Abstract
Fibroblasts play an important role in the wound-healing process by generating extracellular matrix (ECM) and undergoing differentiation into myofibroblasts, but these cells can also be involved in pathologic remodeling of tissue. Nascent ECM provides a substrate for re-epithelialization to occur, restoring damaged tissue to a functional state. Dysregulation of this process can result in fibrosis—stiffening and scarring of the tissue. Current treatments cannot halt or reverse this process. The molecular mechanisms underlying fibrotic dysregulation are poorly understood, providing an untapped pool of potential therapeutic targets. Transforming growth factor-β (TGF- β) and adhesion signaling are involved in inducing fibroblast differentiation into α-smooth muscle actin (α SMA) expressing myofibroblasts, while prostaglandin E2 (PGE2) has been shown to antagonize TGF β signaling; however, the temporal and mechanistic details of this relationship have not yet been fully characterized. We measured α SMA, a marker of fibroblast to myofibroblast differentiation, as a function of: TGF-β1 receptor–ligand complex internalization, PGE2 binding, and adhesion signaling and developed a mathematical model capturing the molecular mechanisms of fibroblast differentiation. Using our model, we predict the following: Periodic dosing with PGE2 temporarily renders fibroblasts incapable of differentiation and refractory to additional TGF-β1 stimulation; conversely, periodic dosing with TGF-β1 in the presence of PGE2 induces a reduced signal response that can be further inhibited by the addition of more PGE2. Controlled fibroblast differentiation is necessary for effective wound healing; however, excessive accumulation of α SMA-expressing myofibroblasts can result in fibrosis. Homeostasis of α SMA in our model requires a balance of positive and negative regulatory signals. Sensitivity analysis predicts that PGE2 availability, TGF-β1 availability, and the rate of TGF-β1 receptor recycling each highly influence the rates of α SMA production. With this model, we are able to demonstrate that regulation of both TGF-β1 and PGE2 signaling levels is essential for preventing fibroblast dysregulation.
Keywords: TGF-β1, PGE2, Mathematical model
1 Author Summary
Fibrosis is a common disease feature characterized by the stiffening and scarring of tissue. When fibrosis occurs in lungs, it can result in a poor prognosis for patients and a 2–4years life expectancy, resulting in approximately 40,000 deaths annually. Fibrosis results from dysregulation of specific cell types that are normally involved in promoting wound closure and healing. When these cells are not properly regulated, deposition of scar tissue and reduction in lung flexibility occur making it difficult for patients to breathe. Although the specific action of this cellular dysregulation has not been defined, fibrosis has been shown to be related to cellular communication. There is a great need to understand the underlying mechanisms responsible for fibrotic disease in order to provide therapeutic options for patients. We use a combination of mathematical modeling and wetlab experimentation to identify key regulatory factors regulating this process, and predict potential therapeutic targets. Our work suggests that in order to control fibrotic disease, we need to establish a balanced chemical environment of positive and negative signaling in affected areas of lung.
2 Introduction
Fibroblasts are necessary for tissue regeneration and wound healing (Velnar et al. 2009; Guo and Dipietro 2010). Fibroblasts exist in tissue in a quiescent state (Desmouliere et al. 1993) until disruption of the tissue structure triggers differentiation into myofibroblasts (Desmouliere et al. 1993). This differentiation is characterized by secretion of extracellular matrix (ECM) proteins and production of α-smooth muscle actin (α SMA), a cytoskeletal protein that enables contraction and tissue remodeling (Velnar et al. 2009; Guo and Dipietro 2010). Previous work has identified several critical functions performed by myofibroblasts including ECM protein secretion (Diegelmann and Evans 2004; Ramasastry 2005; Goldman 2004) and remodeling of damaged tissue. Dysregulation of fibroblast to myofibroblast differentiation can result in severe pathology that can compromise function of affected tissue; however, excessive ECM secretion can lead to detrimental tissue remodeling and fibrosis (Strieter 2008). Fibrosis, the stiffening and scarring of tissue, can result in poor clinical outcomes depending on the extent of the affected tissue, and patients with fibrosis often have poor prognoses (Tomioka et al. 2007). Idiopathic pulmonary fibrosis (IPF), for example, results in decreased inspiration and expiration capacity and has an average prognosis of 2–4years (Ley et al. 2011; Taniguchi et al. 2011; King et al. 2014; Richeldi et al. 2014; Xaubet et al. 2014). There are currently two treatments available for pulmonary fibrosis in the USA, both of which provide only a moderate extension of prognosis (about 6months) (Ley et al. 2011; Taniguchi et al. 2011; King et al. 2014; Richeldi et al. 2014; Xaubet et al. 2014). There are no available treatments that halt or reverse fibrosis. We aim to understand why fibroblast differentiation becomes dysregulated and which signaling mechanisms drive this outcome in order to develop new therapeutics for this and other fibrotic diseases.
The cytokine transforming growth factor-β1 (TGF-β1) has been shown to play a critical role in fibrosis-associated pathologies. TGF-β1 is a major contributing factor to pulmonary complications and fibrosis following bone marrow transplant (Coomes et al. 2011, 2010). TGF-β1 sustains myofibroblast function and can exacerbate IPF (Camelo et al. 2014; Lilja-Maula et al. 2014). It has been clearly established that TGF-β1 is key in driving development of fibrosis; however, there are still open questions as to the mechanisms involved in TGF-β1-induced dysregulation of fibroblast differentiation and myofibroblast function. To identify which factors contribute to fibrotic dysregulation and predict how best to inhibit this process, we construct a mathematical model describing the contribution of molecular mechanisms driving fibroblast to myofibroblast differentiation.
TGF-β1 is a growth factor and member of the transforming growth factor-β superfamily of cytokines known to drive differentiation of fibroblasts as well as secretion of ECM proteins (Bettinger et al. 1996; Broekelmann et al. 1991; Fine and Goldstein 1987). TGF-β1 is secreted in a latent form as a homodimer of TGF-β1 bound to a homodimer of the latency-associated peptide (LAP) referred to as the small latent TGF-β1 complex (42kDa) (Annes et al. 2003). It is stored in the ECM as a large latent TGF-β1 complex (290 kDa) which includes the small complex and an additional protein referred to as the latent TGF-β1 binding protein (LTBP1) (Horiguchi et al. 2012). Release of TGF-β1 from the LAP produces an active TGF-β1 molecule. Latent TGF-β1 can be activated by proteolytic cleavage, mechanical extraction, and changes in pH of the surrounding environment (Annes et al. 2003). TGF-β1 is bound in its active form by a specific membrane receptor complex of ALK5 (TGF-β1 receptor 1) and TGF β 1RII (TGF-β1 receptor II) (Finnson et al. 2013; Rider and Mulloy 2010). The effect of TGF-β1 on cells is tissue- and cell-specific. For example, TGF-β1 can induce differentiation of fibroblasts through downstream canonical SMAD2/3 and non-canonical rho/ROCK signaling cascades (Thannickal et al. 2003). In keratinocytes, TGF-β1 causes cell cycle inhibition (Coffey et al. 1988; Pietenpol et al. 1990), and it has been shown to play several opposing roles in T cell differentiation and maturation (Li et al. 2007; Marie et al. 2006; Leveen et al. 2005; Kronenberg and Rudensky 2005; Liston and Rudensky 2007; Liu et al. 2008; Bendelac et al. 1997). Because of the cell specificity associated with TGF-β1 signaling, we focus on identifying fibroblast-specific mechanisms of action.
Several mathematical models have been developed to complement experimental approaches in exploring TGF-β1 receptor–ligand signaling dynamics in other cell types (Vilar et al. 2006; Vizan et al. 2013; Zi and Klipp 2007). Vilar et al. showed that the ratio of constitutive degradation of TGF-β1 receptors to degradation induced by ligand binding dictates whether cellular responses to TGF-β1 are transient or permanently elevated for keratinocytes and pancreatic cancer cell lines (Vilar et al. 2006). Zi et al. identified that the duration of a cellular response to TGF-β1 is dependent on whether the receptor–ligand complex is internalized into a clatherin-coated or clatherin-independent endosomal compartment. In keratinocytes, if the predominant form of endocytosis is clatherin-independent, the response will be transient. However, if the predominate form of endocytosis is clatherin-dependent, the response will be prolonged (Zi et al. 2011). Recently, Vizan et al. demonstrated that keratinocytes experience a refractory state following a TGF-β1 signaling event, where cells are temporarily insensitive to further TGF-β1 stimulation. They showed that the duration of this refractory period is dependent on the rate of receptor turnover and the ratio of ligand-induced to constitutive receptor degradation for keratinocytes (Vizan et al. 2013). Together, these models emphasize the importance of endocytosis and constitutive versus ligand-induced receptor degradation in determining the cellular response to TGF-β1 signaling.
