Abstract
Nanomaterials constitute a class of structures that have unique physiochemical properties and are excellent scaffolds for presenting carbohydrates, important biomolecules that mediate a wide variety of important biological events. The fabrication of carbohydrate-presenting nanomaterials, glyconanomaterials, is of high interest and utility, combining the features of nanoscale objects with biomolecular recognition. The structures can also produce strong multivalent effects, where the nanomaterial scaffold greatly enhances the relatively weak affinities of single carbohydrate ligands to the corresponding receptors, and effectively amplifies the carbohydrate-mediated interactions. Glyconanomaterials are thus an appealing platform for biosensing applications. In this review, we discuss the chemistry for conjugation of carbohydrates to nanomaterials, summarize strategies, and tabulate examples of applying glyconanomaterials in in vitro and in vivo sensing applications of proteins, microbes, and cells. The limitations and future perspectives of these emerging glyconanomaterials sensing systems are furthermore discussed.
Keywords: glyconanomaterials, biosensing, carbohydrates, nanotechnology, glycoscience
1. Introduction
Carbohydrates are essential in living systems, and collectively have the highest abundance of all biomolecules in nature. They serve for example as energy storage and metabolic intermediates, and carbohydrates conjugated to proteins and lipids mediate molecular recognition, signal transduction, molecular trafficking, cell adhesion, cellular differentiation, inflammation and immune responses (Crocker et al., 2007; Dube and Bertozzi, 2005; Liu and Rabinovich, 2005; Szymanski and Wren, 2005). However, individual carbohydrate-based interactions are often of low affinity, and to overcome this limitation, nature takes advantages of the multivalency effect, where carbohydrates are clustered together to interact with receptors cooperatively (Lee and Lee, 1995).
Glyconanomaterials, where nanomaterials are used as scaffolds to present carbohydrates, have recently emerged as important structures, showing great potential in many applications including sensing and detection (Adak et al., 2014; Bernardi et al., 2013; Chen et al., 2014; El-Boubbou and Huang, 2011; García et al., 2010; Huang, 2013; Marradi et al., 2013; Reichardt et al., 2013; Wang et al., 2009a, 2010a). Compared to other types of scaffolds, nanomaterials offer a number of attractive features as carbohydrate carriers, such as high specific surface area for accommodating high density ligands, tunable size and shape for modulating ligand density and presentation, nanosized dimensions for exploring the interactions with organisms, and unique optical, electronic, photonic, or magnetic properties for transducing the molecular recognition signals for sensing and detection.
In this review, we begin with a brief discussion on coupling chemistry for glyconanomaterials, including a photoconjugation approach developed in our laboratory. We next summarize the synthesis of gold-, iron oxide-, carbon-, quantum dot- (QD-), silica-, liposome-, polymer-, and dendrimer- based glyconanomaterials, and their in vitro and in vivo applications in sensing and imaging of proteins, microbes, and cells. Finally, we discuss current limitations and future perspectives of this field.
2. Carbohydrate conjugation to nanomaterials
Glyconanomaterials are typically prepared following two general conjugation strategies of either non-covalent interactions or covalent bonds, both of which associated with different advantages and drawbacks. In comparison to non-covalent methods, covalent approaches are more frequently used due to the higher stabilities of the covalent adducts. The photocoupling strategy developed in our group, which utilizes suitably functionalized perfluorophenyl azides (PFPAs) for molecular conjugation, provides in this context an efficient alternative.
2.1 Non-covalent conjugation
The non-covalent approach relies in conjugation of the carbohydrate structures to the nanomaterials via typical non-covalent interactions, such as charge interactions, hydrogen bonding, van der Waals' forces, or solvophobic effects. This approach can typically be used under relatively mild conditions, and often requires no, or minimal, chemical derivatization of the carbohydrate ligands or the nanomaterial substrates. The interactions can occasionally be very strong, for example based on biotin-streptavidin recognition, but the bond strengths may also be weaker, which could lead to detachment and thereby increased nonspecific interactions with the target. The process can also be non-selective and less controllable compared to covalent linkages. These effects must be taken into account, as they can affect the sensitivity and specificity in sensing applications. For certain systems, however, this approach is highly useful, efficiently applied to large carbohydrate structures like polysaccharides, glycoproteins, and glycolipids.
2.2 Covalent conjugation
Mono- and oligosaccharides are commonly conjugated to nanomaterials covalently, either directly or via post-modification coupling reactions. This approach holds the advantage of generating stable linkages and robust surface structures. Typical examples include thiol/disulfide chemisorption on gold and quantum dots, phosphates on iron oxides, and silanes on silica. Of the different covalent systems evaluated, thiol/gold is the most studied and used. This system is well established, relatively stable and reproducible. Post-modification coupling is based on typical conjugation chemistries where complementary functional groups react to form covalent linkages such as amides or triazoles. However, this approach generally requires chemical derivatization of the carbohydrates, the synthesis of which may present considerable challenges, especially for oligosaccharide structures.
In order to achieve high spatial and temporal control over the conjugation process, we developed a photocoupling chemistry that is based on fluorinated aryl azides (Liu and Yan, 2006, 2010; Liu et al., 2010a; Park and Yan, 2013; Wang et al., 2009a, 2010a; Yan and Harnish, 2003; Yan and Ren, 2004). Upon light irradiation and nitrogen extrusion from the aryl azide group, highly reactive singlet nitrene entities are formed, which can insert into CH bonds or add to C=C bonds. This method has been successfully applied to conjugation of carbohydrates (Wang et al., 2009b, 2010b), small molecules (Al-Bataineh et al., 2009), polymers (Gann and Yan, 2008; Wang et al., 2011), carbon materials (Liu et al., 2010a, 2010b), and discrete nanoparticles (Park et al., 2015) to different nanomaterials. Interestingly, this photocoupling approach can be efficiently used for un-derivatized carbohydrates, and is perhaps especially useful for oligosaccharides, which are often difficult to derivatize. Furthermore, the reaction is fast and straightforward, often occurring within minutes at room temperature. Maalouli et al. compared the photocoupling with the classic copper-catalyzed alkyne-azide cycloaddition reaction (CuAAC), and found that carbohydrate surfaces prepared by perfluorophenyl azide photocoupling had higher ligand density and also generated stronger surface plasmon resonance (SPR) signals than those prepared by the click reaction (Maalouli et al., 2013).
3. Synthesis of glyconanomaterials
In this section, we focus on several key types of glyconanomaterials, including gold-, iron oxide-, carbon-, quantum dot-, silica-, liposome-, polymer-, and dendrimer-based glyconanomaterials. The specific synthetic approaches to the glyconanomaterials are summarized.
3.1 Gold glyconanomaterials
Gold nanoparticles constitute the most extensively employed and studied type of nanomaterials, basically due to the straightforward preparation and surface chemistry, the high stability, and the attractive optoelectronic properties. The particles give rise to localized surface plasmon resonance (LSPR) upon light irradiation, an effect that is highly sensitive to the dielectric environment close to the nanoparticle surface. This phenomenon renders the structures useful for transducing recognition events at the metal surface (Kelly et al., 2003). Colorimetric sensing has thus been established based on the LSPR shift (Aslan et al., 2004; Saha et al., 2012; Wang and Ma, 2009). When the carbohydrate-receptor interactions cause additional aggregation of gold nanomaterials, larger LSPR shifts will occur, leading to intense color changes that are often visible by the naked eye (Liu et al., 2007). This unique optical property allows for highly sensitive sensing and detection. In addition, the LSPR effect is not subject to blinking, an effect that is associated with quantum dots (QDs), or fluorophore photobleaching of organic structures (Wang et al., 2009a).
Several methods have been employed to synthesize gold glyconanomaterials. A straightforward approach uses reducing sugars as both reducing agents and capping ligands during the formation of gold nanoparticles (Guo and Yan, 2008; Kemp et al., 2009). As the gold precursors become reduced by the carbohydrates to generate gold nanoparticles, hydroxyl/gold interactions lead to a protective carbohydrate layer on the gold nanoparticles. Another in situ method involves the addition of thiol-functionalized carbohydrates to the gold precursors (Chen et al., 2005; De La Fuente et al., 2001; Halkes et al., 2005). The thiol-carbohydrate serves as the capping ligand for gold nanoparticles as they are formed. One drawback of this method is that the particle size could vary significantly depending on the ligand structure as well as the experimental conditions, which are difficult to predict and control (Sundgren and Barchi, 2008). In the ligand exchange method (Chien et al., 2008; Hone et al., 2003; Mahon et al., 2010a; Thygesen et al., 2009), gold nanoparticles are first prepared, and the original ligand is then replaced by thiol-terminated carbohydrates. This protocol could reproducibly generate nanoparticles with predicable sizes. Similarly, in our photocoupling protocol, gold nanoparticles are first subjected to thiol/disulfide-terminated perfluoroaryl azides, and are then subjected to light activation in the presence of carbohydrates (Jayawardena et al., 2013b). With this protocol, a wide range of mono-, oligo-, and polysaccharides have been conjugated to different nanomaterials.
3.2 Magnetic glyconanomaterials
Magnetic nanomaterials constitute an important type of nanomaterials that display magnetic properties when subjected to external magnetic fields. Magnetite (Fe3O4) nanoparticles are the most widely used type in sensing applications. Major attributes of Fe3O4 nanoparticles include: i) straightforward preparation methods yielding particles of 5-30 nm size; ii) excellent biocompatibility, as exemplified by the FDA-approved formulations Feridex for liver imaging (Wang et al., 2001) and Feraheme for iron deficiency anaemia (Lu et al., 2010); iii) high magnetic relaxivities, rendering the materials well suited as contrast agents for in vitro/in vivo magnetic resonance imaging (MRI); and iv) ease of surface functionalization (Gao et al., 2006).
The most direct method to attach carbohydrates to iron oxide nanoparticles is to take advantage of their stabilizing properties during the nanoparticle synthesis (Horak et al., 2007). However, excess carbohydrate is generally required for this approach, and the method is therefore used for easily accessible polysaccharides. An alternative strategy is post-modification of pre-formed nanoparticles by covalent conjugation, for example conjugation of carboxylated carbohydrates to amine-functionalized iron oxide nanoparticles (Shi et al., 2009; Srinivasan and Huang, 2008). Carbohydrates were also conjugated by the CuAAC reaction, where alkyne-derivatized carbohydrates were allowed to react with azide-functionalized nanoparticles (El-Boubbou et al., 2007, 2010; Lin et al., 2007). This coupling configuration was also reported to give better conjugation efficiency compared to coupling azide-derivatized carbohydrates to alkynylated nanoparticles (Lin et al., 2007). In addition, biotinylated carbohydrates were synthesized and conjugated to streptavidin-coated magnetic nanoparticles (Hatch et al., 2008; Pera et al., 2010). Our photocoupling chemistry can also be applied to make magnetic glyconanomaterials. In this case, iron oxide nanoparticles were treated with phosphate-derivatized PFPAs, to which carbohydrates were conjugated by light activation (Jayawardena et al., 2013a; Liu et al., 2009).
3.3 Carbon glyconanomaterials
Carbon nanomaterials include materials ranging from the amorphous carbon to the more recently discovered fullerenes, carbon nanotubes (CNTs) and graphene. The many attractive physical properties of these materials render them highly useful for biosensing. However, these materials are fairly chemically inert, lack reactive functionality, have poor water solubility, and are potentially toxic to cells (Liu et al., 2012). This can however be overcome by carbohydrate functionalization, which improves solubility, biocompatibility and sensing capability.
Because carbon materials are generally non-polar and hydrophobic, non-covalent conjugation approaches often rely on van der Waals' forces, π-π stacking, and hydrophobic effects. Derivatization of carbohydrates with lipophilic groups are thus generally required, including lipids (Chen et al., 2004, 2006; Feng et al., 2011; Khiar et al., 2009; Murthy et al., 2012), polyaromatic hydrocarbons (Chen et al., 2011; Sudibya et al., 2009; Wu et al., 2008) or porphyrins (Chen et al., 2012) before treatment with the carbon substrate. Because the carbon glyconanomaterials are not chemically functionalized in these cases, their physical properties can be preserved.
Covalent modification requires either the carbon nanomaterials or the carbohydrates, or both, to be chemically functionalized. Among the carbon nanomaterials, the most homogeneous structures are obtained with fullerenes (Prato, 1997; Wudl, 2002). For graphene and carbon nanotubes, the oxidized materials are most commonly employed, further functionalized with, e.g., amine-derivatized carbohydrate structures (Chen et al., 2013; Gorityala et al., 2010). Pristine graphene and carbon nanotubes are fairly inert, and require the use of reactive species to chemically functionalize them. These include aryl radicals from aryl diazonium salts (Pinson and Podvorica, 2005; Ragoussi et al., 2013), 1,3-dipolar cycloaddition of azomethine ylides to form pyrrolidine (Hong et al., 2010), and cheletropic cycloaddition of nitrenes to form aziridines (Holzinger et al., 2003; Prato et al., 1993). Studies in our group have for example shown that singlet perfluorophenyl nitrenes are especially useful, and highly reactive towards carbon nanomaterials (Park and Yan, 2013). Once the materials are functionalized, functional groups can be introduced which can be readily used for carbohydrate conjugation (Kong et al., 2015).
3.4 QD glyconanomaterials
Inorganic quantum dots (QDs) are luminescent semiconductor nanomaterials with attractive physical properties for biosensing. QDs can for example show broad optical excitation and narrow emission with good quantum yields, and are less susceptible to photobleaching (Alivisatos, 2004; Michalet et al., 2005). Combined with the possibility for multivalent ligand presentation, these characteristics are appealing for sensing and imaging applications.
QD glyconanomaterials can be prepared by capping QDs with carbohydrates through non-covalent interactions, including hydrophobic effects (Osaki et al., 2004) and electrostatic interactions by mixing negatively charged CdSe/ZnS core-shell QDs, capped with carboxymethyl dextran and sulfanylsuccinate groups, with positively charged polylysine (Chen et al., 2003). Covalent approaches include in situ protocols, for example, based on the addition of thiol-functionalized neoglycoconjugates to QD precursor solutions (De la Fuente and Penadés, 2005; Mukhopadhyay et al., 2009). The ligand exchange protocol is similar to the synthesis of gold glyconanomaterials, where the original capping agents can be replaced by thiol-derivatized glycoconjugates (Babu et al., 2007; Chen et al., 2008; Niikura et al., 2007, 2008; Robinson et al., 2005). QD glyconanomaterials have also been prepared by post-modification protocols similar to gold nanoparticles (Higuchi et al., 2008; Kikkeri et al., 2009b; Sandros et al., 2007).
3.5 Silica glyconanomaterials
Silica nanomaterials are highly tunable, show high thermal and mechanical stability, good water dispersability, and are easy to functionalize (He et al., 2010). Among different silica formats, mesoporous silica nanomaterials are especially attractive, displaying large pore sizes, high internal volumes and surface areas (Hao et al., 2012, 2014a, 2014b, 2015; Zhou et al., 2015b). Although silica nanomaterials do not inherently possess optical or magnetic properties, these properties can be easily introduced by entrapment of fluorescent dyes or encapsulation of gold/QDs/magnetic nanoparticles, enabling the use of these materials in sensing and imaging (Slowing et al., 2007; Trewyn et al., 2007).
Carbohydrates are typically conjugated to silica nanomaterials through post-modification strategies, including, for example, amide/triazole formation and photocoupling. Examples of the popular CuAAC method include the conjugation of alkyne- or azide-functionalized carbohydrate derivatives (Peng et al., 2007; Zhao et al., 2012), in the latter case resulting in galactose-presenting silica nanoparticles for sensing live hepatic cancer cells. Similarly, solid tumors were targeted with mannose-functionalized mesoporous silica nanoparticles (Gary-Bobo et al., 2011). Our photocoupling method has furthermore been applied to aryl azide-functionalized silica nanoparticles, resulting in glyconanomaterials that were successfully used to sense proteins (Tong et al., 2012; Wang et al., 2011b, 2011c, 2013), and detect bacteria (Jayawardena et al., 2013a, 2015; Wang et al., 2011c) and cancer cells (Jiang et al., 2015).
3.6 Liposome/micelle glyconanomaterials
Amphiphilic molecules consisting of hydrophilic carbohydrate head groups and lipophilic hydrocarbon chain segments are generally able to self-assemble in aqueous solutions, some of which forming structures that can ultimately result in vesicular liposome- or micelle-glyconanomaterials depending on the relative lengths, sizes and structures of the involved segments. A large variety of such amphiphilic structures occur naturally (glycolipids) as part of cell membranes, and serve a variety of different functions in living systems. These and other structures can be prepared from suitable lipophilic entities, such as fatty alcohols, phospholipids, and cholesterol. Following self-assembly, the resulting liposome- or micelle- glyconanomaterials may possess several attractive features, such as high biocompatibility and loading capacity. The structures have furthermore the potential to be efficiently taken up by cells from interactions with the cell membranes, for example owing to fusion with the lipid bilayers of the membranes (Jayaraman et al., 2013). In view of the similarities with glycolipid-presenting cell surfaces, these self-assembled structures show potential for a variety of applications, such as for instance inhibitor development and biosensors.
