities of antigen-specific responses across all cell-subsets for each subject (26). Functionality score analysis clearly demonstrates a hierarchy in which GMM is dominant among the glycolipid antigens tested (Fig.5A). Next, we compared the average functionality score among lipids antigens with the average functionality score among peptide antigens and found them to be poorly correlated (Spearman ρ=-0.01; p=0.95) (Fig 5B). We considered the possibility that this low correlation might be due to the inclusion of CD1b-restricted T-cell responses with unknown antigen specificity within the lipid data. However, when we limited the lipid-specific T-cell responses to only those that included CD40L as a proxy for specificity to mycobacterial lipids, there was still a poor correlation (data not shown). This was also true when we considered total IFN-γ production independent of COMPASS (Fig.5C). Thus, our data demonstrate immunodominance among mycobacterial lipid antigens and a remarkably low correlation between T-cell responses to lipids and peptides.
Figure 5. GMM is immunodominant among lipid antigens and there is a poor correlation between CD4 T-cell responses to mycobacterial lipids and peptides.
(A). Functionality score boxplots reveal a hierarchy among CD4 T-cell responses to lipid antigens. The functionality score is calculated by COMPASS for each subject as the proportional of antigen-specific responses within the set of all possible responses after accounting for the posterior probability of detection. ANOVA F-statistic 27.88, p<0.001. GMM has the highest functionality score. (B). Correlation between CD4 T-cell responses to lipid and peptide antigens. The average functionality score across all lipid antigens was plotted against the average functionality score for all peptides for 34 subjects that underwent both assays. Simple linear regression revealed β = 0.04 plotted in blue (95% CI −0.3 – 0.4 plotted in gray). Spearman’s rank correlation ρ=−0.01, p=0.95. (C) Correlation between the average CD4+ IFN-γ magnitude for lipids and peptides. Simple linear regression b=−0.002 plotted in blue (95% CI −0.36 − 0.33 plotted in gray). Spearman’s rank correlation r=0.03, p=0.90.
Discussion
We developed an assay that enables the study of lipid-specific T cells ex-vivo and used this assay to perform a systematic analysis of T-cell responses to mycobacterial glycolipids in a population where tuberculosis is endemic. We incorporated a recently developed computational approach to describe the functional profiles of CD1b-restricted CD4+, CD8+, and CD4-CD8- T cells and their association with M.tb infection. Our results highlight properties of lipid-specific CD4 T-cell responses that have not been previously appreciated, such as the lack of association with M.tb infection, the phenomenon of immunodominance, and a poor correlation with T-cell responses to mycobacterial proteins.
MHC-restricted T cells circulate extensively as the move from the site of infection to secondary lymphoid organs and back. Accordingly, our data are consistent with many prior studies showing increased numbers of T-cell responses to protein antigens during M.tb infection. For CD1-reactive cells, it is unclear if infection causes durable memory responses. Several published studies have documented that T-cell responses to mycobacterial lipid antigens are increased after infection, supporting the existence of memory and systemic action (11, 12, 18, 30). Here, we did not find an association between lipid-specific CD4 T-cell responses and M.tb infection at the time points measured, and there are several potential explanations for these discordant results. First, our study is the first to be conducted in an endemic setting, so it is possible that intense exposure to M.tb among uninfected subjects induces lipid-specific T cell immunity even if it does not prime peptide-specific responses. It is also possible that these responses are the result of exposure to mycobacteria ubiquitously present in the environment or previous vaccination with BCG. Second, T cells recognizing lipid antigens might preferentially home to sites of infection, reducing the chance of detecting them in the blood of infected subjects. This would be similar to MR1-restricted T cells, which are not known to have memory or a requirement for trafficking to secondary lymphoid organs and show diminished numbers in peripheral blood during acute infection(8). Third, we used a statistical modeling approach to quantify antigen-specific T cells present at very low frequency ex-vivo. This is in contrast with published studies in which lymphocytes were incubated with DCs up to six days, which could amplify small differences that are not initially apparent (11, 12). Fourth, our experimental approach was designed to study CD1b-restricted T-cell responses only, while the results of published studies are confounded by the expression of all human CD1 isoforms on DCs. Finally, we did not specifically assess the contribution of CD8+ and CD4-CD8- T-cell responses, while these studies examined IFN-γ production from bulk lymphocyte preparations.