Fibroblast responses to TGF-β1 differ from keratinocyte and pancreatic cell responses. As a result, there are mechanisms specific to fibroblasts that were not considered in these prior mathematical models. Fibroblast responses to TGF-β1 are highly dependent on the simultaneous presence of adhesion signaling (Thannickal et al. 2003). Adhesion signaling through integrin binding is necessary for TGF-β1-induced fibroblast to myofibroblast differentiation to occur (Thannickal et al. 2003). Previous work has demonstrated that fibroblasts plated onto plastic surfaces, such as a tissue culture plate, respond to TGF-β1 to induce differentiation (measured by synthesis of α -SMA). In contrast, when fibroblasts are suspended in liquid culture, they show no response to treatment with TGF-β1 (Thannickal et al. 2003). These data indicate that fibroblasts require adhesion in order to respond to TGF-β1 stimulation (Thannickal et al. 2003). The stiffness of the substrate adhered to is influential in the strength of the fibroblast response to TGF-β1; the stiffer the substrate, the greater the adhesion signaling, and stronger the response (Liu et al. 2010; Huang et al. 2012; Tamada et al. 2004; Sawada et al. 2006; de Ulrich et al. 2009; Peyton et al. 2008; Giannone and Sheetz 2006). Another important factor in the regulation of TGF-β1 signaling in fibroblasts is a negative regulator present in the system, namely prostaglandin E2 (PGE2). PGE2 indirectly inhibits TGF-β1 signaling by inhibition of FAK in the adhesion signaling cascade (Thomas et al. 2007) rather than acting directly on TGF-β1 by limiting SMAD phosphorylation (Thomas et al. 2007). No other forms of cross talk between PGE2 and the canonical TGF-β1 signaling cascade have been demonstrated. PGE2 has been shown to inhibit adhesion signaling and in turn inhibit fibroblast responses to TGF-β1 (Thomas et al. 2007; Kolodsick et al. 2003; Fine et al. 1989; Tian and Schiemann 2010). Fibroblasts are exposed in vivo to PGE2 secreted by epithelial cells, and it has been proposed that this constitutive signaling induces fibroblast quiescence, maintaining homeostasis of the tissue environment (Lama et al. 2002; Moore et al. 2003). High levels of PGE2 have been linked to increased fibroblast apoptosis (Huang et al. 2009). Previous work has shown that fibroblasts can lose sensitivity to PGE2 in vitro by down-regulating EP2 receptor synthesis (Saltzman et al. 1982). This is a phenomenon that is also seen during fibrotic responses in the lung (Moore et al. 2005). PGE2 can mediate functions of multiple cells via binding to four unique receptors (Sugimoto and Narumiya 2007). Because it is a strong inhibitor of adhesion signaling, however, understanding the mechanistic relationships between TGF-β1, adhesion signaling, and PGE2 allows us to identify environmental conditions favorable to healthy wound resolution as well as signaling mechanisms that are key to establishing those conditions.
Thus, to gain further insight into the role of TGF-β1 in regulation of fibroblast differentiation into a myofibroblast, we take a systems’ biology approach to identify the influence of molecular-scale mechanisms of TGF β signaling on regulation of this transition. We use a combination of in vitro experimentation, mathematical modeling, and statistical analyses to identify key mechanisms driving fibroblast differentiation and dysregulation. We developed a nonlinear ordinary differential equation model that tracks the temporal concentrations of key species (Table 1) in receptor–ligand binding, trafficking, and signaling cascades to evaluate how these events drive α SMA synthesis (Fig. 1). We build and test the model with data derived from fibroblast differentiation experimental studies performed herein, and we analyze the model to predict which mechanisms are affecting fibroblast regulation. These factors are potential therapeutic targets for fibrosis.
Table 1.
Model variables
| Symbol | Definition | Units |
|---|---|---|
| TGF- β 1 receptor–ligand variables | ||
| TGF β 1lat | Latent TGF- β 1 | |
| TGF β 1act | Activated TGF- β 1 | |
| R surf | Free receptor on the cell surface | |
| R int | Internalized free receptors in the cytoplasm | |
| TRCsurf | Receptor–ligand complexes on the surface of the cell | |
| TRCint | Internalized receptor–ligand complexes in the cytoplasm | |
| PGE2 input variables | ||
| PGE2Sol | Soluble prostaglandin E2 | |
| PGE2int | Internalized prostaglandin E2 | |
| α-Smooth muscle actin output variables | ||
| PRDS | Post-receptor simplified downstream signaling events | |
| αSMA | α-Smooth muscle actin | |
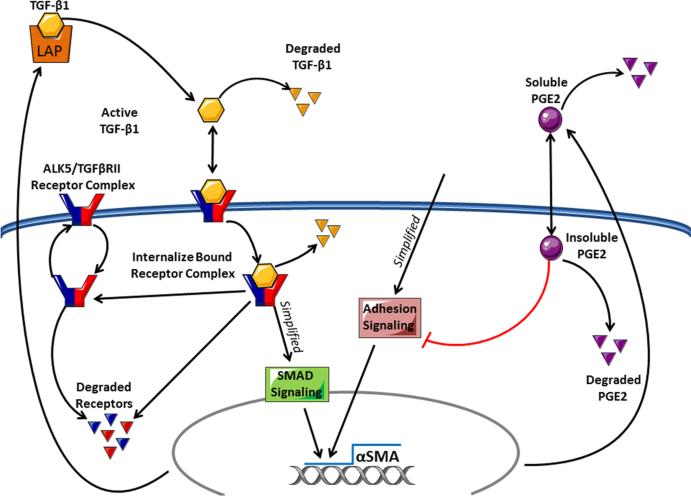
Fig. 1.
Processes relevant to fibroblast differentiation. Latent TGF-β1 can be activated and then is able to bind surface receptors. Receptor–ligand complexes are internalized, initiating a downstream signaling cascade which, in combination with adhesion signaling, induces the synthesis of α-smooth muscle actin. PGE2 can inhibit the adhesion signaling, preventing the completion of the signal cascade and protein synthesis. Our model tracks latent TGF-β1, active TGF-β1, free surface receptors, free cytoplasmic receptors, bound surface receptors, bound cytoplasmic receptors, the number of bound receptor internalization events, extracellular PGE2, intracellular PGE2, and α-smooth muscle actin synthesis. We have coarse-grained the adhesion, SMAD, and PGE2 signaling pathways (Color figure online)
3 Results
3.1 PGE2 is Necessary to Explain In Vitro Data of α SMA Synthesis
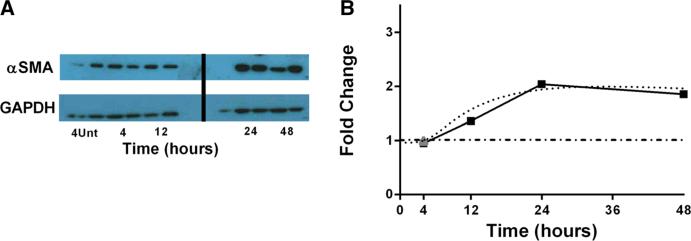
We first evaluated the level of α SMA synthesized in the presence of TGF-β1. We compared fold changes in the concentration of α SMA at 4, 12, 24, or 48h in the presence of an initial concentration of 1ng/ml of TGF-β1 to 4h untreated. Data show no increase at
4h post-treatment over the untreated and an approximately 2.5-fold increase in α SMA at 12h post-treatment that is maintained at 48h post-treatment (Fig. 2A). These results mirror our earlier findings when stimulating fibroblasts with 2ng/ml TGF-β1, indicating that TGF-β1 is not limiting. We also simulated fibroblasts with our mathematical model under the same experimental conditions (untreated or 1ng/ml of TGF-β1) and measured the α SMA concentration at 4, 12, 24, 48h. We parameterized the model using values derived from previously published data or estimated using uncertainty analysis (Table 2) and then validated the model against experimental data generated in our laboratory (Fig. 3). Our simulations show an approximately twofold increase in α SMA by 4h post-treatment and a sixfold increase by 12h post-treatment which was maintained at 48h (data not shown). These levels are much higher than the experimental data, indicating a lack of negative regulation in the system. We predicted that the absence of negative regulation is responsible for the discrepancy in experimental and simulation results. We introduced negative regulation by PGE2 into our model and simulated fibroblasts (untreated or 2ng/ml of TGF-β1) and measured the α SMA concentration at 4, 12, 24, 48 hours in the presence of 100μM PGE2 and found no increase in α SMA at 4h post-treatment over the untreated and an approximately 2.5-fold increase at 12 hours post-treatment which is maintained at 48h post-treatment. Simulation results closely match the experimental data consistent with the idea that a negative regulator is necessary to explain in vitro data regarding fibroblast differentiation (Fig. 2B).
Fig. 2.
(Colour figure online) α SMA time course studies and simulations. a α SMA measured in 3T12 fibroblasts at either 4h untreated or 4, 12, 24, or 48h treated with 1 ng/ml of activated TGF-β1 using Western blot and densitometry analysis. N = 2/condition. b Simulation of α SMA production. Solid line with filled squares representing the experimental data described in part (a) and gray triangle representing 4-h untreated sample using 3T12 fibroblast cell line and 1 ng/ml of activated exogenous TGF-β1, dotted curve representing simulation results following treatment with 1 ng/ml of activated exogenous TGF-β1, and PGE2 inhibition, and dot-dashed line representing simulation control with no TGF-β1 treatment. We have previously published similar kinetics using 2 ng/ml TGF β, suggesting that TGF-β1 is in excess in this system (7)
Table 2.
Model parameters
| Symbol | Parameter | Value | Range | Unit | Source |
|---|---|---|---|---|---|
| General parameters | |||||
| Vol | Volume of media | 2.0 | N/A | mL | N/A |
| Cells | Total number of cells | 5.0 × 105 | N/A | # | N/A |
| TGF-β1 receptor-ligand parameters | |||||
| Latent TGF-β1 Synthesis | 2.4 × 10–20 | 1.0 × 10–20–1.0 × 10–10 | moles/min × volume | est. | |
| Latent TGF-β1 Degradation | 0.006 | N/A | 1/min | Wakefield et al. (1990) | |
| Latent TGF-β1 Activation | 0.06 | 1.0 × 10–4–1.0 × 103 | 1/min | est. | |
| Latent TGF-β1-induced synthesis | 0.06 | 1.0 × 10–4–1.0 × 103 | 1/min | est. | |
| Active TGF-β1 Receptor-Ligand Binding | 1.38 × 109 | N/A | 1/min | De Crescenzo et al. (2003) | |
| Active TGF-β1 Receptor-Ligand Dissociation | 9.0 × 10–3 | N/A | 1/min | De Crescenzo et al. (2003) | |
| Active TGF-β1 Degradation | 0.258 | N/A | 1/min | Wakefield et al. (1990) | |
| TGF-β1 Receptor Synthesis | 4.0 | N/A | #/(min) × cell | Di Guglielmo et al. (2003) | |
| TGF-β1 Receptor Internalization | 0.333 | N/A | 1/(min) | Vilar et al. (2006) | |
| TGF-β1 Receptor Recycling | 0.333 | N/A | 1/(min) | Vilar et al. (2006) | |
| TGF-β1 Receptor-Ligand Complex Internalization | 0.333 | N/A | 1/(min) | Vilar et al. (2006) | |
| TGF-β1 Receptor Degradation | 0.0004 | 1 × 10–4–1 × 10–2 | 1/(min) | est. | |
| TGF-β1 receptor-ligand-induced degradation | 1 × 10–20 | 0-0.25 | 1/min | est. | |
| PGE2 Parameters | |||||
| PGE2 Synthesis | 3.48 × 10–20 | 5.0 × 10–21–5.0 × 10–19 | #/(min) × cell | Lin et al. (1992) | |
| PGE2 Receptor-Ligand Binding Rate | 1.8 × 10–6 | 1.0 × 10–7–1.0 × 102 | 1/min | est. | |
| PGE2 Degradation | 0.001 | N/A | 1/min | Ishihara et al. (1991) | |
| α-Smooth Muscle Actin Parameters | |||||
| PRDS Degradation Rate | 0.24 | 3.0 × 10–3–1.0 × 10 | 1/min | est. | |
| Adhesion Signal Strength | 1 | 0 or 1 | N/A | est. | |
| Matrix Stiffness Effect | 100 | 0-1000 | N/A | est. | |
| Strength of PGE2 Inhibition | 10 | 1.0 × 10–7–1.0 × 103 | 1/min | est. | |
| α 1 | Nonzero so denominator ≠ 0 | 1 × 10–10 | 0 < n ≤ 1 | #/cell | est. |
| α SMA Degradation | 6 × 10–5 | 1.0 × 10–5–1.0 | 1/min | est. | |
Fig. 3.