Many strategies to prepare vesicular liposome- or micelle- glyconanomaterials via self-assembly of amphiphilic structures have been developed (Chabre and Roy, 2010; Jayaraman, 2009; Kiessling et al., 2006; Yan et al., 2005). Principal synthetic approaches to multivalent aggregates include: i) direct self-assembly of appropriate glycosylated amphiphilic molecules, for example based on polyethylene glycol, peptides and/or alkyl linkers with long carbon chains; ii) incorporation of glycosylated amphiphilic molecules with suitable lipid matrices at optimal molar ratios (usually ∼5-10%); and iii) functionalization of pre-formed liposomes or micelles with specific carbohydrate structures (Harada et al., 2005). Owing to their relatively straightforward fabrication, liposome- or micelle- based glyconanomaterials have continuously been developed for biomedical applications, and also as tools for biosensing (Assali et al., 2013; Hildebrand et al., 2002; Mahon et al., 2010b). However, a number of challenges need to be addressed for efficient applications in sensing technology. For example, high densities of surface-exposed carbohydrates are difficult to obtain due to the increased risk of vesicular collapse from high concentrations of glycolipid elements. In addition, the orientation and mobility of the carbohydrate moieties can be difficult to control, resulting in reduced carbohydrate surface accessibility for efficient recognition. The structures may furthermore display relatively low stabilities and characterization can be challenging (Jayaraman et al., 2013).
3.7 Polymer glyconanomaterials
Some of the limitations met with liposomes can be addressed with polymer- or dendrimer-based glyconanomaterials, i.e. synthetic polymeric/dendrimeric structures carrying core or pendant carbohydrate groups. Thus, the carbohydrate surface densities can in principle be improved, without the risk of severe disruption of the nanoparticulate structure. In addition, since multivalency is an attractive feature for sensing and recognition applications, increased interest in polymers and dendrimers as frameworks for carbohydrate presentation has emerged to improve the interactions between the binding partners. Together with developments in high structural control and biocompatibility, these features offer high potential for the materials to be employed in in vitro and in vivo sensing and detection.
The glycocode of living organisms being highly structure-dependent, the carbohydrate presentation on polymer glyconanomaterials has to be designed with high accuracy where small structural differences may considerably influence target binding. The correct carbohydrate arrangement along the polymeric skeleton is essential in order to accomplish specific recognition and cell communication effects. In principle, two methods can be adopted for polymer glyconanomaterials synthesis: polymerization with carbohydrate-functionalized monomers, and grafting carbohydrate entities to a polymer backbone. Traditionally, both methods have been carried out in a stochastic manner, resulting in relatively low control over the detailed structure. However, recent developments in controlled (radical) polymerization and solid-phase synthesis have resulted in improved structural fidelity (Yilmaz and Becer, 2013). These efforts, based on modern polymer chemistry in combination with glycoscience, has lead to enhanced understanding of polymer- and dendrimer- based glyconanomaterials, enabling more complex and well-defined architectures of different shapes with high biocompatibilities and affinities. The resulting glyconanomaterials show significant potential for many biological applications, perhaps especially for biosensors (Sunasee and Narain, 2013; Voit and Appelhans, 2010). However, some of their properties may result in limitations, in particular with respect to the structural homogeneity from the polymer synthesis. These effects may be less important for certain applications such as (qualitative) binding and imaging, whereas detailed, quantitative analysis requires highly homogeneous entities.
3.8 Dendrimer glyconanomaterials
In contrast to regular polymers, dendrimers are in principle monodisperse macromolecules with well-defined, usually spherical, architectures. The structures are generally of high symmetry, comprise a core scaffold from which branching segments protrude, and are decorated with external functional groups (Astruc et al., 2010; Bosman et al., 1999). In addition, dendrons, i.e. non-spherical, dendritic structures based on single focal points rather than cores with branching points in all directions, can be applied. The dendrimer structures typically contain internal cavities for potential encapsulation, while the external groups define the solubility and chemical performances. Reproducible synthesis of structurally defined entities is therefore accessible, where the structures can be tailored for specific applications. These features have led to the development of complex, yet highly defined, nanoscale carbohydrate-functionalized structures that are more robust than liposomes and which can be further modified (Ashton and Boyd, 1997). High control of ligand densities can also be obtained, owing to efficient synthesis strategies developed. The resulting dendrimer glyconanomaterials have also increasingly found important uses in glycoscience and technology. Many examples of such structures have been reported over last two decades with the main purpose to enhance the binding efficiencies via the multivalency effect.
Three types of dendrimer glyconanomaterials can be distinguished, based on either carbohydrate core structures, pendant carbohydrate entities; or dendrimers built entirely from carbohydrates (Chabre and Roy, 2010; Turnbull et al., 2002), generally synthesized following different divergent (from core and outwards) or convergent (assembly of dendrons) methodologies. Both synthetic methods are based on repeated reaction sequences, where each repetition leads to a new dendritic ‘generation’. The divergent approach requires highly efficient and orthogonal chemistry in order to avoid incomplete reactions, but minimizes potential steric complications. The convergent approach, on the other hand, normally yields dendrimer glyconanomaterials of high purity, but may suffer from steric constraints with hindered cores. Dendrimer glyconanomaterials show several advantages as carbohydrate-presenting entities when used in sensing applications. They are generally well-defined structures that display high chemical stabilities (Boas and Heegaard, 2004), resulting in ease of characterization and evaluation. They permit geometrical control over carbohydrate positioning and density and can be constructed to show high degrees of multivalency (Seebach et al., 1998). These advantages, however, come with a potential drawback of relatively high synthetic efforts, and high production costs.
4. Glyconanomaterials for sensing and imaging
For sensing and imaging applications, it is essential that the carbohydrates retain their recognition effects following their conjugation to the nanomaterials. Compared to other biological recognition elements such as antibodies and enzymes, this is less challenging since carbohydrates often display higher stabilities and a wider range of conditions can be applied. Nevertheless, the recognition effects are generally sensitive to the surface presentation, such as ligand density, linker length, surface chemistry, etc. In this section, we overview recent developments in in vitro/in vivo sensing and imaging of proteins, microbes, and cells using various glyconanomaterials.
4.1 Sensing proteins
Many physiological and pathophysiological processes, such as cell-cell communication, cell adhesion, and cell infection, start with carbohydrates recognizing their cognate binding proteins (Holgersson et al., 2005) (Fig. 1). Understanding the carbohydrate-protein interactions are important, and can generate new leads in developing diagnostic and therapeutic tools (Ernst and Magnani, 2009). Glyconanomaterials can in this context serve as cell mimics, where the recognition event forms the basis for sensing proteins. Studies from our laboratory and others have shown that glyconanomaterials can amplify the binding affinity of carbohydrates to proteins by several orders of magnitude (La Belle et al., 2007; Liu and Yan, 2010; McLean et al., 2005; Wang et al., 2010b, 2011a, 2012a, 2013). This laid a strong foundation for glyconanomaterials in sensing proteins with high sensitivity. Among the proteins tested, concanavalin A (ConA), is ubiquitously applied, especially used for basic studies to develop new sensing systems. In 2001, Kataoka's and Shinohara's groups used gold- and carbon-based glyconanomaterials to study carbohydrate-lectin interactions (Kato et al., 2001; Otsuka et al., 2001). These pioneering studies demonstrated the feasibility of multivalent glyconanomaterials in sensing proteins.
Fig. 1.
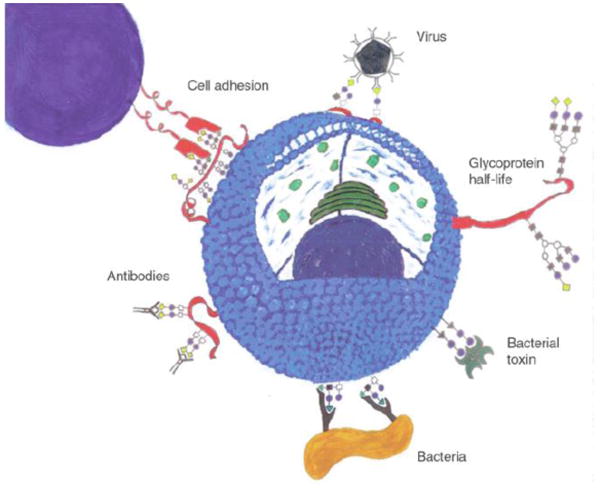
Schematic illustration of typical carbohydrate-mediated interactions at cell surfaces. Reproduced from (Holgersson et al., 2005) by permission from Nature Publishing Group.
Table 1 summarizes examples when gold-, carbon-, polymer-, and dendrimer-based glyconanomaterials have been applied to sensing lectins. Well-established sensing and imaging techniques have generally been adopted, taking advantages of the unique properties of the proteins, as well as the specific physiochemical properties of the nanomaterials. Figures 2 - 4 are selected examples where glyconanomaterials are used for sensing lectins. The most used transduction and imaging techniques include: 1) UV-Vis spectroscopy/optical microscopy, which relies on the interactions of glyconanomaterials with proteins leading to light absorption/scattering, causing absorbance changes. In addition, oligomeric carbohydrate-binding proteins can interact with multivalent nanoparticles to form aggregates, which can be visualized under the optical microscope or by naked eyes when the agglomerates are large enough. When gold glyconanomaterials are used, interaction/agglomeration also induces a bathochromic shift of the LSPR signal, together with a decrease in the absorption intensity. 2) Fluorescence spectroscopy/microscopy, which generally takes advantage of fluorescently-labeled proteins and/or glyconanomaterials in a variety of formats. 3) Surface plasmon resonance (SPR). SPR signals are generated upon the interaction of glyconanomaterials with protein-functionalized SPR sensor chips, or vice versa. 4) Dynamic light scattering (DLS). Binding of glyconanomaterials to the proteins increases the hydrodynamic volume of the complex, which can be sensed by DLS. 5) Quartz crystal microbalance (QCM). QCM has proven especially suitable for nanomaterials, owing to their large molecular mass and thereby enhanced signals.
Table 1.
Sensing and imaging proteins using gold-, carbon-, polymer-, and dendrimer-glyconanomaterials.
| Carbohydrat e |
Particle size/type |
Coupling method (Function al group) |
Carbohydr ate density |
Protein types |
Detectio n methods/ assay |
Detecti on range/l imit |
Selectivity | Reference s |
|---|---|---|---|---|---|---|---|---|
| Gold glyconanomaterials | ||||||||
| Lac; d-Gal | 8.9 nm | Covalent (-SH) | 20-50 wt% | RCA120 | UV-Vis; visualization | 5-50 μg/mL ; 1 μg/mL (1 ppm) | Lac/RCA120 | (Otsuka et al., 2001) |
| α-d-Man | 16 nm | Covalent (-SH) | ConA | UV-Vis | 0.192-0.385 μM; <0.1μM | α-d-Man/ConA | (Hone et al., 2003) | |
| α-d-Man; d-Glc; d-Gal | 6-20 nm | Covalent (-SH) | 11-128/particle | ConA | SPR | 0.095-6 μM | α-d-Man/ConA | (Lin et al., 2003) |
| α-d-Man; d-Glc | 1.79-3.84 nm | Covalent (-SH) | 23-98/particle | ConA | SPR; UV-Vis | 400-7000 RU | α-d-Man/ConA | (Halkes et al., 2005) |
| d-GalNAc | 1-12 nm | Covalent (-SH) | 90/particle | PNA; PSA | UV-Vis | GalNAc/PNA | (Svarovsky et al., 2005) | |
| α-d-Man | 32 nm | Covalent (-SH) | ConA; BS-I; SBA; MAL | SPR; UV-Vis; visualization | 5 nM | α-d-Man/ConA | (Tsai et al., 2005) | |
| β-d-Gal | 50-150 nm | Covalent (-SH) | 4/nm2 | RCA120 | UV-Vis | 100 μg/mL | β-Lac/RCA120 | (Bergen et al., 2006) |
| Heparan sulfate | 15-16 nm | Covalent (-sulfate) | Heparanase | UV-Vis; visualization | 0.7-5.6 μg/mL | Heparan/Heparanase | (Ban et al., 2008) | |
| α-d-Man | ∼8 nm | Covalent (-SH) | ConA | QCM | 1.3×10-10 M | α-d-Man/ConA | (Lyu et al., 2008) | |
| β-d-Gal; α-d-Man | 16 nm | Covalent (-SH) | RCA120; ConA; BSA | UV-Vis | 0.01-0.8 μM (Gal); 0.7-2.4 μM (Man) | Lac/RCA120; α-d-Man/ConA | (Schofield et al., 2008) | |
