Abstract
Oxidative stress has been implicated in many common age-related diseases and is hypothesized to play a role in posttraumatic stress disorder (PTSD)-related neurodegeneration (Miller and Sadeh, 2014). This study examined the influence of the oxidative stress-related genes ALOX 12 and ALOX 15 on the association between PTSD and cortical thickness. Factor analyses were used to identify and compare alternative models of the structure of cortical thickness in a sample of 218 veterans. The best-fitting model was then used for a genetic association analysis in White non-Hispanic participants (n = 146) that examined relationships between 33 single nucleotide polymorphisms (SNPs) spanning the two genes, 8 cortical thickness factors, and each SNP × PTSD interaction. Results identified a novel ALOX12 locus (indicated by two SNPs in perfect linkage disequilibrium: rs1042357 and rs10852889) that moderated the association between PTSD and reduced thickness of the right prefrontal cortex. A whole-cortex vertex-wise analysis showed this effect to be localized to clusters spanning the rostral middle frontal gyrus, superior frontal gyrus, rostral anterior cingulated cortex, and medial orbitofrontal cortex. These findings illustrate a novel factor-analytic approach to neuroimaging-genetic analyses and provide new evidence for the possible involvement of oxidative stress in PTSD-related neurodegeneration.
Keywords: Posttraumatic stress disorder, oxidative stress, ALOX12, cortical thickness, factor analysis, genetic association analysis
1. Introduction
The cortex is the thin, convoluted outer layer of the brain composed of four to six layers of neurons, neuroglia, and nerve fibers aligned in columns. The number, size, and arrangement of these cells determine cortical thickness (CT) at a given point. Twin studies have found thickness variation to be coordinated by a heritable latent structure that corresponds approximately to the four primary lobes and known patterns of gene expression (Chen et al., 2008; Chen et al., 2012; Chen et al., 2013; Lerch et al., 2006; O'Leary et al., 2007; Rimol et al., 2010). Though heritability studies have arrived at different conclusions about how to best model this structure, principles of phylogenetics and ontogenetics point to the primacy of the symmetrical four-lobe organization within each hemisphere. CT is also influenced by a host of developmental processes, environmental factors, and diseases, including psychiatric conditions such as posttraumatic stress disorder (PTSD). Structural neuroimaging studies have linked PTSD to reduced volume and/or thickness of several regions of the cortex and a recent meta-analysis concluded that the most consistent associations have been found in the anterior cingulate cortex, left temporal pole/middle temporal gyrus, and ventromedial prefrontal cortex/anterior cingulate cortex (Kühn and Gallinat, 2013). Though the extent to which observed differences reflect pre-existing vulnerabilities, consequences of the illness, or some combination thereof remain open questions, neuroimaging-genetic studies can potentially shed light on their answers by identifying genetic loci that influence brain morphology directly and/or influence molecular processes that might link PTSD to changes in brain morphology.
The plethora of data involved in studies that combine neuroimaging and genetic methods, however, yields formidable analytic challenges. Vertex-wise analyses of the cortical mantle can involve comparisons of upwards of 10,000 vertices, and the number of genetic loci can range from dozens or more in candidate gene studies to millions in a genome-wide association study. Hence, it is essential in studies combining these approaches to reduce the number of comparisons on one or both sides of the equation to avoid either false positives or overly punitive multiple-testing correction. In this study, building on prior research on the genetic organization of the cortex, we approached this problem using factor analysis to model the latent structure of cortical thickness and identified a multidimensional structure that we then submitted to a genetic association analysis.
1.1 Oxidative Stress, ALOX12, ALOX15, and Neurodegeneration in PTSD
Oxidative stress (OXS) is a cellular condition that occurs when pro-oxidant molecules exceed the capacity of available antioxidants to counteract their effects. It is a process fundamental to aging and widely implicated in many common diseases. Psychological stress and stress-related mental illnesses have been linked to elevated levels of OXS-related biomarkers in both human and animals studies (for reviews, see Hovatta et al., 2010; Palta et al., 2014; Schiavone et al., 2013) that, in turn, have also been implicated in mechanisms of neurodegeneration (Li et al., 2013). For example, using a novel animal model of PTSD, Wilson and colleagues (2013) studied effects of acute and chronic stress on OXS in rats by measuring biomarkers of OXS and antioxidant activity during stress exposure and later in post-mortem analysis of brain and adrenal tissue. Compared to controls, rats in the stress condition exhibited slower growth, higher plasma corticosterone levels, greater anxiety-like behavior on an elevated plus-maze task, and a dose-response relationship between the duration of the protocol and levels of ROS. Post-mortem analysis of brain tissue revealed elevated levels of reactive oxygen species and other byproducts of OXS in the hippocampus and pre-frontal cortex.
We conceptualize chronic PTSD as a stress-perpetuating condition that promotes OXS through sleep disturbance, the repeated activation of stress-related physiology that accompanies reexperiencing symptoms, and other mechanisms, and we hypothesize that the condition results in accelerated cellular aging and neurodegeneration (for a review, see Miller and Sadeh, 2014). One of the pathways by which OXS damages neural integrity is via the enzyme 12/15-lipoxygenase (12/15-LOX). When levels of the antioxidant glutathione are depleted during OXS, 12/15-LOX becomes neurotoxic by attacking mitochondria and producing destructive reactive oxygen species (Pallast et al., 2009; Seiler et al., 2008). Given these characteristics, investigators have identified this enzyme as “the central executioner in an OXS-related neuronal death program” (Pallast et al., 2009, p882). 12/15-LOX is transcribed by the genes ALOX12 and ALOX15. The two sit in close physical proximity to each other on chromosome 17, share 86% sequence homology, and their enzymes are classified as “dual-specificity” due to similarities in their structure and function (Huret et al., 2013). In this study, based on evidence for the possible involvement of OXS in PTSD and the role of12/15-LOX in OXS-related neurodegeneration, we tested the hypothesis that genetic variation in ALOX12 and/or ALOX15 moderates the association between PTSD and CT and we used a novel model of the latent structure of CT to examine these associations.