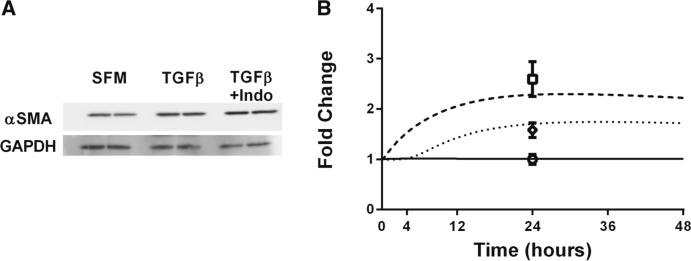
α SMA studies and simulations with and without PGE2 signaling. a α SMA measured in 3T12 fibroblasts at 24h either untreated, treated with 2ng/ml of TGF-β1, or treated with 2ng/ml of TGF-β1 and 10μM of indomethacin using Western blot and densitometry analysis. N = 2/condition. b Simulation of α SMA production. Dashed curve represents simulation results in the absence of PGE2 inhibition. Dotted curve represents simulation results in the presence of PGE2 inhibition, and solid line represents simulation control with no TGF-β1 treatment. Experimental data for α SMA at 24h following treatment with 2 ng/ml of TGF-β1 and 10μM of indomethacin, 2 ng/ml of TGF-β1, or serum-free media (SFM) are represented by open square, open diamond, and open circle, respectively. N = 2
In order to test our hypothesis that PGE2 signaling was needed for our model to recapitulate experimental data, we performed the experiment described above with fibroblasts in the absence of PGE2 signaling by treating them simultaneously with 2ng/ml of TGF-β1 and indomethacin (an inhibitor of PGE2) (Fig. 3A). We found that in the absence of PGE2 signaling, the concentration of α SMA in fibroblasts was significantly increased (p value < 0.05).
3.2 Transient TGF-β1 Signaling in the Presence of PGE2
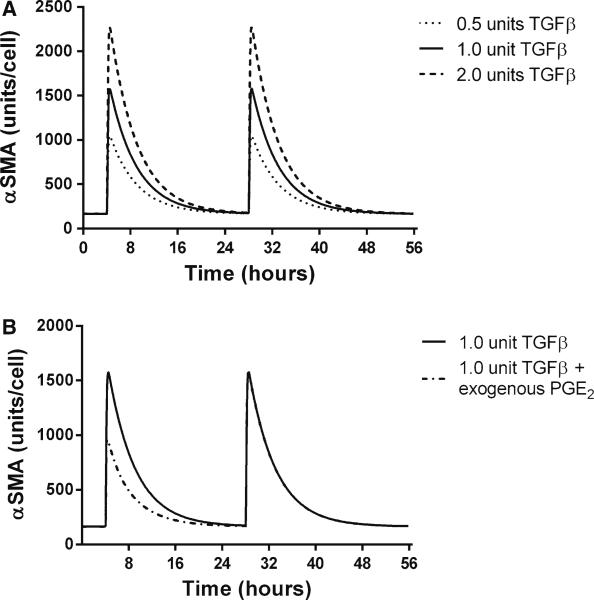
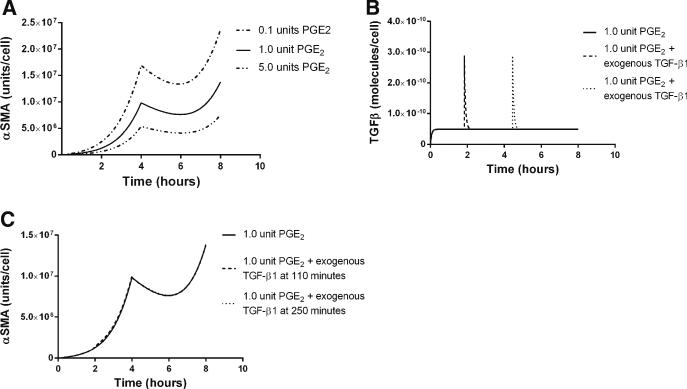
Previously published work identifies PGE2 as an important regulator of TGF-β1 signaling (Thomas et al. 2007), and we show above that including PGE2 gives simulation that is consistent with experimental data of TGF-β1-induced α SMA synthesis. In order to better understand the dynamics of PGE2 inhibition of fibroblast differentiation, we characterized the fibroblast response to TGF-β1 in the presence of a fixed concentration of PGE2 using our in silico model. During this simulation, we identified two phenomena which we refer to as response and refraction. Response describes periods of increasing α SMA concentrations in the fibroblast. Refraction refers to periods of declining concentrations of α SMA, indicating that fibroblasts are not able to respond to TGF-β1 and that α SMA is degrading in the cell. We define the magnitude and duration of a response as the maximal concentration of α SMA achieved following treatment and the time it takes for this concentration to return to pre-stimulatory levels, respectively. In the transient TGF-β1 signaling simulations, we fixed the concentration of PGE2 and simulated periodic dosing with exogenous active TGF-β1 (Fig. 4).
Fig. 4.
In silico TGF-β1-induced α SMA synthesis in the presence of constant concentration of PGE2. The x-axis represents time in hours, and the y-axis represents the concentration of α SMA in units/cell. a α SMA synthesis in a constant concentration (1 nmol) of PGE2 and dosing with 0.5ng/ml TGF-β1 (dotted) 1 ng/ml TGF-β1 (solid), and 2 ng/ml (dashed) at 4 and 28h. b α SMA synthesis in a constant concentration (1 nmol) of PGE2 and dosing with 1 ng/ml TGF-β1 at 4 and 28h. Dashed line simulation receives an additional dose of PGE2 at 440min
Prior to TGF-β1 treatment, the cells are in a refractory state with no change in the concentration of α SMA (Fig. 4a). Following treatment with TGF-β1, an induction of α SMA synthesis occurs but is transient, and α SMA levels eventually return to a refractory state indicating that the signal has dissipated. We observed that the magnitude of a signal response increases as the TGF-β1 dose is increased; however, the duration of this response does not change. Having observed a dose-dependent signal response to TGF-β1 in the presence of a constant concentration of PGE2, we wanted to see whether this response could be abrogated. We next repeated the simulation, but in addition to periodic doses of TGF-β1, we added an additional dose of exogenous PGE2 during the response phase (Fig. 4b). We observe that the addition of exogenous PGE2 following TGF-β1 treatment does reduce the magnitude of the signal. However, this reduction is transient and restricted to the current dose response. It does not affect the ability of the cell to respond fully to a later dose of TGF-β1. This suggests that intermittent treatment of differentiated cells with negative regulators is only temporarily effective at suppressing α SMA synthesis.
3.3 PGE2-Induced TGF-β1 Signaling Refractory State
We showed above that PGE2 can reduce the magnitude of response to a single dose of TGF-β1. We tested how PGE2 affects fibroblast responses to a constant concentration of TGF-β1 in order to evaluate the effects of PGE2 dosing on fibroblasts in the context of excessive TGF-β1 synthesis. We examined α SMA concentration in conjunction with constant TGF-β1 concentrations and periodic PGE2 dosing. We predicted that dosing with PGE2 induces a refractory period where a cell is unresponsive to TGF-β1 (Fig. 5a). This refractory period is transient, and eventually, the cellular response to TGF-β1 is restored. We varied the size of the dose of PGE2 and examined how that affects the dynamics of the TGF-β1 signal response. We found that higher doses of PGE2 induced a longer refractory period and a decreased magnitude of signal response following the refractory period (Fig. 5a). As we observed (Fig. 4), additional treatment with exogenous PGE2 during the response phase could reduce the magnitude of the fibroblast response to TGF-β1. We tested the inverse of the phenomenon to see whether a fibroblast could be rescued from refraction by adding exogenous TGF-β1 during the refractory period. We simulated treatment of our in silico cells with PGE2 and gave an additional dose of exogenous TGF-β1 (Fig. 5b) during the refractory period. We observed that additional TGF-β1 could not rescue cells from refraction (Fig. 5b). We also treated cells with additional exogenous TGF-β1 during the response phase to test whether we could induce a stronger signal response; however, we predict that treatment does not alter the magnitude or duration of the signal response in these cells (Fig. 5c). This suggests that consistent exposure to negative regulators such as PGE2 reduces the cells’ sensitivity to fluctuations in the concentration of positive differentiation signals such as TGF-β1.
Fig. 5.
