Abstract
The triggering receptor expressed on myeloid cells (TREM) family, which is abundantly expressed in myeloid lineage cells, plays a pivotal role in innate and adaptive immune response. In this study, we aimed to identify a novel receptor expressed on hematopoietic stem cells (HSCs) by using in silico bioinformatics and to characterize the identified receptor. We thus found the TREM-like transcript (TLT)-6, a new member of TREM family. TLT-6 has a single immunoglobulin domain in the extracellular region and a long cytoplasmic region containing 2 immunoreceptor tyrosine-based inhibitory motif-like domains. TLT-6 transcript was expressed in HSCs, monocytes and macrophages. TLT-6 protein was up-regulated on the surface of bone marrow-derived and peritoneal macrophages by lipopolysaccharide stimulation. TLT-6 exerted anti-proliferative effects in macrophages. Our results demonstrate that TLT-6 may regulate the activation and proliferation of macrophages.
Keywords: TLT-6, ITIM motif, Anti-proliferation, Macrophages, TREM family
INTRODUCTION
Triggering receptor expressed on myeloid cells (TREM) and TREM-like receptors are a structurally linked protein family, which contains a variable single immunoglobulin domain that is located on chromosome 17C in mice (1). These receptors are expressed on a variety of myeloid lineages cells such as neutrophils, monocytes, macrophages, and dendritic cells (1). TREM family receptors play important roles in the regulation of both innate and adaptive immune response. For example, TREM-1 is mainly associated with inflammation, expressed by the stimulation of lipopolysaccharides (LPS), and up-regulated in Gr-1 (+) F4/80 (+) monocytes from tumor mice (2,3). Soluble TREM-1, which is secreted in innate immune cells, is involved in intra-amniotic infection/inflammation (4). TREM-2 plays a key role in negative regulation of autoimmunity (1,5). In the TREM-like family, TREM-like transcript (TLT)-1 is found in the naïve platelet and megakaryocyte α-granules and its expression is upregulated on surface of activated cells (6). TLT-1 had an important role for the protection of inflammation-associated hemorrhage (7). Unlike other TREM family, TLT-2 is expressed in B cells as well as peripheral lymphoid organs. In myeloid cells, TLT-2 was not expressed in naïve monocytes in the bone marrow and peripheral blood but was up-regulated in differentiated macrophages and granulocytes upon activation or inflammatory stimulation (8,9). TLT-4 is predominantly expressed on dendritic cells and macrophages, especially in red pulp and marginal metallophilic macrophages in the spleen. Moreover, TLT-4 in dendritic cells showed the ability to bind to Annexin V+ PI+ late apoptotic or necrotic cells and was not affected by maturation induced by Toll-like receptor ligands (10).
Several TREM including TREM-1, TREM-2, TREM-3, and TREML-4 can participate in the signaling through their association with an adaptor molecule DNAX activation protein 12 kDa (DAP12) that contains an immunoreceptor tyrosine-based activation motif (ITAM), which is phosphorylated upon activation of DAP12-linked receptors. The phosphorylated ITAM recruits or activates Syk and NTAL/LAB, leading to activation of their subsignaling molecules such as Akt, calcium, and mitogen-activated protein kinase, for various cellular responses (1,11). In contrast, TREML-1 and TREML-2, which encode TLT-1 and TLT-2, respectively, contain a cytoplasmic tail with immunoreceptor tyrosin inhibitory motifs (ITIMs) that can associate with the SHP-1 and SHP-2 proteins (12,13).
In this study, we attempted to identify the novel receptor-type genes expressed in hematopoietic stem cells (HSCs) or hematopoietic lineage cells using an in silico differential display method. We found several receptortype unigenes expressed in HSCs and mature hematopoietic cells. Among them, we identified a novel TREM family gene, TLT-6, and demonstrated its molecular character, expression, and biological activity in macrophages.
MATERIALS AND METHODS
Materials
Doxycycline, LPS, propidium iodide and pervanadate were purchased from Sigama (St. Louis, MO, USA). Dulbecco's modified Eagle's medium (DMEM), bovine calf serum, fetal bovine serum (FBS) and penicillin/streptomycin were obtained from Gibco (Grand Island, NY, USA). Trizol was from Invitrogen Life Technologies™ (Carlsbad, NM, USA). Macrophage colony-stimulating factor (M-CSF) was purchased from R&D Systems (Minneapolis, MN, USA). Antibodies against hemagglutinin (HA), SHIP and SHP-2 were from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Cells
The investigation was performed in accordance with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH publication No. 85-23, revised 1996, latest revision in 2011) and was approved by the Animal Subjects Committee and by the Institutional Guidelines of Konkuk University, Korea. C57BL/6 mice (6- to 8-weeks old) were purchased from Orient Bio (Seoul, Korea) and maintained in a pathogen-free environment. Hematopoietic lineage cells were isolated from the bone marrow, thymus, lymph nodes, and peripheral blood of mice, and they were analyzed by staining with specific antibodies using FACSVantage SE (BD Biosciences, San Jose, CA, USA). Peritoneal macrophages were isolated from peritoneal exudates of mice (14). Bone marrow-derived macrophages were obtained from the total bone marrows from femurs and tibiae of mice (15,16). Bone marrow-derived macrophages were cultured in DMEM supplemented with 10% bovine calf serum and 5 ng/ml M-CSFfor 5~6 days. RAW264.7 and HeLa cells were purchased from ATCC (Manassas, VA, USA) and were maintained in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin.