| Lac; maltose | 1.8 nm | Covalent (-SH) | 15-30 wt% | β-galactosidase | UV-vis | 0.01-0.1 mg/mL | β-Lac/β-galactosidase | (Barrientos et al., 2009) |
| maltose; d-Man; d-Glc; Lac | 12.33 nm | Covalent (native) | ConA | UV-Vis | 0.03-100 nM | α-d-Man/ConA | (Chuang et al., 2009) | |
| α-d-Man | 2.9 nm | Covalent (-SH) | ConA; BSA | Fluorescence | 0.015-100 nM; 10 pM | α-d-Man/ConA | (Huang et al., 2009) | |
| α-d-Glc; maltose; maltotriose | 12 nm | Covalent (-SH) | 300-1000 molligand/mol particles | ConA; glucoamylase | UV-Vis | 1 μM | α-d-Glc/ConA | (Thygesen et al., 2009) |
| α-d-Man; Manα1-4Man | ∼20 nm | Photocoupling (native) | 1200/particle | ConA; GS-II; PNA | UV-Vis; SPR | 10 μg/mL | α-d-Man/ConA | (Wang et al., 2009b) |
| α-d-Man; d-GlcNAc | 4-6 nm | Covalent (-SH) | 4-52/particle | ConA; WGA | Diffractive optical technology | α-d-Man/ConA; GlcNAc/WGA | (Jiang et al., 2010) | |
| d-Man | 10 nm | Covalent (-SH) | ConA; human IgG | Anodic stripping voltammetry | 0.084-50 μg/mL; 0.07 μg/mL | Man/ConA | (Min et al., 2010) | |
| d-Man; d-Glc | 7-30 nm | Photocoupling (native) | 297-4486/particle | ConA | UV-Vis; fluorescence microscopy | 0.01-0. 32 μM; 16 nM | Man/ConA | (Wang et al., 2010b) |
| α-d-Man | 22 nm | Photocoupling (native) | 1037-1516/particle | Cyanovirin | UV-Vis; fluorescence microscopy | 100 nM | α-d-Man/Cyanovirin | (Wang et al., 2011a) |
| d-Man; d-Gal | 13-16 nm | Photocoupling (naive) | ConA; RCA120 | DLS | 2.9 nM | d-Man/ConA | (Wang et al., 2011b) | |
| β-d-Gal; β-d-Glc; α-d-Man | 1.2-1.7 nm | Covalent (-SH) | 11-67/particle | PA-IL | SPR; ITC | 300 RU | β-d-Gal/PA-IL | (Reynolds et al., 2012) |
| d-Man; d-Gal | 22 nm | Photocoupling (native) | 3600/particle | ConA | ITC | 10 μM | d-Man/ConA | (Wang et al., 2012) |
| Dextran | Covalent (native) | ConA | SPR | 1-20 μg/mL; 0.39 μg/mL | Dex/ConA | (Huang et al., 2013) | ||
| d-Man; d-GlcNAc; Lac; d-Gal; Lac; sucrose; d-Ara; d-Glc | 20 nm | Photocoupling (native) | ConA; GS-II; PNA; SBA | UV-Vis | 10 μg/mL | d-Man/ConA; d-GlcNAc/GS-I I; Lac/PNA; d-Gal/SBA | (Jayawardena et al., 2013b) | |
| d-Man | 10 nm | Covalent (-SH) | ConA | UV-Vis | 10-50 μg/mL | d-Man/ConA | (Lim et al., 2013) | |
| α-d-Man | 12.5 nm | Covalent (-SH) | ConA | QCM | 1 μM | α-d-Man/ConA | (Mahon et al., 2013) | |
| Lac; β-CD | 12.1±1 nm | Covalent (-SH) | PNA; human galectin-3 | UV-Vis | 2 μM | Lac/PNA | (Aykaç et al., 2014) | |
| α-d-Man | Covalent (-SH) | ConA; SNA; PNA; UEA; WGA | QCM | 0.5-17.5 nM | α-d-Man/ConA | (Zeng et al., 2014) | ||
| β-d-Gal | 10-100 nm | Covalent (-N3) | PNA; WGA; ConA; LCA120; BSA; pepsin | UV-Vis | 0.01-0.05μM; 3.5 nM | β-d-Gal/PNA | (Hu et al., 2015) | |
| Lac; α-d-Man; β-d-GlcNAc | 15 nm | Covalent (-SH) | RCA120; ConA; WGA | UV-Vis; DLS | 0.5-10 nM; 300 pM | Lac/RCA120; α-d-Man/ConA; β-d-GlcNAc/WGA | (Huang et al., 2015) | |
| Carbon glyconanomaterials | ||||||||
| α-d-Man; Dextran | N/A (Fullerene) | Covalent (-N3) | ConA | SPR | 0.13 mM | α-d-Man/ConA | (Kato et al., 2001) | |
| α-d-Gal; β-d-Gal | 65-70 nm (SWNTs) | Non-covalent (-lipid) | 10-25 nm thickness | HPA | Fluorescence | 100 μg/mL | α-d-Gal/HPA | (Chen et al., 2004) |
| Lac | 0.8-1.4 nm (SWNTs) | Covalent (native) | RCA120; ConA; WGA | Confocal microscopy | 0.28 mg/mL | Lac/RCA120 | (Hasegawa et al., 2004) | |
| α-d-Man; Lac; β-d-Gal | 5-10 nm (SWNTs) | Non-covalent (-pyrene) | ConA; PNA; PTA | Fluorescence | 100 μg/mL | α-d-Man/ConA; Lac/PNA; β-d-Gal/PTA | (Wu et al., 2008) | |
| d-GlcNAc; β-d-Glc; α-d-Man | 10-30 nm (SWNTs) | Non-covalent (-pyrene) | HPA; ConA | Fluorescence | 5 mM | α-d-Man/ConA | (Sudibya et al., 2009) | |
| Maltose | 1.12±0.04 nm (Graphene) | Non-covalent (-pyrene) | ConA; BSA | Fluorescence | 0.02-1 mM; 0.8 nM | Maltose/ConA | (Chen et al., 2011) | |
| Lac | 20-50 nm (MWNTs) | Non-covalent (-lipid) | BSA | Circular dichroism | 0.04 mg/mL | Lac/BSA | (Feng et al., 2011) | |
| α-d-Man; α-d-Glc | N/A (Fullerene) | Covalent (-N3) | 24/particle | ConA | ITC | 0.017-0.055 mM | α-d-Man/ConA | (Sánchez-Navarro et al., 2011) |
| β-d-Gal; α-L-Fuc; α-d-Man | 0.73-10.4 nm (Graphene); 3.5-23.4 nm (SWNTs) | Non-covalent (-pyrene) | PA-IL; PA-IIL; ConA | Electrolyte-gated FET; ITC | 2 μM | β-d-Gal/PA-IL; α-l-Fuc/PA-IIL; α-d-Man/ConA | (Chen et al., 2012) | |
| d-Glc | 15±3 nm (MWNTs) | Covalent (-CHO) | ConA | Electrochemical impedance spectroscopy | 3.3 pM-9.3 nM; 1.0 pM | d-Glc/ConA | (Hu et al., 2012) | |
| α-d-Man | 1.2-2.5 nm (SWNTs) | Non-covalent (-lipid) | ConA; WGA | Confocal microscopy; SPR | ∼6000 RU | α-d-Man/ConA | (Murthy et al., 2012) | |
| α-d-Man | 4-8 nm (Graphene); 3.5 nm (SWNTs) | Covalent (-N3) | 6 wt%; 35 wt% | ConA; BSA | UV-Vis | 0.2 mg/mL | α-d-Man/ConA | (Ragoussi et al., 2013) |
| Polymer glyconanomaterials | ||||||||
| α-d-Man | N/A (Acrylamide) | Covalent (-acrylic amide) | 5% of monomers | ConA | QCM; SPR | 5×10-10 M | α-d-Man/ConA | (Yu et al., 2007) |
| Neu5Acα2-6 Galβ1-4Glc | N/A (Acrylamide) | Covalent (-OCN) | MAA; SNA; PNA | SPR; microarray | 0.075-7.5 nM; 10-6 M | β-d-Gal/PNA | (Narla and Sun, 2012) | |
| α-d-Man | 7-10 nm (Acrylamide) | Covalent (-N3) | 1-3/chain | ConA | SPR; ITC | 8 nM | α-d-Man/ConA | (Ponader et al., 2012) |
| α-d-Man | 2.7 nm (Acrylamide) | Covalent (-acrylic amide) | 36, 4.8 μmol/m2 | ConA; BSA | QCM | 5 mg/L | α-d-Man/ConA | (Seto et al., 2012) |
| α-d-Man; β-d-Gal; β-d-GlcNAc | N/A (Styrene) | Covalent (-N3) | ConA; PNA; WGA | QCM | 0.2 mg/mL | α-d-Man/ConA; β-d-Gal/PNA; β-d-GlcNAc/WGA | (Baradel et al., 2013) | |
| α-d-Man | 50 nm (Man-trimethoxysilane) | Covalent (-acrylic amide) | 39 nm | ConA; BSA | QCM; SPR | 10−9 -10−6 M; 1 ng/mm2 | α-d-Man/ConA | (Seto et al., 2014) |
| α-d-Man; Lac | N/A (Acrylamide) | Covalent (-acrylic amide) | 5.3-20.4 %/polymer | ConA; PNA | QCM; UV-Vis | 200 nM | α-d-Man/ConA; Lac/PNA | (Tanaka et al., 2014a) |
| Lac; Neu5Acα2-6 Galβ1-4Glc; N-glycan | N/A (Acrylamide) | Covalent (-N3) | 7.2-7.7 %/polymer | PNA; BSA; SSA | QCM | 20 nM | Lac/PNA; Neu5Acα2-6Galβ1-4Glc/BSA; N-glycan/SSA | (Tanaka et al., 2014b) |
| α-d-Man | 80-200 nm (Methacrylate) | Covalent (-acrylic amide) | ConA; BSA | QCM; SPR; UV-Vis | 10-5-10-1 g/L; 6.0 ng/mL | α-d-Man/ConA | (Terada et al., 2014) | |
| Lac; β-d-Glc; d-Gal | N/A (Methacrylate) | Covalent (-acrylic amide) | RCA120 | QCM | 10 μg/mL | Lac/RCA120 | (Wang et al., 2014) | |
| α-d-Man; β-d-Gal | 250 × 2 nm | Covalent (-NH2) | ConA; SBA | fluorescence | α-d-Man/ConA; β-d-Gal/SBA | (Zhou et al., 2014) | ||
| α-d-Man; β-d-Gal | 265 × 5 nm | Covalent (-NH2) | 0.6 mmol/g | ConA; RCA120 | fluorescence | α-d-Man/ConA; β-d-Gal/RCA120 | (Zhou et al., 2015a) | |
| Dendrimer glyconanomaterials | ||||||||
| α-d-Man; β-d-Glc | N/A (CD) | Covalent (-SH) | 7-14/compound | ConA | ITC | 0.07-0.4 μM | α-d-Man/ConA | (Ortega-Caballero et al., 2001) |
| d-GlcNAc | N/A (CD) | Covalent (-SH) | 7/compound | E-selectin; BSA | SPR | 0.9 μM | d-GlcNAc/E-selectin | (Furuike et al., 2005) |
| d-Glc | 4.5 nm (Amidoamine) | Covalent (-alkene) | 10/compound | ConA | Fluorescence | 0-33 mM; 5.5 mM | d-Glc/ConA | (Ibey et al., 2005) |
| Lac | 16 nm | Covalent (-SH) | Cholera toxin | UV-Vis | 54 nM (3μg/mL) | Lac/Cholera toxin | (Schofield et al., 2007) | |
| Globotriose | 4-20 nm | Covalent (-SH) | 60-1970/particle | Shiga-like Toxin | SPR; UV-Vis | 1 mg/mL | Globotriose/Shiga-like Toxin | (Chien et al., 2008) |
| α-d-Man; β--d-Gal; β-d-Glc | N/A (Amidoamine) | Covalent (-NH2) | 6-18/compound | ConA | Turbidimetry | 1 mg/mL | α-d-Man/ConA | (Kikkeri et al., 2008) |
| Lac | 9.0-18.1 nm (silole-core carbosilane) | Covalent (-SAc) | 6/compound | PNA; WGA | Fluorescence | 20 μM | Lac/PNA | (Hatano et al., 2009) |
| α-d-Man | N/A (Ruthenium-Amidoamine) | Covalent (-NH2) | 6-18/compound | GNA; ConA | Fluorescence | 25-347 nM; 25 nM | α-d-Man/GNA | (Kikkeri et al., 2009a) |
| α-d-Man | 0.7-1.6 nm (Gallic acid-triethylene glycol) | Covalent (-N3) | 3-27/compound | ConA | SPR | 12-24 nM | α-d-Man/ConA | (Munoz et al., 2009) |
| α-d-Man | N/A (Amidoamine) | Covalent (-NCS) | 16-172/compound | ConA | Fluorescence | 100 μg/mL | α-d-Man/ConA | (Schlick et al., 2009) |
| Lac; Neu5Acα2-6 Galβ1-4Glc | N/A (Tetraphenylethylene) | Covalent (-N3) | 4/compound | RCA120; SSA | Fluorescence | 20 μM | Lac/RCA120 | (Kato et al., 2010) |
| α-d-Man; β-d-Gal | N/A (Ruthenium-Amidoamine) | Covalent (-NH2) | 2-18/compound | ConA; Asialoglycoprotein; GNA | Fluorescence | 25-38 nM; 2.5 nM | α-d-Man/ConA | (Kikkeri et al., 2010a) |
| α-d-Man; β-d-Gal; d-Glc; maltose | N/A (Ruthenium-Amidoamine) | Covalent (-NH2) | 6-18/compound | ConA | Microarray | 2.5 nM | α-d-Man/ConA | (Kikkeri et al., 2010b) |
| d-Glc | N/A (Ethyleneglycol) | Covalent (-dichlorotriazine) | 12/compound | ConA | Confocal microscopy; fluorescence | 50-200 mg/dL | d-Glc/ConA | (Cummin et al., 2011) |
| α-d-Man | N/A (Ruthenium-CD) | Non-covalent (host-guest interaction) | 14-56/compound | ConA (high density); ConA (low density) | SPR | 0.138-4.573 μM; 0.1380 μM | α-d-Man/ConA (high density) | (Grünstein et al., 2011) |
| α-d-Man | 2 nm (Amidoamine) | Covalent (-NCS) | 10/compound | ConA; WGA | Fluorescence | 1 mg/mL | α-d-Man/ConA | (Bogdan et al., 2012) |
| α-d-Man | N/A (Amidoamine) | Covalent (-alkyne) | 4-16/com pound | ConA; BSA | ITC; DPV | 10 μM | α-d-Man/ConA | (Martos-Maldonado et al., 2013) |
| α-d-Man | N/A (Ether) | Covalent (-N3) | 9-81/compound | ConA | SPR | 1 pg/mm2 | α-d-Man/ConA | (Munoz et al., 2013) |
Abbreviations: Carbohydrates Ara, arabinose; CD, cyclodextrin; Dex, dextran; Fuc, fucose; Gal, galactose; Glc, glucose; GlcNAc, N-acetyl-D-glucosamine; Lac, lactose; Man, mannose; Mal, maltose; Suc, sucrose. Proteins BSA, bovine serum albumin; BS-I, bandeiraea simplicifolia lectin I; ConA, Concanavalin A; GNA, Galanthus nivilis agglutinin; GS-II, Griffonia simplicifolia II; HPA, Helix pomatia agglutinin; MAA, Macckia amurensi agglutinin; MAL, maackia amurensis; PA-IL, Pseudomonas aeruginosa I lectin; PA-IIL, Pseudomonas aeruginosa II lectin; PNA, peanut agglutinin; PSA, Pisum sativum agglutinin; PTA, Psophocarpus tetragonolobus agglutinin; RCA120, Ricinus communis agglutinin; SBA, soybean agglutinin; SNA, Sambucus nigra agglutinin; SSA, Sambucus sieboldiana agglutinin; UEA, Ulex europaeus agglutinin; VAA, Viscum album agglutinin; WGA, wheat germ agglutinin. Instruments DLS, dynamic light scattering; DPV, differential pulse voltammetry; ITC, isothermal titration calorimetry; QCM, quartz crystal microbalance; SPR, surface plasmon resonance spectroscopy.
Fig. 2.
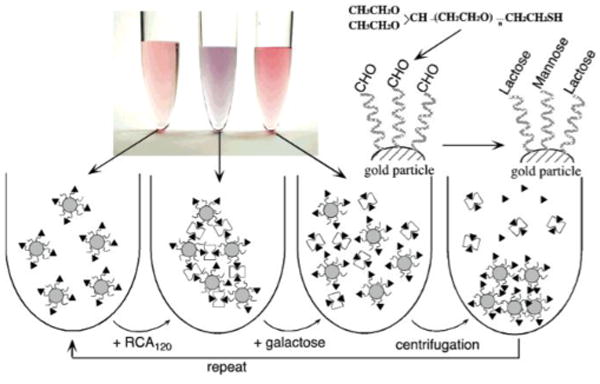
Gold glyconanoparticles for lectin recognition. Amino-terminated lactose was conjugated to the aldehyde-presenting gold nanoparticles by reductive amination. The resulting glyconanomaterials were subsequently applied to Ricinus communis (castor bean) agglutinin (RCA120), resulting in significant aggregation and color changes. Reproduced from (Otsuka et al., 2001) with permission from the American Chemical Society.
Figure 4.
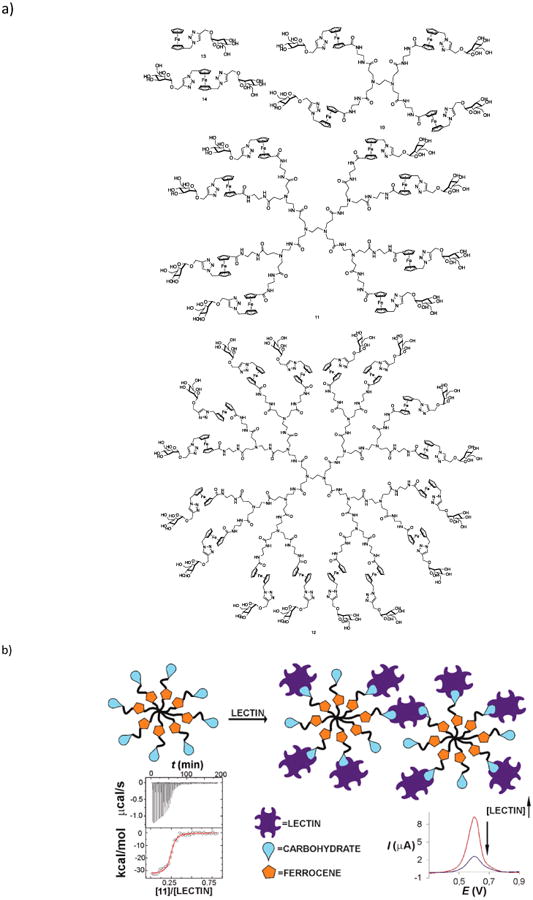
Electrochemical sensing of lectins using glycodendrimers. (a) Structures of mannosylated, ferrocene-containing, PAMAM-based glycodendrimers of different generations. (b) Differential pulse voltammetry showing decrease in the peak current upon binding of Man-dendrimer with ConA. Adapted from (Martos-Maldonado et al. 2013) with permission from American Chemical Society.
In addition, microarray technologies can be applied to several of the techniques. Multiple ligands are thus typically tethered to solid surfaces, and the parallel binding events with glyconanomaterials monitored. This technique is often preferred when high-throughput screening and rapid binding analysis are required.
4.2 Sensing microbes and cells
Infections by pathogens are often mediated by carbohydrate recognition at cell/virus particle surfaces (Fig. 1) (Finlay and Cossart, 1997). Multivalent lectin-carbohydrate interactions generate strong adhesive forces, and can in this context be used as an effective means of sensing and detecting pathogens. Thus, glyconanomaterials can enable rapid and sensitive detection of pathogen and toxins without time-consuming procedures, such as multiple incubations and washings, or use of nucleic acid amplification/detection. Optical spectroscopy and microscopy (UV-Vis, fluorescence), often combined with cytometry or staining procedures, are the most typically used. SPR and nuclear magnetic resonance technique (MRI) have also been employed. Literature survey of sensing and detection of microbes and cells using glyconanomaterials is tabulated in Table 2. Figure 5 shows an earlier example.
Table 2.