In silico PGE2-induced refraction in TGF-β1-induced α SMA synthesis. The x-axis represents time in hours (a–c). The y-axis represents the concentration of α SMA for panels (a) and (c). The y-axis represents the concentration of active TGF-β1 for panel (b). a α SMA synthesis in the presence of a constant dose (1ng/ml) of TGF-β1. Dashed curve represents dosing with 10nmol PGE2, solid curve represents dosing with 100nmol PGE2, and dotted curve represents dosing with 5μmol of PGE2. b Concentration of TGF-β1 over time corresponding to output in panel (c). c All were dosed with 100nmol of PGE2 at 0 and 4h. Filled curve simulation was also treated with and additional dose of 1ng/ml of TGF-β1 at 100min during the refractory period. Dotted curve simulation was treated with an additional dose of 1ng/ml of TGF-β1 at 220min during the response period
3.4 Identifying States of Controlled Myofibroblast Function
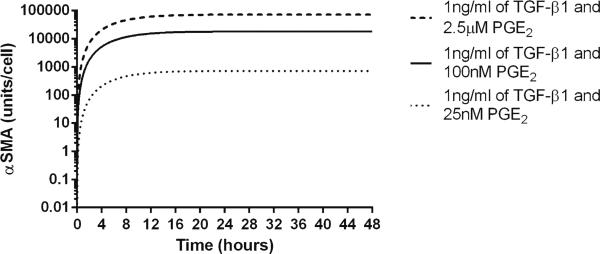
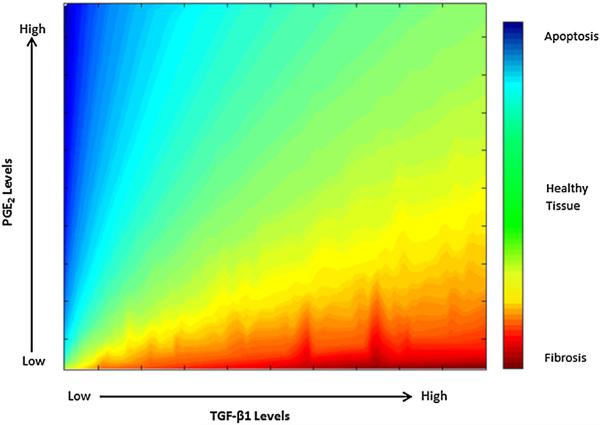
We predicted that constitutive levels of PGE2 result in predominantly quiescent fibroblasts (decreasing the concentration of α SMA) with transient responsiveness to periodic dosing with TGF-β1, and that constitutive levels of TGF-β1 result in increasing fibroblast differentiation (based on increasing the concentrations of α SMA) with transient refraction following periodic PGE2 dosing (Figs. 4, 5). Based on these studies, we know that PGE2 and TGF-β1 influence the state of fibroblast differentiation. The question remains as to whether fibroblasts can achieve a steady state in terms of the concentration of α SMA. We simulated treating cells with constant concentrations of both PGE2 and TGF-β1 and observed that under these conditions, α SMA concentration achieved a steady state that was maintained (Fig. 6). The magnitude of this steady state is determined by the ratio of TGF-β1 to PGE2 (Fig. 7).
Fig. 6.
Three examples of PGE2 and TGF-β1-induced steady-state fibroblast α SMA levels in the model. X-axis represents time in hours, and Y-axis represents the concentration of α SMA
Fig. 7.
Predicted response outcomes across different TGF-β1-to-PGE2 ratios. We compare α SMA concentrations of a fibroblast under different ratios of TGF-β1 to PGE2 for 24h in the model (1)–(10). The x-axis represents the concentration of TGF-β1, and the y-axis represents the concentration of PGE2. The color gradient represents the level of α SMA produced in response to these levels of TGF-β1 to PGE2. Blue corresponds to low concentrations of α SMA likely leading to fibrosis, and red corresponds to high concentration of α SMA likely leading to apoptosis (Tomioka et al. 2007; Ley et al. 2011). Plotted are the steady-state concentrations of α SMA at 24h for these TGF-β1 to PGE2 levels. A balance of these mediators leads to the best outcome: When moderate levels of αSMA are produced (green) (Color figure online)
3.5 Identifying Key Molecular Mechanisms Regulating Fibroblast Differentiation
To identify molecular mechanisms critical to the dynamics of fibroblast differentiation, we performed uncertainty and sensitivity analyses (see Sect. 5). When all model parameters are varied over wide ranges (Table 2) and partial rank correlation coefficients (PRCCs) are calculated, we can identify mechanisms that are significantly correlated with α-smooth muscle actin concentration over time (Table 3). Because ECM stiffness () strongly influences α SMA synthesis (Liu et al. 2010; Huang et al. 2012), we also performed our analysis holding this parameter constant to reflect the biological scenario where we are not able to vary the stiffness of the ECM. In this scenario, we can examine the roles of other mechanisms/parameters not influenced by the effects of stiffness (Table 4). Several key mechanisms identified by the model analysis to be driving fibroblast activation are: PGE2-induced inhibition, TGF-β1 receptor recycling rate, and active TGF-β1 degradation rate, indicating the importance of both positive and negative regulators of fibroblast differentiation. Further study of PGE2-induced inhibition is necessary to characterize specific therapeutic strategies that take advantage of existing negative regulatory pathways. Therapeutic treatments that inhibit receptor recycling or restrict the availability of active TGF-β1 could aid in inducing a shift in the environment by down-regulating positive feedback in the TGF-β1 signaling cascade.
Table 3.
PRCC values varying ECM stiffness
| Time hours | α1 | αSMA degradation rate | Active TGF-β1 degradation rate | Signal recovery rate | Receptor recycling rate | PGE2-induced inhibition | Adhesion-dependent signaling | ECM stiffness |
|---|---|---|---|---|---|---|---|---|
| 4 | –0.63 | –0.23 | –0.17 | –0.10 | 0.20 | 0.65 | 0.98 | 0.99 |
| 12 | –0.63 | –0.49 | –0.28 | –0.16 | 0.18 | 0.65 | 0.98 | 0.99 |
| 24 | –0.63 | –0.61 | –0.53 | –0.15 | 0.18 | 0.64 | 0.98 | 0.99 |
| 48 | –0.79 | –0.64 | –0.60 | –0.13 | 0.17 | 0.63 | 0.97 | 0.99 |
Parameters with positive values are positively correlated to αSMA synthesis, and negative values are negatively correlated to synthesis
Table 4.
PRCC values with fixed matrix stiffness
| Time hours | α1 | αSMA degradation rate | Active TGF-β1 degradation rate | Signal recovery rate | Receptor recycling rate | PGE2-Induced inhibition |
|---|---|---|---|---|---|---|
| 4 | –0.95 | –0.61 | –0.54 | –0.21 | 0.50 | 0.95 |
| 12 | –0.93 | –0.86 | –0.62 | –0.49 | 0.44 | 0.93 |
| 24 | –0.91 | –0.90 | –0.85 | –0.43 | 0.39 | 0.91 |
| 48 | –0.95 | –0.88 | –0.87 | –0.37 | 0.33 | 0.87 |
4 Discussion
Fibroblasts and myofibroblasts play a key role in promoting wound healing. They restore critical extracellular matrix and remodel architecture of damaged tissue (Velnar et al. 2009; Guo and Dipietro 2010; Diegelmann and Evans 2004; Ramasastry 2005; Goldman 2004). Their actions are tightly regulated to prevent disruption of healthy tissues but dysregulation can occur resulting in fibrotic diseases that are detrimental to the functionality of surrounding tissues. One such disease, IPF, leads to poor prognosis for patients with very limited treatment options, and lung transplant is currently the only long-term solution. Identifying key mechanisms driving fibroblast to myofibroblast differentiation and dysregulation enables us to predict targets for therapeutic treatment for fibrotic diseases.
We developed a mathematical model that captures the dynamics of TGF-β1-induced fibroblast differentiation under isolated conditions and characterized by the synthesis of α SMA. We utilized previously published (Di Guglielmo et al. 2003; Ishihara et al. 1991; Kalter et al. 1991) and our own in vitro data generated herein together with uncertainty and sensitivity analyses to build and test our model. The model details receptor–ligand binding and trafficking of TGF-β1, PGE inhibition, and α SMA synthesis. Our model includes a simple representation of adhesion-dependent signaling, which has been shown to be necessary for TGF-β1-induced fibroblast differentiation, and for PGE2 signaling, a major inhibitor of TGF-β1-induced fibroblast differentiation.
Prior models of TGF-β1 receptor–ligand signaling have been developed based on keratinocytes and pancreatic cancer cell lines (Vilar et al. 2006; Vizan et al. 2013; Zi and Klipp 2007). In these cell types, TGF-β1 is a negative regulator of cell growth and proliferation. These models are able to describe how temporal loss of sensitivity to TGF-β1 plays a key role in keratinocyte and pancreatic cancer cell regulation (Zimmermann et al. 2007). They demonstrate that desensitization is highly dependent on the ratio of constitutive and ligand-induced receptor degradation rates. In our model, we examine the regulatory mechanisms of fibroblast differentiation with TGF-β1 as our focus. TGF-β1 has been shown to induce fibroblast differentiation in the presence of adhesion signaling. As a result, we see a completely opposite cellular response to TGF-β1 signaling in fibroblasts compared to previously modeled cell types. Dysregulation of fibroblasts during wound healing is characterized by excessive differentiation, characteristic of a positive response to TGF-β1. For this reason, a new model of TGF-β1 signaling specifically for fibroblasts is necessary to identify key molecular factors that dictate in dysregulation.
Our results suggest that experimental outputs were only able to be recapitulated in the presence of PGE2 inhibition emphasizing that inhibition of TGF-β1 signaling is important for regulation of fibroblast behavior, and in the absence of inhibition factors like PGE2, we observe excessive production of α SMA, a marker for myofibroblast function.
