Abstract
CD99 signaling is crucial to a diverse range of biological functions including survival and proliferation. CD99 engagement is reported to augment activator protein-1 (AP-1) activity through mitogen-activated protein (MAP) kinase pathways in a T-lymphoblastic lymphoma cell line Jurkat and in breast cancer cell lines. In this study, we report that CD99 differentially regulated AP-1 activity in the human myeloma cell line RPMI8226. CD99 was highly expressed and the CD99 engagement led to activation of the MAP kinases, but suppressed AP-1 activity by inducing the expression of basic leucine zipper transcription factor, ATF-like (BATF), a negative regulator of AP-1 in RPMI8226 cells. By contrast, engagement of CD99 enhanced AP-1 activity and did not change the BATF expression in Jurkat cells. CD99 engagement reduced the proliferation of RPMI8226 cells and expression of cyclin 1 and 3. Overall, these results suggest novel CD99 functions in RPMI8226 cells.
Keywords: CD99, BATF, AP-1, Proliferation, MAP kinase
INTRODUCTION
CD99 is a 32 kDa type-I transmembrane glycoprotein that is exclusively found in primates (1) and is strongly expressed in in cortical thymocytes, bone marrow precursor cells and some plasma cells (2,3). Engagement of CD99 on these cells has revealed that CD99 signaling is crucial to diverse biological functions in these cells, such as adhesion (4,5,6), apoptosis (7,8,9), proliferation, and differentiation (10,11). Previous studies have uncovered some signaling pathways that operate downstream of CD99 engagement (4,5,6,7,8,11,12,13,14,15,16). Specifically, CD99 engagement is reported to augment activator protein-1 (AP-1) activity through mitogen-activated protein (MAP) kinase pathways in a T-cell line (11) and in human breast cancer cell lines (13).
Basic leucine zipper transcription factor, ATF-like (BATF), is an endogenous repressor of AP-1-dependent transcription (17,18). Unlike other AP-1 proteins, BATF lacks a transactivation domain. BATF forms transcriptionally inert complexes with Jun proteins that compete with Jun/Fos complexes for the same DNA-binding sites. As shown by studies that utilized reporter assays, BATF inhibits AP-1-dependent luciferase activity in vitro and in vivo (17,18,19,20). Inhibition of AP-1-dependent transcription by the overexpression of BATF effectively blocks cell growth (19,20,21,22).
In this study, we examined the signal transduction pathway of CD99 in the human myeloma cell line RPMI8226. Surprisingly, CD99 diminished AP-1 activity by enhancing the expression of BATF. This contrasts with the findings of previous studies that investigated other CD99-expressing cells. Engagement of CD99 also reduced the proliferation of RPMI8226 cells. Overall, our results reveal a novel pathway of CD99-mediated signaling and cellular function.
MATERIALS AND METHODS
Cells and antibodies (Abs)
RPMI8226 and Jurcat cells (American Tissue Culture Collection, Rockville, MD, USA) were cultured in RPMI-1640 media supplemented with 10% (v/v) fetal bovine serum, penicillin G and streptomycin. Anti-CD99 monoclonal Abs (YG32 and DN16-PE) were purchased from Dynona Inc. (Seoul, Republic of Korea). For the engagement of CD99, cells were stimulated with 10 µg/ml YG32 Ab and 17 µg/ml anti-mouse immunoglobulin G (IgG) monovalent F(ab) fragments (Jackson ImmunoResearch, Westgrove, PA, USA) for crosslinking. All anti-MAP kinase antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA).
Reverse transcription PCR and quantitative real-time PCR
Total RNA was extracted from cells using the NucleoSpin kit (Macherey-Nagel, Düren, Germany) and reverse-transcribed using the iScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA, USA). The cDNA was amplified using specific primers for CD99 type-I and -II, as described previously (5,13). Levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA were used to normalize expression. Real-time PCR was performed using an iCycler iQ system with the iQ SYBR Green Supermix (Bio-Rad Laboratories). Fold induction was calculated with the comparative Ct method (23), using the expression of ribosomal protein S18 as the reference. The primers used in real-time PCR analyses are listed in Table S1.