2. Materials and Methods
2.1 Participants
Participants were 218 United States veterans of conflicts in Iraq and/or Afghanistan with structural magnetic resonance imaging (MRI) scans completed and processed for analysis. Individuals were excluded for history of seizures, prior serious medical illness (e.g. cerebrovascular accident, myocardial infarction, or diabetes), current suicidal and/or homicidal ideation, current diagnosis of bipolar disorder, schizophrenia or other psychotic disorder, cognitive disorder due to a general medical condition, history of moderate to severe traumatic brain injury, metal implant, shrapnel, aneurysm clip, pacemaker, or pregnancy. 89.0% of the veterans were male and the mean age was 32.09 (SD = 8.42, Range: 19.00 – 58.00 years). The majority (71.1%) self-identified as White, 15.6% as Hispanic or Latino, 9.6% as Black or African American, 2.3% as Asian, and 0.5% as American Indian; two did not provide self-reported race or ethnicity. Most (66.1%) served in the Army, 19.7 in the Marines, 9.2% in the Air Force, 4.1% in the Navy and 0.5% in the Coast Guard.
2.2 Procedures and Measures
Participants underwent a series of clinical interviews, MRI scans, and had blood samples drawn for genotyping. All relevant Institutional Review Boards and regulatory committees approved the study and participants gave written informed consent.
2.2.1 Clinical Assessment
PTSD was assessed using the Clinician Administered PTSD Scale (CAPS; Blake et al., 1993). Each DSM-IV PTSD criterion was queried using two items, one for the symptom frequency and one for intensity, which were then summed to yield a dimensional measure of severity. We employed a modified version of the CAPS to assess PTSD symptoms (and diagnosis) across three life intervals. First, symptoms present within the past 30 days (current) were assessed. Next, participants were interviewed about their worst post-deployment symptoms using an interval defined as “your worst 30-day period of PTSD symptoms since returning stateside or leaving the combat theater.” This is similar to the standard method for assessing “lifetime” PTSD but limited in this case to the post-deployment interval. If the last 30 days was identified as the worst period, then the veteran was asked when his/her symptoms initially became that severe (e.g., “How long ago did you start feeling this way? When did things get this bad?”) and that interval was used for worst post-deployment symptoms. Finally, if participants endorsed exposure to pre-military trauma, then pre-deployment symptoms were also assessed. The highest lifetime CAPS score from across these three intervals was used for the primary analyses described below. Lifetime alcohol abuse/dependence was assessed using the Structured Clinical Interview for DSM-IV (SCID-IV; First et al., 1994). CAPS descriptive statistics are provided in Table 1.
Table 1.
PTSD severity across the 3 assessment intervals.
| Minimum | Maximum | Mean | Std. Deviation | |
|---|---|---|---|---|
| CAPS Total Pre-military | 0 | 74 | 15.62 | 21.537 |
| CAPS Total Post-deployment | 0 | 130 | 56.58 | 35.196 |
| CAPS Total Current | 0 | 102 | 43.92 | 28.625 |
2.2.2 Cortical Thickness
CT was measured using a Siemens 3T TIM Trio system with a 12-radiofrequency channels head coil. Detailed descriptions of the scan acquisition and preprocessing methods are available in Corbo et al. (2014) and Lindemer et al. (2013). Thickness estimates were extracted using the FreeSurfer suite (Fischl and Dale, 2000), which generates mean CT values for 34 parcellations in each hemisphere of the cortex. CT factor analyses were based on the mixed ancestry sample of 218 participants with processed MRI scans.
2.2.3 Genotyping
DNA was isolated from peripheral blood samples on a Qiagen AutoPure instrument using Qiagen reagents. Sample DNA concentrations were determined using PicoGreen assays (Invitrogen). DNA quality and quantity was evaluated by a TaqMan® RNase P Detection assay (Applied Biosystems Assay, Life Technologies, Carlsbad, CA) with fluorescence detection on a 7900 Fast Real Time PCR System (Applied Biosystems, Life Technologies, Carlsbad, CA) according to the manufacturer's protocol. Each DNA sample was whole-genome amplified, fragmented and hybridized to an Illumina HumanOmni2.5-8 microarray according to the manufacturer's protocol (Illumina, San Diego, CA). Beadchips were imaged using the Illumina iScan System and analyzed with GenomeStudio V2011.1 software containing v1.9.4 Genotyping Module. Samples for which the call rate was <0.994 were repeated. This included all samples from a defective array. After repeats on new microarrays were completed, a GenomeStudio project was created from a custom genotyping cluster file, and all samples had call rates >0.994. After duplicate genotypes were removed, there were 307 unique samples. All samples passed a check for concordance between reported sex and X chromosome homozygosity (271 males and 36 females) performed in PLINK v1.07 (Purcell et al., 2007).