TLT-6 cloning
Mouse TLT-6 full-length cDNA was amplified by polymerase chain reaction (PCR), with following primer pair: 5' primer (5'-GGATCCATGGCCTGGGA GCCCACATAC-3') and a 3' primer (5'-GAATTCTCAC TGCCCTGGGAG CTCAGC-3'). PCR was performed at 94℃ for 45 s, 56℃ for 30 sec, and 72℃ for 1 min, for a total of 30 cycles. The amplified PCR product was subcloned into a pGEM-T easy vector (Promega, Madison, WI, USA) and confirmed the sequences of cDNA by automated sequencing. Murine TLT-6 sequence was analyzed using the BLAST algorithm of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST). Alignment of sequences was performed using the DNASTAR program (17,18).
Expression of the TLT-6 transcript
Cells were harvested from bone marrow, blood, brain, heart, kidney, ovary, spleen, lung, liver and thymus for determination of organ specific expression analysis. To confirm hematopoietic lineage specific expression analysis, cells were isolated from the bone marrow, thymus, spleen, and peripheral blood and each hematopoietic lineage cells were purified using FACSVantage SE; BD Biosciences). Cell total RNA was isolated with Trizol according to the manufacturer's instructions, and 5µg of RNA was reverse transcribed to cDNA using SuperScript™ (Invitrogen Life Technologies™). The primers for RT-PCR analysis of TLT-6 included a 5' primer (5'-CTCGCTTCAA CTT CTTCACT-3') and a 3' primer (5'-GCTGTAGATGGAGT CCTCAG-3'). The conditions for RT-PCR analysis were 94℃ for 30 s, 60℃ for 30 s, and 72℃ for 1 min for a total of 30 cycles.
TLT-6 transfectant cells
To generate an inducible stable cell line, HA-tagged TLT-6 cDNA was inserted into pRevTRE-EGFP-RSVp-rt TA2S-M2-WPRE retroviral inducible vector by doxycycline treatment (18). TLT-6 transfectant cells were obtained by sorting green fluorescent protein (GFP)-positive cells by using FACSVantage SE (BD Biosciences). To confirm the biological roles, RAW264.7 cells were transfected with TLT-6 construct were treated with doxycyclin (1 ng/ml).
Coimmunoprecipitaion and Western blotting analysis
To confirm the molecular weight of the TLT-6 protein, HA-tagged full-length TLT-6 cDNA was directly inserted into the pcDNA 3.1 expression vector and expressed in vitro with TnT Quick Coupled Transcription/Translation Systems (Promega) following the technical manual. In order to determine the interaction of TLT-6 with SHIP and SHP-2, an HA-tagged TLT-6 inducible retroviral vector was introduced into HeLa cells, and then, the cells were induced with doxycycline treatment. After 48 h of induction, co-immunoprecipitation was performed with an anti- HA antibody, and western blot analysis was performed with anti-SHIP or anti-SHP-2.
Production of monoclonal antibody
To generate a TLT-6-specific monoclonal antibody, the extracelluar domain of TLT-6 was constructed into the pMT/V5-His vector. This construct was used to generate recombinant protein in Schneider (S2) cells by using the Drosophila Expression System. Purification of His-TLT-6 was performed using a HisTrap™ HP (Amersham Biosciences). His-TLT-6 protein was injected into male SD rats (Orient Bio; 7 weeks old) twice at 1-week intervals. After the last injection, lymphocytes from popliteal lymph nodes of the immunized rats were obtained and fused with SP-2 myeloma cells. To screen the positive hybridoma clones, we conducted ELISA by using an His- TLT-6 protein-coated plate and confirmed with transfected cell lines with full-length TLT-cDNA by flow cytometry (FACSVantage SE; BD Biosciences).
Flow cytometry
To investigate the expression of TLT-6 in primary cells, cells were harvested from the bone marrow and bone marrow-derived and peritoneal macrophages were collected, and washed twice in staining buffer (PBS, 0.01% NaN3, and 2% FBS). Cells were stained with fluorescence (FITC, PE, or APC)-, biotin-conjugated hematopoietic-specific monoclonal antibodies (CD4, CD3, CD8, B220, Gr-1, Mac-1, c-Kit, Sca-1, CD11b, Ter-119, and CD11c), or matched isotype controls purchased from eBiosciences (Sand Diego, CA, USA). Macrophages derived from the peritoneum or bone marrow were activated by LPS. Red cells were lysed, and nonspecific binding was blocked by preincubation with nonconjugated antibody to CD16/32. Dead cells were excluded by propidium iodide staining. Whole cells stained were analyzed using FACSVantage SE (BD Biosciences).