Construction of expression plasmid
The human BATF gene was PCR-amplified from cDNA of RPMI8226 cells using the following primers: 5'-GGC GCTAGCGCCACCATGCCTCACAGCTCCGAC-3' and 5'-GCCCTCGAGTCAGGGCTGGAAGCGC-3'. The amplified products were digested using the NheI and XhoI restriction enzymes, and were ligated into the pCDNA3.1- hygro expression vector (Invitrogen, Carlsbad, CA, USA). The resulting expression construct was sequenced to verify the cDNA sequence vectors were used in the reporter assays.
Transfection and luciferase assay
The luciferase assay was performed using the luciferase assay system and the β-galactosidase assay system (Promega, Madison, WI). RPMI8226 and Jurkat cells were transfected using Lipofectamine LTX (Invitrogen) in accordance with the manufacturer's protocol. A standard transfection reaction used 1 µg of DNA, including AP1-Luc (Clontech, Mountain View, CA), pM1-β-Gal (Roche Diagnostics, Indianapolis, IN) and expression plasmid (pCDNA3.1 or pCDNA3.1-BATF). Luciferase activities were normalized with respect to β-galactosidase activities.
Electroporation of small interfering RNA
BATF-targeting and control siRNAs were purchased from Genolution Pharmaceuticals Inc. (Seoul, Korea) (Table S2). RPMI8226 cells (2.0×106) were electroporated using 1 µM BATF-targeting siRNA or control siRNA using a microporator (Invitrogen, Carlsbad, CA). BATF expression was determined using real-time PCR at 48 hours after electroporation.
Carboxyfluorescein succinimidyl ester (CFSE) labeling
1.0×107 cells were labeled with 0.2 µM CFSE and incubated at 37℃ for 10 minutes. Cold fetal calf serum was added to stop the staining reaction. After 3 days of culture, the fluorescence intensity of CFSE was measured using a FACSCalibur flow cytometer and analyzed using FlowJo software (Ashland, OR, USA).
Apoptosis assays
Apoptosis was analyzed by the apoptosis detection kit (BD Biosciences, San Jose, CA, USA) and tetramethylrhodamine ethyl ester perchlorate (TMRE; Molecular Probes, Eugene, OR, USA) staining. For annexin V staining, cells 1×106 cells were suspended in binding buffer and incubated with Annexin V-FITC and propidium iodide for 15 min at room temperature in the dark. For TMRE staining, 1×106 cells were incubated with 50 nM TMRE for 20 minutes at 37℃. The samples were then measured using a FACSCalibur flow cytometer and analyzed using CellQuest-Pro software (BD Biosciences).
RESULTS
The human myeloma cell line RPMI8226 strongly expresses CD99
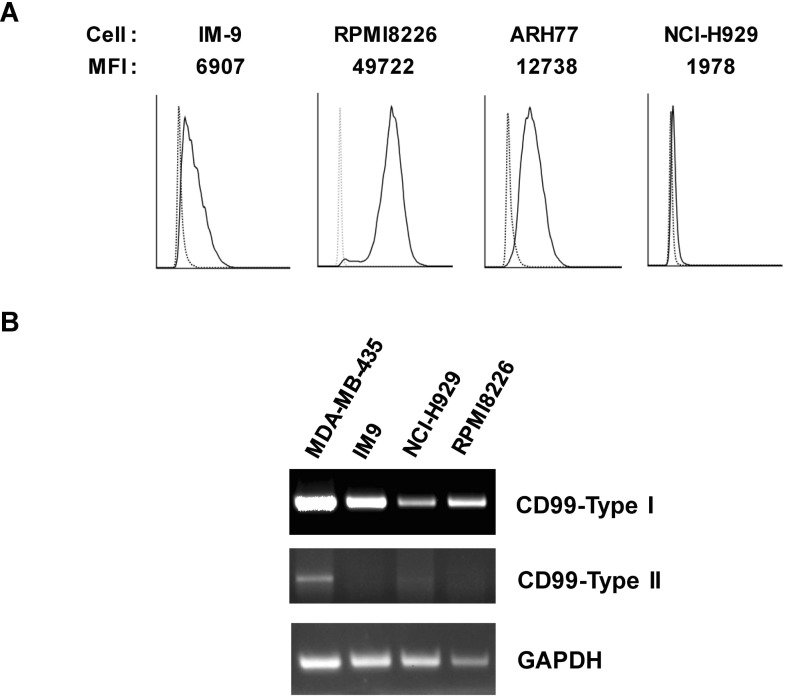
To select a B-cell line for studies of CD99 function, CD99 surface expression was measured in the B-cell lines RPMI8226, ARH77, NCI-H929, and IM-9 by flow cytometry (Fig. 1A). Among the cell lines tested, RPMI8226 cells had the highest surface expression of CD99. In particular, the expression level of CD99 on RPMI8226 cells was higher than on IM-9 cells, in which CD99 engagement triggers intracellular signaling (5). There are two major isoforms of CD99, termed type-I and -II, which have distinct roles in various cell types (5,13). RT-PCR analysis with primers designed to specifically amplify each isoform showed that only the type-I isoform was detected in RPMI8226 cells, whereas both the type-I and -II isoforms were detected in MDA-MB-435 breast cancer cells, as reported previously (13).