Ancestry in these samples was determined based on self-report and confirmed by a principal component (PC) analysis of 100 000 random common (minor allele frequency >0.05) SNPs in a joint analysis with reference-population data from the 1000 Genomes Project (1000 Genomes Project Consortium, 2012). The PC analysis was computed using EIGENSTRAT (Price et al., 2006). Of the 307 genotyped samples, 218 were of self-reported white non-Hispanic (WNH) ancestry. Four of these subjects were dropped because they had PC values > 6 standard deviations away from the mean for WNH subjects, leaving 214 genotype-verified WNH participants. Finally, WNH-only PCs were computed for the purposes of examining substructure within that sample. PCs for these 214 subjects were again computed using 100 000 SNPs. Of the 214 confirmed WNH subjects, 146 of these subjects had both CT and PTSD data available for analysis. In sum, the CT analysis was based on all 218 participants with CT data irrespective of ancestry. The genetic association analysis based on the 146 genome-confirmed WNH subjects. All 22 ALOX12 SNPs and 11 ALOX15 SNPs from the chip with minor allele frequency > 5% were included in the genetic association analysis.
2.2.4 Data Analyses
Exploratory and confirmatory factor analyses (EFA and CFA, respectively) were used to model the latent structure of CT. These approaches are well-suited for applications in which a set of observed indicators (in this case, CT parcellations) are theorized to covary as a function of a common underlying influence. EFA is purely descriptive. In contrast, CFA requires the investigator to specify essential aspects of the model a priori and can be used to test the relative fit of hypothesized alternatives. We began by conducting an EFA of the 34 right hemisphere parcellations. We then evaluated whether the best-fitting EFA model generalized to left hemisphere using CFA and compared the fit of the EFA-derived solution to viable alternatives. These analyses supported a best-fitting model composed of 4 correlated factors in each hemisphere. Additional details are available in the Supplementary Material.
We then extracted the 8 factor scores from these analyses and submitted them to an omnibus linear regression analysis in which PTSD severity, age, and sex were entered in an initial step, the 33 ALOX SNPs (coded additively) were evaluated in turn in a second step, and the 33 PTSD severity × SNP interactions (with the corresponding SNP main effect in the model) were each examined in a third step. Significance for SNP main effects and interactions was determined using Monte-Carlo null simulation with 10,000 replicates in which SNP data was randomly permuted between subjects. This analysis imposed multiple-testing control while taking into account the correlations between the CT factor scores and the correlations between the SNPs.
Finally, for a higher-resolution analysis of the specific cortical region(s) implicated in the omnibus genetic association analysis, we performed a vertex-wise analysis across the entire cortex in FreeSurfer's Qdec using the top SNP. In this analysis the covariates, main effects of SNP and PTSD severity, and the SNP × PTSD severity interaction were again entered in successive steps. The vertex-wise significance threshold was set at p < .005 and, given the direction of the association found in the omnibus analysis, negative contrasts were tested. Monte Carlo simulations (10,000 iterations) were again used to correct for multiple comparisons and the cluster-corrected threshold was set at p < .05.
3. Results
3.1 Exploratory Factor Analysis of Right Hemisphere CT Parcellations
EFA of the right hemisphere CT data suggested a four-factor solution. The primary loading of each parcellation in this model and its relative strength is depicted in Figure 1 (see Supplementary Figure S1 and Tables S1 & S3 for detailed results). Factor 1 (Frontal) included the traditional frontal parcellations plus the insula and the posterior cingulate cortex. Factor 2 (Temporal) was composed of the traditional temporal regions plus the isthmus of the cingulate cortex. Factor 3 (Parietal) was defined by traditional parietal regions plus two frontal regions (caudal middle frontal gyrus and precentral gyrus). Factor 4 (Occipital) was defined exclusively by occipital regions. The four factors were moderately inter-correlated (rs ranging from .32 to .52) and the proportion of variance in each indicator explained by its factor ranged from 12% (isthmus of the cingulate cortex) to 77% (superior frontal cortex). Adding age, alcohol abuse/dependence, sex, and PTSD severity to the model as correlates of each factor revealed significant negative correlations between age and all four CT factors (range of rs = -.25 to -.33) as well as a significant negative correlation between maximum PTSD severity and the right frontal factor (r = -.18, p < .05). No significant effects for alcohol abuse/dependence or sex were found. Then, to examine the generalizability of the PTSD severity effect to the clinical diagnosis of PTSD (rather than the dimensional severity score), we ran a linear regression using a dichotomous lifetime PTSD diagnosis variable and found the same patterns of association with right frontal factor CT, i.e., age (β = -.34, p < .001), sex (β = .13, p = .046), and PTSD (β = -.18, p = .007) were all significant.
Figure 1.
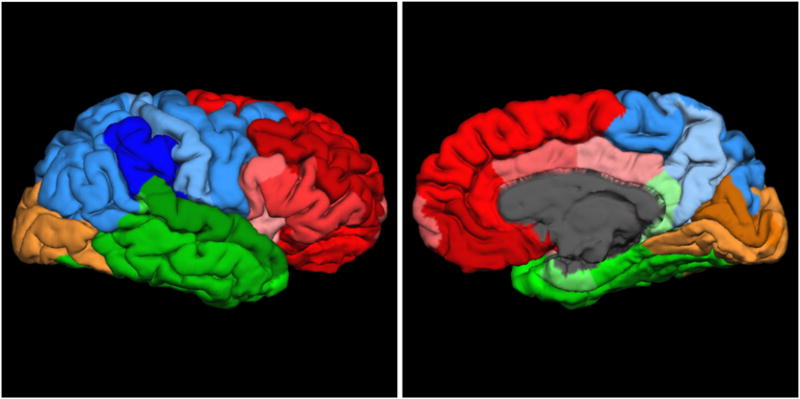
Lateral (left) and medial (right) views of the exploratory factor analysis-derived structure of right hemisphere cortical thickness. Red: frontal; blue: parietal; green: temporal, beige: occipital. Darker shading indicates parcellations with stronger loadings within a factor.