Summary of sensing and imaging microbes and cells in vitro using glyconanomaterials.
| Carbohydrate | Particle size/type |
Couplin g method s (Functi onal group) |
Carbohydra te density |
Microbe/ cell types |
Detection methods/ass ay |
Dete ction limit (/mL ) |
Selectivity | Referenc es |
|---|---|---|---|---|---|---|---|---|
| Gold glyconanomaterials | ||||||||
| Lac; β-d-Glc; maltose | 1.6-2.1 nm | Covalent (-SH) | hTERT-BJ1 cells | Fluorescence microscopy; optical microscopy | 1×104 | Lac/hTERT-BJ1 cell; β-d-Glc/hTERT-BJ1 cell | (De la Fuente et al., 2007) | |
| Dextran | 22±3 nm | One-pot (native) | E. coli 8739 | UV-Vis | 1×106 | Dextran/E. coli 8739 | (Nath et al., 2008) | |
| α-d-Man | 2.9 nm | Covalent (-SH) | E. coli K12 | Fluorescence | 7.2× 105 | α-d-Man/E. coli K12 | (Huang et al., 2009) | |
| β-d-Gal | 4 nm | Covalent (-SH) | ∼65/particle | Vero cells | Luminometry | 1×104 | β-d-Gal/Vero cell | (Kulkarni et al., 2010) |
| Polysialic acid; d-Man; d-Gal | 13 nm | Covalent (native) | 55/particle | BGC cells | Confocal microscopy; fluorescence | 210 | Polysialic acid/BGC cell | (Han et al., 2011b) |
| d-Glc | 5 nm | Covalent (-SH) | KB cells; A549 cells | Flow cytometry; confocal microscopy | 2×104 | Glc/KB cell | (Li et al., 2011) | |
| Lac | Covalent (-SH) | C33 cells | Confocal microscopy | Lac/C33 cell | (Gallo et al., 2012) | |||
| d-Man | 200 nm | Covalent (native) | E. coli ORN178; E. coli ORN208; E. coli 13762 | Fluorescence microscopy | Man/E. coli ORN178 | (Vedantam et al., 2012) | ||
| α-Neu5Ac | 16.4±1.6 nm | Covalent (-SH) | influenza virus H3N2; H5N1 | UV-Vis | 2.55 μg/mL | α-Neu5Ac/H3N2 | (Marín et al., 2013) | |
| d-Galf; d-Glc; d-Gal | 3 nm | Covalent (-SH) | Human monocyte -derived dendritic cells | Flow cytometry | 1×106 | d-Galf/dendritic cell | (Chiodo et al., 2014) | |
| β-d-Glc | 1.5 nm | Covalent (-SH) | HepG2 cells | Confocal microscopy; fluorescence correlation spectroscopy | 1×105 | β-d-Glc/HepG 2 cell | (Murray et al., 2014) | |
| Magnetic glyconanomaterials | ||||||||
| Vancomycin | 3-4 nm | Covalent (-NH2) | Staphylococcus aureus; S. epidermidis | Optical microscopy | 4 | Vancomycin/Both | (Gu et al., 2003a) | |
| Vancomycin | 4 nm | Covalent (-NH2) | E. coli | Optical microscopy | 15 | Vancomycin/E. coli | (Gu et al., 2003b) | |
| α-d-Man; β-d-Gal | 10-20 nm | Covalent (-N3/-NH2) | 300/particle | E. coli ORN178; E. coli ORN208 | Fluorescent microscopy | 104 | α-d-Man/E. coli ORN178 | (El-Boubbou et al., 2007) |
| α-d-Man | 1.6-6.6 nm | Covalent (native) | 30 wt% | rMSCs | MRI; Prussian blue staining | 31100 | α-d-Man/rMSCs | (Horak et al., 2007) |
| Hyaluronic acid | 40-50 nm | Non-covalent (native) | HEK293 cells; A549 cells | Fluorescence microscopy | 40000 | Hyaluronic acid/Both | (Kumar et al., 2007) | |
| β-d-Gal | ∼10 nm | Covalent (-amphiphile) | Hepatocytes | Confocal microscopy | 105 | β-d-Gal/Hepatocytes | (Yoo et al., 2007) | |
| α-d-Man; Lac; α-Neu5Ac | 2-3 μm | Covalent (-biotin) | E. coli ORN178; E. coli ORN208; J96; J96-PiIE; PT22ΔTox; B41; CFT073 | Optical microscopy | 105 | α-d-Man/E. coli ORN178 | (Hatch et al., 2008) | |
| Vancomycin | 10 nm | Covalent (-NH2) e | 9-12/particle | E. coli; E. faecalis; S. epidermidis; S. aureus | Microagglutination | Vancomycin/All | (Kell et al., 2008) | |
| Hyaluronic acid | 12±0.1 nm | Non-covalent (native) | HCT116 cells; NIH3T3 cells | MRI | 2×105 | Hyaluronic acid/Both | (Lee et al., 2008) | |
| α-d-Man | ∼10 nm | Covalent (native) | Macrophage cells | MRI | α-d-Man/Macrophage cells | (Yoo et al., 2008) | ||
| Chitosan | 6-10 nm | Covalent (-COOH) | hMSCs | MRI; Prussian blue staining | 40 | Chitosan/hMSCs | (Shi et al., 2009) | |
| α-d-Man; β-d-Gal; β-L-Fuc; β-Neu5Ac; β-d-GlcNAc | 6 nm | Covalent (-N3/-NH2) | 8 wt% | 184B5; A498; A549; HT29; SKOV-3; B16-F10; B16-F1; MCF-7; TA3-HA; TA3-ST cells | MRI; Prussian blue staining | 105 | β-Fuc/A549&HT29; α-d-Man/184B 5&MCF-7; β-d-Gal/B16-F 10&MCF-7; β-Neu5Ac/All; β-d-GlcNAc/S KOV-3 | (El-Boubbou et al., 2010) |
| Hyaluronic acid | 6 nm | Covalent (native) | THP-1 cells; EA.hy926 cells; LNCaP cells | MRI; Prussian blue staining; flow cytometry; confocal microscopy | 4×105 | Hyaluronic acid/THP-1 cell | (Kamat et al., 2010) | |
| Galα1-4Gal | 250 nm | Covalent (-N3) | Streptococcus suis bacteria | Luminescence | 105 | Galα1-4Gal/Streptococcus suis | (Pera et al., 2010) | |
| Lac | 9 nm | Covalent (-SH) | ∼60/particl e | Peripheral blood mononuclear cells | MRI; fluorescence microscopy | 105 | Lac/mononuclear cell | (Gallo et al., 2011) |
| Lac | 6 nm | Covalent (-SH) | C33 cells; Raji cells | MRI; flow cytometry; fluorescence microscopy | 5×103 | Lac/C33 cell | (García et al., 2011) | |
| β-d-Gal | 14 nm | Covalent (-SH) | A549 cells | Epifluorescence microscopy; confocal microscopy | 104 | β-d-Gal/A549 cell | (Pfaff et al., 2011) | |
| Hyaluronic acid | 5 nm | Covalent (native) | 44 wt% | SKOV-3 cells | MRI; confocal microscopy | 2×105 | Hyaluronic acid/SKOV-3 cell | (El-Dakdouki et al., 2012) |
| Carbon glyconanomaterials | ||||||||
| β-d-Gal; α-d-Man | ∼20 nm (SWNTs) | Covalent (-NH2) | E. coli O157:H7 | Optical microscopy | β-d-Gal/E. coli O157:H7 | (Gu et al., 2005) | ||
| α-d-Gal; β-d-Gal | 65-70 nm (SWNTs) | Non-covalent (-lipid) | 10-25 nm thickness | CHO cells | Fluorescence microscopy; flow cytometry | α-d-Gal/CHO cell | (Chen et al., 2006) | |
| d-GlcN | 20 nm (SWNTs) | Covalent (-NH2) | 3T3 fibroblasts cells | Optical microscopy | 104 | d-GlcN/3T3 cell | (Nimmagadda et al., 2006) | |
| β-d-Gal; α-d-Man | N/A (SWNTs) | Covalent (-NH2) | B. anthracis spores | Optical microscopy | α-d-Man/B. anthracis spores | (Wang et al., 2006) | ||
| β-d-Gal; α-d-Man | ∼20 nm (SWNTs) | Covalent (-NH2) | 37-45 wt%; 35-47 wt% | E. coli O157:H7; B. subtilis spores | Fluorescence microscopy; optical microscopy | 5×107 | β-d-Gal/E. coli O157:H7; α-d-Man/B. anthracis spores | (Gu et al., 2008) |
| α-d-Man; β-d-Gal | 5-10 nm (SWNTs) | Non-covalent (-pyrene) | CHO cells | Fluorescence microscopy | α-d-Man/CHO cell | (Wu et al., 2008) | ||
| d-GlcNAc; β-d-Glc; α-d-Man | 10-30 nm (SWNTs) | Non-covalent (-pyrene) | PC12 cells | Semiconductor device analyzer | d-GlcNAc/PC 12 cell | (Sudibya et al., 2009) | ||
| d-Man; d-Glc | N/A (Diamond) | Covalent (-alkene /-NH2) | E. coli PKL1162 | Agglutination-filtration assay | 37 | d-Man/E. coli PKL1162 | (Hartmann et al., 2012) | |
| α-d-Man; β-d-Gal | 1 nm (Fullerene) | Covalent (-N3) | 12-36/particle | Ebola pseudovirus | 5000 | α-d-Man/Ebola pseudovirus | (Luczkowiak et al., 2013) | |
| QD glyconanomaterials | ||||||||
| Cellobiose; Lac; Maltose Maltoheptose | 5 nm (CdSe) | Non-covalent (-amphiphile) | HeLa cells | Fluorescence microscopy | Cellobiose/HeLa cell | (Osaki et al., 2004) | ||
| β-d-GlcNAc | 5 nm (CdSe/ZnS) | Covalent (-SH) | 210/particle | Mouse sperm; pigsperm; sea-urchin sperm | Confocal microscopy; flow cytometry | 2.7× 106 | β-d-GlcNAc/All | (Robinson et al., 2005) |
| d-GlcNAc; β-d-Gal; α-d-Man | 12 nm (CdTe) | Covalent (-SH) | HeLa cells | Confocal microscopy | 105 | d-GlcNAc/HeLa cell | (Niikura et al., 2007) | |
| Chitosan | 29 nm (InGaP/ZnS) | Covalent (native) | PC12 cells | Flow cytometry; Explore Optix imaging | Chitosan/PC12 cell | (Sandros et al., 2007) | ||
| β-d-Gal | 5±0.5 nm (CdSe/ZnS) | Covalent (-N3) | A549; H467; HeLa; COS-7; CL1-1 cells | Confocal microscopy | β-d-Gal/A549 cell | (Chen et al., 2008) | ||
| d-Man | 20-30 μm (Qdot® 655) | Non-covalent (-PEG) | Macrophage cells | Fluorescence microscopy | 1.3 × 105 | d-Man/Macrophage cell | (Higuchi et al., 2008) | |
| Maltotriose; panose; maltose | 12 nm (CdTe) | Covalent (-SH) | 28/particle | HeLa cells | Confocal microscopy | Maltotriose/HeLa cell | (Niikura et al., 2008) | |
| Hyaluronic acid | 5.7 nm (CdSe/CdS/Z nS) | Non-covalent (native) | HeLa cells; Human dermal fibroblast cells | Fluorescence microscopy | 104 | Hyaluronic acid/HeLa cell | (Bhang et al., 2009) | |
| β-d-Gal; α-d-Man | 15-20 nm (CdSe/ZnS) | Covalent (-COOH) | HepG2 cells | Flow cytometry; fluorescence microscopy | β-d-Gal/HepG2 cell | (Kikkeri et al., 2009b) | ||
| α-d-Man | 15 nm (CdS) | Covalent (-SH) | E. coli ORN178; E. coli ORN208 | Confocal microscopy | 104 | α-d-Man/E. coli ORN178 | (Mukhopadhyay et al., 2009) | |
| β-d-Gal; β-d-Man; β-d-Glc | 2.5 nm (CdTe) | Non-covalent (native) | Saccharomyces cerevisiae; Kluyveromyces bulgaricus | Epifluorescence microscopy; Optical microscopy | β-d-Gal/Saccharomyces cerevisiae; β-Man/Kluyveromyces bulgaricus | (Coulon et al., 2010) | ||
| Hyaluronic acid | 42.3 nm (Qdot® 800) | Covalent (-NH2) | HepG2; HSC-T6; FL83B cells | Flow cytometry; confocal microscopy; immunofluorescence staining | 3×104 | Hyaluronic acid/HepG2 | (Kim et al., 2010) | |
| Lac | 4.5±0.5 nm (CdSeS/ZnS) | Covalent (-SH) | 134-140/particle | HUVEC cells | SPR; fluorescence microscopy | Lac/HUVEC cell | (Yang et al., 2010a) | |
| β-d-Gal; β-Lac | 4.5±0.5 nm (CdSeS/ZnS) | Covalent (-SH) | 35.3-48.7 wt% | HepG2 cells | Flow cytometry; fluorescence microscopy | 105 | β-d-Gal/HepG2 cell | (Yang et al., 2010b) |
| Mannan | 3.1 nm (CdS) | Covalent (-NH2) | BGC cells | Flow cytometry | 1.2× 103 | Mannan/BGC cell | (Han et al., 2011a) | |
| β-d-Gal | 4 nm (CdS) | Covalent (-NH2) | 2-10 wt% | HepG2; MCF-7 cells | Fluorescence; confocal microscopy | β-d-Gal/HepG2 cell | (Cai et al., 2012) | |
| Silica glyconanomaterials | ||||||||
| β-d-Gal | 60±5 nm (solid) | Covalent (-COOH) | 5.045 wt% | Liver cancer cells; MCF-7 cells; blood cells | Fluorescence microscopy; flow cytometry | β-d-Gal/Liver cancer cell | (Peng et al., 2007) | |
| α-d-Man | ∼100 nm (solid and mesoporous) | Covalent (-Diethyl squarate) | 0.18 mmol/g | MDA-MB-231 cells | Confocal microscopy | α-d-Man/MDA-MB-231 cell | (Hocine et al., 2010) | |
| α-d-Man | 118 nm (mesoporous) | Covalent (-Diethyl squarate) | MCF-7; MDA-MB-231; HCT-116 cells | Optical microscopy | α-d-Man/MCF-7& MDA-MB-231 cell | (Gary-Bobo et al., 2011) | ||
| d-Gal; d-Glc | 54±4 nm (solid) | Covalent (-alkynyl) | -0.6 μmol/mg | Oligodendrocytes | Confocal microscopy | d-Gal/Oligodendrocytes | (Zhao et al., 2012) | |
| β-d-Man | ∼6 nm (solid) | Covalent (-COOH) | MCF-7 cells | Fluorescence microscopy | β-d-Man/MCF-7 cell | (Ahire et al., 2013) | ||
| d-Maltoheptaose; β-CD; d-Man | 81.2±7.3 nm (solid) | Photocoupling (native) | 11517-68623/particle | E. coli | Confocal microscopy | d-Maltoheptaose/E. coli | (Jayawardena et al., 2013a) | |
| α-d-Man | 148-161 nm (solid) | Covalent (-Diethyl squarate) | 1.95 mmol/g | MDA-MB-231 cells | Confocal microscopy | α-d-Man/MDA-MB-231 cell | (Perrier et al., 2013) | |
| Vancomycin | 90±37 nm (mesoporous) | Covalent (-COOH) | 50 wt% | S. aureus; RAW 264.7 cells; E. coli | Confocal microscopy | 105 | Vancomycin/S. aureus& RAW 264.7 cell | (Qi et al., 2013) |
| d-Tre; β-CD; d-Glc; d-Maltoheptaose | 42.1±1.9 nm (solid) | Photocoupling (native) | 7.25-16.0 × 10-16 μg/nm2 | M. smegmatis strain mc2 155 | Confocal microscopy | d-Tre/M. smegmatis strain mc2 155 | (Jayawardena et al., 2015) | |
| Dendrimer glyconanomaterials | ||||||||
| Galα1-4Gal | N/A (Benzenedimethanethiol) | Covalent (-NH2) | 1-4/compound | Streptococcus suis | Hemagglutination | 108 | Galα1-4Gal/Streptococcus suis | (Hansen et al., 1997) |
| α-d-Man | N/A (Amide series) | Covalent (-NH2) | 2-16/compound | E. coli K12 | Hemagglutination | 108 | α-d-Man/E. coli K12 | (Nagahori et al., 2002) |
| α-d-Man | N/A (BoltornH30) | Covalent (-NH2) | 32/compound | Dendritic cells; Ebola virus | α-d-Man/Dendritic cell& Ebola virus | (Lasala et al., 2003) | ||
| Galα1-4Gal | N/A (Amidoamine) | Covalent (-COOH) | 8/compound | Streptococcus suis | Hemagglutination; SPR | Galα1-4Gal/Streptococcus suis | (Joosten et al., 2004) | |
| d-GlcNAc | N/A (Amidoamine) | Covalent (-NCS) | 8/compound | Mononuclear cells | Fluorescence microscopy; confocal microscopy | d-GlcNAc/Mononuclear cells | (Krist et al., 2004) | |
| d-GlcN; glucosamine 6-sulfate | N/A (Amidoamine) | Covalent (native) | 14% | Dendritic cells; monocyte-derived macrophages | Trypan blue exclusion; hemagglutination | 106 | d-GlcN/Both | (Shaunak et al., 2004) |
| Galα1-4Gal; d-Man | N/A (Amidoamine) | Covalent (-COOH) | 1-8/compound | E. coli HB101; T24 cells | SPR; hemagglutination | Galα1-4Gal/Both | (Salminen et al., 2007) | |
| α-d-Man; mannan | N/A (Tetra-compound) | Covalent (-N3/alkyne) | 4/compound | E. coli UTI89 | Hemagglutination; SPR | α-d-Man/E. coli UTI89 | (Touaibia et al., 2007) | |
| Galα1-4Gal | N/A (Phenolic acid) | Covalent (-N3) | 1-8/compound | Streptococcus suis | Hemagglutination | Galα1-4Gal/Streptococcus suis | (Branderhorst et al., 2008) | |
| d-GlcNAc | N/A (Amidoamine) | Covalent (-NCS) | 8/compound | Mononuclear cells | Flow cytometry | d-GlcNAc/Mononuclear cells | (Hulikova et al., 2009) | |
| Neu5Acα2-6 Galβ1-4Glc; Lac | N/A (Tetraphenylethylene) | Covalent (-N3) | 4/compound | Influenza virus A/WSN/33 | Fluorescence | 5×104 | Neu5Acα2-6Galβ1-4Glc /Influenza virus A/WSN/33 | (Kato et al., 2010) |
| α-d-Man | N/A (Ruthenium-CD) | Non-covalent (host-guest interaction) | 14-56/compound | E. coli ORN178; E. coli ORN208 | Confocal microscopy | α-d-Man/E. coli ORN178 | (Grünstein et al., 2011) | |
| d-GlcNAc | N/A (Amidoamine) | Covalent (-NCS) | 8/compound | Mononuclear cells | Flow cytometry | 106 | d-GlcNAc/Mononuclear cell | (Hulikova et al., 2011) |
Fig. 5.