Figure 1. Expression of CD99 in B-cell lines. (A) Surface expression of CD99 in B-cell lines. Cells were stained using an anti-CD99 Ab (DN16-PE, solid line) or isotype control Ab (dotted line) and measured using flow cytometry. (B) PCR analysis with primers designed to specifically amplify CD99 type-I and -II. As described previously, the 583 bp fragment is characteristic of CD99 type-I and the 515 bp fragment is characteristic of CD99 type-II (5,13). Levels of GAPDH mRNA were used to normalize expression.
CD99 engagement reduces AP-1 activity and induces BATF expression
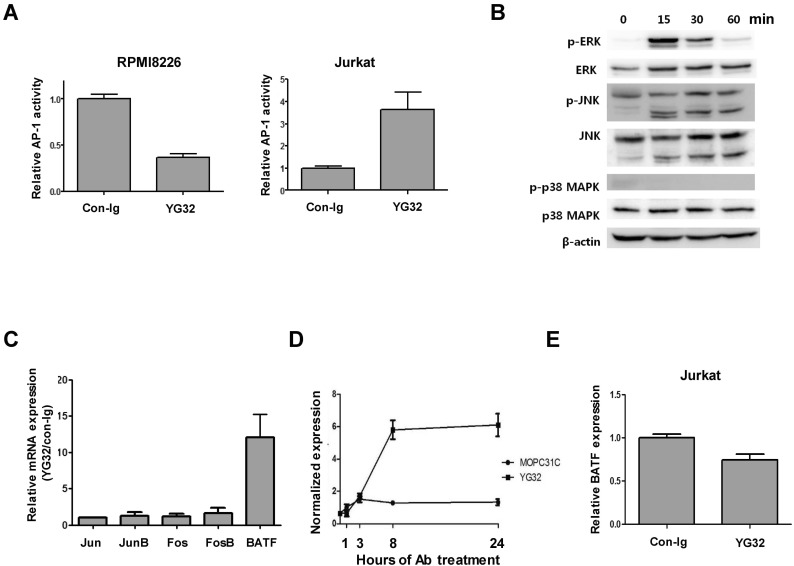
CD99 engagement with the functional anti-CD99 antibody (Ab) is the typical way to trigger CD99-mediated intracellular signaling that culminates in the aggregation, apoptosis, and proliferation of immune cells (4,5,6,7,8,9,10,11,24). To elucidate the function of CD99-mediated signaling in RPMI8226 cells, we employed an agonistic anti-CD99 Ab, YG32, to trigger intracellular signaling (12,25). We measured changes in AP-1 activity upon CD99 engagement by YG32 in RPMI8226 cells because CD99 has been reported to stimulate AP-1 signaling in other cells (11,13). Surprisingly, CD99 engagement reduced AP-1 activity in RPMI8226 cells, whereas it enhanced AP-1 activity in Jurkat cells, as reported previously (Fig. 2A) (11). We next examined MAP kinase activity because MAP kinases are upstream of AP-1, and CD99-mediated signaling activates MAP kinases in Jurkat cells (11,12). However, phosphorylation of both extracellular signal-regulated kinase (ERK) and c-Jun N-terminal kinase (JNK) was increased by CD99 engagement in RPMI8226 cells (Fig. 2B). We postulated that AP-1 activity could be regulated by a negative regulator of AP-1 in RPMI8226 cells, rather than by altered MAP kinase signaling mediated by CD99. We measured changes in the expression of AP-1 transcription factors, including BATF, using real-time PCR (Fig. 2C). BATF was identified as being potentially responsible for the reduction in AP-1 levels because it is a negative endogenous regulator of AP-1-mediated transcription. BATF expression was 12-fold higher in CD99-engaged cells than in control cells, whereas the expression levels of Jun, JunB, Fos, and FosB were all less than 2-fold higher in CD99-engaged cells than in control cells (Fig. 2C). Time-course experiments showed that BATF expression was rapidly induced within 8 hours after YG32 Ab treatment, and was sustained at a high level up to 24 hours after CD99 engagement (Fig. 2D). This result is completely different to the effects observed in Jurkat cells, where BATF expression did not increase upon YG32 Ab treatment (Fig. 2E). These results suggest that the induction of BATF expression following CD99 engagement suppresses AP-1 activity in RPMI8226 cells.