We identified GMM as an immunodominant glycolipid antigen recognized by CD4 T cells. GMM-specific CD4 T cells were polyfunctional, simultaneously producing IFN-γ, TNF-α, and IL-2 upon stimulation with antigen, and present at frequencies between 0.001 and 0.01%. These data confirm and extend published data using GMM-loaded tetramers to estimate precursor frequency in peripheral blood, characterization of GMM-specific T cell clones, and experimental immunization of cattle with GMM(15, 16, 31, 32). We also showed induction of CD40L by GMM and MA-specific CD4 T cells, suggesting these T cells are capable of interacting with cognate B cells to facilitate the production of high affinity antibodies mycobacterial surface lipids. Polyfunctional T cells responses to M. tuberculosis are associated with the clinical stage of disease as well as protection after vaccination in animal models(29, 33–35). It is not known if lipid-specific polyfunctional T-cell responses or antibodies are important for protective immunity against M.tb, but these question can be addressed experimentally in ongoing human trials of whole mycobacterial vaccines(36, 37).
Our study is the first to comprehensively assess T-cell effector function and compare responses to five mycobacterial lipid antigens with responses to five immunodominant protein antigens in a cross-sectional study. Thus, we are able to quantify the relative contribution of T cells targeting structurally distinct antigens produced by mycobacteria. The poor correlation we observed support a model in which the functions of T cells targeting lipids and proteins are different with regard to timing or localization. This is consistent with comparative genomic studies, in which CD1 gene families have been identified in virtually every mammalian species, suggesting they perform an important and non-redundant function(38). Our data have several implications for the development of T-cell based diagnostics and therapies for tuberculosis. The definition of M.tb infection is based on detecting T-cell responses to protein antigens, but less than 10% of those deemed infected will eventually develop active tuberculosis. Thus, including other immune parameters could improve the prognostic utility of an immunodiagnostic. The poor correlation between CD4 T-cell responses to lipids and proteins suggest that measuring T-cell responses to lipid antigens would provide non-redundant information that could be studied for its prognostic utility in a prospective clinical trial. Second, our results predict that whole cell vaccines that included both lipid and protein antigens might activate T cells with a broader set of functions than either class alone. Functional breadth of T cells could partly explain the observed clinical efficacy of whole cell vaccines, such as BCG and M. vaccae(39, 40). Third, a formidable challenge to performing population-based studies of mycobacterial T-cell immunity is the highly polymorphic nature of MHC Class I and Class II molecules. By contrast, virtually all humans express the same CD1b molecules, so assays like the one we have developed can be used to measure T-cell signatures independent of genetic background.
Supplementary Material
Acknowledgments
This work was supported by the NIH (K08-AI89938 to CS, U01-AI068618 to MJM) and the Firland Foundation (CS).
The authors would like to thank Blessing Kadira, Rebecca J. Smith, Bryce A. Manso, Saheli Datta, and Krystle K. Quan for technical assistance as well as the SATVI Study Team.
Footnotes
Author Contributions
C.S., G.P., D.F., N.F., S.R.D, M.J.M., and T.R.H. designed the experiments. T.J.S., H.M., and W.A.H. established the clinical cohort. D.B.M., J.P, and M.G. provided new reagents. C.S. and G.P. conducted the experiments. C.S., D.F., S.R.D., N.F., S.D., L.L., W.J., G.F., and R.G. analyzed the data. C.S. and T.R.H. wrote the manuscript with contributions from all authors.
Competing Financial Interests
All authors state that they do not have any competing financial interests with the work presented here.