3.2 Confirmatory Factor Analyses of Left Hemisphere Parcellations
Next, we applied the primary loadings from the right hemisphere EFA model to the left hemisphere parcellations using CFA (see Table S4 for indicator assignments for this and each competing CFA model). This model yielded acceptable fit (Table S2 with all indicators loading significantly on their respective factors (Table S5). Due to the simpler structure of CFA, we found higher factor inter-correlations compared to the EFA model (rs = .59 to .81), though variance explained was comparable, ranging from 11% (isthmus of the cingulate cortex) to 70% (superior frontal cortex). Adding the covariates and PTSD severity as correlates of the four left hemisphere factors revealed global effects of age (rs = - .38 to -.44) but no significant association with PTSD severity or sex.
Our next step was to compare the fit of the EFA-derived four-factor model to a four-factor alternative in which each parcellation loaded on a factor corresponding to its traditional lobar location. Given the conventional distinction between the four primary lobes and a set of highly interconnected regions seated below the cortical surface (Mesulam, 2000), we also evaluated a five-factor model that included a limbic factor defined by subdivisions of the cingulate cortex, parahippocampal gyrus, and insula. Finally, we evaluated a six-factor solution corresponding to Chen et al.'s (2008) graph theoretical network analysis-based model of CT. Results showed that the alternative four- and six-factor models showed poorer fit relative to the EFA-derived model. The five-factor limbic model showed largely equivalent fit to the EFA-derived four-factor model but included a very high correlation between the limbic and frontal factors (r = .92). This redundancy combined with evidence from the right-side EFA indicating the superior fit of the original four-factor model led us to retain this as our final model. Factor scores from the best-fitting EFA (right) and CFA (left) models were then extracted for use in the genetic association analysis.
3.3 Genetic Association Analysis
In the genotype-confirmed White non-Hispanic subsample (n = 146), we began by examining the possible influence of genetic substructure on CT by regressing each CT factor on the first two genetic PCs. No significant associations were found (smallest p-value = 0.255) so the PCs were omitted from subsequent analyses. Next, we submitted the CT factor scores to an omnibus linear regression analysis that corrected for repeated testing across the 8 factors and 33 SNPs. The first step included the covariates PTSD severity, age, and sex. As before, PTSD severity showed a significant negative association with the right frontal factor (p < .04), age was associated with seven of the eight factors (ps < .001), and sex was non-significant. The SNPs were entered in the second block with none showing a significant main effect. When the PTSD severity × SNP interaction terms were entered in a third step, results showed significant multiple-testing corrected PTSD severity × SNP associations with the right frontal factor for two ALOX12 SNPs in perfect linkage disequilibrium (LD; rs1042357 and rs10852889; see Table 2 for detailed regression results and Supplementary Figure S3 for LD). Several other ALOX12 SNPs interacted with PTSD severity in predicting right frontal factor CT at a level that fell just short of multiple-testing corrected significance (see Table 3).
Table 2. Results of Hierarchical Regression Analysis of Main Effects and PTSD × ALOX12 Locusa Interaction on Right Frontal Factor Cortical Thickness.
| PTSD and Covariates Only | |||
|---|---|---|---|
|
| |||
| B | SE | p | |
| Int | 0.958 | 0.453 | 0.036 |
| Age | -0.034 | 0.009 | <0.001 |
| Sex | 0.363 | 0.293 | 0.217 |
| PTSD | --0.005 | 0.002 | 0.039 |
|
| |||
| Model R2 = 0.102 | |||
| With SNP Main Effect | |||
|
| |||
| B | SE | p | |
|
| |||
| Int | 1.007 | 0.455 | 0.029 |
| Age | -0.033 | 0.009 | <0.001 |
| Sex | 0.370 | 0.293 | 0.210 |
| PTSD | -0.005 | 0.002 | 0.061 |
| SNP | -0.110 | 0.111 | 0.326 |
|
| |||
| Model R2= 0.108 | |||
| With PTSD × SNP Interaction | |||
|
| |||
| B | SE | p | |
|
| |||
| Int | 0.566 | 0.456 | 0.216 |
| Age | -0.033 | 0.009 | <0.001 |
| Sex | 0.342 | 0.282 | 0.228 |
| PTSD | 0.003 | 0.003 | 0.228 |
|
| |||
| SNP | .671 | 0.247 | 0.008 |
| PTSD × SNP | -0.013 | 0.004 | 0.00062 |
| pcorb 0.049 | |||
|
| |||
| Model R2 = 0.180 | |||
Int, intercept; PTSD, posttraumatic stress disorder; SNP, single nucleotide polymorphism; SE, standard error.
The ALOX12 locus in this model was rs1042357/rs1085288 which were in perfect linkage disequilibrium.
pcor refers to significance estimated through simulation correcting across the 33 SNPs and 8 factor scores in the omnibus analysis. Significant p-values are bolded.
Table 3.
ALOX12 SNPs with uncorrected p-values less than .01 in the SNP × PTSD Interaction with right frontal factor scoresa.
| SNP | Estimate | Std Error | t-value | p-value | p-corrected |
|---|---|---|---|---|---|
| rs10852889 | -0.0126 | 0.003599 | -3.50198 | 0.00062 | 0.0499 |
| rs1042357 | -0.0126 | 0.003599 | -3.50198 | 0.00062 | 0.0499 |
| rs15966 | -0.01249 | 0.00361 | -3.46133 | 0.000713 | 0.0583 |
| rs2070590 | -0.01232 | 0.003603 | -3.41902 | 0.000824 | 0.0673 |
| rs1042356 | -0.01232 | 0.003603 | -3.41902 | 0.000824 | 0.0673 |
| rs1126667 | -0.01232 | 0.003603 | -3.41902 | 0.000824 | 0.0673 |
| rs434473 | -0.01232 | 0.003603 | -3.41902 | 0.000824 | 0.0673 |
| rs312466 | -0.0123 | 0.003627 | -3.39143 | 0.000914 | 0.0736 |
| rs202156062 | -0.0122 | 0.003678 | -3.31854 | 0.001153 | 0.0901 |
| rs2920421 | -0.0108 | 0.00388 | -2.78415 | 0.006126 | 0.3357 |
| rs11571326 | 0.015291 | 0.005499 | 2.780831 | 0.006174 | 0.3373 |
| rs2073438 | -0.01087 | 0.003952 | -2.74995 | 0.006748 | 0.3569 |
No ALOX15 SNPs showed uncorrected p-values less than .01 in the SNP × PTSD association with right frontal factor scores, so none are listed here.