Figure 2. CD99 engagement reduces AP-1 activity and induces BATF expression in RPMI8226 cells. (A) AP-1 activity in RPMI8226 and Jurkat cells upon CD99 engagement. Both AP-1 luciferase constructs and a β-galactosidase expression plasmid were transfected into RPMI8226 and Jurkat cells. Luciferase activity was measured 24 hours after CD99 engagement and normalized against β-galactosidase activity. (B) Activation of ERK and JNK upon CD99 engagement in RPMI8226 cells. RPMI8226 cells were stimulated with 10 µg/ml of an anti-CD99 Ab (YG32) at 37℃ for 15, 30, and 60 min. The activities of ERK, JNK, and p38 kinase were measured by western blotting with antibodies specific for the phosphorylated form of each kinase, as well as suitable controls. Results are from a representative experiment performed in triplicate. (C) Expression of AP-1 transcription factors was measured using real-time PCR at 72 hours after CD99 engagement. Expression levels of the transcript in CD99-engaged cells are shown relative to those in control Ab (mouse IgG)-treated cells. (D) BATF expression was measured using real-time PCR at 1, 3, 8, and 24 hours after CD99 engagement in RPMI8226 cells. (E) BATF expression in CD99-engaged and control Jurkat cells. BATF expression was measured 24 hours after CD99 engagement by real-time PCR.
BATF regulates AP-1 activity in RPMI8226 cells
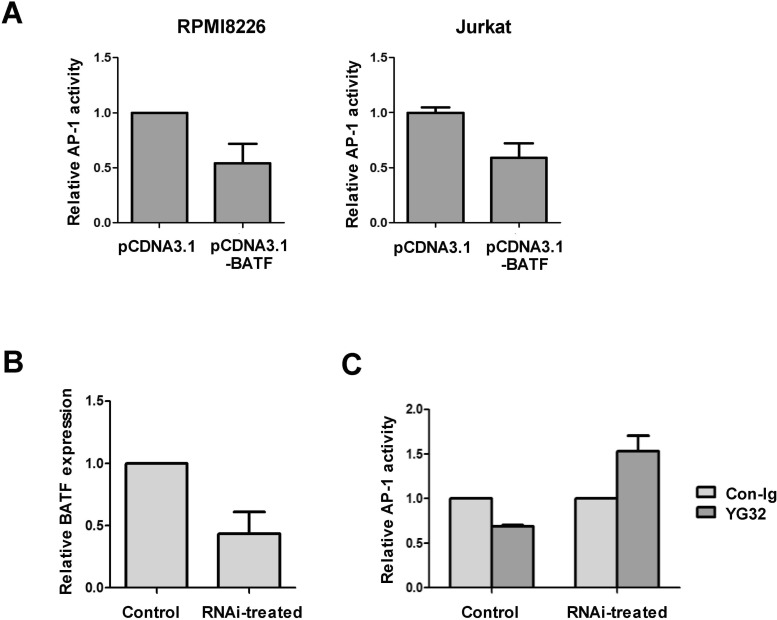
To investigate the function of BATF in RPMI8226 cells, we measured AP-1 activity when BATF was ectopically overexpressed. As previously reported in other cells (17,18,19), overexpression of BATF reduced AP-1 activity in RPMI8226 cells, as well as in 293T cells (Fig. 3A). To investigate the extent of BATF involvement in the CD99-induced reduction of AP-1 activity, BATF expression was knocked down by siRNA transfection. Transfection of BATF-targeting siRNA reduced BATF expression by about 50% in comparison to that in cells transfected with non-silencing control siRNA (Fig. 3B). In cells transfected with control siRNA, CD99 engagement reduced AP-1 activity. However, in cells transfected with BATF-targeting siRNA, the CD99-mediated reduction in AP-1 activity was abolished (Fig. 3C). These results demonstrate that induction of BATF expression by CD99 engagement suppresses AP-1 activity in RPMI8226 cells.