Proliferation assay
RAW 264.7 cells were transfected with TLT-GFP fusion retroviral vector or GFP vector as the control, and the GFP-positive cells were sorted by flow cytometry. Bone marrow-derived macrophages were obtained by M-CSF stimulation for 4 days and then treated with anti-TLT-6 monoclonal antibody for 2 days. The proliferation effects were calculated using a Cell Counting Kit-8 (DOJINDO Lab, Kumamoto, Japan).
Statistical analysis
The data are expressed as means±SD. Statistical significance of the differences between experimental groups was assessed by Student's t-test. Statistical significance was determined at the level of p<0.05.
RESULTS
Cloning and character of TLT-6
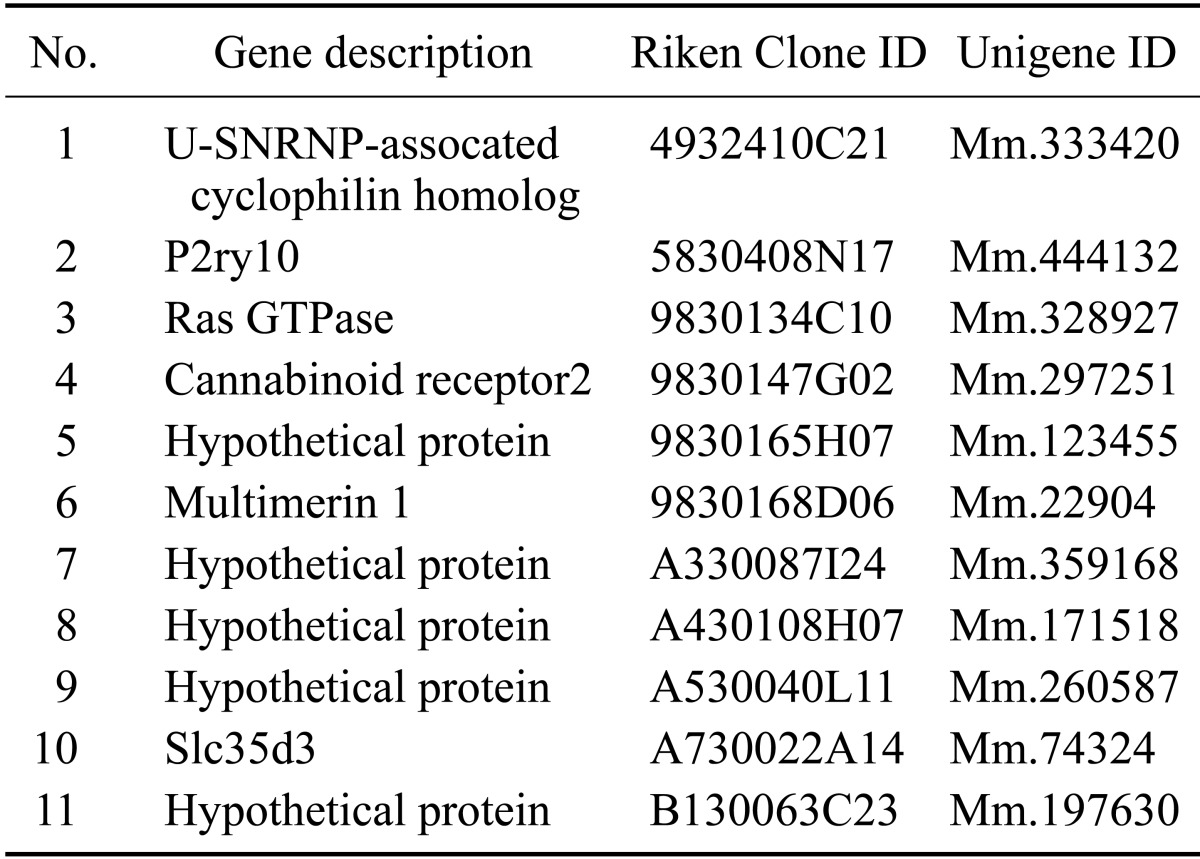
To identify novel candidate genes, which are predominantly expressed in HSCs, we performed a digital differentiation display by using expressed sequence tag (EST) unigenes libraries (http://www.ncbi.nlm.nih.gov/unigene) between mouse HSCs (c-Kit+Sca-1+Lin-) and mature hematopoietic cells (Table 1). For one of these genes, TLT-6, we found a higher sequence homology with TREM receptor families. The predicted amino acid sequence showed a 22-aa leader sequence, 56-aa extracellular domain, 23-aa transmembrane domain, and 112-aa cytoplasmic region. The extracellular region of TLT-6 contains an immunoglobulin domain and a cytoplasmic region with 2 representative ITIM-like motifs (ICYASL and VEYASI) (Fig. 1A). To confirm the molecular weight of TLT-6, we translated the protein with the Flag-tagged TLT-6 cDNA using an in vitro translation method and identified approximately 42-kD proteins (Fig. 1B). As next trials, we confirmed whether two ITIM motifs of TLT-6 in the cytoplasmic might recruit a protein phosphatase such as SHIP or SHP-2 when phosphorylated. To test this possibility, an HA-tagged TLT-6 transgene was introduced into HeLa cells expressing the SHIP and SHP-2 proteins. After stimulation with 100 µM pervanadate for 20 min, we performed immunoprecipitation with anti-HA antibody and then confirmed anti-SHIP or SHP-2 protein by western blotting. As shown in Fig. 1C, TLT-6 was associated with SHP-2, but not with SHIP (Fig. 1C).