Decomposition of the rs1042357/rs10852889 × PTSD interaction by genotype using partial correlations controlling for age and sex suggested an additive effect of the minor allele on the association between PTSD and right frontal factor scores (0 copies [n = 45]: r = .22, p = .16; 1 copy [n = 73]: r = -.29, p = .014; 2 copies [n = 28]: r = -.51, p = .008; Figure 2). Then, as before, we examined the generalizability of the PTSD severity effect to the clinical diagnosis of PTSD by comparing the association between genotype and right frontal factor scores in participants with versus without a lifetime diagnosis of PTSD. This decomposition revealed a significant effect of genotype in cases with PTSD (F[2,99] = 4.195, p = .0180) but not controls (F[2,47] = 1.853, p = .169). Finally, we performed several analyses examining possible effects of different types of trauma exposure on these associations but found no main effects or SNP interactions involving these variables (see Supplementary Materials).
Figure 2.
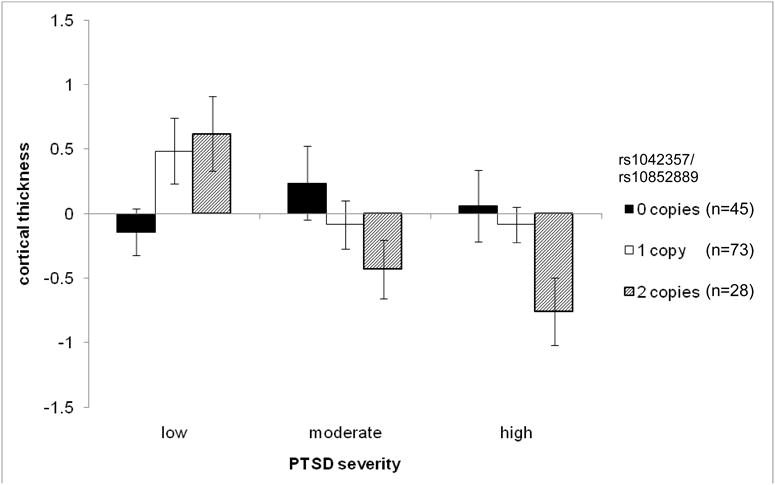
Mean right frontal factor cortical thickness (with standard errors) as a function of PTSD severity (tertiles) and rs1042357/rs10852889 genotype. ns in each cell were as follows [0 copies: low = 22, moderate = 9 high = 14] [1 copy: low = 24, moderate = 24, high = 25] [2 copies: low = 4, moderate = 16, high = 8]. Post-hoc analysis showed that the association between PTSD severity and right frontal cortical thickness was significant only in subjects homozygous for the minor allele (2 copies: r = -.46, p = .014; 1 copy: r = -.228, p < .052; 0 copies r = .210, p < .165).
3.4 Whole-cortex Vertex-wise Analysis
The whole-cortex analysis yielded a significant multiple-testing corrected PTSD severity × ALOX12 locus (rs1042357/rs10852889) association in two clusters of right pre-frontal cortex (Figure 3; Supplementary Table S6). The first spanned the rostral middle frontal gyrus and superior frontal gyrus; the second included the rostral anterior cingulate cortex and medial orbitofrontal cortex. There were no significant main effects of the ALOX12 peak SNPs or PTSD.
Figure 3.
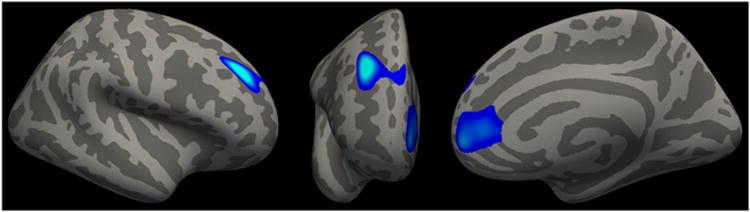
Lateral (left), anterior (middle), and medial (right) views of right hemisphere cortical thickness clusters associated with the rs1042357/rs10852889 × PTSD severity effect.
4. Discussion
This study used a genetically-informed model of the latent structure of the cortex to examine the influence of the OXS-related genes ALOX 12 and ALOX 15 on the association between PTSD and CT. Results identified a novel ALOX12 locus (rs1042357/rs10852889) that moderated the association between maximum lifetime PTSD severity and reduced thickness of the right prefrontal cortex and this effect explained approximately seven percent of the variance of CT in this region. We focused on these two genes based on prior evidence for the role of their corresponding enzymes in the link between OXS and neurodegeneration. 12-LOX (the enzyme transcribed by ALOX12) is the predominant LOX enzyme in the brain and is present in neurons, oligodendrocytes, and astrocytes (Bendani et al., 1995). During OXS, it initiates a cascade of cell death by attacking mitochondria and producing destructive reactive oxygen species (Li et al., 2009; Li et al., 1997; Pallast et al., 2009; Seiler et al., 2008). Evidence suggests that it is also involved in the modulation of tau metabolism (Giannopoulos et al., 2013) and its metabolites have been found in post-mortem brain and cerebral spinal fluid of patients with Alzheimer's disease (Practicò et al., 2004; Yao et al., 2005) and in neurons and endothelial cells surrounding stroke damage (Yigitkanli et al., 2013).