Figure 3. BATF-mediated regulation of AP-1 activity in RPMI8226 cells. (A) Overexpression of BATF reduces AP-1 activity in RPMI8226 and 293T cells. Cells were transiently transfected with reporter plasmids and expression plasmids, and treated with YG32. AP-1 activity was measured using a luciferase assay. Luciferase activity in cells co-transfected with pcDNA3.1-BATF is shown relative to that in cells co-transfected with pCDNA3.1 (set to 1). (B) Expression of BATF in RPMI8226 cells was reduced by siRNA treatment. Expression of BATF was measured using real-time PCR at 2 days after electroporation of siRNA oligonucleotides. (C) BATF knockdown reverses the reduction in AP-1 activity upon CD99 engagement. Cells were transfected with reporter plasmids 2 days after electroporation with siRNA oligonucleotides. Luciferase activities were normalized with respect to -galactosidase activities. The values show this normalized activity in Ab-treated cells relative to that in cells treated with control IgG. The error bars indicate the SD of three independent experiments.
CD99 engagement reduces the proliferation of RPMI8226 cells
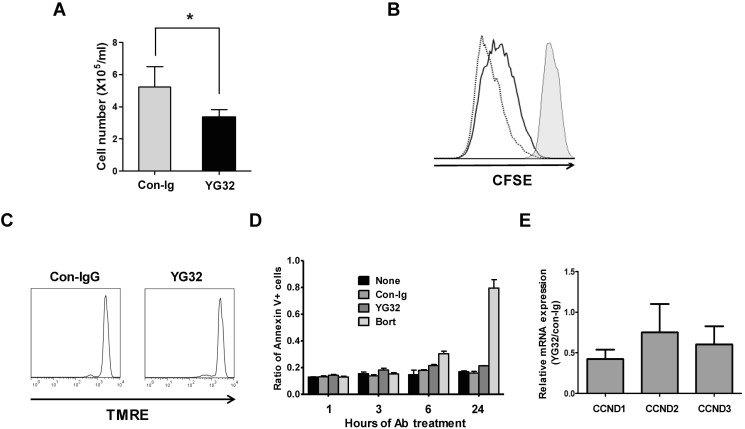
Given that CD99 engagement suppressed AP-1 activity in RPMI8226 cells, we investigated the effect of CD99 signaling on the proliferation of these cells. Engagement of CD99 by its agonistic Ab YG32 significantly reduced the number of cells recovered after 3 days of culture (Fig. 4A). To investigate whether growth suppression triggered by CD99 engagement was owing to a reduction in proliferation or an increase in apoptosis, apoptosis was evaluated by the TMRE assay and annexin V staining, and proliferation was evaluated by CFSE labeling. After 3 days of culture, the decrease in CFSE fluorescence intensity observed in control cells was evidently retarded in YG32-treated cells (Fig. 4B). In the TMRE assay, there was no marked difference between YG32-treated and control cells (Fig. 4C). Over 24 h of culture, the ratio of annexin V-positive cells to the total number of cells did not significantly differ between YG32-treated and control samples, whereas this ratio significantly increased in bortezomib-treated samples (Fig. 4D). These results suggest that the reduction in the number of cells recovered after CD99 engagement was mostly caused by the suppression of cell division, and not by a marked increase in the rate of apoptosis. We measured the expression of cyclin D1, D2, and D3 using real-time PCR after CD99 engagement because cyclin D proteins are major regulators of proliferation and cyclin D1 is a target gene of active AP-1 (26). The expression levels of cyclin D1 and D3 were significantly lower in CD99-engaged RPMI8226 cells than in untreated cells (Fig. 4E). These results suggest that the decrease in the recovery of RPMI8226 cells following treatment with an anti-CD99 Ab is mainly owing to a reduction in cell proliferation, possibly through reduced AP-1 activity via BATF. Nonetheless, the possible contribution of pro-apoptotic mechanisms cannot be entirely excluded.