Table I. In silico bioinformatics unigenes expressing strongly in HSCs.
Figure 1. Sequence analysis and character of TLT-6. (A) Predicted amino acid sequences, domains, and motifs of TLT-6. The leader sequence and transmembrane domain are indicated by dotted lines, and the variable type of immunoglobulin-like domain in the extracellular region is denoted by a solid line. The ITIM-like motifs in the intracellular region of TLT-6 are shown in the box. (B) Analysis of deduced molecular weight for Flag-tagged TLT-6 using an in vitro transcription/translation system. (C) Identification of interacting protein with TLT-6 using co-immunoprecipitation. HeLa cells, endogenously expressing SHIP and SHP-2, were used for transfection with HA-tagged TLT-6, immunoprecipitated with anti-Flag antibody, and then immunoblotted with anti-phospho SHIP or SHP-2 monoclonal antibodies. IB, Immunobloting; IP, immunoprecipitation.
TLT-6 expression
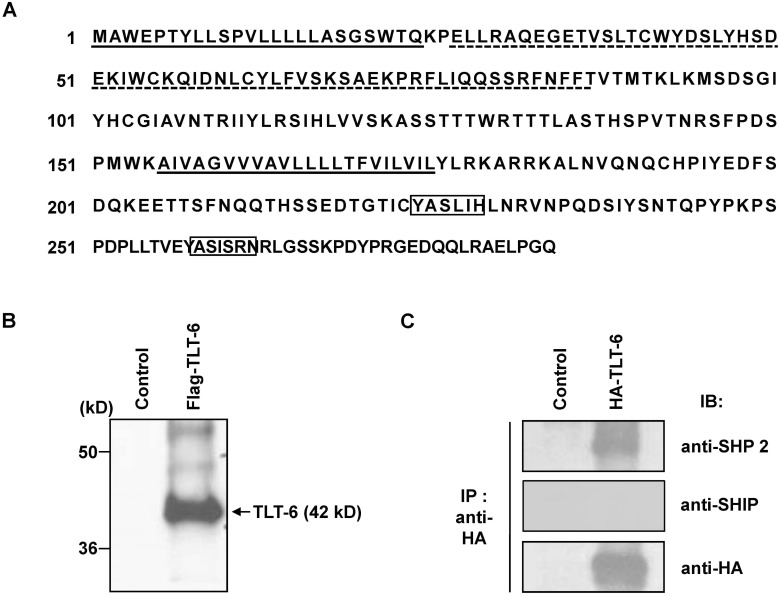
To confirm organ specific expression pattern of TLT-6, RT-PCR analysis was tested. TLT-6 mRNA transcript was observed high expression in bone marrow, peripheral blood and liver and also low expression in brain, spleen, lung and thymus, but not in heart, kidney and ovary (Fig. 2A). In addition, to further characterize the expression profile of TLT-6 in hematopoietic lineage cells, RT-PCR analysis was performed in purified hematopoietic lineage cells. TLT-6 was strongly expressed in B cells, immature erythrocytes, CD4+ T cells, CD8+ T cells, CD11b+ monocytes, granulocytes, and CD11c+ cells (Fig. 2B). In addition, TLT-6 transcripts showed relatively strong expression in HSCs (c-Kit+Sca-1+Lin-) or progenitor cells (Fig. 2C).
Figure 2. RT-PCR analyses of TLT-6 expression. (A) Organ specific expression of TLT-6 and (B) hematopoietic lineage cells specific expression of TLT-6 purified in the bone marrow, spleen, and peripheral blood, as described in the methods section. cDNA concentration was standardized and quantified using β-actin. (C) TLT-6 expression in HSCs (K+S+L-) and progenitor cells (K+S+L-, K-S+L-, and K-S-L-) isolated from the mouse bone marrow using RT-PCR. For RT-PCR, cDNA concentration was normalized with β-actin. PBL, leukocytes isolated from peripheral blood.