The two ALOX12 SNPs implicated in this study are located 4,186 base pairs apart on opposite sides of a block of SNPs in high LD spanning the second through eighth exons of the gene. rs10852889 is an intron variant positioned between the second and third exons; rs1042357 is in exon 8 and makes an exonic splicing enhancer with its A allele (Kim et al., 2010) implying that the A (risk) allele in this study could contribute directly to an abnormal enzyme through inaccurate splicing. Furthermore, rs1042357 is also in almost perfect LD with rs434473 and rs1126667, which are the only two common missense SNPs in ALOX12. Though the functional importance of these SNPs has yet to be examined in depth, all three were implicated previously in a study of schizophrenia (Kim et al., 2010).
We approached our examination of possible associations between ALOX12 and ALOX 15, PTSD, and CT using a data reduction approach based on modeling the latent structure of thickness variations across the cortex. Results of factor analyses supported the broad correspondence between the latent structure of CT and its manifest four lobe organization. Interestingly, these analyses also revealed several regions where parcellation loadings deviated from their traditional functional subdivisions and/or crossed boundaries defined by macroanatomical landmarks. For example, in our best-fitting model, the precentral gyrus and caudal middle frontal gyrus of the frontal cortex loaded more strongly on the parietal factor than on the frontal factor. These regions comprise part of the primary motor/premotor cortex and differ substantially in function and cytoarchitecture from the other primarily prefrontal regions that defined the frontal factor. Specifically, the primary motor/premotor cortex has highly specialized interconnections with the parietal cortex that subserve functions such as prehension and visuomotor processing (Grafton et al., 1992; Mesulam, 2000). In addition, whereas the prefrontal cortex is characterized by its granular, prominent six layer cell structure, the motor area lacks cell layer IV and the premotor area has only a poorly developed layer IV. That such subtle and intricate anatomical and functional commonalities were detected by factor analysis highlights the sensitivity of the approach and its potential value for future studies of CT.
Previous investigations have used principal components analysis for cortical data reduction (Narr et al., 2005) but that method does not distinguish the common variance underlying a set of indicators from error variance. Instead, error remains in the extracted components reducing their reliability. In contrast, analyses based on the common factor model, like those used in this study— whether exploratory or confirmatory—model error variance separately from common variance and can therefore be expected to provide greater statistical power for association analysis. Furthermore, multiple indicator models like these are well-suited for identifying effects that are small but widely distributed and/or characterized by abnormalities in the relationships between factors. Finally, twin studies indicate that multivariate representations of CT correspond more closely to their genetic substrate than do the individual parcellations that define them (Chen et al., 2013, 2012; Lerch et al., 2006; Rimol et al., 2010).
We followed our multivariate modeling with a vertex-wise analysis designed to pin-point the locus of the ALOX12 SNP × PTSD interaction and found two whole-cortex multiple-testing corrected clusters of reduced CT. The first was located primarily in the right dorsolateral prefrontal cortex—an area known to play a key role in working memory and emotion regulation processes (Delgado et al., 2008). The second spanned the rostral anterior cingulate cortex and medial orbital-frontal cortex, which have been implicated in numerous prior structural and functional MRI studies of PTSD (Kühn and Gallinat, 2013). These regions have neuroanatomical and functional connections with the amygdala and other subcortical components of emotional response systems (Bush et al., 2000; Etkin et al., 2006) and hypo-activation in these areas is thought to play a role in the emotion regulation and fear extinction deficits observed in PTSD (Patel et al., 2012).
These findings should be considered in light of several limitations. First, the cross-sectional design precludes conclusions about the direction of association between PTSD severity and CT. Given prior evidence pointing to the possible involvement of OXS in PTSD (Miller and Sadeh, 2014) and the known role of12/15-LOX in OXS-related neurodegeneration, we believe it is plausible that ALOX12 variants moderate the effects of PTSD on CT though their influence on 12-LOX; however, other mechanisms are also conceivable and only experimental and longitudinal studies can provide definitive answers. Second, the sample size, while large for a PTSD neuroimaging-genetic study, was modest for factor analysis and genetic association analysis, so replication analyses using independent datasets are needed. Third, the sample was based on a predominantly male cohort of military veterans so it is unclear to what extent results will generalize to other PTSD samples. Fourth, given the degree of LD in ALOX12, we were unable to identify the specific locus underlying the observed association. To do so would require functional work, analyses in larger samples, and/or samples from different ancestral populations with different LD structures.
4.1 Conclusions
To conclude, the findings of this study implicated a novel locus in the OXS gene ALOX12 that moderated the association between PTSD and reduced thickness of the right prefrontal cortex. The study also illustrated a unique approach to neuroimaging-genetic analyses using a model of the latent structure of CT. These analytic methods are well-established in certain fields (most notably, in psychometric scale development and construct validation) but as we have shown, they are also potentially well-suited to modeling neural structures and networks for genetic association analysis and other applications.
Supplementary Material
Highlights.
We examined the relationship of ALOX12 and ALOX15 SNPs to PTSD and cortical thickness.
ALOX12 and ALOX15 transcribe an enzyme involved in oxidative stress-related neuronal damage.
We used a novel factor analytic approach to model the structure of the cortex.
An ALOX12 locus moderated the association between PTSD and reduced cortical thickness.
Whole-cortex vertex-wise analysis showed this effect to be localized to right prefrontal cortex.