We characterized fibroblast responses to different environmental conditions in order to isolate conditions that are favorable for controlled wound healing. These conditions may be artificially created in instances of fibroblast dysregulation in order to limit damaging effects of fibrosis. We predicted conditions resulting in quiescence (no α SMA), increasing differentiation (increasing the concentrations of α SMA), and steady-state differentiation (constant concentrations of α SMA). We simulated how fibroblasts respond to periodic dosing of TGF- β1 in the presence of a constant concentration of PGE2, in order to understand the ability of cells to perform wound-healing activities in the presence of continual negative regulation. We found that under these conditions, fibroblasts were able to respond modestly to dosing with TGF-β1 but quickly returned to a quiescent state when the signal was resolved. Cells are not able to maintain a constant concentration of α SMA, with only periodic stimulation from TGF-β1. The magnitude of the response, but not the duration, is dependent on the magnitude of the dose of TGF-β1 administered. We found that while the magnitude of the response in the presence of constant PGE2 was dependent on the TGF-β1 dose, the concentration of PGE2 dictated the duration of that response. Higher levels of negative regulators such as PGE2 present at the site of a wound may inversely correlate with the extent of damage present and thus inversely correlate with the amount of time required for repair mechanisms to take place. It is also possible that rather than shortening the duration of the repair, PGE2 and other potential negative regulators of TGF-β1 may decrease the amount of time each individual fibroblast responds to TGF-β1 signaling reducing the overall amount of fibroblast differentiation occurring during a repair event. Additional treatment with PGE2 during the response phase of fibroblasts to TGF-β1 was able to induce a short refractory period and temporarily decrease the strength of the signal response to TGF-β1 following the additional PGE2 treatment. This finding suggests that the cells are not refractory to PGE2 signaling in the presence of TGF-β1, and that PGE2 plays an important role in regulating TGF-β1 signaling. These results imply that fibroblasts are able to produce a limited amount of α SMA in response to TGF-β1 and potentially perform modest wound-healing activities in the presence of constant negative regulators, but they are not able to maintain a prolonged state of continuous α SMA synthesis. An environment with sustained concentrations of negative regulatory signals like PGE2 could be permissive to wound healing but also preventative of fibroblast dysregulation. It is also possible that under these conditions, TGF-β1 levels are insufficient for wound healing. Steady-state secretion of PGE2 by alveolar epithelial cells has been demonstrated and is suspected of keeping fibroblasts quiescent in the absence of injury (Lama et al. 2002; Moore et al. 2003).
We also tested how fibroblasts respond to periodic dosing with PGE2 in constant concentrations of TGF-β1 (Fig. 5), to evaluate the efficacy of inhibitory factors in the presence of substantial stimulation. We found that in the absence of PGE2, the fibroblasts were continuously responsive to the presence of TGF-β1, presenting steadily increasing concentrations of α SMA. Following treatment with PGE2, the cells were refractory to further stimulation of TGF-β1 for a limited period of time, and the duration of the refractory state was dependent on the dose of PGE2 administered. We tested whether cells could be relieved of refraction early if dosed with additional TGF-β1 during that time in order to determine whether treatment with PGE2 could sustain inhibition of the cellular differentiation in the presence of rapidly changing levels of TGF-β1. We found that treatment with additional TGF-β1 could not rescue fibroblasts from the refractory period, suggesting that PGE2 can block TGF-β1 signaling even during a flux in TGF-β1 concentration. These findings illustrate the ability of negative regulators such as PGE2 to inhibit fibroblast differentiation during response phase and in the presence of constant TGF-β1. This regulation, however, is transient resolving as soon as the inhibitor is degraded. Periodic dosing with PGE2 cannot sustain a long-term steady state of α SMA production. These results suggest that if PGE2 levels are insufficient or inconsistent during a wound-healing response, myofibroblasts may not maintain a steady-state level of α SMA production, but rather experience constant stimulation to TGF-β1 except for short transient periods of time when PGE2 levels are high enough to induce temporary quiescence. Our model therefore predicts that therapeutic treatments with fluctuating concentrations of negative regulators may be insufficient to restrict fibroblast differentiation for extended periods of time. It calls instead for treatment strategies that provide prolonged alterations to the environmental concentrations of positive and negative regulators. Our predictions remain to be tested experimentally, not only in vitro but in vivo as well.
Having identified environmental conditions that result in constant induction and quiescence of fibroblasts, we predict that constitutive concentration of both TGF-β1 and PGE2 will result in steady-state levels of fibroblast differentiation. Our model allows us to compare the contribution of each of these molecular factors to the overall outcome of fibroblast differentiation characterized by α SMA synthesis and predict patterns of signal availability that are currently untestable in vitro. We tested how fibroblasts respond to simultaneous constant levels of PGE2 and TGF-β1 (Fig. 6), and found that in the presence of both molecules, fibroblasts maintained a steady concentration of α SMA. This result indicates that a continuous presence of both molecules is necessary for sustained and controlled fibroblast response. This suggests that periodic inhibition of TGF-β1 is insufficient to prevent fibroblast dysregulation. Periodic dosing with TGF-β1 may or may not induce levels of fibroblast α SMA sufficient for successful wound healing, depending on the size and severity of the wound as well as the number of fibroblasts available to respond. Further characterization of the functionality of these environmental conditions in restricting and reversing the effects of fibroblast dysregulation will require a multiscale model capturing events at molecular, cellular, and tissue scale, which we are currently building.
Sensitivity analysis indicates that the parameters responsible for controlling strength of PGE2-induced inhibition (, α1) are the most significant factors for regulating fibroblast differentiation. In addition to the strength of PGE2-induced inhibition, rates of active TGF-β1 degradation and receptor recycling were also found to be very important. These parameters contribute to either the actual concentration of TGF-β1 and PGE2 in the media or to the level of concentration at which the receptor–ligand interaction becomes saturated. These findings indicate that PGE2 and TGF-β1 receptor–ligand signaling dynamics simultaneously contribute to fibroblast regulation and together dictate the differentiation of the cell. Because PGE2 inhibition was found to be one of the most important parameters in regulating fibroblast differentiation, but fibroblasts lose sensitivity to PGE2 over time (Moore et al. 2005), our findings call for therapeutics that mimic the effects of PGE2 through inhibition of adhesion signaling. Therapeutics that provide constant inhibition of adhesion signaling are needed to establish favorable environmental conditions for regulated wound healing. Inhibition of TGF-β1 receptor recycling () and decreased availability of active TGF-β1 (TGF β 1act) may increase the efficacy of PGE2 mimics in establishing favorable environmental conditions.
The findings of our model indicate that restricting positive differentiation signals like TGF-β1 alone cannot account for the regulation of fibroblast differentiation. We identify a need for balanced environmental conditions for fibroblasts with consistent levels of positive and negative regulators (Fig. 7). Extremely high ratios of TGF-β1 to PGE2 produce outcomes of excessive α SMA synthesis which we hypothesize correlate to fibrosis. Extremely low ratios of TGF-β1 to PGE2 result in very minimal fibroblast differentiation which may be insufficient for effective wound healing. In order to better understand this system and identify specific targets for therapeutic treatment, there is great need to explore more detailed dynamics of PGE2 inhibition of adhesion signaling in conjunction with TGF-β1 receptor ligand signaling dynamics.
5 Methods
5.1 In Vitro Studies of TGF-β1-Induced α SMA Synthesis
3T12 mouse fibroblast cell line was obtained from American Type Culture Collection (ATCC; CCL-164). Approximately 7.5 × 105 cells/400k cells/well are plated onto six-well plates and either left untreated, treated with 1–2ng/ml of activated TGF-β1, or treated simultaneously with 2 ng/ml of activated TGF-β1 and 10μM indomethacin. Untreated cells were harvested at 4h post-treatment. Treated cells were harvested at 4, 12, 24, or 48h post-treatment. Cells were lysed in radioimmunoprecipitation assay buffer with protease inhibitor cocktail (Sigma) for 15min at 4 °C and centrifuged. Total protein concentrations in the supernatants were determined by the bicinchoninic acid assay (Pierce). Equal amounts of protein from each sample were separated on a 4–20% gradient SDS-polyacrylamide gel and transferred to a PVDF membrane (Amersham/GE Healthcare, Pittsburgh, PA). PVDF membrane was then probed with a monoclonal antibody (Clone 1A4; Dako, Carpinteria, CA) for 1h to detect α SMA protein. This process was repeated to detect GAPDH (Santa Cruz).
5.2 Mathematical Model
5.2.1 TGF-β1 Receptor–Ligand Dynamics
To construct a model of fibroblast differentiation that captures molecular mechanisms necessary to study fibroblast dysregulation, we first describe TGF-β1 receptor–ligand binding and trafficking kinetics using mass action kinetics (Lauffenburger and Linderman 1993), building on studies in a variety of human and mouse cell lines and primary cells (Vilar et al. 2006; Vizan et al. 2013; Zi and Klipp 2007). We account for the TGF-β1 receptors ALK5 and TGF-β1RII as a single receptor complex. Because we are exclusively studying TGF-β1 and no other members of the TGF- β family, we disregard competition for subunits in establishing receptor complexes and assume that all stable receptor complexes include the ALK5 and TGF-β1RII. Modeling these receptors together as a single receptor unit is relevant because both receptors are required for induction of downstream signaling cascades. Because receptor subunits have different parameter values, we use the rate-limiting value for each parameter to describe the dynamics of the receptor complex. About 10% of the TGF-β1 receptors are present on cell surfaces in the absence of stimulation; the remaining 90% are sequestered intracellularly (Di Guglielmo et al. 2003).
Equations 1–6 track the rates of change over time for concentrations of six TGF-β1 signal cascade species. These species include the following: latent TGF-β1 [TGF β 1lat], active unbound TGF-β1 [TGF β 1act], unbound receptors on the fibroblast surface [Rsurf], unbound receptors in cytoplasm [Rint], bound receptor/TGF-β1 complexes on the fibroblast surface [TRCsurf], and bound receptor/TGF-β1 complexes in cytoplasm [T RCint] (Table 1; Fig. 1). The rates of change in the concentration of these species are dictated by parameter values (see Table 2).
The rate of change in the concentration of latent TGF-β1 is captured by:
| (1) |
The first term on the right-hand side (RHS) of the equation is , the constitutive TGF-β1 synthesis by fibroblasts, and is estimated by uncertainty analysis (described below) (Marino et al. 2008). The next term in the RHS of the equation represents the loss of latent TGF-β1 to degradation () and activation (). The final term on the RHS of the equation is a positive feedback term for additional latent TGF-β1 synthesis in the presence of TGF-β1 signaling. The rate constant is also estimated by uncertainty analysis. Nav represents Avogadro's number, and V ol represents the volume of the experimental environment. Cells represents the number of fibroblasts in a given simulation.