Expression character of TLT-6 protein
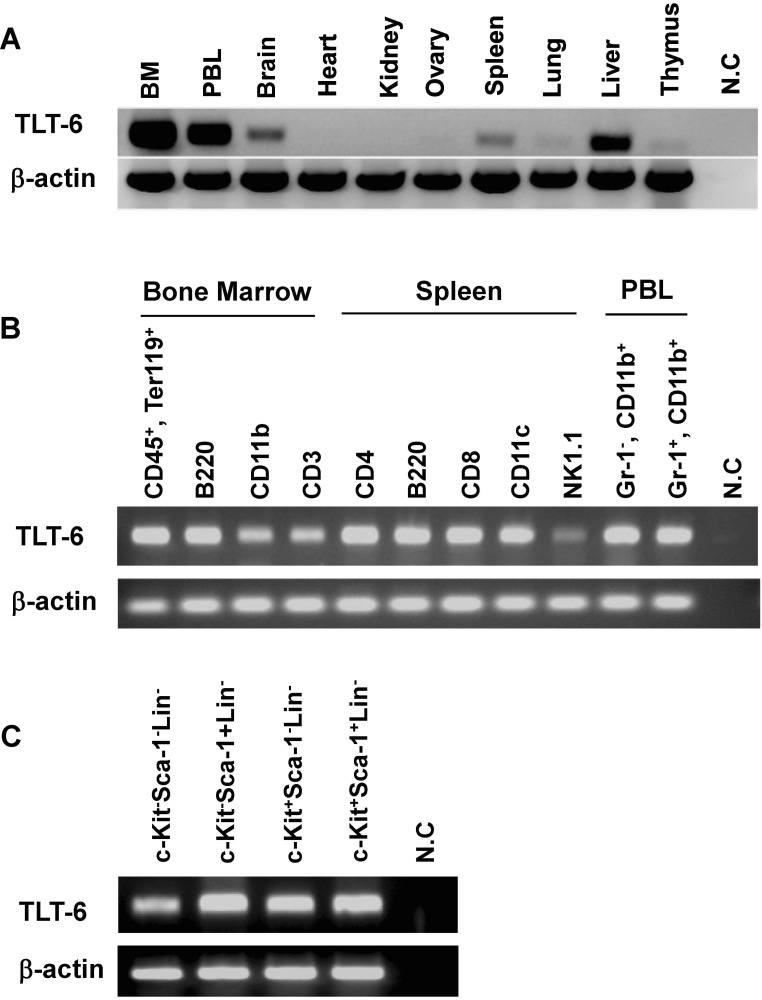
To determine the expression levels of the TLT-6 protein in hematopoietic lineage cells, we generated a monoclonal antibody against the extracellular domain of TLT-6 protein. Using the monoclonal antibody, we analyzed the expression character of TLT-6 on the surface of HSCs or CD11b+ cells from the bone marrow. As shown in Figure 3A, flow cytometry analysis showed that few TLT-6 protein-positive cells were observed on the surface of CD11b+ cells, but not on that of HSCs (c-Kit+Sca-1+Lin- cells) from the bone marrow.
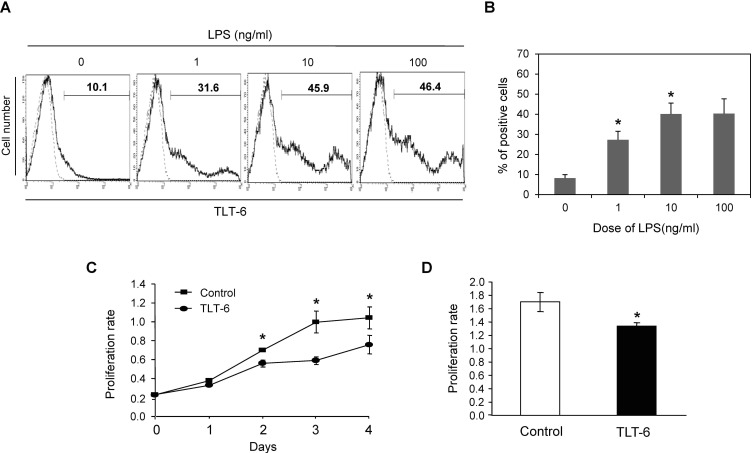
Figure 3. TLT-6 expressions in hematopoietic stem cells, monocyts and macrophages. (A) Expression patterns of TLT-6 in the surface of hematopoietic lineage cells from the bone marrow of mice analyzed by flow cytometry. Flow cytometry analysis was performed as described in the methods section, on the surface region of CD11b+ cells and HSCs (c-Kit+Sca-1+Lin-cells) from the bone marrow of mice. Expression of TLT-6 on the surface region of peritoneum (B, C)- and bone marrow (D, E)-derived macrophages. Each isolated macrophage sample was stimulated with LPS (10 ng/ml) during the indicated days. After treatment with anti-TLT-6 monoclonal antibody for 2 days, flow cytometry analysis was carried out as described in the methods section. Numbers in the upper parts of histogram curves indicate the positive intensity values of TLT- 6 expression in cells. Dotted line, control; Solid line, LPS-stimulated condition.