References
- 1.WHO. Global Tuberculosis Report 2013. 2013. [Google Scholar]
- 2.Zumla A, Raviglione M, Hafner R, von Reyn CF. Tuberculosis. N Engl J Med. 2013;368:745–755. doi: 10.1056/NEJMra1200894. [DOI] [PubMed] [Google Scholar]
- 3.Mogues T, Goodrich ME, Ryan L, LaCourse R, North RJ. The relative importance of T cell subsets in immunity and immunopathology of airborne Mycobacterium tuberculosis infection in mice. J Exp Med. 2001;193:271–280. doi: 10.1084/jem.193.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behar SM, Dascher CC, Grusby MJ, Wang CR, Brenner MB. Susceptibility of mice deficient in CD1D or TAP1 to infection with Mycobacterium tuberculosis. J Exp Med. 1999;189:1973–1980. doi: 10.1084/jem.189.12.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin PL, Rutledge T, Green AM, Bigbee M, Fuhrman C, Klein E, Flynn JL. CD4 T cell depletion exacerbates acute Mycobacterium tuberculosis while reactivation of latent infection is dependent on severity of tissue depletion in cynomolgus macaques. AIDS Res Hum Retroviruses. 2012;28:1693–1702. doi: 10.1089/aid.2012.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted alpha beta+ T cells. Nature. 1994;372:691–694. doi: 10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka Y, Sano S, Nieves E, De Libero G, Rosa D, Modlin RL, Brenner MB, Bloom BR, Morita CT. Nonpeptide ligands for human gamma delta T cells. Proc Natl Acad Sci U S A. 1994;91:8175–8179. doi: 10.1073/pnas.91.17.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gold MC, Cerri S, Smyk-Pearson S, Cansler ME, Vogt TM, Delepine J, Winata E, Swarbrick GM, Chua WJ, Yu YY, Lantz O, Cook MS, Null MD, Jacoby DB, Harriff MJ, Lewinsohn DA, Hansen TH, Lewinsohn DM. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. 2010;8:e1000407. doi: 10.1371/journal.pbio.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Rhijn I, Zajonc DM, Wilson IA, Moody DB. T-cell activation by lipopeptide antigens. Curr Opin Immunol. 2005;17:222–229. doi: 10.1016/j.coi.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Moody DB, Reinhold BB, Guy MR, Beckman EM, Frederique DE, Furlong ST, Ye S, Reinhold VN, Sieling PA, Modlin RL, Besra GS, Porcelli SA. Structural requirements for glycolipid antigen recognition by CD1b-restricted T cells. Science. 1997;278:283–286. doi: 10.1126/science.278.5336.283. [DOI] [PubMed] [Google Scholar]
- 11.Gilleron M, Stenger S, Mazorra Z, Wittke F, Mariotti S, Bohmer G, Prandi J, Mori L, Puzo G, De Libero G. Diacylated sulfoglycolipids are novel mycobacterial antigens stimulating CD1-restricted T cells during infection with Mycobacterium tuberculosis. J Exp Med. 2004;199:649–659. doi: 10.1084/jem.20031097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Layre E, Collmann A, Bastian M, Mariotti S, Czaplicki J, Prandi J, Mori L, Stenger S, De Libero G, Puzo G, Gilleron M. Mycolic acids constitute a scaffold for mycobacterial lipid antigens stimulating CD1-restricted T cells. Chem Biol. 2009;16:82–92. doi: 10.1016/j.chembiol.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 13.Sieling PA, Chatterjee D, Porcelli SA, Prigozy TI, Mazzaccaro RJ, Soriano T, Bloom BR, Brenner MB, Kronenberg M, Brennan PJ, Modlin RL. CD1-restricted T cell recognition of microbial lipoglycan antigens. Science. 1995;269:227–230. doi: 10.1126/science.7542404. [DOI] [PubMed] [Google Scholar]
- 14.Kasmar AG, van Rhijn I, Cheng TY, Turner M, Seshadri C, Schiefner A, Kalathur RC, Annand JW, de Jong A, Shires J, Leon L, Brenner M, Wilson IA, Altman JD, Moody DB. CD1b tetramers bind alphabeta T cell receptors to identify a mycobacterial glycolipid-reactive T cell repertoire in humans. J Exp Med. 2011;208:1741–1747. doi: 10.1084/jem.20110665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Rhijn I, Kasmar A, de Jong A, Gras S, Bhati M, Doorenspleet ME, de Vries N, Godfrey DI, Altman JD, de Jager W, Rossjohn J, Moody DB. A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nat Immunol. 