The rate of change in the concentration of active TGF-β1 is described by:
| (2) |
The first term on the RHS of the equation represents the rate of activation of latent TGF-β1 (from Eq. 1). The second term in the equation is the rate of active TGF-β1 binding to the receptor. The next term in the equation is the dissociation rate of active TGF-β1 from cell surface receptors. The final term in this equation represents the rate of degradation of active TGF-β1.
Equation 3 represents the rate of change over time of the concentration of unbound surface receptor complexes.
| (3) |
() represents the constitutive rate of receptor synthesis. The second and third terms on the RHS of the equation represent the rates of active TGF-β1 binding to and dissociation from receptors on the surface of the cell (from Eq. 2). The fourth term is the rate of internalization of unbound receptors into the cytoplasm and is proportional to the concentration of unbound surface receptors ([Rsurf]). The last term in the equation is the rate of receptor recycling from the cytoplasm to the surface of the cell (Di Guglielmo et al. 2003).
Equation 4 represents the rate of change over time in the concentration of internalized unbound receptors.
| (4) |
The first term on the RHS of the equation represents the rate of receptor internalization as described for Eq. 3. The second term in the equation is the loss of internal unbound receptors to recycling and degradation. The rate constant represents receptor degradation and is defined to satisfy the experimental data as described for above.
Equation 5 represents the rate of change over time in the concentration of bound surface receptors.
| (5) |
The first and second terms on the RHS of the equation represent the rate of TGF-β1 binding to unbound surface receptors and the dissociation of active TGF-β1 from surface receptor–ligand complexes (from Eq. 2). The last term in the equation represents the rate of internalization of bound receptor ligand complexes.
Equation 6 represents the rate of change over time in the number of internalized bound receptor–ligand complexes per cell.
| (6) |
The first term on the RHS of the equation represents the rate of internalization of surface receptor–ligand complexes (from Eq. 5). The second term in the equation represents the loss of internalized receptor–ligand complexes to recycling, ligand-independent degradation, and ligand-induced degradation and is proportional to the number of internalized receptor–ligand complexes ([T RCint]).
5.2.2 PGE2 Dynamics
Equations 7 and 8 track the concentration of extracellular PGE2 as well as the intracellular concentration of PGE2 (Table 1; Fig. 1). Recent studies have shown that PGE2 is an effective inhibitor of TGF-β1-induced fibroblast differentiation (Thomas et al. 2007). PGE2 can be synthesized and bound by fibroblasts making it a component of fibroblast autocrine signaling. Thus, to fully understand factors involved in fibroblast regulation and differentiation, we include equations for soluble and internalized PGE2 that allow for tracking of inhibition of TGF-β1-induced differentiation.
| (7) |
[PGE2Sol] represents the concentration of soluble PGE2. The first term on the RHS of the equation represents the constitutive rate of PGE2 synthesis by the fibroblast. The second term in the equation represents the rate of PGE2 being internalized into the cell. The final term in the equation is the rate of degradation of extracellular PGE2.
| (8) |
[PGE2int] represents the concentration of internalized PGE2. The first term on the RHS of the equation represents the rate of PGE2 being internalized into the cell (from Eq. 7). The second term in the equation is the rate of degradation of internalized PGE2.
5.2.3 α SMA Synthesis
We track α SMA concentration, a known indicator of fibroblast differentiation (3). α SMA serves to increase the contractile strength of fibroblasts a phenotype that is associated with their differentiated state (Hinz et al. 2001). This phenotype is important for wound contraction and tissue remodeling. TGF-β1 receptor binding and internalization induces a signaling cascade through either the canonical SMAD2/3 pathway or the non-canonical Rho/ROCK pathway (Derynck and Zhang 2003). Because experimental data detailing the rates of SMAD2/3 and Rho/ROCK signaling cascades are limited in fibroblasts, we focused on the receptor–ligand dynamics of the TGF-β1 signaling cascade and simplified, i.e., we replaced the entire signaling cascade with a term that tracks the effect of the kinase signaling cascade (see more below), the downstream phosphorylation cascades into a single event. We can still learn much about the system using this approach, and our analyses point to which elements can be elaborated later for further study.
We have two equations in our model that enable us to track the synthesis of α SMA. Equation 9 tracks the number of receptor–ligand internalization events over time [PRDS]. Because we do not explicitly measure all the kinase signaling cascades downstream of receptor–ligand complex internalization, we use this equation as a surrogate for those events. This equation contains a loss term which is the degradation of signaling complexes. These complexes serve as a simplification of the complex biological signal cascade far downstream of the initial internalization event. We use this equation to bridge the temporal gap between receptor–ligand events which occur very fast (on a timescale of minutes) and the protein synthesis events that occur much slower (on a timescale of hours), without the need to explicitly model every signal in the cascade. It is one way to coarse grain (simplify) these intracellular signaling events. Course graining the kinase signaling cascades, for which many parameter values are not known, allows us to reduce ambiguity from many unknown parameter values and processes. However, as key factors are identified, our model can point to features that can be fine-grained in future studies to further elucidate key signaling mechanisms driving fibroblast differentiation.
| (9) |
The first term on the RHS of the equation represents the rate of internalization of receptor complexes (from Eq. 5). The second term represents the rate of degradation of the signal.
Equation 10 captures the rate of change over time for the concentration of α SMA per cell [aSM A].
| (10) |
The first term on the RHS of the equation represents the rate of α SMA synthesis and is proportional to the number of receptor–ligand complex internalization events ([PRDS]) which promote α SMA synthesis and inversely proportional to the concentration of internalized PGE2 ([PGE2int]) which antagonizes α SMA synthesis. The rate constant represents the stiffness of the matrix to which the cell is adhering which positively correlates to adhesion signaling (Huang et al. 2012; Peyton et al. 2008; Discher et al. 2005; Levental et al. 2009). represents the rate of PGE2 inhibition of adhesion signaling. This term dictates how well PGE2 is able to antagonize the induction of α SMA synthesis by TGF-β1. α1 represents a small, nonzero number to bound the denominator away from zero. The final term in the equation represents the rate of degradation of α SMA.
5.3 Parameter Derivation and Estimation
We use uncertainty analysis (below) to estimate parameters in our model. When a parameter value is available from the literature, it is used directly, and others are obtained during model calibration (Fig. 2, also see ranges in Table 2). For the parameters that were available in the literature, we computed their values as follows. The rate constants for latent TGF-β1 degradation () and active TGF-β1 degradation() are derived from the half-life of latent and active TGF-β1 in rat plasma (9.2 ± 1.4min and 2.7 ± 0.4min, respectively) and published by Wakefield et al. (1990) assuming first-order kinetics. The rate constants for TGF-β1 binding and dissociation from the receptor and were published by De Crescenzo et al. (2003). The rate is fit to satisfy the experimental data, suggesting that approximately 90 of the total TGF-β1 receptors reside in the cytoplasm of the cell with the remaining 10% localizing to the surface under steady conditions (Penheiter et al. 2002), given that there are approximately 10,550 ± 1400 receptors per cell (Kalter et al. 1991). Rate constants for receptor recycling () is derived from Vilar et al. (2006). The rate is derived from Lin et al. (1992). Ishihara et al. (1991) assuming first-order kinetics.
The parameter represents a switch for the presence or absence of adhesion signaling and has a value of either 0 or 1. represents the magnitude of PGE2 inhibition of adhesion signaling and is estimated by uncertainty analysis. α1 represents a small nonzero number. In the absence of PGE2, α1 is given the same numerical value as , these terms reduce to 1 resulting in no inhibition by PGE2 and avoid dividing by zero.
For the remaining parameters, quantitative values for these rate constants were not available in the literature and would be difficult to measure in vitro (, , , , , , and ) so we estimate their values using uncertainty analysis and model calibration techniques.
5.4 Uncertainty Analysis
We use uncertainty analysis to quantify how variations in parameter values lead to variations in model outputs. Uncertainty analysis allows us to examine outcomes based on a wide value range for each unknown parameter value individually and simultaneously. We vary numerous parameters in the model over a wide range (Table 2) and compare how these variations affect model outputs. Latin hypercube sampling (LHS) is a method for varying multiple parameters simultaneously and then sampling the parameter space (Marino et al. 2008). When used for parameter estimation, uncertainty analysis allows model calibration to available data and identification of values for unknown parameters that allow for this when varied simultaneously.
5.5 Sensitivity Analysis
We use sensitivity analyses to identify which model parameters have a significant effect on model output and the extent of this effect. Sensitivity analysis identifies which parameters have a significant effect and the extent of the effect. Partial rank correlation coefficients (PRCCs) are used to quantify the effects of varying each parameter on the model output and therefore discerning which parameters have the strongest influence on a given output, or in other words the sensitivity of an output to a given parameter. PRCC values range from −1 to +1. A value of −1 signifies a perfect negative correlation between the parameter and the output, whereas a value of +1 signifies a perfect positive correlation between the parameter and the output. The closer a PRCC value is to 0, the weaker the correlation, whether positive or negative. PRCC values are differentiated with a Student's t test of significance. However, since PRCC quantifies nonlinear correlations, even small PRCC values can be significant. In this work, we use the LHS algorithm to generate 1000 unique parameter sets. This number of simulations gives high accuracy when identifying PRCC values (Marino et al. 2008). PRCC values are considered significant and with a p value of 0.01.
5.6 Model Solution, Calibration, and Validation
We solve Eqs. (1)–(10) with parameters as listed in Tables 1 and 2 using MATLAB ODE15s solver. While the equations are given in units/time, we simulate both calibration and validation studies using quantities in terms of fold change. This is how experimental data for these studies are measured, and it allows us to compare directly with data. We also compare the relative outputs in responses to treatment as fold change compared to untreated simulations.