To test whether the activation of macrophages from bone marrow or peritoneum by LPS stimulation induces TLT-6 expression on the cell surface, both macrophages treated with LPS showed an increased number of TLT-6-expression-positive responses on their surface (Fig. 3B-E).
TLT-6 is upregulated and regulates proliferation in macrophages in response to LPS
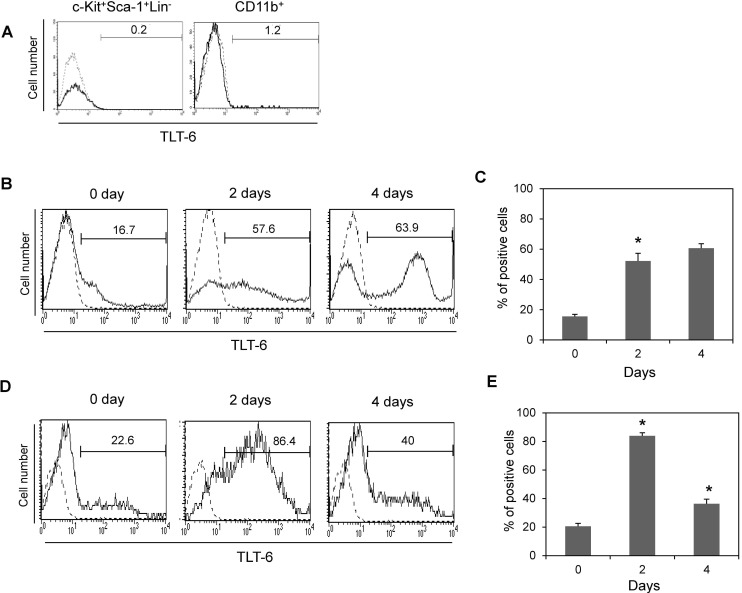
We next investigated the expression profile or biological activity of TLT-6 in macrophages. TLT-6 was up-regulated by LPS stimulation in macrophages derived from RAW264.7 cells. LPS stimulation increased the TLT-6 expression on the cell surface in dose-dependent manner in RAW264.7 cells (Fig. 4A and B). Moreover, TLT-6 overexpression in RAW264.7 cells significantly inhibited cell proliferation from day 2, compared with control cells transfected with TLT-free vector (Fig. 4C). Similar to this response, TLT-6 monoclonal antibody-treated bone marrow-derived macrophages also showed decreased proliferation when compared with control cells treated with IgG isotype (Fig. 4D).
Figure 4. TLT-6 expression and its effect on macrophages. (A, B) TLT-6 expression in activated RAW264.7 cells. The cells were stimulated with each indicated concentration of LPS for 3 days and the positive cells expressing TLT-6 were observed by flow cytometry analysis. Numbers in the upper parts of histogram curves indicate the positive intensity values of TLT-6 expression in cells. Dotted line, control; Solid line, LPS-stimulated condition. (C) Effect of TLT-6 overexpression on RAW264.7 cell proliferation. RAW 264.7 cells were transfected with the TLT-6 cDNA transgene, and cell proliferation was tested using the Cell Counting Kit-8 during the indicated days. (D) Effect of antibody treatment on bone marrow-derived macrophage proliferation. Cells were simultaneously treated with 10 ng/ml M-CSF and 1 µg/ml monoclonal TLT-6 antibody for 2 days and cell proliferation was analyzed using the Cell Counting Kit-8. Data in panel B and C are expressed as mean±standard deviation (SD), respectively.*p<0.05 compared with cells transfected with GFP vector (control in panel B), or with cells treated with IgG isotype (control in panel C).
DISCUSSION
HSCs have multipotential differentiation and self-renewal capacity to maintain blood homeostasis (19). Several investigators have thus conducted studies to identify the key regulator involved in controlling the self-renewal or multi-differentiation process (20,21). However, the key regulators remain to be identified. In this study, we identified a new TREM family gene, TLT-6, with unknown expression properties and functional roles, having a structure similar to other TREM family genes by using in silico bioinformatics. In silico bioinformatics provides a powerful tool for discovering simple cloning of unigenes that show a differential expression pattern between 2 representative samples (22). In the present study, we used this tool to identify a novel regulator that is predominantly expressed in HSCs compared to mature blood cells and identified several receptor-type unigenes candidates. Among these candidate unigenes, TLT-6 was identified as a novel TREM family molecule expressed in various hematopoietic lineage cells and its overexpression also inhibited macrophage proliferation. To the best of our knowledge, the present study is the first report on the expression property and function of a new TREM family gene, TLT-6.