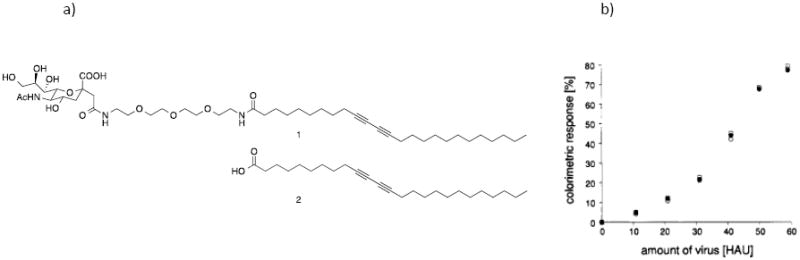
(a) Sialic acid-presenting glycolipid formed glycoliposome which was then crosslinked by irradiation to give a blue- or purple-colored liposome. (b) When influenza virus was added to the liposome, the solution changed to a pink or orange color. The colorimetric response was quantified by measuring the percent change in the absorption at 626 nm relative to the total absorption maxima, and the response increased with the amount of influenza virus added. Adapted from (Reichert et al. 1995) with permission from American Chemical Society.
As cellular surfaces are rich in carbohydrates, it is conceivable that glyconanomaterials could serve as cell mimics and interact with different biological entities. Compared to monovalent ligands, multivalent glyconanomaterials bind to cell receptors with greater avidity and specificity, and could lead to fine-tuned sensing combined with modern single particle- and cell detection techniques. The cell status could thus be analyzed more precisely and efficiently, providing in-depth understanding of the interactions between glyconanomaterials and cells. Among different sensing systems, UV-Vis- and fluorescence spectroscopy/microscopy, SPR, and QCM, are for example amenable to quantitative analysis of the binding performance of glyconanomaterials to cell surfaces. Effective nanoprobes that can detect, image, and profile microbes and cells will not only aid the understanding of the roles carbohydrates played in disease process, but also the development of new theranostic tools in disease prevention and treatment.
4.3 Tissue- and in vivo- sensing/imaging
Early examples of glyconanomaterials research have focused on carbohydrate-mediated in vitro interactions of proteins, viruses and cells. Recent development has however emerged where carbohydrate ligands are used as targeting entities to direct the nanomaterials to receptor sites in vivo for imaging and tracking specific cells, tissues, and organs based on the selective carbohydrate/protein-carbohydrate interactions. Similar to sensing and imaging of cells, fluorescence spectroscopic/microscopic techniques are the most commonly adopted. In addition, typical medical imaging methods, such as PET and MRI are also applied, where the glyconanomaterials can serve as contrast agents.
In 2004, an early in vivo study was reported to show that glyconanomaterials could behave as anti-adhesion agents against progression of lung metastasis in mice (Rojo et al., 2004) (Fig. 6). Table 3 summarizes examples of detecting and imaging specific disease states in animals using gold-, magnetic-, and QD-based glyconanomaterials. These results highlight the potential of glyconanomaterials for disease diagnostics and eventually as therapeutics to combat infection and cancer. The materials constitute in this sense a particular promising in vivo sensing and imaging platform, relatively easily modified to display high biocompatibilities, and avoiding immune responses and nonspecific interactions (García et al., 2015). However, when designing glyconanomaterials as theranostic platforms in vivo, clearance of the materials prior to reaching the therapeutic targets, and enzymatic degradation are important factors that need to be taken into consideration and more studies are warranted in this respect.
Fig. 6.
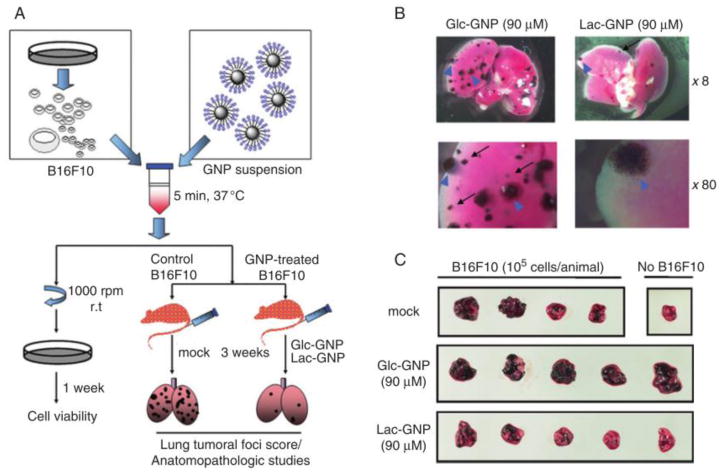
Antimetastatic effect of lactose-functionalized gold glyconanoparticles (GNPs) on lung tumor development in mice. (A) Evaluation of nanomaterials antimetastatic performance. (B) Lung images displaying lung tumoral foci. (C) Mouse lung tissue treated with murine melanoma cells (B16F10) and GNPs. Reproduced from (Rojo et al., 2004) by permission of Wiley-VCH.
Table 3.
Tissue- and in vivo- sensing/imaging using gold-, magnetic-, and QD-glyconanomaterials.
| Carbohydrate | Particle size/type |
Coupli ng method s (Functi onal group) |
Carbohy drate density |
Animal type |
Detection methods/a ssay |
Target | Selectivity | Referenc es |
|---|---|---|---|---|---|---|---|---|
| Gold glyconanomaterials | ||||||||
| Lac; maltose; β-d-Glc | < 2 nm | Covalent (-SH) | 70/particle | C57BL/6 mice | Histological analysis | Lung tumor | Lac/Lung tumor | (Rojo et al., 2004) |
| β-d-Gal | 50-150 nm | Covalent (-SH) | 4/nm2 | C57BL/6 (female) | Instrumental neutron activation technique | Liver | β-d-Gal/Liver | (Bergen et al., 2006) |
| Hyaluronic acid | 16 nm | Covalent (-SH) | 30.9±1.7/particle | DBA-1J mice; BALB/c nude mice | Near-infrared fluorescent imaging | Arthritis; tumor | Hyaluronic acid/Both | (Lee et al., 2008) |
| β-d-Glc; Lac; β-d-Gal | 1.9-4.4 nm | Covalent (-SH) | C57BL/6 | MRI | Brain tumor | β-d-Glc/Brain tumor | (Marradi et al., 2009) | |
| Heparin; d-Glc | 14±4 nm | Covalent (-NH2) | Chick embryos; C57BL/6 NCr (male) | Stereomicroscopy | Chorioallantoic membrane | Heparin/Chorioallantoic membrane | (Kemp et al., 2009) | |
| Sialyl LewisX | 18 nm | Covalent (-NH2) | 10.5±1.2 nmol/mg | C57BL/6 (male) | MRI; Prussian blue staining | Brain; spleen; liver | Sialyl LewisX/All | (Farr et al., 2014) |
| β-d-Glc | 1.8-3.2 nm | Covalent (-SH) | 59-174 | Sprague– Dawley rats (male) | PET/CT | Brain | β-d-Glc/Brain | (Frigell et al., 2014) |
| Magnetic glyconanomaterials | ||||||||
| β-d-Gal | ∼10 nm | Covalent (-amphiphile) | Rats (SD, female) | MRI | Liver | β-d-Gal/Liver | (Yoo et al., 2007) | |
| Sialyl LewisX; β-d-GlcNAc; LacNAc; Neu5Acα2-3Gal β1-4GlcNAc | ∼1 μm | Covalent (-CN) | 105-107/particle | Wistarrats (male) | MRI | Brain | Sialyl LewisX/Brain | (van Kasteren et al., 2008) |
| Hyaluronic acid | 6 nm | Covalent (native) | 43 wt% | C57BL/6 mice | MRI; Prussian blue staining | Liver; kidney | Hyaluronic acid/Both | (El-Dakdouki et al., 2011) |
| Lac | 4 nm | Covalent (-COOH) | FVB mice; C57B1/6 mice | Stereomicroscopy; confocal microscopy; MRI | Brain tumor | Lac/Brain tumor | (Elvira et al., 2012) | |
| β-CD | 6 nm | Covalent (native) | 19 wt% | Atherosclerotic rabbit | Optical microscopy; MRI; Prussian blue staining | Aorta tissues | β-CD/Aorta tissues | (Li et al., 2012) |
| Neu5Ac | 5 nm | Covalent (-COOH) | 4 wt% | C57BL/6 mice | MRI; Prussian blue staining | Brain | Neu5Ac/Brain | (Kouyoumdjian et al., 2013) |
| Hyaluronic acid | 6 nm | Covalent (native) | 46 wt% | Atheroscl erotic rabbit | MRI; optical microscopy; Prussian blue staining | Aorta tissues | Hyaluronic acid/Aorta tissues | (El-Dakdouki et al., 2014) |
| Lac | 4 nm | Covalent (-COOH) | Mice | MRI; flow cytometry | Brain tumor | Lac/Brain tumor | (Elvira et al., 2015) | |
| QD glyconanomaterials | ||||||||
| Chitosan | 29 nm (InGaP/ZnS) | Covalent (native) | Mice | Flow cytometry; Near-infrared fluorescent imaging | Brain | Chitosan/Brain | (Sandros et al., 2007) | |
| Hyaluronic acid | 5.7 nm (CdSe/CdS/ZnS) | Non-covalent (native) | Nude mice | Fluorescence microscopy | Ear | Hyaluronic acid/Ear | (Bhang et al., 2009) | |
| β-d-Gal; α-d-Man | 15-20 nm (CdSe/ZnS) | Covalent (-COOH) | C57BL/6 mice (female) | Flow cytometry; fluorescence microscopy | Liver | β-d-Gal/Liver | (Kikkeri et al., 2009b) | |
| Hyaluronic acid | 42.3 nm (Qdot® 800) | Covalent (-NH2) | Balb/c mice (female) | Flow cytometry; Confocal microscopy; luminescent imaging | Liver; Spleen; kidney | Hyaluronic acid/Liver | (Kim et al., 2010) | |
| α-Neu5Ac; Lac; Neu5Acα2-3Galβ1-4GlcNAc | 5.8-9.3 nm (CdSe/Z nS) | Covalent (-SH) | Mice | Near-infrared fluorescent imaging | Liver; brain; heart; stomach; spleen kidney; bladder | Neu5Acα2-3Galβ1-4GlcNAc/Liver | (Ohyanagi et al., 2011) |
5. Summary and perspective
Clearly, the merge of nanotechnology with glycoscience has resulted in a wide range of important new applications, especially during the past decade. The emerging field of glyconanomaterials has witnessed rapid growth, and already shown strong potential in sensing and detection. A rich variety of glyconanomaterials has furthermore been developed, taking advantage of the different physicochemical properties of specific structures. This has had special impact on the biosensor area, where these materials are now used as useful sensing platforms.
Essential to glyconanomaterials synthesis is the coupling chemistries that can yield efficient conjugation of carbohydrates to nanomaterials. Either non-covalent or covalent approaches have been applied, resulting in functionalized nanomaterials that take advantage of their unique intrinsic properties. The nanomaterials furthermore result in multivalent presentation of the carbohydrate entities at their surfaces, thereby in a sense mimicking certain cells and virus particles. This feature often leads to dramatically increased affinities between the materials and the target receptors, with large impact on the sensing performance of the glyconanomaterials.
Until now, the carbohydrate displays developed have been fairly simple, especially in comparison to the complex carbohydrate-coating (sometimes called the glycocalyx) of different cells. New strategies are thus still needed, particularly regarding the synthesis of glyconanomaterials with high carbohydrate diversity and more sophisticated carbohydrate display, where the ligand density, spatial arrangement, and accessibility can be precisely controlled. This will ultimately result in detailed modulation of the affinities and specificities of the glyconanomaterials to suit different theranostic and biosensing requirements.
Nevertheless, a multitude of successful sensing applications using glyconanoparticles have been demonstrated, many of which mentioned in this review. Many different entities have thus been targeted, both in vitro and in vivo, ranging from discrete carbohydrate-binding proteins, such as lectins, through viruses, bacteria and mammalian cells, to sensing of tissues in live organisms. This development is in rapid progress, leading to sensing and imaging of specific binding partners and eventually to monitor various disease loci and states. Although such biomedical applications are of very high potential, the biodistribution, clearance, and biocompatibility of the glyconanomaterials need to be established for in vivo sensing. This development, together with the progress in glycoscience and glycobiology, will result in the realization of fine-tuned glyconanomaterials for efficient biosensing, diagnostics and therapeutics applications.
Fig. 3.
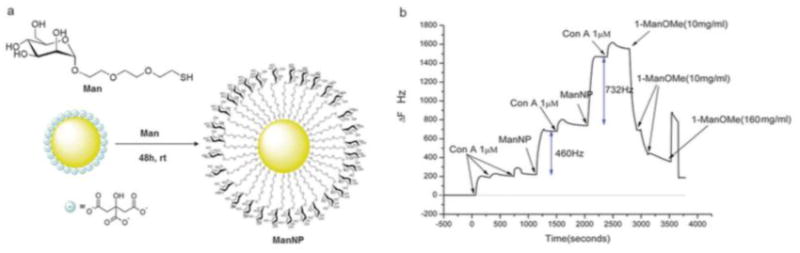
QCM sensor using gold glyconanoparticles. (a) Glyconanoparticles were prepared by treating gold nanoparticles with thiolated mannose. (b) Gold-plated QCM sensors were coated with mannan, to which ConA was adsorbed. The Man-conjugated gold nanoparticles were subsequently introduced, and bound to the protein surface through multivalent interactions. Repeated administration of protein and glyconanoparticles resulted in multilayer formation. Reproduced from (Mahon et al., 2013) by permission from the Royal Society of Chemistry.
Highlights.
Glyconanomaterials have witnessed rapid development in the past decade
A wide variety of glyconanomaterials have been developed and synthesized
Glyconanomaterials have demonstrated high potential in sensing and imaging proteins, microbes, and cells in vitro and in vivo
Acknowledgments
This work was in part supported by the National Institutes of Health (R01GM080295 and R21AI109896, to M.Y.), the Royal Institute of Technology, and the People Programme (Marie Curie Actions) of the European Union's Seventh Framework Programme FP7/2007-2013/under REA grant agreement no 264645.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Olof Ramström, Email: ramstrom@kth.se.
Mingdi Yan, Email: mingdi_yan@uml.edu.