Figure 4. CD99 engagement reduces the proliferation of RPMI8226 cells. (A) The number of RPMI8226 cells was counted using a hemocytometer after 3 days of treatment with an anti-CD99 Ab (YG32) or control IgG. The asterisk denotes a significant difference (p<0.05) between the two groups. (B) CFSE-labeled RPMI8226 cells at day 0 (filled histogram) were incubated for 3 days with control IgG (dotted line) or YG32 (solid line), and analyzed by flow cytometry. (C) RPMI8226 cells were cultured with YG32 or control IgG for 24 hours, stained with TMRE, and quantified using flow cytometry. (D) Annexin V-positive cells were quantified using flow cytometry after culture for the indicated amount of time in the presence of control IgG, YG32, or bortezomib. The ratio of the number of annexin V-positive cells to the total number of cells is shown (E) Expression of cyclin D1, D2, and D3 was measured by performing real-time PCR using the cDNA of RPMI8226 cells harvested after 3 days of incubation with YG32.
DISCUSSION
Variation in the AP-1 response to identical stimuli in different cell types and different microenvironments is well-documented (27). Overall, AP-1 signal transduction is controlled by a variety of factors, including the expression level of individual AP-1 family members, post-transcriptional modification, and the composition of AP-1 molecules on DNA-binding sites of target genes. Inhibition of AP-1 blocks tumor promotion, transformation, progression, and invasion (28). Inhibition of AP-1-dependent transcription by the overexpression of BATF effectively blocks cell growth (19,20,21,22). A small number of exogenous stimulating factors, including interleukin-6, leukemia inhibitory factor (29), interferon-β (30), and programmed cell death-1 (31), have been reported to be capable of inducing BATF expression. The present study demonstrated that CD99 is a BATF-inducing factor that reduces AP-1 activity in RPMI8226 cells.
CD99-mediated intracellular signaling involves activation of MAP kinases and increased AP-1 activity in Jurkat acute leukemia cells (11) and breast cancer cells (13). CD99 engagement augmented ERK and JNK activation in RPMI8226 cells. Therefore, the reduction in AP-1 activity by CD99 engagement in RPMI8226 cells prompted us to examine changes in the composition of AP-1 transcription factors following CD99 engagement. In various hematopoietic cells, increased BATF expression inhibits proliferation by reducing AP-1 activity (21,22,31). A reduction in AP-1 activity caused by the c-Jun-dominant negative mutant decreases the expression of D-type cyclins (32). In the present study, CD99 engagement inhibited proliferation and reduced the expression of cyclin D1 and D3 in RPMI8226 cells. Altogether, augmented expression of BATF by CD99 is a plausible mechanism for the inhibition of AP-1 activity, the reduced proliferation, and the downregulation of cyclin D in RPMI8226 cells. Under our experimental conditions, CD99 engagement did not appear to induce extensive apoptosis, in contrast to the findings of previous studies in other cells (7,8,9).
The present study showed that CD99 engagement reduces AP-1 activity and the proliferation of RPMI8226 cells. These results suggest a novel pathway of CD99 signaling that controls cell proliferation, and raise the possibility that an anti-CD99 Ab could be used as a novel therapeutic molecule.
ACKNOWLEDGEMENTS
This study was supported by the National Research Foundation of Korea, an MRC grant (no. 2008-0062286), and a grant from the Asan Institute for Life Sciences, Seoul, Korea (no. 2011-0794).
Abbreviations
- BATF
basic leucine zipper transcription factor
- ATF-like
- TMRE
tetramethylrhodamine ethyl ester perchlorate
Footnotes
CONFLICTS OF INTEREST: The authors have declared that no conflicts of interests exist.