The homology search of TLT-6 with TREM family genes revealed up to 50% higher sequence homology with TREM-4 and TREM-5, but relatively lower homology with other TREM family genes with a range of 10.8%~15.5% for TREM-1, TREM-2, TREM-3, TLT-1, TLT-2, TLT-3, and TLT-4 (data not shown). Moreover, TLT-6 gene was located on chromosome 17, which showed structures very similar to TREM family genes that have a single variable-type immunoglobulin domain in the extracellular and long cytoplasmic regions. These results indicate that TLT-6 is a novel of TREM family. It was known that TREM-1, TREM-2, and TREM-3 induced the signaling associated with the co-adapter molecule DAP12, which contains an ITAM (1,11). In contrast to this finding, TLT-6, similar to the TREM-like family genes such as TREML-1 and TREML-2, contained two ITIMs domains in the cytoplasmic tail, which has been shown to recruit SHP-2 protein, but not the SHIP protein. These results imply that TLT-6 may inhibit the early signaling pathway. In this study, we observed that TLT-6 was expressed on the cell surface and might be negatively involved in a complicated cellular reaction through extracellular signaling pathways though two ITIMs domain in monocytes.
In the case of myeloid cells, monocytes from the bone marrow or macrophages derived from the bone marrow or peritoneum showed relatively high expression of TLT-6 within the cell, but not on the cell surface (data not shown). Moreover, when activated with LPS, TLT-6 was up-regulated on cell surface in macrophages from bone marrow or peritoneum, as well as in RAW264.7 cells. This data imply that TLT-6 protein may be translocated rapidly from intracellular to cell surface by activation in macrophages. We also observed that TLT-6 was increased in macrophages stimulated with M-CSF or in thioglycolateelicited inflammatory macrophages compared with resident macrophages, indicating that TLT-6 may be important during the innate immune response stimulated by inflammatory stimuli. Moreover, we also demonstrated that TLT-6 revealed anti-proliferation activity against myeloid cells. These results suggest that TLT-6 strongly inhibits in the process of activation in myeloid cells and macrophages.
In summary, the present study demonstrated that TLT-6 was expressed on the surface of CD11b+ monocytes in the bone marrow and that its expression was increased by inflammatory or activation signaling. Moreover, TLT-6 was involved in the activation or proliferation of macrophages in vitro. These results suggest that TLT-6 may regulate the activation and proliferation of macrophages.
ACKNOWLEDGEMENTS
This work was carried out with the support of "Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ009321032013)" Rural Development Administration, Republic of Korea.
Abbreviations
- HSCs
hematopoietic stem cells
- TLT
TREM-like transcript
- TREM
triggering receptor expressed on myeloid cells
Footnotes
CONFLICTS OF INTEREST: The authors have no financial conflict of interest.
References
- 1.Ford JW, McVicar DW. TREM and TREM-like receptors in inflammation and disease. Curr Opin Immunol. 2009;21:38–46. doi: 10.1016/j.coi.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klesney-Tait J, Turnbull IR, Colonna M. The TREM receptor family and signal integration. Nat Immunol. 2006;7:1266–1273. doi: 10.1038/ni1411. [DOI] [PubMed] [Google Scholar]
- 3.Zanzinger K, Schellack C, Nausch N, Cerwenka A. Regulation of triggering receptor expressed on myeloid cells 1 expression on mouse inflammatory monocytes. Immunology. 2009;128:185–195. doi: 10.1111/j.1365-2567.2009.03091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kusanovic JP, Romero R, Chaiworapongsa T, Mittal P, Mazaki-Tovi S, Vaisbuch E, Erez O, Gotsch F, Than NG, Edwin SS, Pacora P, Jodicke C, Yeo L, Hassan SS. Amniotic fluid sTREM-1 in normal pregnancy, spontaneous parturition at term and preterm, and intra-amniotic infection/inflammation. J Matern Fetal Neonatal Med. 2010;23:34–47. doi: 10.3109/14767050903009248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turnbull IR, Gilfillan S, Cella M, Aoshi T, Miller M, Piccio L, Hernandez M, Colonna M. Cutting edge: TREM-2 attenuates macrophage activation. J Immunol. 2006;177:3520–3524. doi: 10.4049/jimmunol.177.6.3520. [DOI] [PubMed] [Google Scholar]
- 6.Washington AV, Schubert RL, Quigley L, Disipio T, Feltz R, Cho EH, McVicar DW. A TREM family member, TLT-1, is found exclusively in the alpha-granules of megakaryocytes and platelets. Blood. 2004;104:1042–1047. doi: 10.1182/blood-2004-01-0315. [DOI] [PubMed] [Google Scholar]