References
- Adak AK, Li BY, Lin CC. Carbohydr Res. 2014;405:2–12. doi: 10.1016/j.carres.2014.07.026. [DOI] [PubMed] [Google Scholar]
- Ahire JH, Chambrier I, Mueller A, Bao YP, Chao YM. ACS Appl Mater Interfaces. 2013;5:7384–7391. doi: 10.1021/am4017126. [DOI] [PubMed] [Google Scholar]
- Al-Bataineh Sa, Luginbuehl R, Textor M, Yan M. Langmuir. 2009;25:7432–7437. doi: 10.1021/la900334w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alivisatos P. Nat Biotechnol. 2004;22:47–52. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- Ashton P, Boyd S. Angew Chem Int Ed. 1997;36:421–424. [Google Scholar]
- Aslan K, Zhang J, Lakowicz JR, Geddes CD. J Fluorsc. 2004;14:391–400. doi: 10.1023/b:jofl.0000031820.17358.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assali M, Cid JJ, Fernández I, Khiar N. Chem Mater. 2013;25:4250–4261. [Google Scholar]
- Astruc D, Boisselier E, Ornelas C. Chem Rev. 2010;110:1857–1959. doi: 10.1021/cr900327d. [DOI] [PubMed] [Google Scholar]
- Aykaç A, Martos-Maldonado MC, Casas-Solvas JM, Quesada-Soriano I, García-Maroto F, García-Fuentes L, Vargas-Berenguel A. Langmuir. 2014;30:234–242. doi: 10.1021/la403454p. [DOI] [PubMed] [Google Scholar]
- Babu P, Sinha S, Surolia A. Bioconjugate Chem. 2007;18:146–151. doi: 10.1021/bc060204q. [DOI] [PubMed] [Google Scholar]
- Ban ZH, Bosques CJ, Sasisekharan R. Organ Biomol Chem. 2008;6:4290–4292. doi: 10.1039/b813210k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baradel N, Fort S, Halila S, Badi N, Lutz JF. Angew Chem Int Ed. 2013;52:2335–2339. doi: 10.1002/anie.201209052. [DOI] [PubMed] [Google Scholar]
- Barrientos AG, Fuente JMDLa, Jiménez M, Solís D, Cañada FJ, Martín-Lomas M, Penadés S. Carbohydr Res. 2009;344:1474–1478. doi: 10.1016/j.carres.2009.04.029. [DOI] [PubMed] [Google Scholar]
- Bergen JM, von Recum Ha, Goodman TT, Massey AP, Pun SH. Macromol Biosci. 2006;6:506–516. doi: 10.1002/mabi.200600075. [DOI] [PubMed] [Google Scholar]
- Bernardi A, Jiménez-Barbero J, Casnati A, De Castro C, Darbre T, Fieschi F, Finne J, Funken H, Jaeger KE, Lahmann M, Lindhorst TK, Marradi M, Messner P, Molinaro A, Murphy PV, Nativi C, Oscarson S, Penadés S, Peri F, Pieters RJ, Renaudet O, Reymond JL, Richichi B, Rojo J, Sansone F, Schäffer C, Turnbull WB, Velasco-Torrijos T, Vidal S, Vincent S, Wennekes T, Zuilhof H, Imberty A. Chem Soc Rev. 2013;42:4709–4727. doi: 10.1039/c2cs35408j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhang SH, Won N, Lee T, Jin H, Nam J, Park J, Chung H, Park H, Sung Y, Hahn SK, Kim BS, Kim S. ACS Nano. 2009;3:1389–1398. doi: 10.1021/nn900138d. [DOI] [PubMed] [Google Scholar]
- Boas U, Heegaard P. Chem Soc Rev. 2004;33:43–63. doi: 10.1039/b309043b. [DOI] [PubMed] [Google Scholar]
- Bogdan N, Roy R, Morin M. RSC Adv. 2012;2:985–991. [Google Scholar]
- Bosman AW, Janssen HM, Meijer EW. Chem Rev. 1999;99:1665–1688. doi: 10.1021/cr970069y. [DOI] [PubMed] [Google Scholar]
- Branderhorst HM, Kooij R, Salminen A, Jongeneel LH, Arnusch CJ, Liskamp RMJ, Finne J, Pieters RJ. Org Biomol Chem. 2008;6:1425–1434. doi: 10.1039/b800283e. [DOI] [PubMed] [Google Scholar]
- Cai XJ, Li XH, Liu YW, Wu GN, Zhao YC, Chen F, Gu ZW. Pharm Res. 2012;29:2167–2179. doi: 10.1007/s11095-012-0745-1. [DOI] [PubMed] [Google Scholar]
- Chabre YM, Roy R. Adv Carbohydr Chem Biochem. 2010;63:165–393. doi: 10.1016/S0065-2318(10)63006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CT, Munot YS, Salunke SB, Wang YC, Lin RK, Lin CC, Chen CC, Liu YH. Adv Funct Mater. 2008;18:527–540. [Google Scholar]
- Chen QS, Wei WL, Lin JM. Biosens Bioelectron. 2011;26:4497–4502. doi: 10.1016/j.bios.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Chen X, Lee GS, Zettl A, Bertozzi CR. Angew Chem Int Ed. 2004;43:6111–6116. doi: 10.1002/anie.200460620. [DOI] [PubMed] [Google Scholar]
- Chen X, Ramström O, Yan M. Nano Res. 2014;7:1381–1403. doi: 10.1007/s12274-014-0507-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Tam UC, Czlapinski JL, Lee GS, Rabuka D, Zettl A, Bertozzi CR. J Am Chem Soc. 2006;128:6292–6293. doi: 10.1021/ja060276s. [DOI] [PubMed] [Google Scholar]
- Chen YF, Ji T, Rosenzweig Z. Nano Lett. 2003;3:581–584. [Google Scholar]
- Chen YJ, Chen SH, Chien YY, Chang YW, Liao HK, Chang CY, Jan MD, Wang KT, Lin CC. ChemBioChem. 2005;6:1169–1173. doi: 10.1002/cbic.200500023. [DOI] [PubMed] [Google Scholar]
- Chen YN, Star A, Vidal S. Chem Soc Rev. 2013;42:4532–4542. doi: 10.1039/c2cs35396b. [DOI] [PubMed] [Google Scholar]
- Chen YN, Vedala H, Kotchey GP, Audfray A, Cecioni S, Imberty A, Vidal S, Star A. ACS Nano. 2012;6:760–770. doi: 10.1021/nn2042384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien YY, Jan MD, Adak AK, Tzeng HC, Lin YP, Chen YJ, Wang KT, Chen CT, Chen CC, Lin CC. ChemBioChem. 2008;9:1100–9. doi: 10.1002/cbic.200700590. [DOI] [PubMed] [Google Scholar]
- Chiodo F, Marradi M, Park J, Ram AFJ, Penadés S, van Die I, Tefsen B. ACS Chem Biol. 2014;9:383–9. doi: 10.1021/cb4008265. [DOI] [PubMed] [Google Scholar]
- Chuang YJ, Zhou XC, Pan ZW, Turchi C. Biochem Biophys Res Commun. 2009;389:22–27. doi: 10.1016/j.bbrc.2009.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon J, Thouvenin I, Aldeek F, Balan L, Schneider R. J Fluoresc. 2010;20:591–597. doi: 10.1007/s10895-009-0590-8. [DOI] [PubMed] [Google Scholar]
- Crocker PR, Paulson JC, Varki A. Nat Rev Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- Cummin BM, Lim J, Simanek EE, Pishko MV, Coté GL. Biomed Opt Exp. 2011;2:1243–1257. doi: 10.1364/BOE.2.001243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Fuente M, Alcántara D, Penadés S. IEEE Trans NanoBioscience. 2007;6:275–281. doi: 10.1109/tnb.2007.908981. [DOI] [PubMed] [Google Scholar]
- De la Fuente M, Barrientos AG, Rojas TC, Rojo J, Cañada J, Fernández A, Penadés S. Angew Chem Int Ed. 2001;40:2257–2261. [PubMed] [Google Scholar]
- De la Fuente M, Penadés S. Tetrahedron Asymmetry. 2005;16:387–391. [Google Scholar]
- Dube DH, Bertozzi CR. Nat Rev Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- El-Boubbou K, Gruden C, Huang XF. J Am Chem Soc. 2007;129:13392–13393. doi: 10.1021/ja076086e. [DOI] [PubMed] [Google Scholar]
- El-Boubbou K, Huang XF. Curr Med Chem. 2011;18:2060–2078. doi: 10.2174/092986711795656144. [DOI] [PubMed] [Google Scholar]
- El-Boubbou K, Zhu DC, Vasileiou C, Borhan B, Prosperi D, Li W, Huang XF. J Am Chem Soc. 2010;132:4490–4499. doi: 10.1021/ja100455c. [DOI] [PubMed] [Google Scholar]
- El-Dakdouki MH, El-Boubbou K, Kamat M, Huang RP, Abela GS, Kiupel M, Zhu DC, Huang XF. Pharm Res. 2014;31:1426–1437. doi: 10.1007/s11095-013-1021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Dakdouki MH, El-Boubbou K, Zhu DC, Huang X. RSC Adv. 2011;1:1449–1452. doi: 10.1039/C1RA00737H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Dakdouki MH, Zhu DC, El-Boubbou K, Kamat M, Chen JJ, Li W, Huang XF. Biomacromolecules. 2012;13:1144–1151. doi: 10.1021/bm300046h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvira G, García I, Benito M, Gallo J, Desco M, Penadés S, Garcia-Sanz JA, Silva A. PLoS One. 2012;7:e44466. doi: 10.1371/journal.pone.0044466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvira G, García I, Gallo J, Benito M, Montesinos P, Holgado-Martin E, Ayuso-Sacido A, Penadés S, Desco M, Silva A, Garcia-Sanz JA. Stem Cell Res. 2015;14:114–129. doi: 10.1016/j.scr.2014.11.006. [DOI] [PubMed] [Google Scholar]
- Ernst B, Magnani JL. Nat Rev Drug Discov. 2009;8:661–677. doi: 10.1038/nrd2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr TD, Lai CH, Grünstein D, Orts-Gil G, Wang C, Boehm-Sturm P, Seeberger PH, Harms C. Nano Lett. 2014;14:2130–2134. doi: 10.1021/nl500388h. [DOI] [PubMed] [Google Scholar]
- Feng W, Luo RM, Xiao J, Ji PJ, Zheng ZG. Chem Eng Sci. 2011;66:4807–4813. [Google Scholar]
- Finlay BB, Cossart P. Science. 1997;276:718–725. doi: 10.1126/science.276.5313.718. [DOI] [PubMed] [Google Scholar]
- Frigell J, García I, Gómez-Vallejo V, Llop J, Penadés S. J Am Chem Soc. 2014;136:449–457. doi: 10.1021/ja411096m. [DOI] [PubMed] [Google Scholar]
- Furuike T, Sadamoto R, Niikura K, Monde K, Sakairi N, Nishimura SI. Tetrahedron. 2005;61:1737–1742. [Google Scholar]
- Gallo J, García I, Genicio N, Padro D, Penadés S. Biomaterials. 2011;32:9818–9825. doi: 10.1016/j.biomaterials.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Gallo J, Genicio N, Penadés S. Adv Healthc Mater. 2012;1:302–307. doi: 10.1002/adhm.201200051. [DOI] [PubMed] [Google Scholar]
- Gann JP, Yan M. Langmuir. 2008;24:5319–5323. doi: 10.1021/la7029592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Li L, Ho PL, Mak GC, Gu H, Xu B. Adv Mater. 2006;18:3145–3148. [Google Scholar]
- García I, Gallo J, Genicio N, Padro D, Penadés S. Bioconjugate Chem. 2011;22:264–273. doi: 10.1021/bc1003923. [DOI] [PubMed] [Google Scholar]
- García I, Marradi M, Penadés S. Nanomedicine. 2010;5:777–792. doi: 10.2217/nnm.10.48. [DOI] [PubMed] [Google Scholar]
- García I, Sánchez-Iglesias A, Henriksen-Lacey M, Grzelczak M, Penadés S, Liz-Marzán LM. J Am Chem Soc. 2015;137:3686–3692. doi: 10.1021/jacs.5b01001. [DOI] [PubMed] [Google Scholar]
- Gary-Bobo M, Mir Y, Rouxel C, Brevet D, Basile I, Maynadier M, Vaillant O, Mongin O, Blanchard-Desce M, Morère A, Garcia M, Durand JO, Raehm L. Angew Chem Int Ed. 2011;50:11425–11429. doi: 10.1002/anie.201104765. [DOI] [PubMed] [Google Scholar]
- Gorityala BK, Ma JM, Wang X, Chen P, Liu XW. Chem Soc Rev. 2010;39:2925–2934. doi: 10.1039/b919525b. [DOI] [PubMed] [Google Scholar]
- Grünstein D, Maglinao M, Kikkeri R, Collot M, Barylyuk K, Lepenies B, Kamena F, Zenobi R, Seeberger PH. J Am Chem Soc. 2011;133:13957–13966. doi: 10.1021/ja2036767. [DOI] [PubMed] [Google Scholar]
- Gu HW, Ho PL, Tsang KWT, Wang L, Xu B. J Am Chem Soc. 2003a;125:15702–15703. doi: 10.1021/ja0359310. [DOI] [PubMed] [Google Scholar]
- Gu HW, Ho PL, W T, Tsang K, Yu CW, Xu B. Chem Commun. 2003b:1966–1967. doi: 10.1039/b305421g. [DOI] [PubMed] [Google Scholar]
- Gu LR, Elkin T, Jiang XP, Li HP, Lin Y, Qu LW, Tzeng TRJ, Joseph R, Sun YP. Chem Commun. 2005:874–876. doi: 10.1039/b415015e. [DOI] [PubMed] [Google Scholar]
- Gu LR, Luo PG, Wang H, Meziani MJ, Lin Y, Veca LM, Cao L, Lu FS, Wang X, Quinn RA, Wang W, Zhang PY, Lacher S, Sun YP. Biomacromolecules. 2008;9:2408–2418. doi: 10.1021/bm800395e. [DOI] [PubMed] [Google Scholar]
- Guo YL, Yan HT. J Carbohydr Chem. 2008;27:309–319. [Google Scholar]
- Halkes KM, Carvalho de Souza A, Maljaars CEP, Gerwig GJ, Kamerling JP. Eur J Org Chem. 2005;2005:3650–3659. [Google Scholar]
- Han E, Ding L, Jin S, Ju HX. Biosens Bioelectron. 2011a;26:2500–2505. doi: 10.1016/j.bios.2010.10.044. [DOI] [PubMed] [Google Scholar]
- Han E, Ding L, Ju HX. Anal Chem. 2011b;83:7006–7012. doi: 10.1021/ac201488x. [DOI] [PubMed] [Google Scholar]
- Hansen HC, Haataja S, Finne J, Magnusson G. J Am Chem Soc. 1997;119:6974–6979. [Google Scholar]
- Hao NJ, Li LL, Zhang Q, Huang XL, Meng XW, Zhang YQ, Chen D, Tang FQ, Li LL. Microporous Mesoporous Mater. 2012;162:14–23. [Google Scholar]
- Hao NJ, Jayawardana KW, Chen X, Yan M. ACS Appl Mater Interfaces. 2015;7:1040–1045. doi: 10.1021/am508219g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao NJ, Li LF, Tang FQ. J Biomed Nanotechnol. 2014a;10:2508–2538. doi: 10.1166/jbn.2014.1940. [DOI] [PubMed] [Google Scholar]
- Hao NJ, Li LF, Tang FQ. J Mater Chem A. 2014b;2:11565–11568. [Google Scholar]
- Harada YH, Murata TM, Totani KT, Kajimoto TK, Masum SM, Tamba YT, Yamazaki MY, Usui TU. Biosci Biotechnol Biochem. 2005;69:166–178. doi: 10.1271/bbb.69.166. [DOI] [PubMed] [Google Scholar]
- Hartmann M, Betz P, Sun Y, Gorb SN, Lindhorst TK, Krueger A. Chem Eur J. 2012;18:6485–6492. doi: 10.1002/chem.201104069. [DOI] [PubMed] [Google Scholar]
- Hasegawa T, Fujisawa T, Numata M, Umeda M, Matsumoto T, Kimura T, Okumura S, Sakurai K, Shinkai S. Chem Commun. 2004:2150–2151. doi: 10.1039/b407409b. [DOI] [PubMed] [Google Scholar]
- Hatano K, Saeki H, Yokota H, Aizawa H, Koyama T, Matsuoka K, Terunuma D. Tetrahedron Lett. 2009;50:5816–5819. [Google Scholar]
- Hatch DM, Weiss Aa, Kale RR, Iyer SS. ChemBioChem. 2008;9:2433–2442. doi: 10.1002/cbic.200800188. [DOI] [PubMed] [Google Scholar]
- He Y, Fan CH, Lee ST. Nano Today. 2010;5:282–295. [Google Scholar]