2013;14:706–713. doi: 10.1038/ni.2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Rhijn I, Gherardin NA, Kasmar A, de Jager W, Pellicci DG, Kostenko L, Tan LL, Bhati M, Gras S, Godfrey DI, Rossjohn J, Moody DB. TCR Bias and Affinity Define Two Compartments of the CD1b-Glycolipid-Specific T Cell Repertoire. J Immunol. 2014;192:4054–4060. doi: 10.4049/jimmunol.1400158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulrichs T, Moody DB, Grant E, Kaufmann SH, Porcelli SA. T-cell responses to CD1-presented lipid antigens in humans with Mycobacterium tuberculosis infection. Infect Immun. 2003;71:3076–3087. doi: 10.1128/IAI.71.6.3076-3087.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montamat-Sicotte DJ, Millington KA, Willcox CR, Hingley-Wilson S, Hackforth S, Innes J, Kon OM, Lammas DA, Minnikin DE, Besra GS, Willcox BE, Lalvani A. A mycolic acid-specific CD1-restricted T cell population contributes to acute and memory immune responses in human tuberculosis infection. J Clin Invest. 2011;121:2493–2503. doi: 10.1172/JCI46216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Jong A, Pena-Cruz V, Cheng TY, Clark RA, Van Rhijn I, Moody DB. CD1a-autoreactive T cells are a normal component of the human alphabeta T cell repertoire. Nat Immunol. 2010;11:1102–1109. doi: 10.1038/ni.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahomed H, Hawkridge T, Verver S, Geiter L, Hatherill M, Abrahams DA, Ehrlich R, Hanekom WA, Hussey GD. Predictive factors for latent tuberculosis infection among adolescents in a high-burden area in South Africa. Int J Tuberc Lung Dis. 2011;15:331–336. [PubMed] [Google Scholar]
- 21.de la Salle H, Mariotti S, Angenieux C, Gilleron M, Garcia-Alles LF, Malm D, Berg T, Paoletti S, Maitre B, Mourey L, Salamero J, Cazenave JP, Hanau D, Mori L, Puzo G, De Libero G. Assistance of microbial glycolipid antigen processing by CD1e. Science. 2005;310:1321–1324. doi: 10.1126/science.1115301. [DOI] [PubMed] [Google Scholar]
- 22.Gau B, Lemetais A, Lepore M, Garcia-Alles LF, Bourdreux Y, Mori L, Gilleron M, De Libero G, Puzo G, Beau JM, Prandi J. Simplified deoxypropionate acyl chains for Mycobacterium tuberculosis sulfoglycolipid analogues: chain length is essential for high antigenicity. Chembiochem : a European journal of chemical biology. 2013;14:2413–2417. doi: 10.1002/cbic.201300482. [DOI] [PubMed] [Google Scholar]
- 23.De Rosa SC, Carter DK, McElrath MJ. OMIP-014: validated multifunctional characterization of antigen-specific human T cells by intracellular cytokine staining. Cytometry Part A : the journal of the International Society for Analytical Cytology. 2012;81:1019–1021. doi: 10.1002/cyto.a.22218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horton H, Thomas EP, Stucky JA, Frank I, Moodie Z, Huang Y, Chiu YL, McElrath MJ, De Rosa SC. Optimization and validation of an 8-color intracellular cytokine staining (ICS) assay to quantify antigen-specific T cells induced by vaccination. J Immunol Methods. 2007;323:39–54. doi: 10.1016/j.jim.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finak G, Frelinger J, Jiang W, Newell EW, Ramey J, Davis MM, Kalams SA, De Rosa SC, Gottardo R. OpenCyto: an open source infrastructure for scalable, robust, reproducible, and automated, end-to-end flow cytometry data analysis. PLoS Comput Biol. 2014;10:e1003806. doi: 10.1371/journal.pcbi.1003806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin L, Finak G, Ushey K, Seshadri C, Hawn TR, Frahm N, Scriba TJ, Mahomed H, Hanekom W, Bart PA, Pantaleo G, Tomaras GD, Rerks-Ngarm S, Kaewkungwal J, Nitayaphan S, Pitisuttithum P, Michael NL, Kim JH, Robb ML, O’Connell RJ, Karasavvas N, Gilbert P, CDRS, McElrath MJ, Gottardo R. COMPASS identifies T-cell subsets correlated with clinical outcomes. Nature biotechnology. 2015 doi: 10.1038/nbt.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chattopadhyay PK, Yu J, Roederer M. A live-cell assay to detect antigen-specific CD4+ T cells with diverse cytokine profiles. Nat Med. 2005;11:1113–1117. doi: 10.1038/nm1293. [DOI] [PubMed] [Google Scholar]