- 7.Washington AV, Gibot S, Acevedo I, Gattis J, Quigley L, Feltz R, De La MA, Schubert RL, Gomez-Rodriguez J, Cheng J, Dutra A, Pak E, Chertov O, Rivera L, Morales J, Lubkowski J, Hunter R, Schwartzberg PL, McVicar DW. TREM-like transcript-1 protects against inflammation-associated hemorrhage by facilitating platelet aggregation in mice and humans. J Clin Invest. 2009;119:1489–1501. doi: 10.1172/JCI36175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King RG, Herrin BR, Justement LB. Trem-like transcript 2 is expressed on cells of the myeloid/granuloid and B lymphoid lineage and is up-regulated in response to inflammation. J Immunol. 2006;176:6012–6021. doi: 10.4049/jimmunol.176.10.6012. [DOI] [PubMed] [Google Scholar]
- 9.Hashiguchi M, Kobori H, Ritprajak P, Kamimura Y, Kozono H, Azuma M. Triggering receptor expressed on myeloid cell-like transcript 2 (TLT-2) is a counter-receptor for B7-H3 and enhances T cell responses. Proc Natl Acad Sci U S A. 2008;105:10495–10500. doi: 10.1073/pnas.0802423105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hemmi H, Idoyaga J, Suda K, Suda N, Kennedy K, Noda M, Aderem A, Steinman RM. A new triggering receptor expressed on myeloid cells (Trem) family member, Trem-like 4, binds to dead cells and is a DNAX activation protein 12-linked marker for subsets of mouse macrophages and dendritic cells. J Immunol. 2009;182:1278–1286. doi: 10.4049/jimmunol.182.3.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharif O, Knapp S. From expression to signaling: roles of TREM-1 and TREM-2 in innate immunity and bacterial infection. Immunobiology. 2008;213:701–713. doi: 10.1016/j.imbio.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 12.Washington AV, Quigley L, McVicar DW. Initial characterization of TREM-like transcript (TLT)-1: a putative inhibitory receptor within the TREM cluster. Blood. 2002;100:3822–3824. doi: 10.1182/blood-2002-02-0523. [DOI] [PubMed] [Google Scholar]
- 13.Barrow AD, Astoul E, Floto A, Brooke G, Relou IA, Jennings NS, Smith KG, Ouwehand W, Farndale RW, Alexander DR, Trowsdale J. Cutting edge: TREM-like transcript-1, a platelet immunoreceptor tyrosine-based inhibition motif encoding costimulatory immunoreceptor that enhances, rather than inhibits, calcium signaling via SHP-2. J Immunol. 2004;172:5838–5842. doi: 10.4049/jimmunol.172.10.5838. [DOI] [PubMed] [Google Scholar]
- 14.Davies JQ, Gordon S. Isolation and culture of murine macrophages. Methods Mol Biol. 2005;290:91–103. doi: 10.1385/1-59259-838-2:091. [DOI] [PubMed] [Google Scholar]
- 15.Levesque SA, Kukulski F, Enjyoji K, Robson SC, Sevigny J. NTPDase1 governs P2X7-dependent functions in murine macrophages. Eur J Immunol. 2010;40:1473–1485. doi: 10.1002/eji.200939741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sordet O, Rébé C, Plenchette S, Zermati Y, Hermine O, Vainchenker W, Garrido C, Solary E, Dubrez-Daloz L. Specific involvement of caspases in the differentiation of monocytes into macrophages. Blood. 2002;100(13):4446–4453. doi: 10.1182/blood-2002-06-1778. [DOI] [PubMed] [Google Scholar]
- 17.Kim MS, Kim YK, Kim YS, Seong M, Choi JK, Baek KH. Deubiquitinating enzyme USP36 contains the PEST motif and is polyubiquitinated. Biochem Biophys Res Commun. 2005;30:797–804. doi: 10.1016/j.bbrc.2005.03.051. [DOI] [PubMed] [Google Scholar]
- 18.Gu BC, Kwon MS, Kim TW. Control of expression for GFP gene using tetracycline inducible retrovirus vector system. Reprod Dev Biol. 2005;29:57–62. [Google Scholar]
- 19.Deneault E, Cellot S, Faubert A, Laverdure JP, Frechette M, Chagraoui J, Mayotte N, Sauvageau M, Ting SB, Sauvageau G. A functional screen to identify novel effectors of hematopoietic stem cell activity. Cell. 2009;137:369–379. doi: 10.1016/j.cell.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seita J, Weissman IL. Hematopoietic stem cell: self-renewal versus differentiation. Wiley Interdiscip Rev Syst Biol Med. 2010;2:640–653. doi: 10.1002/wsbm.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma S, Gurudutta GU, Satija NK, Pati S, Afrin F, Gupta P, Verma YK, Singh VK, Tripathi RP. Stem cell c-KIT and HOXB4 genes: critical roles and mechanisms in self-renewal, proliferation, and differentiation. Stem Cells Dev. 2006;15:755–778. doi: 10.1089/scd.2006.15.755. [DOI] [PubMed] [Google Scholar]
- 22.Nonas SA, Finigan JH, Gao L, Garcia JG. Functional genomic insights into acute lung injury: role of ventilators and mechanical stress. Proc Am Thorac Soc. 2005;2:188–194. doi: 10.1513/pats.200501-005AC. [DOI] [PubMed] [Google Scholar]