Supplementary Materials
Primers used for the real-time PCR
SiRNA sequences
References
- 1.Levy R, Dilley J, Fox RI, Warnke R. A human thymus-leukemia antigen defined by hybridoma monoclonal antibodies. Proc Natl Acad Sci U S A. 1979;76:6552–6556. doi: 10.1073/pnas.76.12.6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dworzak MN, Fritsch G, Buchinger P, Fleischer C, Printz D, Zellner A, Schollhammer A, Steiner G, Ambros PF, Gadner H. Flow cytometric assessment of human MIC2 expression in bone marrow, thymus, and peripheral blood. Blood. 1994;83:415–425. [PubMed] [Google Scholar]
- 3.Park CK, Shin YK, Kim TJ, Park SH, Ahn GH. High CD99 expression in memory T and B cells in reactive lymph nodes. J Korean Med Sci. 1999;14:600–606. doi: 10.3346/jkms.1999.14.6.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard G, Zoccola D, Deckert M, Breittmayer JP, Aussel C, Bernard A. The E2 molecule (CD99) specifically triggers homotypic aggregation of CD4+ CD8+ thymocytes. J Immunol. 1995;154:26–32. [PubMed] [Google Scholar]
- 5.Hahn JH, Kim MK, Choi EY, Kim SH, Sohn HW, Ham DI, Chung DH, Kim TJ, Lee WJ, Park CK, Ree HJ, Park SH. CD99 (MIC2) regulates the LFA-1/ICAM-1-mediated adhesion of lymphocytes, and its gene encodes both positive and negative regulators of cellular adhesion. J Immunol. 1997;159:2250–2258. [PubMed] [Google Scholar]
- 6.Kasinrerk W, Tokrasinwit N, Moonsom S, Stockinger H. CD99 monoclonal antibody induce homotypic adhesion of Jurkat cells through protein tyrosine kinase and protein kinase C-dependent pathway. Immunol Lett. 2000;71:33–41. doi: 10.1016/s0165-2478(99)00165-0. [DOI] [PubMed] [Google Scholar]
- 7.Jung KC, Kim NH, Park WS, Park SH, Bae Y. The CD99 signal enhances Fas-mediated apoptosis in the human leukemic cell line, Jurkat. FEBS Lett. 2003;554:478–484. doi: 10.1016/s0014-5793(03)01224-9. [DOI] [PubMed] [Google Scholar]
- 8.Pettersen RD, Bernard G, Olafsen MK, Pourtein M, Lie SO. CD99 signals caspase-independent T cell death. J Immunol. 2001;166:4931–4942. doi: 10.4049/jimmunol.166.8.4931. [DOI] [PubMed] [Google Scholar]
- 9.Bernard G, Breittmayer JP, de MM, Trampont P, Hofman P, Senik A, Bernard A. Apoptosis of immature thymocytes mediated by E2/CD99. J Immunol. 1997;158:2543–2550. [PubMed] [Google Scholar]
- 10.Waclavicek M, Majdic O, Stulnig T, Berger M, Sunder-Plassmann R, Zlabinger GJ, Baumruker T, Stockl J, Ebner C, Knapp W, Pickl WF. CD99 engagement on human peripheral blood T cells results in TCR/CD3-dependent cellular activation and allows for Th1-restricted cytokine production. J Immunol. 1998;161:4671–4678. [PubMed] [Google Scholar]
- 11.Yoon SS, Kim HJ, Chung DH, Kim TJ. CD99 costimulation up-regulates T cell receptor-mediated activation of JNK and AP-1. Mol Cells. 2004;18:186–191. [PubMed] [Google Scholar]
- 12.Hahn MJ, Yoon SS, Sohn HW, Song HG, Park SH, Kim TJ. Differential activation of MAP kinase family members triggered by CD99 engagement. FEBS Lett. 2000;470:350–354. doi: 10.1016/s0014-5793(00)01330-2. [DOI] [PubMed] [Google Scholar]
- 13.Byun HJ, Hong IK, Kim E, Jin YJ, Jeoung DI, Hahn JH, Kim YM, Park SH, Lee H. A splice variant of CD99 increases motility and MMP-9 expression of human breast cancer cells through the AKT-, ERK-, and JNK-dependent AP-1 activation signaling pathways. J Biol Chem. 2006;281:34833–34847. doi: 10.1074/jbc.M605483200. [DOI] [PubMed] [Google Scholar]
- 14.Choi EY, Park WS, Jung KC, Kim SH, Kim YY, Lee WJ, Park SH. Engagement of CD99 induces up-regulation of TCR and MHC class I and II molecules on the surface of human thymocytes. J Immunol. 1998;161:749–754. [PubMed] [Google Scholar]
- 15.Husak Z, Printz D, Schumich A, Potschger U, Dworzak MN. Death induction by CD99 ligation in TEL/AML1-positive acute lymphoblastic leukemia and normal B cell precursors. J Leukoc Biol. 2010;88:405–412. doi: 10.1189/jlb.0210097. [DOI] [PubMed] [Google Scholar]