Acknowledgments
This research and preparation of this manuscript was supported by a National Institute of Mental Health (NIMH) grant awarded to MWM (R21MH102834, “Neuroimaging Genetics of PTSD”) and the Translational Research Center for TBI and Stress Disorders (TRACTS), a U.S. Department of Veterans Affairs (VA) Rehabilitation Research and Development (RR&D) Traumatic Brain Injury Center of Excellence (B9254-C) at VA Boston Healthcare System. This research is the result of work supported with resources and the use of facilities at the Pharmacogenomics Analysis Laboratory, Research and Development Service, Central Arkansas VA Healthcare System, Little Rock, Arkansas. This work was also supported by a Career Development Award to EJW from the VA Clinical Sciences Research and Development Program. The contents of this manuscript do not represent the views of the VA, NIH or the United States Government.
Footnotes
Appendix A. Supplementary Data: Supplementary information associated with this article can be found in the online version at the Psychoneuroendocrinology website.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bendani MK, Palluy O, Cook-Moreau J, Beneytout JL, Rigaud M, Vallat JM. Localization of 12-lipoxygenase mRNA in cultured oligodendrocytes and astrocytes by in situ reverse transcriptase and polymerase chain reaction. Neurosci Lett. 1995;189:159–162. doi: 10.1016/0304-3940(95)11482-c. [DOI] [PubMed] [Google Scholar]
- Blake DD, Weathers FW, Nagy LM. A clinician rating scale for assessing current and lifetime PTSD: the CAPS-I. Behav Ther. 1993;18:187–188. [Google Scholar]
- Bush G, Luu P, Posner MI. Cognitive and emotional influences in anterior cingulate cortex. Trends Cogn Sci. 2000;4:215–222. doi: 10.1016/s1364-6613(00)01483-2. [DOI] [PubMed] [Google Scholar]
- Chen CH, Fiecas M, Gutiérrez ED, Panizzon MS, Eyler LT, Vuoksimaa E, Thompson WK, Fennema-Notestine C, Haglar DJ, Jr, Jernigan TL, Neale MC, Franz CE, Lyons MJ, Fischl B, Tsuang MT, Dale AM, Kremen WS. Genetic topography of brain morphology. Proc Natl Acad Sci U S A. 2013;110:17089–17094. doi: 10.1073/pnas.1308091110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Gutierrez ED, Thompson W, Panizzon MS, Jernigan TL, Eyler LT, Fennema-Notestine C, Jak AJ, Neale MC, Franz CE, Lyons MJ, Grant MD, Fischl B, Seidman LJ, Tsuang MT, Kremen WS, Dale AM. Hierarchical genetic organization of human cortical surface area. Science. 2012;335:1634–1636. doi: 10.1126/science.1215330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, He Y, Rosa-Neto P, Germann J, Evans AC. Revealing modular architecture of human brain structural networks by using cortical thickness from MRI. Cereb Cortex. 2008;18:2374–2381. doi: 10.1093/cercor/bhn003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbo V, Salat DH, Amick MM, Leritz EC, Milberg WP, McGlinchey RE. Reduced cortical thickness in veterans exposed to early life trauma. Psychiatry Res: Neuroimaging. 2014;223:53–60. doi: 10.1016/j.pscychresns.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado MR, Nearing KI, LeDoux JE, Phelps EA. Neural circuitry underlying the regulation of conditioned fear and its relation to extinction. Neuron. 2008;59:829–838. doi: 10.1016/j.neuron.2008.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etkin A, Egner T, Peraza DM, Kandel ER, Hirsch J. Resolving emotional conflict: a role for the rostral anterior cingulate cortex in modulating activity in the amygdala. Neuron. 2006;51:871–882. doi: 10.1016/j.neuron.2006.07.029. [DOI] [PubMed] [Google Scholar]
- First MB, Spitzer RL, Gibbon M, Williams J. Structured Clinical Interview for Axis I DSM-IV Disorders. New York Psychiatric Institute, Department of Biometrics Research; New York: 1994. [Google Scholar]
- Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A. 2000;97:11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannopoulos PF, Joshi YB, Chu J, Pratico D. The 12-15-lipoxygenase is a modulator of Alzheimer's related tau pathology in vivo. Aging Cell. 2013;12:1082–1090. doi: 10.1111/acel.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafton ST, Mazziotta JC, Woods RP, Phelps ME. Human functional anatomy of visually guided finger movements. Brain. 1992;115:565–587. doi: 10.1093/brain/115.2.565. [DOI] [PubMed] [Google Scholar]
- Hovatta I, Juhila J, Donner J. Oxidative stress in anxiety and comorbid disorders. Neurosci Res. 2010;68:261–275. doi: 10.1016/j.neures.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Huret JL, Ahmad M, Arsaban M, Bernheim A, Cigna J, Desangles F, Guignard JC, Jacquemot-Perbal MC, Labarussias M, Leberre V, Malo A, Morel-Pair C, Mossafa H, Potier JC, Texier G, Viguié F, Yau Chun Wan-Senon S, Zasadzinski A, Dessen P. Atlas of genetics and cytogenetics in oncology and haematology in 2013. Nucleic Acids Res. 2013;41(Database issue):D920–D924. doi: 10.1093/nar/gks1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Kim HJ, Park JK, Kim JW, Chung JH. Association between polymorphisms of arachidonate 12-lipoxygenase (ALOX12) and schizophrenia in a Korean population. Behav Brain Funct. 2010;6:1–4. doi: 10.1186/1744-9081-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn S, Gallinat J. Gray matter correlates of posttraumatic stress disorder: a quantitative meta-analysis. Biol Psychiatry. 2013;73:70–74. doi: 10.1016/j.biopsych.2012.06.029. [DOI] [PubMed] [Google Scholar]