- 28.Lewinsohn DA, Winata E, Swarbrick GM, Tanner KE, Cook MS, Null MD, Cansler ME, Sette A, Sidney J, Lewinsohn DM. Immunodominant tuberculosis CD8 antigens preferentially restricted by HLA-B. PLoS Pathog. 2007;3:1240–1249. doi: 10.1371/journal.ppat.0030127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Day CL, Mkhwanazi N, Reddy S, Mncube Z, van der Stok M, Klenerman P, Walker BD. Detection of polyfunctional Mycobacterium tuberculosis-specific T cells and association with viral load in HIV-1-infected persons. J Infect Dis. 2008;197:990–999. doi: 10.1086/529048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moody DB, Ulrichs T, Muhlecker W, Young DC, Gurcha SS, Grant E, Rosat JP, Brenner MB, Costello CE, Besra GS, Porcelli SA. CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature. 2000;404:884–888. doi: 10.1038/35009119. [DOI] [PubMed] [Google Scholar]
- 31.Van Rhijn I, Nguyen TK, Michel A, Cooper D, Govaerts M, Cheng TY, van Eden W, Moody DB, Coetzer JA, Rutten V, Koets AP. Low cross-reactivity of T-cell responses against lipids from Mycobacterium bovis and M. avium paratuberculosis during natural infection. Eur J Immunol. 2009;39:3031–3041. doi: 10.1002/eji.200939619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen TK, Koets AP, Santema WJ, van Eden W, Rutten VP, Van Rhijn I. The mycobacterial glycolipid glucose monomycolate induces a memory T cell response comparable to a model protein antigen and no B cell response upon experimental vaccination of cattle. Vaccine. 2009;27:4818–4825. doi: 10.1016/j.vaccine.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aagaard C, Hoang T, Dietrich J, Cardona PJ, Izzo A, Dolganov G, Schoolnik GK, Cassidy JP, Billeskov R, Andersen P. A multistage tuberculosis vaccine that confers efficient protection before and after exposure. Nat Med. 2011;17:189–194. doi: 10.1038/nm.2285. [DOI] [PubMed] [Google Scholar]
- 34.Harari A, Dutoit V, Cellerai C, Bart PA, Du Pasquier RA, Pantaleo G. Functional signatures of protective antiviral T-cell immunity in human virus infections. Immunol Rev. 2006;211:236–254. doi: 10.1111/j.0105-2896.2006.00395.x. [DOI] [PubMed] [Google Scholar]
- 35.Pollock KM, Whitworth HS, Montamat-Sicotte DJ, Grass L, Cooke GS, Kapembwa MS, Kon OM, Sampson RD, Taylor GP, Lalvani A. T-cell immunophenotyping distinguishes active from latent tuberculosis. J Infect Dis. 2013;208:952–968. doi: 10.1093/infdis/jit265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grode L, Ganoza CA, Brohm C, Weiner J, 3rd, Eisele B, Kaufmann SH. Safety and immunogenicity of the recombinant BCG vaccine VPM1002 in a phase 1 open-label randomized clinical trial. Vaccine. 2013;31:1340–1348. doi: 10.1016/j.vaccine.2012.12.053. [DOI] [PubMed] [Google Scholar]
- 37.Arbues A, Aguilo JI, Gonzalo-Asensio J, Marinova D, Uranga S, Puentes E, Fernandez C, Parra A, Cardona PJ, Vilaplana C, Ausina V, Williams A, Clark S, Malaga W, Guilhot C, Gicquel B, Martin C. Construction, characterization and preclinical evaluation of MTBVAC, the first live-attenuated M. tuberculosis-based vaccine to enter clinical trials. Vaccine. 2013;31:4867–4873. doi: 10.1016/j.vaccine.2013.07.051. [DOI] [PubMed] [Google Scholar]
- 38.Dascher CC, Brenner MB. Evolutionary constraints on CD1 structure: insights from comparative genomic analysis. Trends Immunol. 2003;24:412–418. doi: 10.1016/s1471-4906(03)00179-0. [DOI] [PubMed] [Google Scholar]
- 39.Colditz GA, Berkey CS, Mosteller F, Brewer TF, Wilson ME, Burdick E, Fineberg HV. The efficacy of bacillus Calmette-Guerin vaccination of newborns and infants in the prevention of tuberculosis: meta-analyses of the published literature. Pediatrics. 1995;96:29–35. [PubMed] [Google Scholar]
- 40.von Reyn CF, Mtei L, Arbeit RD, Waddell R, Cole B, Mackenzie T, Matee M, Bakari M, Tvaroha S, Adams LV, Horsburgh CR, Pallangyo K G. DarDar Study. Prevention of tuberculosis in Bacille Calmette-Guerin-primed, HIV-infected adults boosted with an inactivated whole-cell mycobacterial vaccine. AIDS. 2010;24:675–685. doi: 10.1097/QAD.0b013e3283350f1b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.