Initial conditions are chosen to calibrate the model based on previously identified values specific for fibroblasts to achieve a steady state (Di Guglielmo et al. 2003), or selected to replicate experimental conditions (Fig. 2). Kalter et al. observed that fibroblasts have approximately 10,550±1400 TGF-β1 receptors per cell, and approximately 90% of those receptors are in the cytoplasm at steady state (Kalter et al. 1991). Based on this observation and rates derived in Vilar et al. for receptor recycling and synthesis, we select parameter values using uncertainty analysis that allow us to achieve these steady states. Variables for which the steady state is unknown are given initial values of 0 (Table 2). Using our model and the set of identified baseline parameter values that reflect differentiation of fibroblasts (Table 2), we validate our model by comparing output to additional experimental data derived in our laboratory and not used in the calibration of the model (Fig. 3). First, we simulate exposing our model fibroblasts to an initial concentration of 0.6nM of TGF-β1 and compare the concentration of α SMA at 4, 12, 24, and 48h as well as a 4h untreated control to in vitro experimental data described below (Fig. 2). Second, we simulate how output from different model constructions compares to experimental data published previously by our group. For example, we introduce into the model PGE2 inhibition of adhesion signaling and compare to simulation experimental results in the presence and absence of PGE2 (Fig. 2) (Thomas et al. 2007). Time course in vitro experiments were performed on mouse 3T12 transformed fibroblasts. Additional in vitro experiments were performed using primary mouse lung fibroblasts to effectively capture the sensitivity of lung fibroblasts to PGE2. Comparative simulations were done using different initial concentrations of TGF-β1 to reflect differences in effective concentration of and sensitivity to TGF-β1 between these two cell types. The difference in magnitude of α SMA synthesis in response to TGF-β1 treatment between in vitro experiments (Fig. 2) is likely due to differences between transformed cells and primary cells.
5.7 Calculating the Balance Between TGF-β1 and PGE2 Levels
We use LHS sampling as described above to generate 1000 simulations of our model with different TGF-β1 and PGE2 concentrations and then compare the concentration of α SMA at 48h. From this simulated data, we generated a heat map of the concentration of α SMA with respect to TGF-β1 and PGE2 using the meshgrid, TriSCatteredInterp, and contour functions in MATLAB (Fig. 7). The outcomes vary from severe fibrosis to healthy tissue to apoptosis (Huang et al. 2009).
Acknowledgments
This research was supported by the following Grants: R01 EB012579, R01 HL 110811 (both awarded to D.E.K. and J.J.L.), and R01 HL 115618 (awarded to B.B.M.).
References
- Annes JP, Munger JS, Rifkin DB. Making sense of latent TGFbeta activation. J Cell Sci. 2003;116(Pt 2):217–224. doi: 10.1242/jcs.00229. [DOI] [PubMed] [Google Scholar]
- Bendelac A, Rivera MN, Park SH, Roark JH. Mouse CD1-specific NK1 T cells: development, specificity, and function. Annu Rev Immunol. 1997;15:535–562. doi: 10.1146/annurev.immunol.15.1.535. [DOI] [PubMed] [Google Scholar]
- Bettinger DA, Yager DR, Diegelmann RF, Cohen IK. The effect of TGF-beta on keloid fibroblast proliferation and collagen synthesis. Plast Reconstr Surg. 1996;98(5):827–833. doi: 10.1097/00006534-199610000-00012. [DOI] [PubMed] [Google Scholar]
- Broekelmann TJ, Limper AH, Colby TV, McDonald JA. Transforming growth factor beta 1 is present at sites of extracellular matrix gene expression in human pulmonary fibrosis. Proc Natl Acad Sci USA. 1991;88(15):6642–6646. doi: 10.1073/pnas.88.15.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camelo A, Dunmore R, Sleeman MA, Clarke DL. The epithelium in idiopathic pulmonary fibrosis: breaking the barrier. Front Pharmacol. 2014;4:173. doi: 10.3389/fphar.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey RJ, Jr, Bascom CC, Sipes NJ, Graves-Deal R, Weissman BE, Moses HL. Selective inhibition of growth-related gene expression in murine keratinocytes by transforming growth factor beta. Mol Cell Biol. 1988;8(8):3088–3093. doi: 10.1128/mcb.8.8.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coomes SM, Wilke CA, Moore TA, Moore BB. Induction of TGF-beta 1, not regulatory T cells, impairs antiviral immunity in the lung following bone marrow transplant. J Immunol. 2010;184(9):5130–5140. doi: 10.4049/jimmunol.0901871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coomes SM, Farmen S, Wilke CA, Laouar Y, Moore BB. Severe gammaherpesvirus-induced pneumonitis and fibrosis in syngeneic bone marrow transplant mice is related to effects of transforming growth factor-beta. Am J Pathol. 2011;179(5):2382–2396. doi: 10.1016/j.ajpath.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Crescenzo G, Pham PL, Durocher Y, O'Connor-McCourt MD. Transforming growth factor-β (TGF-β) binding to the extracellular domain of the type II TGF-β receptor: receptor capture on a biosensor surface using a new coiled-coil capture system demonstrates that avidity contributes significantly to high affinity binding. J Mol Biol. 2003;328(5):1173–1183. doi: 10.1016/s0022-2836(03)00360-7. [DOI] [PubMed] [Google Scholar]
- de Ulrich TA, Juan Pardo EM, Kumar S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009;69(10):4167–4174. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425(6958):577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- Desmouliere A, Geinoz A, Gabbiani F, Gabbiani G. Transforming growth factor-beta 1 induces alpha-smooth muscle actin expression in granulation tissue myofibroblasts and in quiescent and growing cultured fibroblasts. J Cell Biol. 1993;122(1):103–111. doi: 10.1083/jcb.122.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Guglielmo GM, Le Roy C, Goodfellow AF, Wrana JL. Distinct endocytic pathways regulate TGF-beta receptor signalling and turnover. Nat Cell Biol. 2003;5(5):410–421. doi: 10.1038/ncb975. [DOI] [PubMed] [Google Scholar]
- Diegelmann RF, Evans MC. Wound healing: an overview of acute, fibrotic and delayed healing. Front Biosci. 2004;9:283–289. doi: 10.2741/1184. [DOI] [PubMed] [Google Scholar]
- Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310(5751):1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- Fine A, Poliks CF, Donahue LP, Smith BD, Goldstein RH. The differential effect of prostaglandin E2 on transforming growth factor-beta and insulin-induced collagen formation in lung fibroblasts. J Biol Chem. 1989;264(29):16988–16991. [PubMed] [Google Scholar]
- Fine A, Goldstein RH. The effect of transforming growth factor-beta on cell proliferation and collagen formation by lung fibroblasts. J Biol Chem. 1987;262(8):3897–3902. [PubMed] [Google Scholar]
- Finnson KW, McLean S, Di Guglielmo GM, Philip A. Dynamics of transforming growth factor beta signaling in wound healing and scarring. Adv Wound Care (New Rochelle) 2013;2(5):195–214. doi: 10.1089/wound.2013.0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone G, Sheetz MP. Substrate rigidity and force define form through tyrosine phosphatase and kinase pathways. Trends Cell Biol. 2006;16(4):213–223. doi: 10.1016/j.tcb.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Goldman R. Growth factors and chronic wound healing: past, present, and future. Adv Skin Wound Care. 2004;17(1):24–35. doi: 10.1097/00129334-200401000-00012. [DOI] [PubMed] [Google Scholar]
- Guo S, Dipietro LA. Factors affecting wound healing. J Dent Res. 2010;89(3):219–229. doi: 10.1177/0022034509359125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz B, Celetta G, Tomasek JJ, Gabbiani G, Chaponnier C. Alpha-smooth muscle actin expression upregulates fibroblast contractile activity. Mol Biol Cell. 2001;12(9):2730–2741. doi: 10.1091/mbc.12.9.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi M, Ota M, Rifkin DB. Matrix control of transforming growth factor-beta function. J Biochem. 2012;152(4):321–329. doi: 10.1093/jb/mvs089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SK, White ES, Wettlaufer SH, Grifka H, Hogaboam CM, Thannickal VJ, et al. Prostaglandin E(2) induces fibroblast apoptosis by modulating multiple survival pathways. FASEB J. 2009;23(12):4317–4326. doi: 10.1096/fj.08-128801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Yang N, Fiore VF, Barker TH, Sun Y, Morris SW, et al. Matrix stiffness-induced myofibroblast differentiation is mediated by intrinsic mechanotransduction. Am J Respir Cell Mol Biol. 2012;47(3):340–348. doi: 10.1165/rcmb.2012-0050OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara O, Sullivan MH, Elder MG. Differences of metabolism of prostaglandin E2 and F2 alpha by decidual stromal cells and macrophages in culture. Eicosanoids. 1991;4(4):203–207. [PubMed] [Google Scholar]
- Kalter VG, Brody AR. Receptors for transforming growth factor-beta (TGF-beta) on rat lung fibroblasts have higher affinity for TGF-beta 1 than for TGF-beta 2. Am J Respir Cell Mol Biol (USA) 1991;4(5):397–407. doi: 10.1165/ajrcmb/4.5.397. [DOI] [PubMed] [Google Scholar]
- King TE, Jr, Bradford WZ, Castro-Bernardini S, Fagan EA, Glaspole I, Glassberg MK, et al. A phase 3 trial of pirfenidone in patients with idiopathic pulmonary fibrosis. N Engl J Med. 2014;370(22):2083–2092. doi: 10.1056/NEJMoa1402582. [DOI] [PubMed] [Google Scholar]
- Kolodsick JE, Peters-Golden M, Larios J, Toews GB, Thannickal VJ, Moore BB. Prostaglandin E2 inhibits fibroblast to myofibroblast transition via E. prostanoid receptor 2 signaling and cyclic adenosine monophosphate elevation. Am J Respir Cell Mol Biol. 2003;29(5):537–544. doi: 10.1165/rcmb.2002-0243OC. [DOI] [PubMed] [Google Scholar]