- Lerch JP, Worsley K, Shaw WP, Greenstein DK, Lenroot RK, Giedd J, Evans AC. Mapping anatomical correlations across cerebral cortex (MACACC) using cortical thickness from MRI. Neuroimage. 2006;31:993–1003. doi: 10.1016/j.neuroimage.2006.01.042. [DOI] [PubMed] [Google Scholar]
- Li J, O W, Li W, Jiang ZG, Ghanbari HA. Oxidative stress and neurodegenerative disorders. Int J Mol Sci. 2013;14:24438–24475. doi: 10.3390/ijms141224438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Maher P, Schubert D. A role for 12-lipoxygenase in nerve cell death caused by glutathione depletion. Neuron. 1997;19:453–463. doi: 10.1016/s0896-6273(00)80953-8. [DOI] [PubMed] [Google Scholar]
- Li J, Wang H, Rosenberg PA. Vitamin K prevents oxidative cell death by inhibiting activation of 12-lipoxygenase in developing oligodendrocytes. J Neurosci Res. 2009;87:1997–2005. doi: 10.1002/jnr.22029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemer ER, Salat DH, Leritz EC, McGlinchey RE, Milberg WP. Reduced cortical thickness with increased lifetime burden of PTSD in OEF-OIF veterans and the impact of comorbid TBI. Neuroimage Clin. 2013;2:601–611. doi: 10.1016/j.nicl.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesulam M. Principles of behavioral and cognitive neurology. Oxford University Press; New York: 2000. [Google Scholar]
- Miller MW, Sadeh N. Traumatic stress, oxidative stress and posttraumatic stress disorder: neurodegeneration and the accelerated-aging hypothesis. Mol Psychiatry. 2014;19:1156–1162. doi: 10.1038/mp.2014.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narr KL, Bilder RM, Toga AW, Woods RP, Rex DE, Szeszko PR, Robinson D, Sevy S, Gunduz-Bruce H, Wang YP, DeLuca H, Thompson PM. Mapping cortical thickness and gray matter concentration in first episode schizophrenia. Cereb Cortex. 2005;15:708–719. doi: 10.1093/cercor/bhh172. [DOI] [PubMed] [Google Scholar]
- O'Leary DD, Chou SJ, Sahara S. Area patterning of the mammalian cortex. Neuron. 2007;56:252–269. doi: 10.1016/j.neuron.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Pallast S, Arai K, Ziaoying W, Lo EH, Leyen K. 12/15-Lipoxygenase targets neuronal mitochondria under oxidative stress. J Neurochem. 2009;111:882–889. doi: 10.1111/j.1471-4159.2009.06379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palta P, Samuel LJ, Miller ER, III, Szanton SL. Depression and oxidative stress: results from a meta-analysis of observational studies. Psychosom Med. 2014;76:12. doi: 10.1097/PSY.0000000000000009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R, Spreng RN, Shin LM, Girard TA. Neurocircuitry models of posttraumatic stress disorder and beyond: a meta-analysis of functional neuroimaging studies. Neurosci & Biobehav Rev. 2012;36:2130–2142. doi: 10.1016/j.neubiorev.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Practicò D, Zhukareva V, Yao Y, Uryu K, Funk CD, Lawson JA, Trojanowski JQ, Lee VMY. 12/15-lipoxygenase is increased in Alzheimer's disease: possible involvement in brain oxidative stress. Am J Pathol. 2004;164:1655–1662. doi: 10.1016/S0002-9440(10)63724-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimol LM, Panizzon MS, Fennema-Notestine C, Eyler LT, Fischl B, Franz CE, Hagler DJ, Lyons MJ, Neale MC, Pacheco J, Perry ME, Schmitt JE, Grant MD, Seidman LJ, Thermenos HW, Tsuang MT, Eisen SA, Kremen WS, Dale AM. Cortical thickness is influenced by regionally-specific genetic factors. Biol Psychiatry. 2010;67:493–499. doi: 10.1016/j.biopsych.2009.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavone S, Jaquet V, Trabace L, Krause K. Severe life stress and oxidative stress in the brain: from animal models to human pathology. Antioxid Redox Signal. 2013;18:1475–1490. doi: 10.1089/ars.2012.4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler A, Schneider M, Förster H, Roth S, Wirth EK, Culmsee C, Plesnila N, Kremmer E, Rådmark O, Wurst W, Bornkamm GW, Schweizer U, Conrad M. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent-and AIF-mediated cell death. Cell Metab. 2008;8:237–248. doi: 10.1016/j.cmet.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Wilson CB, McLaughlin LD, Nair A, Ebenezer PJ, Dange R, Francis J. Inflammation and oxidative stress are elevated in the brain, blood, and adrenal glands during the progression of post-traumatic stress disorder in a predator exposure animal model. PLoS One. 2013;8:e76146. doi: 10.1371/journal.pone.0076146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Clark CM, Trojanowski JQ, Lee VM, Practico D. Elevation of 12/15 lipoxygenase products in AD and mild cognitive impairment. Ann Neurol. 2005;58:623–626. doi: 10.1002/ana.20558. [DOI] [PubMed] [Google Scholar]
- Yigitkanli K, Pekcec A, Karatas H, Pallast S, Mandeville E, Joshi N, Smirnova N, Gazaryan I, Ratan RR, Witztum JL, Montaner J, Holman TR, Lo EH, van Leyen K. Inhibition of 12/15-lipoxygenase as therapeutic strategy to treat stroke. Ann Neurol. 2013;73:129–135. doi: 10.1002/ana.23734. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