- Higuchi Y, Oka M, Kawakami S, Hashida M. J Control Release. 2008;125:131–136. doi: 10.1016/j.jconrel.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Hildebrand A, Schaedlich A, Rothe U, Neubert RHH. J Colloid Interf Sci. 2002;249:274–281. doi: 10.1006/jcis.2002.8272. [DOI] [PubMed] [Google Scholar]
- Hocine O, Gary-Bobo M, Brevet D, Maynadier M, Fontanel S, Raehm L, Richeter S, Loock B, Couleaud P, Frochot C. Int J Clin Pharm. 2010;402:221–230. doi: 10.1016/j.ijpharm.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Holgersson J, Gustafsson A, Breimer ME. Immunol Cell Biol. 2005;83:694–708. doi: 10.1111/j.1440-1711.2005.01373.x. [DOI] [PubMed] [Google Scholar]
- Holzinger M, Abraham J, Whelan P, Graupner R, Ley L, Hennrich F, Kappes M, Hirsch A. J Am Chem Soc. 2003;125:8566–8580. doi: 10.1021/ja029931w. [DOI] [PubMed] [Google Scholar]
- Hone DC, Haines AH, Russell DA. Langmuir. 2003;19:7141–7144. [Google Scholar]
- Hong SY, Tobias G, Al-Jamal KT, Ballesteros B, Ali-Boucetta H, Lozano-Perez S, Nellist PD, Sim RB, Finucane C, Mather SJ, Green MLH, Kostarelos K, Davis BG. Nat Mater. 2010;9:485–490. doi: 10.1038/nmat2766. [DOI] [PubMed] [Google Scholar]
- Horak D, Babic M, Jendelová P, Herynek V, Trchová M, Pientka Z, Pollert E, Hájek M, Syková E. Bioconjugate Chem. 2007;18:635–644. doi: 10.1021/bc060186c. [DOI] [PubMed] [Google Scholar]
- Hu FX, Chen SH, Wang CY, Yuan R, Xiang Y, Wang C. Biosens Bioelectron. 2012;34:202–207. doi: 10.1016/j.bios.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Hu XLe, Jin HY, He XP, James TD, Chen GR, Long YT. ACS Appl Mater Interfaces. 2015;7:1874–1878. doi: 10.1021/am5076293. [DOI] [PubMed] [Google Scholar]
- Huang CC, Chen CT, Shiang YC, Lin ZH, Chang HT. Anal Chem. 2009;81:875–882. doi: 10.1021/ac8010654. [DOI] [PubMed] [Google Scholar]
- Huang CF, Yao GH, Liang RP, Qiu JD. Biosens Bioelectron. 2013;50:305–310. doi: 10.1016/j.bios.2013.07.002. [DOI] [PubMed] [Google Scholar]
- Huang LDe, Adak AK, Yu CC, Hsiao WC, Lin HJ, Chen ML, Lin CC. Chem Eur J. 2015;21:3956–3967. doi: 10.1002/chem.201405747. [DOI] [PubMed] [Google Scholar]
- Huang GL. Curr Med Chem. 2013;20:782–788. [PubMed] [Google Scholar]
- Hulikova K, Benson V, Svoboda J, Sima P, Fiserova A. Int Immunopharmacol. 2009;9:792–799. doi: 10.1016/j.intimp.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Hulikova K, Svoboda J, Benson V, Grobarova V, Fiserova A. Int Immunopharmacol. 2011;11:955–961. doi: 10.1016/j.intimp.2011.02.009. [DOI] [PubMed] [Google Scholar]
- Ibey BL, Beier HT, Rounds RM, Coté GL, Yadavalli VK, Pishko MV. Anal Chem. 2005;77:7039–7046. doi: 10.1021/ac0507901. [DOI] [PubMed] [Google Scholar]
- Jayaraman N. Chem Soc Rev. 2009;38:3463–3483. doi: 10.1039/b815961k. [DOI] [PubMed] [Google Scholar]
- Jayaraman N, Maiti K, Naresh K. Chem Soc Rev. 2013;42:4640–4656. doi: 10.1039/c3cs00001j. [DOI] [PubMed] [Google Scholar]
- Jayawardena HSN, Jayawardana KW, Chen X, Yan M. Chem Commun. 2013a;49:3034–3036. doi: 10.1039/c3cc40491a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayawardena HSN, Wang X, Yan M. Anal Chem. 2013b;85:10277–10281. doi: 10.1021/ac402069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayawardana KW, Jayawardena HSN, Wijesundera SA, De Zoysa T, Sundhoro M, Yan M. Chem Commun. 2015 doi: 10.1039/C5CC04251H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Wang X, Geng H, Beer TM, Qian DZ, Ramström O, Yan M. ScienceJet. 2015;4:132. [PMC free article] [PubMed] [Google Scholar]
- Jiang XZ, Housni A, Gody G, Boullanger P, Charreyre MT, Delair T, Narain R. Bioconjugate Chem. 2010;21:521–530. doi: 10.1021/bc900431p. [DOI] [PubMed] [Google Scholar]
- Joosten JAF, Loimaranta V, Appeldoorn CCM, Haataja S, El Maate FA, Liskamp RMJ, Finne J, Pieters RJ. J Med Chem. 2004;47:6499–6508. doi: 10.1021/jm049476+. [DOI] [PubMed] [Google Scholar]
- Kamat M, El-Boubbou K, Zhu DC, Lansdell T, Lu XW, Li W, Huang XF. Bioconjugate Chem. 2010;21:2128–2135. doi: 10.1021/bc100354m. [DOI] [PubMed] [Google Scholar]
- Kato H, Yashiro A, Mizuno A, Nishida Y, Kobayashi K, Shinohara H. Bioorg Med Chem Lett. 2001;11:2935–2939. doi: 10.1016/s0960-894x(01)00583-2. [DOI] [PubMed] [Google Scholar]
- Kato T, Kawaguchi A, Nagata K, Hatanaka K. Biochem Biophys Res Commun. 2010;394:200–204. doi: 10.1016/j.bbrc.2010.02.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kell AJ, Stewart G, Ryan S, Peytavi R, Boissinot M, Huletsky A, Bergeron MG, Simard B. ACS Nano. 2008;2:1777–1788. doi: 10.1021/nn700183g. [DOI] [PubMed] [Google Scholar]
- Kelly KL, Coronado E, Zhao LL, Schatz GC. J Phys Chem B. 2003;107:668–677. [Google Scholar]
- Kemp MM, Kumar A, Mousa S, Dyskin E, Yalcin M, Ajayan P, Linhardt RJ, Mousa SA. Nanotechnology. 2009;20:455104. doi: 10.1088/0957-4484/20/45/455104. [DOI] [PubMed] [Google Scholar]
- Khiar N, Leal MP, Baati R, Ruhlmann C, Mioskowski C, Schultz P, Fernández I. Chem Commun. 2009:4121–4123. doi: 10.1039/b904717d. [DOI] [PubMed] [Google Scholar]
- Kiessling LL, Gestwicki JE, Strong LE. Angew Chem Int Ed. 2006;45:2348–2368. doi: 10.1002/anie.200502794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkeri R, García-Rubio I, Seeberger PH. Chem Commun. 2009a:235–237. doi: 10.1039/b814146k. [DOI] [PubMed] [Google Scholar]
- Kikkeri R, Grünstein D, Seeberger PH. J Am Chem Soc. 2010a;132:10230–10232. doi: 10.1021/ja103688s. [DOI] [PubMed] [Google Scholar]
- Kikkeri R, Hossain LH, Seeberger PH. Chem Commun. 2008:2127–2129. doi: 10.1039/b802177e. [DOI] [PubMed] [Google Scholar]
- Kikkeri R, Kamena F, Gupta T, Hossain LH, Boonyarattanakalin S, Gorodyska G, Beurer E, Coullerez G, Textor M, Seeberger PH. Langmuir. 2010b;26:1520–1523. doi: 10.1021/la9038792. [DOI] [PubMed] [Google Scholar]
- Kikkeri R, Lepenies B, Adibekian A, Laurino P, Seeberger PH. J Am Chem Soc. 2009b;131:2110–2112. doi: 10.1021/ja807711w. [DOI] [PubMed] [Google Scholar]
- Kim KS, Hur W, Park SJ, Hong SW, Choi JE, Goh EJ, Yoon SK, Hahn SK. ACS Nano. 2010;4:3005–3014. doi: 10.1021/nn100589y. [DOI] [PubMed] [Google Scholar]
- Kong N, Shimpi MR, Ramström O, Yan M. Carbohydr Res. 2015;405:33–38. doi: 10.1016/j.carres.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouyoumdjian H, Zhu DC, El-Dakdouki MH, Lorenz K, Chen JJ, Li W, Huang XF. ACS Chem Neurosci. 2013;4:575–584. doi: 10.1021/cn3002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krist P, Vannucci L, Kuzma M, Man P, Sadalapure K, Patel A, Bezouska K, Pospísil M, Petrus L, Lindhorst TK, Kren V. ChemBioChem. 2004;5:445–452. doi: 10.1002/cbic.200300669. [DOI] [PubMed] [Google Scholar]
- Kulkarni AA, Fuller C, Korman H, Weiss AA, Iyer SS. Bioconjugate Chem. 2010;21:1486–1493. doi: 10.1021/bc100095w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Sahoo B, Montpetit A, Behera S, Lockey RF, Mohapatra SS. Nanomedicine. 2007;3:132–137. doi: 10.1016/j.nano.2007.03.001. [DOI] [PubMed] [Google Scholar]
- La Belle JT, Gerlach JQ, Svarovsky S, Joshi L. Anal Chem. 2007;79:6959–6964. doi: 10.1021/ac070651e. [DOI] [PubMed] [Google Scholar]
- Lasala F, Arce E, Otero JR, Rojo J, Delgado R. Antimicrob Agents Chemother. 2003;47:3970–3972. doi: 10.1128/AAC.47.12.3970-3972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Lee K, Kim IK, Park TG. Biomaterials. 2008;29:4709–4718. doi: 10.1016/j.biomaterials.2008.08.038. [DOI] [PubMed] [Google Scholar]
- Lee Y, Lee H, Kim YB, Kim J, Hyeon T, Park H, Messersmith PB, Park TG. Adv Mater. 2008;20:4154–4157. doi: 10.1002/adma.200800756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YC, Lee RT. Acc Chem Res. 1995;28:321–327. [Google Scholar]
- Li HG, El-Dakdouki MH, Zhu DC, Abela GS, Huang XF. Chem Commun. 2012;48:3385–3387. doi: 10.1039/c2cc17852d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhou HY, Yang L, Du GQ, Pai-Panandiker AS, Huang XF, Yan B. Biomaterials. 2011;32:2540–2545. doi: 10.1016/j.biomaterials.2010.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KR, Park JM, Choi HN, Lee WY. Microchem J. 2013;106:154–159. [Google Scholar]
- Lin CC, Yeh YC, Yang CY, Chen GF, Chen YC, Wu YC, Chen CC. Chem Commun. 2003:2920–2921. doi: 10.1039/b308995a. [DOI] [PubMed] [Google Scholar]
- Lin PC, Ueng Shua, Yu SC, Jan Mdan, Adak AK, Yu CC, Lin CC. Org Lett. 2007;9:2131–2134. doi: 10.1021/ol070588f. [DOI] [PubMed] [Google Scholar]
- Liu FT, Rabinovich GA. Nat Rev Cancer. 2005;5:29–41. doi: 10.1038/nrc1527. [DOI] [PubMed] [Google Scholar]
- Liu L, Yan M. Angew Chem Int Ed. 2006;45:6207–6210. doi: 10.1002/anie.200602097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LH, Dietsch H, Schurtenberger P, Yan M. Bioconjugate Chem. 2009;20:1349–1355. doi: 10.1021/bc900110x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LH, Lerner MM, Yan M. Nano Lett. 2010a;10:3754–3756. doi: 10.1021/nl1024744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LH, Yan M. Acc Chem Res. 2010;43:1434–1443. doi: 10.1021/ar100066t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LH, Zorn G, Castner DG, Solanki R, Lerner MM, Yan M. J Mater Chem. 2010b;20:5041–5046. doi: 10.1039/C0JM00509F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu RR, Liew RS, Zhou J, Xing BG. Angew Chem Int Ed. 2007;46:8799–8803. doi: 10.1002/anie.200702773. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhao YL, Sun BY, Chen CY. Acc Chem Res. 2012;46:702–713. doi: 10.1021/ar300028m. [DOI] [PubMed] [Google Scholar]
- Lu M, Cohen MH, Rieves D, Pazdur R. Am J Hematol. 2010;85:315–319. doi: 10.1002/ajh.21656. [DOI] [PubMed] [Google Scholar]
- Luczkowiak J, Muñoz A, Sánchez-Navarro M, Ribeiro-Viana R, Ginieis A, Illescas BM, Martín N, Delgado R, Rojo J. Biomacromolecules. 2013;14:431–437. doi: 10.1021/bm3016658. [DOI] [PubMed] [Google Scholar]
- Lyu YK, Lim KR, Lee BY, Kim KS, Lee WY. Chem Commun. 2008:4771–4773. doi: 10.1039/b807438k. [DOI] [PubMed] [Google Scholar]
- Maalouli N, Barras A, Siriwardena A, Bouazaoui M, Boukherroub R, Szunerits S. Analyst. 2013;138:805–812. doi: 10.1039/c2an36272d. [DOI] [PubMed] [Google Scholar]
- Mahon E, Aastrup T, Barboiu M. Chem Commun. 2010a;46:5491–5493. doi: 10.1039/c002652b. [DOI] [PubMed] [Google Scholar]
- Mahon E, Aastrup T, Barboiu M. Chem Commun. 2010b;46:2441–2443. doi: 10.1039/b924766a. [DOI] [PubMed] [Google Scholar]
- Mahon E, Mouline Z, Silion M, Gilles A, Pinteala M, Barboiu M. Chem Commun. 2013;49:3004–3006. doi: 10.1039/c3cc41074a. [DOI] [PubMed] [Google Scholar]
- Marín MJ, Rashid A, Rejzek M, Fairhurst SA, Wharton SA, Martin SR, McCauley JW, Wileman T, Field RA, Russell DA. Org Biomol Chem. 2013;11:7101–7107. doi: 10.1039/c3ob41703d. [DOI] [PubMed] [Google Scholar]
- Marradi M, Alcántara D, de la Fuente JM, García-Martín ML, Cerdán S, Penadés S. Chem Commun. 2009:3922–3924. doi: 10.1039/b900957d. [DOI] [PubMed] [Google Scholar]
- Marradi M, Chiodo F, García I, Penadés S. Chem Soc Rev. 2013;42:4728–4745. doi: 10.1039/c2cs35420a. [DOI] [PubMed] [Google Scholar]
- Martos-Maldonado MC, Casas-Solvas JM, Quesada-Soriano I, García-Fuentes L, Vargas-Berenguel A. Langmuir. 2013;29:1318–1326. doi: 10.1021/la304107a. [DOI] [PubMed] [Google Scholar]
- McLean JA, Stumpo KA, Russell DH. J Am Chem Soc. 2005;127:5304–5305. doi: 10.1021/ja043907w. [DOI] [PubMed] [Google Scholar]
- Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min IH, Choi L, Ahn KS, Kim BK, Lee BY, Kim KS, Choi HN, Lee WY. Biosens Bioelectron. 2010;26:1326–1331. doi: 10.1016/j.bios.2010.07.038. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay B, Martins MB, Karamanska R, Russell DA, Field RA. Tetrahedron Lett. 2009;50:886–889. [Google Scholar]
- Munoz EM, Correa J, Fernandez-Megia E, Riguera R. J Am Chem Soc. 2009;131:17765–17767. doi: 10.1021/ja9074826. [DOI] [PubMed] [Google Scholar]
- Munoz EM, Correa J, Riguera R, Fernandez-Megia E. J Am Chem Soc. 2013;135:5966–5969. doi: 10.1021/ja400951g. [DOI] [PubMed] [Google Scholar]
- Murray RA, Qiu Y, Chiodo F, Marradi M, Penadés S, Moya SE. Small. 2014;10:2602–2610. doi: 10.1002/smll.201303604. [DOI] [PubMed] [Google Scholar]
- Murthy BN, Zeile S, Nambiar M, Nussio MR, Gibson CT, Shapter JG, Jayaraman N, Voelcker NH. RSC Adv. 2012;2:1329–1333. [Google Scholar]