- 16.Yoon SS, Jung KI, Choi YL, Choi EY, Lee IS, Park SH, Kim TJ. Engagement of CD99 triggers the exocytic transport of ganglioside GM1 and the reorganization of actin cytoskeleton. FEBS Lett. 2003;540:217–222. doi: 10.1016/s0014-5793(03)00268-0. [DOI] [PubMed] [Google Scholar]
- 17.Williams KL, Nanda I, Lyons GE, Kuo CT, Schmid M, Leiden JM, Kaplan MH, Taparowsky EJ. Characterization of murine BATF: a negative regulator of activator protein-1 activity in the thymus. Eur J Immunol. 2001;31:1620–1627. doi: 10.1002/1521-4141(200105)31:5<1620::aid-immu1620>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 18.Iacobelli M, Wachsman W, McGuire KL. Repression of IL-2 promoter activity by the novel basic leucine zipper p21SNFT protein. J Immunol. 2000;165:860–868. doi: 10.4049/jimmunol.165.2.860. [DOI] [PubMed] [Google Scholar]
- 19.Echlin DR, Tae HJ, Mitin N, Taparowsky EJ. B-ATF functions as a negative regulator of AP-1 mediated transcription and blocks cellular transformation by Ras and Fos. Oncogene. 2000;19:1752–1763. doi: 10.1038/sj.onc.1203491. [DOI] [PubMed] [Google Scholar]
- 20.Dorsey MJ, Tae HJ, Sollenberger KG, Mascarenhas NT, Johansen LM, Taparowsky EJ. B-ATF: a novel human bZIP protein that associates with members of the AP-1 transcription factor family. Oncogene. 1995;11:2255–2265. [PubMed] [Google Scholar]
- 21.Thornton TM, Zullo AJ, Williams KL, Taparowsky EJ. Direct manipulation of activator protein-1 controls thymocyte proliferation in vitro. Eur J Immunol. 2006;36:160–169. doi: 10.1002/eji.200535215. [DOI] [PubMed] [Google Scholar]
- 22.Liao J, Humphrey SE, Poston S, Taparowsky EJ. Batf promotes growth arrest and terminal differentiation of mouse myeloid leukemia cells. Mol Cancer Res. 2011;9:350–363. doi: 10.1158/1541-7786.MCR-10-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Bernard G, Raimondi V, Alberti I, Pourtein M, Widjenes J, Ticchioni M, Bernard A. CD99 (E2) up-regulates alpha4beta1-dependent T cell adhesion to inflamed vascular endothelium under flow conditions. Eur J Immunol. 2000;30:3061–3065. doi: 10.1002/1521-4141(200010)30:10<3061::AID-IMMU3061>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 25.Gil MC, Lee MH, Seo JI, Choi YL, Kim MK, Jung KC, Park SH, Kim TJ. Characterization and epitope mapping of two monoclonal antibodies against human CD99. Exp Mol Med. 2002;34:411–418. doi: 10.1038/emm.2002.58. [DOI] [PubMed] [Google Scholar]
- 26.Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- 27.Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 28.Matthews CP, Colburn NH, Young MR. AP-1 a target for cancer prevention. Curr Cancer Drug Targets. 2007;7:317–324. doi: 10.2174/156800907780809723. [DOI] [PubMed] [Google Scholar]
- 29.Senga T, Iwamoto T, Humphrey SE, Yokota T, Taparowsky EJ, Hamaguchi M. Stat3-dependent induction of BATF in M1 mouse myeloid leukemia cells. Oncogene. 2002;21:8186–8191. doi: 10.1038/sj.onc.1205918. [DOI] [PubMed] [Google Scholar]
- 30.Su ZZ, Lee SG, Emdad L, Lebdeva IV, Gupta P, Valerie K, Sarkar D, Fisher PB. Cloning and characterization of SARI (suppressor of AP-1, regulated by IFN) Proc Natl Acad Sci U S A. 2008;105:20906–20911. doi: 10.1073/pnas.0807975106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quigley M, Pereyra F, Nilsson B, Porichis F, Fonseca C, Eichbaum Q, Julg B, Jesneck JL, Brosnahan K, Imam S, Russell K, Toth I, Piechocka-Trocha A, Dolfi D, Angelosanto J, Crawford A, Shin H, Kwon DS, Zupkosky J, Francisco L, Freeman GJ, Wherry EJ, Kaufmann DE, Walker BD, Ebert B, Haining WN. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat Med. 2010;16:1147–1151. doi: 10.1038/nm.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Lu C, Shen Q, Munoz-Medellin D, Kim H, Brown PH. AP-1 blockade in breast cancer cells causes cell cycle arrest by suppressing G1 cyclin expression and reducing cyclin-dependent kinase activity. Oncogene. 2004;23:8238–8246. doi: 10.1038/sj.onc.1207889. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used for the real-time PCR
SiRNA sequences