- Kronenberg M, Rudensky A. Regulation of immunity by self-reactive T cells. Nature. 2005;435(7042):598–604. doi: 10.1038/nature03725. [DOI] [PubMed] [Google Scholar]
- Lama V, Moore BB, Christensen P, Toews GB, Peters-Golden M. Prostaglandin E2 synthesis and suppression of fibroblast proliferation by alveolar epithelial cells is cyclooxygenase-2-dependent. Am J Respir Cell Mol Biol. 2002;27(6):752–758. doi: 10.1165/rcmb.4857. [DOI] [PubMed] [Google Scholar]
- Lauffenburger DA, Linderman JJ. Receptors: models for binding, trafficking, and signaling. Oxford University Press; New York: 1993. [Google Scholar]
- Leveen P, Carlsen M, Makowska A, Oddsson S, Larsson J, Goumans MJ, et al. TGF-beta type II receptor-deficient thymocytes develop normally but demonstrate increased CD8+ proliferation in vivo. Blood. 2005;106(13):4234–4240. doi: 10.1182/blood-2005-05-1871. [DOI] [PubMed] [Google Scholar]
- Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139(5):891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley B, Collard HR, King TE., Jr Clinical course and prediction of survival in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2011;183(4):431–440. doi: 10.1164/rccm.201006-0894CI. [DOI] [PubMed] [Google Scholar]
- Li MO, Wan YY, Flavell RA. T cell-produced transforming growth factor-beta1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity. 2007;26(5):579–591. doi: 10.1016/j.immuni.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Lilja-Maula L, Syrja P, Laurila HP, Sutinen E, Ronty M, Koli K, et al. Comparative study of transforming growth factor-beta signalling and regulatory molecules in human and canine idiopathic pulmonary fibrosis. J Comp Pathol. 2014;150(4):399–407. doi: 10.1016/j.jcpa.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Lin LL, Lin AY, DeWitt DL. Interleukin-1 alpha induces the accumulation of cytosolic phospholipase A2 and the release of prostaglandin E2 in human fibroblasts. J Biol Chem. 1992;267(33):23451–23454. [PubMed] [Google Scholar]
- Liston A, Rudensky AY. Thymic development and peripheral homeostasis of regulatory T cells. Curr Opin Immunol. 2007;19(2):176–185. doi: 10.1016/j.coi.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang P, Li J, Kulkarni AB, Perruche S, Chen W. A critical function for TGF-beta signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells. Nat Immunol. 2008;9(6):632–640. doi: 10.1038/ni.1607. [DOI] [PubMed] [Google Scholar]
- Liu F, Mih JD, Shea BS, Kho AT, Sharif AS, Tager AM, et al. Feedback amplification of fibrosis through matrix stiffening and COX-2 suppression. J Cell Biol. 2010;190(4):693–706. doi: 10.1083/jcb.201004082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie JC, Liggitt D, Rudensky AY. Cellular mechanisms of fatal early-onset autoimmunity in mice with the T cell-specific targeting of transforming growth factor-beta receptor. Immunity. 2006;25(3):441–454. doi: 10.1016/j.immuni.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Marino S, Hogue IB, Ray CJ, Kirschner DE. A methodology for performing global uncertainty and sensitivity analysis in systems biology. J Theor Biol. 2008;254(1):178–196. doi: 10.1016/j.jtbi.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BB, Peters-Golden M, Christensen PJ, Lama V, Kuziel WA, Paine R, 3rd, et al. Alveolar epithelial cell inhibition of fibroblast proliferation is regulated by MCP-1/CCR2 and mediated by PGE2. Am J Physiol Lung Cell Mol Physiol. 2003;284(2):L342–L349. doi: 10.1152/ajplung.00168.2002. [DOI] [PubMed] [Google Scholar]
- Moore BB, Ballinger MN, White ES, Green ME, Herrygers AB, Wilke CA, et al. Bleomycin-induced E prostanoid receptor changes alter fibroblast responses to prostaglandin E2. J Immunol. 2005;174(9):5644–5649. doi: 10.4049/jimmunol.174.9.5644. [DOI] [PubMed] [Google Scholar]
- Penheiter SG, Mitchell H, Garamszegi N, Edens M, Dore JJ, Jr, Leof EB. Internalization-dependent and -independent requirements for transforming growth factor beta receptor signaling via the Smad pathway. Mol Cell Biol. 2002;22(13):4750–4759. doi: 10.1128/MCB.22.13.4750-4759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyton SR, Kim PD, Ghajar CM, Seliktar D, Putnam AJ. The effects of matrix stiffness and RhoA on the phenotypic plasticity of smooth muscle cells in a 3-D biosynthetic hydrogel system. Biomaterials. 2008;29(17):2597–2607. doi: 10.1016/j.biomaterials.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietenpol JA, Stein RW, Moran E, Yaciuk P, Schlegel R, Lyons RM, et al. TGF-beta 1 inhibition of c-myc transcription and growth in keratinocytes is abrogated by viral transforming proteins with pRB binding domains. Cell. 1990;61(5):777–785. doi: 10.1016/0092-8674(90)90188-k. [DOI] [PubMed] [Google Scholar]
- Ramasastry SS. Acute wounds. Clin Plast Surg. 2005;32(2):195–208. doi: 10.1016/j.cps.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Richeldi L, du Bois RM, Raghu G, Azuma A, Brown KK, Costabel U, et al. Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N Engl J Med. 2014;370(22):2071–2082. doi: 10.1056/NEJMoa1402584. [DOI] [PubMed] [Google Scholar]
- Rider CC, Mulloy B. Bone morphogenetic protein and growth differentiation factor cytokine families and their protein antagonists. Biochem J. 2010;429(1):1–12. doi: 10.1042/BJ20100305. [DOI] [PubMed] [Google Scholar]
- Saltzman LE, Moss J, Berg RA, Hom B, Crystal RG. Modulation of collagen production by fibroblasts. Effects of chronic exposure to agonists that increase intracellular cyclic AMP. Biochem J. 1982;204(1):25–30. doi: 10.1042/bj2040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada Y, Tamada M, Dubin-Thaler BJ, Cherniavskaya O, Sakai R, Tanaka S, et al. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127(5):1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strieter RM. What differentiates normal lung repair and fibrosis? Inflammation, resolution of repair, and fibrosis. Proc Am Thorac Soc. 2008;5(3):305–310. doi: 10.1513/pats.200710-160DR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, Narumiya S. Prostaglandin E receptors. J Biol Chem. 2007;282(16):11613–11617. doi: 10.1074/jbc.R600038200. [DOI] [PubMed] [Google Scholar]
- Tamada M, Sheetz MP, Sawada Y. Activation of a signaling cascade by cytoskeleton stretch. Dev Cell. 2004;7(5):709–718. doi: 10.1016/j.devcel.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, Kondoh Y, Ebina M, Azuma A, Ogura T, Taguchi Y, et al. The clinical significance of 5% change in vital capacity in patients with idiopathic pulmonary fibrosis: extended analysis of the pirfenidone trial. Respir Res. 2011;12:93. doi: 10.1186/1465-9921-12-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thannickal VJ, Lee DY, White ES, Cui Z, Larios JM, Chacon R, et al. Myofibroblast differentiation by transforming growth factor-beta1 is dependent on cell adhesion and integrin signaling via focal adhesion kinase. J Biol Chem. 2003;278(14):12384–12389. doi: 10.1074/jbc.M208544200. [DOI] [PubMed] [Google Scholar]
- Thomas PE, Peters-Golden M, White ES, Thannickal VJ, Moore BB. PGE(2) inhibition of TGF-beta1-induced myofibroblast differentiation is Smad-independent but involves cell shape and adhesion-dependent signaling. Am J Physiol Lung Cell Mol Physiol. 2007;293(2):L417–L428. doi: 10.1152/ajplung.00489.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Schiemann WP. PGE2 receptor EP2 mediates the antagonistic effect of COX-2 on TGF-beta signaling during mammary tumorigenesis. FASEB J. 2010;24(4):1105–1116. doi: 10.1096/fj.09-141341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka H, Imanaka K, Hashimoto K, Iwasaki H. Health-related quality of life in patients with idiopathic pulmonary fibrosis-cross-sectional and longitudinal study. Intern Med. 2007;46(18):1533–1542. doi: 10.2169/internalmedicine.46.6218. [DOI] [PubMed] [Google Scholar]
- Velnar T, Bailey T, Smrkolj V. The wound healing process: an overview of the cellular and molecular mechanisms. J Int Med Res. 2009;37(5):1528–1542. doi: 10.1177/147323000903700531. [DOI] [PubMed] [Google Scholar]
- Vilar JM, Jansen R, Sander C. Signal processing in the TGF-beta superfamily ligand-receptor network. PLoS Comput Biol. 2006;2(1):e3. doi: 10.1371/journal.pcbi.0020003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizan P, Miller DSJ, Gori I, Das D, Schmierer B, Hill CS. Controlling long-term signaling: receptor dynamics determine attenuation and refractory behavior of the TGF-beta pathway. Sci Signal. 2013;6(305):ra106. doi: 10.1126/scisignal.2004416. [DOI] [PubMed] [Google Scholar]
- Wakefield LM, Winokur TS, Hollands RS, Christopherson K, Levinson AD, Sporn MB. Recombinant latent transforming growth factor beta 1 has a longer plasma half-life in rats than active transforming growth factor beta 1, and a different tissue distribution. J Clin Invest. 1990;86(6):1976–1984. doi: 10.1172/JCI114932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xaubet A, Serrano-Mollar A, Ancochea J. Pirfenidone for the treatment of idiopathic pulmonary fibrosis. Expert Opin Pharmacother. 2014;15(2):275–281. doi: 10.1517/14656566.2014.867328. [DOI] [PubMed] [Google Scholar]
- Zi Z, Feng Z, Chapnick DA, Dahl M, Deng D, Klipp E, et al. Quantitative analysis of transient and sustained transforming growth factor-beta signaling dynamics. Mol Syst Biol. 2011;7:492. doi: 10.1038/msb.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi Z, Klipp E. Constraint-based modeling and kinetic analysis of the Smad dependent TGF-beta signaling pathway. PLoS ONE. 2007;2(9):e936. doi: 10.1371/journal.pone.0000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann CS, Carvalho CR, Silveira KR, Yamaguti WP, Moderno EV, Salge JM, et al. Comparison of two questionnaires which measure the health-related quality of life of idiopathic pulmonary fibrosis patients. Braz J Med Biol Res. 2007;40(2):179–187. doi: 10.1590/s0100-879x2007000200004. [DOI] [PubMed] [Google Scholar]