- Nagahori N, Lee RT, Nishimura S, Page D, Roy R, Lee YC. ChemBioChem. 2002;3:836–844. doi: 10.1002/1439-7633(20020902)3:9<836::AID-CBIC836>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Narla SN, Sun X. Biomacromolecules. 2012;13:1675–1682. doi: 10.1021/bm3003896. [DOI] [PubMed] [Google Scholar]
- Nath S, Kaittanis C, Tinkham A, Perez JM. Anal Chem. 2008;80:1033–1038. doi: 10.1021/ac701969u. [DOI] [PubMed] [Google Scholar]
- Niikura K, Nishio T, Akita H, Matsuo Y, Kamitani R, Kogure K, Harashima H, Ijiro K. ChemBioChem. 2007;8:379–384. doi: 10.1002/cbic.200600496. [DOI] [PubMed] [Google Scholar]
- Niikura K, Sekiguchi S, Nishio T, Masuda T, Akita H, Matsuo Y, Kogure K, Harashima H, Ijiro K. ChemBioChem. 2008;9:2623–2627. doi: 10.1002/cbic.200800464. [DOI] [PubMed] [Google Scholar]
- Nimmagadda A, Thurston K, Nollert MU, McFetridge PS. J Biomed Mater Res A. 2006;76:614–625. doi: 10.1002/jbm.a.30577. [DOI] [PubMed] [Google Scholar]
- Ohyanagi T, Nagahori N, Shimawaki K, Hinou H, Yamashita T, Sasaki A, Jin T, Iwanaga T, Kinjo M, Nishimura SI. J Am Chem Soc. 2011;133:12507–12517. doi: 10.1021/ja111201c. [DOI] [PubMed] [Google Scholar]
- Ortega-Caballero F, Giménez-Martínez JJ, García-Fuentes L, Ortiz-Salmerón E, Santoyo-González F, Vargas-Berenguel A. J Org Chem. 2001;66:7786–7795. doi: 10.1021/jo015875q. [DOI] [PubMed] [Google Scholar]
- Osaki F, Kanamori T, Sando S, Sera T, Aoyama Y. J Am Chem Soc. 2004;126:6520–6521. doi: 10.1021/ja048792a. [DOI] [PubMed] [Google Scholar]
- Otsuka H, Akiyama Y, Nagasaki Y, Kataoka K. J Am Chem Soc. 2001;123:8226–8230. doi: 10.1021/ja010437m. [DOI] [PubMed] [Google Scholar]
- Park J, Jayawardena HSN, Chen X, Jayawardana KW, Sundhoro M, Ada E, Yan M. Chem Commun. 2015;51:2882–2885. doi: 10.1039/c4cc07936a. [DOI] [PubMed] [Google Scholar]
- Park J, Yan M. Acc Chem Res. 2013;46:181–189. doi: 10.1021/ar300172h. [DOI] [PubMed] [Google Scholar]
- Peng J, Wang K, Tan W, He X, He C, Wu P, Liu F. Talanta. 2007;71:833–840. doi: 10.1016/j.talanta.2006.05.064. [DOI] [PubMed] [Google Scholar]
- Pera NP, Kouki A, Haataja S, Branderhorst HM, Liskamp RMJ, Visser GM, Finne J, Pieters RJ. Org Biomol Chem. 2010;8:2425–2429. doi: 10.1039/c000819b. [DOI] [PubMed] [Google Scholar]
- Perrier M, Gary-Bobo M, Lartigue L, Brevet D, Morère A, Garcia M, Maillard P, Raehm L, Guari Y, Larionova J, Durand JO, Mongin O, Blanchard-Desce M. J Nanopart Res. 2013;15:1602. [Google Scholar]
- Pfaff A, Schallon A, Ruhland TM, Majewski AP, Schmalz H, Freitag R, Müller AHE. Biomacromolecules. 2011;12:3805–3811. doi: 10.1021/bm201051p. [DOI] [PubMed] [Google Scholar]
- Pinson J, Podvorica F. Chem Soc Rev. 2005;34:429–439. doi: 10.1039/b406228k. [DOI] [PubMed] [Google Scholar]
- Ponader D, Wojcik F, Beceren-Braun F, Dernedde J, Hartmann L. Biomacromolecules. 2012;13:1845–1852. doi: 10.1021/bm300331z. [DOI] [PubMed] [Google Scholar]
- Prato M. J Mater Chem. 1997;7:1097–1109. [Google Scholar]
- Prato M, Li QC, Wudl F, Lucchini V. J Am Chem Soc. 1993;115:1148–1150. [Google Scholar]
- Qi GBin, Li LL, Yu FQ, Wang H. ACS Appl Mater Interfaces. 2013;5:10874–10881. doi: 10.1021/am403940d. [DOI] [PubMed] [Google Scholar]
- Ragoussi ME, Casado S, Ribeiro-Viana R, Torre GDLa, Rojo J, Torres T. Chem Sci. 2013;4:4035–4041. [Google Scholar]
- Reichardt NC, Martín-Lomas M, Penadés S. Chem Soc Rev. 2013;42:4358–4376. doi: 10.1039/c2cs35427f. [DOI] [PubMed] [Google Scholar]
- Reichert A, Nagy JO, Spevak W, Charych D. J Am Chem Soc. 1995;117:829–830. [Google Scholar]
- Reynolds M, Marradi M, Imberty A, Penadés S, Pérez S. Chem Eur J. 2012;18:4264–4273. doi: 10.1002/chem.201102034. [DOI] [PubMed] [Google Scholar]
- Robinson A, Fang JM, Chou PT, Liao KW, Chu RM, Lee SJ. ChemBioChem. 2005;6:1899–1905. doi: 10.1002/cbic.200500112. [DOI] [PubMed] [Google Scholar]
- Rojo J, Díaz V, de la Fuente JM, Segura I, Barrientos AG, Riese HH, Bernad A, Penadés S. ChemBioChem. 2004;5:291–297. doi: 10.1002/cbic.200300726. [DOI] [PubMed] [Google Scholar]
- Saha K, Agasti SS, Kim C, Li X, Rotello VM. Chem Rev. 2012;112:2739–2779. doi: 10.1021/cr2001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A, Loimaranta V, Joosten JaF, Khan aS, Hacker J, Pieters RJ, Finne J. J Antimicrob Chemother. 2007;60:495–501. doi: 10.1093/jac/dkm251. [DOI] [PubMed] [Google Scholar]
- Sánchez-Navarro M, Muñoz A, Illescas BM, Rojo J, Martín N. Chem Eur J. 2011;17:766–769. doi: 10.1002/chem.201002816. [DOI] [PubMed] [Google Scholar]
- Sandros MG, Behrendt M, Maysinger D, Tabrizian M. Adv Funct Mater. 2007;17:3724–3730. [Google Scholar]
- Schlick KH, Lange CK, Gillispie GD, Cloninger MJ. J Am Chem Soc. 2009;131:16608–16609. doi: 10.1021/ja904073p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield CL, Field RA, Russell DA. Anal Chem. 2007;79:1356–1361. doi: 10.1021/ac061462j. [DOI] [PubMed] [Google Scholar]
- Schofield CL, Mukhopadhyay B, Hardy SM, McDonnell MB, Field RA, Russell DA. Analyst. 2008;133:626–634. doi: 10.1039/b715250g. [DOI] [PubMed] [Google Scholar]
- Seebach D, Rheiner BP, Greiveldinger G, Butz T, Sellner H. Top Curr Chem. 1998;197:125–164. [Google Scholar]
- Seto H, Kamba S, Kondo T, Hasegawa M, Nashima S, Ehara Y, Ogawa Y, Hoshino Y, Miura Y. ACS Appl Mater Interfaces. 2014;6:13234–13241. doi: 10.1021/am503003v. [DOI] [PubMed] [Google Scholar]
- Seto H, Ogata Y, Murakami T, Hoshino Y, Miura Y. ACS Appl Mater Interfaces. 2012;4:411–417. doi: 10.1021/am2014713. [DOI] [PubMed] [Google Scholar]
- Shaunak S, Thomas S, Gianasi E, Godwin A, Jones E, Teo I, Mireskandari K, Luthert P, Duncan R, Patterson S, Khaw P, Brocchini S. Nat Biotechnol. 2004;22:977–984. doi: 10.1038/nbt995. [DOI] [PubMed] [Google Scholar]
- Shi ZL, Neoh KG, Kang ET, Shuter B, Wang SC, Poh C, Wang W. ACS Appl Mater Interfaces. 2009;1:328–335. doi: 10.1021/am8000538. [DOI] [PubMed] [Google Scholar]
- Slowing II, Trewyn BG, Giri S, Lin VSY. Adv Funct Mater. 2007;17:1225–1236. [Google Scholar]
- Srinivasan B, Huang XF. Chirality. 2008;20:265–277. doi: 10.1002/chir.20418. [DOI] [PubMed] [Google Scholar]
- Sudibya HG, Ma JM, Dong XC, Ng S, Li LJ, Liu XW, Chen P. Angew Chem Int Ed. 2009;48:2723–2726. doi: 10.1002/anie.200805514. [DOI] [PubMed] [Google Scholar]
- Sunasee R, Narain R. Macromol Biosci. 2013;13:9–27. doi: 10.1002/mabi.201200222. [DOI] [PubMed] [Google Scholar]
- Sundgren A, Barchi JJ. Carbohydr Res. 2008;343:1594–1604. doi: 10.1016/j.carres.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svarovsky SA, Szekely Z, Barchi JJ. Tetrahedron Asymmetry. 2005;16:587–598. [Google Scholar]
- Szymanski CM, Wren BW. Nat Rev Microbiol. 2005;3:225–237. doi: 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Inoue G, Shoda SI, Kimura Y. J Polym Sci Part A Polym Chem. 2014a;52:3513–3520. [Google Scholar]
- Tanaka T, Ishitani H, Miura Y, Oishi K, Takahashi T, Suzuki T, Shoda S, Kimura Y. ACS Macro Lett. 2014b;3:1074–1078. doi: 10.1021/mz500555x. [DOI] [PubMed] [Google Scholar]
- Terada Y, Hashimoto W, Endo T, Seto H, Murakami T, Hisamoto H, Hoshino Y, Miura Y. J Mater Chem B. 2014;2:3324–3332. doi: 10.1039/c4tb00028e. [DOI] [PubMed] [Google Scholar]
- Thygesen MB, Sauer J, Jensen KJ. Chem Eur J. 2009;15:1649–1660. doi: 10.1002/chem.200801521. [DOI] [PubMed] [Google Scholar]
- Tong Q, Wang X, Wang H, Kubo T, Yan M. Anal Chem. 2012;84:3049–3052. doi: 10.1021/ac203455b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touaibia M, Wellens A, Shiao TC, Wang Q, Sirois S, Bouckaert J, Roy R. ChemMedChem. 2007;2:1190–1201. doi: 10.1002/cmdc.200700063. [DOI] [PubMed] [Google Scholar]
- Trewyn BG, Giri S, Slowing II, Lin VSY. Chem Commun. 2007;43:3236–3245. doi: 10.1039/b701744h. [DOI] [PubMed] [Google Scholar]
- Tsai CS, Yu TBin, Chen CT. Chem Commun. 2005:4273–4275. doi: 10.1039/b507237a. [DOI] [PubMed] [Google Scholar]
- Turnbull WB, Kalovidouris Sa, Stoddart JF. Chem Eur J. 2002;8:2988–3000. doi: 10.1002/1521-3765(20020703)8:13<2988::AID-CHEM2988>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Van Kasteren SI, Campbell SJ, Serres S, Anthony DC, Sibson NR, Davis BG. Proc Natl Acad Sci U S A. 2008;106:18–23. doi: 10.1073/pnas.0806787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedantam P, Tzeng TRJ, Brown AK, Podila R, Rao A, Staley K. Plasmonics. 2012;7:301–308. [Google Scholar]
- Voit B, Appelhans D. Macromol Chem Phys. 2010;211:727–735. [Google Scholar]
- Wang H, Ren J, Hlaing A, Yan M. J Colloid Interf Sci. 2011;354:160–167. doi: 10.1016/j.jcis.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HF, Gu LR, Lin Y, Lu FS, Meziani MJ, Luo PG, Wang W, Cao L, Sun YP. J Am Chem Soc. 2006;128:13364–13365. doi: 10.1021/ja065455o. [DOI] [PubMed] [Google Scholar]
- Wang X, Liu LH, Ramström O, Yan M. Exp Biol Med. 2009a;234:1128–1139. doi: 10.3181/0904-MR-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Matei E, Deng L, Koharudin L, Gronenborn AM, Ramström O, Yan M. Biosens Bioelectron. 2013;47:258–264. doi: 10.1016/j.bios.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Matei E, Deng L, Ramström O, Gronenborn AM, Yan M. Chem Commun. 2011a;47:8620–8622. doi: 10.1039/c1cc12981c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Matei E, Gronenborn AM, Ramström O, Yan M. Anal Chem. 2012;84:4248–4252. doi: 10.1021/ac3006632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ramström O, Yan M. J Mater Chem. 2009b;19:8944–8949. doi: 10.1039/B917900C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ramström O, Yan M. Adv Mater. 2010a;22:1946–1953. doi: 10.1002/adma.200903908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ramström O, Yan M. Anal Chem. 2010b;82:9082–9089. doi: 10.1021/ac102114z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ramström O, Yan M. Analyst. 2011b;136:4174–4178. doi: 10.1039/c1an15469a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ramström O, Yan M. Chem Commun. 2011c;47:4261–4263. doi: 10.1039/c0cc05299j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Narain R, Liu Y. Langmuir. 2014;30:7377–7387. doi: 10.1021/la5016115. [DOI] [PubMed] [Google Scholar]
- Wang YX, Hussain SM, Krestin GP. Eur Radiol. 2001;11:2319–2331. doi: 10.1007/s003300100908. [DOI] [PubMed] [Google Scholar]
- Wang ZX, Ma LN. Coord Chem Rev. 2009;253:1607–1618. [Google Scholar]
- Wu P, Chen X, Hu N, Tam UC, Blixt O, Zettl A, Bertozzi CR. Angew Chem Int Ed. 2008;47:5022–5025. doi: 10.1002/anie.200705363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wudl F. J Mater Chem. 2002;12:1959–1963. [Google Scholar]
- Yan F, Xue J, Zhu J, Marchant RE, Guo Z. Bioconjugate Chem. 2005;16:90–96. doi: 10.1021/bc049805c. [DOI] [PubMed] [Google Scholar]
- Yan M, Harnish B. Adv Mater. 2003;15:244–248. [Google Scholar]
- Yan M, Ren J. Chem Mater. 2004;16:1627–1632. [Google Scholar]
- Yang Y, Yu M, Yan TT, Zhao ZH, Sha YL, Li ZJ. Bioorg Med Chem. 2010a;18:5234–5240. doi: 10.1016/j.bmc.2010.05.046. [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhao YT, Yan TT, Yu M, Sha YL, Zhao ZH, Li ZJ. Tetrahedron Lett. 2010b;51:4182–4185. [Google Scholar]
- Yilmaz G, Becer CR. Eur Polym J. 2013;49:3046–3051. [Google Scholar]
- Yoo MK, Kim IY, Kim EM, Jeong HJ, Lee CM, Jeong YY, Akaike T, Cho CS. J Biomed Biotechnol. 2007;2007:94740. doi: 10.1155/2007/94740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo MK, Park IY, Kim IY, Park IK, Kwon JS, Jeong HJ, Jeong YY, Cho CS. J Nanosci Nanotechnol. 2008;8:5196–5202. doi: 10.1166/jnn.2008.1118. [DOI] [PubMed] [Google Scholar]
- Yu L, Huang M, Wang PG, Zeng X. Anal Chem. 2007;79:8979–8986. doi: 10.1021/ac071453q. [DOI] [PubMed] [Google Scholar]
- Zeng HJ, Yu JS, Jiang YD, Zeng XQ. Biosens Bioelectron. 2014;55:157–161. doi: 10.1016/j.bios.2013.11.018. [DOI] [PubMed] [Google Scholar]
- Zhao JS, Liu YF, Park HJ, Boggs JM, Basu A. Bioconjugate Chem. 2012;23:1166–1173. doi: 10.1021/bc2006169. [DOI] [PubMed] [Google Scholar]
- Zhou J, Butchosa N, Jayawardena HSN, Zhou Q, Yan M, Ramström O. Bioconjug Chem. 2014;25:640–643. doi: 10.1021/bc500004c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Butchosa N, Jayawardena HSN, Park J, Zhou Q, Yan M, Ramström O. Biomacromolecules. 2015a;16:1426–1432. doi: 10.1021/acs.biomac.5b00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Hao NJ, De Zoyza T, Yan M, Ramström O. Chem Commun. 2015b;51:9833–9836. doi: 10.1039/c5cc02907d. [DOI] [PMC free article] [PubMed] [Google Scholar]


