Highlights
-
•
This study suggests that there is close association between ApoE polymorphism and MI risk.
-
•
ApoE ε2 allele is a protective factor of MI.
-
•
ε4 allele is a risk factor for MI, especially in Caucasian and Asian populations.
Abbreviations: ApoE, Apolipoprotein E; MI, myocardial infarction; OR, odds ratio; CIs, confidence intervals; HWE, Hardy–Weinberg equilibrium
Keywords: Myocardial infarction, MI, Apolipoprotein E, ApoE, Polymorphism, Meta-analysis
Abstract
Published data regarding the association between Apolipoprotein E (ApoE) genetic variation and myocardial infarction (MI) risk were not always consistent. Therefore, the current meta-analysis was conducted to derive a more precise estimation of the association between ApoE polymorphism and MI risk. PubMed and Web of Science were searched to identify relevant studies. Summary odds ratio (ORs) and 95% confidence intervals (CIs) were calculated using random-effect or fixed-effect models based on the heterogeneity of included studies. All the tests were performed using Stata 11.0. A total of 22 eligible studies were identified in this meta-analysis. The results show that ApoE ε2 and ε4 alleles were associated with MI risk. The study suggests that there is close association between ApoE polymorphism and MI risk. It shows that ApoE ε2 allele is a protective factor of MI, while ε4 allele is a risk factor of MI, especially in Caucasian and Asian population. Nevertheless, well-designed, unbiased and larger sample size studies are required to confirm the results.
1. Introduction
Myocardial infarction (MI) is a complex syndrome affected by multiple predisposing genetic and environmental factors [1]. The association between ApoE polymorphisms and MI has drawn a lot of attention. ApoE is a multifunctional protein which plays a critical role in the metabolism of triglycerides and cholesterol [2], [3], and the corresponding gene is considered as a excellent candidate to investigate the etiology of MI [4]. The gene is located at 19q13.2 and possesses three common alleles (ε2, ε3, ε4) and forms six genotypes (ε2/ε2, ε2/ε3, ε2/ε4, ε3/ε3, ε3/ε4 and ε4/ε4) [5]. As the previous studies, the ApoE polymorphisms were found to affect ApoE transcription and the levels of cholesterol and triglyceride [6], [7], which was the main underlying risk factor of MI. However, the results of the earlier studies were inconsistent. Therefore, a system review and meta-analysis by collecting and sorting the previously published studies was conducted.
2. Materials and methods
2.1. Identification and eligibility of relevant studies
The online medical databases PubMed and Web of Science were used, using the search term “ApoE/Apolipoprotein E”, “polymorphism/genetic variation” and “myocardial infarction/MI”. The last retrieval was conducted in January 2015. The literatures were limited to papers in English. In addition, studies were identified by manual search of the references listed in the retrieved studies. The inclusion criteria were listed as follows: (1) case–control studies with either a population-based or a hospital-based design; (2) studies evaluated association between the ApoE polymorphisms and cancer risk; (3) present sufficient data to calculate an odds ratio (OR) with 95% confidence interval (CI); (4) not republished data. Moreover, the studies without raw data or those that were case-only studies, case reports, editorials and review articles (including meta-analyses) were eliminated.
2.2. Data extraction
The following detail information were extracted from each study enrolled in this study by two investigators (YLW and LZ) independently: the first author’s last name, year of publication, country of subjects, ethnicity, the source of controls, genotyping method, matching numbers of genotyped cases and controls and P for Hardy–Weinberg equilibrium (HWE). Furthermore, the disagreements were discussed among all authors and resolved with consensus.
2.3. Statistical analysis
The association of the ApoE polymorphism and risk of myocardial infarction was estimated by calculating the pooled ORs and 95%CI. The pooled ORs were estimated for seven genetic models (ε2/ε2 vs. ε3/ε3, ε2/ε3 vs. ε3/ε3, ε2/ε4 vs. ε3/ε3, ε3/ε4 vs. ε3/ε3, ε4/ε4 vs. ε3/ε3, ε2 allele vs. ε3 allele and ε4 allele vs. ε3 allele). Stratified analyses were performed by ethnicity (‘other ethnicity’ group was defined as those ethnicities that contained only one study). Heterogeneity across the studies was evaluated by using the Chi-square test based Q-statistic test [8], and it was considered significant when Pheterogeneity(Ph) < 0.05. The data were combined using random-effects (the DerSimonian and Laird method) in the presence of heterogeneity (P < 0.05 or I2 > 50%) and fixed-effects (the Mantel–Haenszel method) models were used in absence of heterogeneity (P > 0.05 or I2 < 50%) [9]. Furthermore, the sensitivity analysis was used to assess the stability of results, and publication bias was analyzed by Begg’s funnel plot and Egger’s regression test [10]. Additionally, HWE was used to assess the genotype frequencies of the polymorphism by the chi-square test. All statistical tests were performed with STATA 11.0 and all the P values were two-sided.
3. Results
3.1. Characteristics of studies
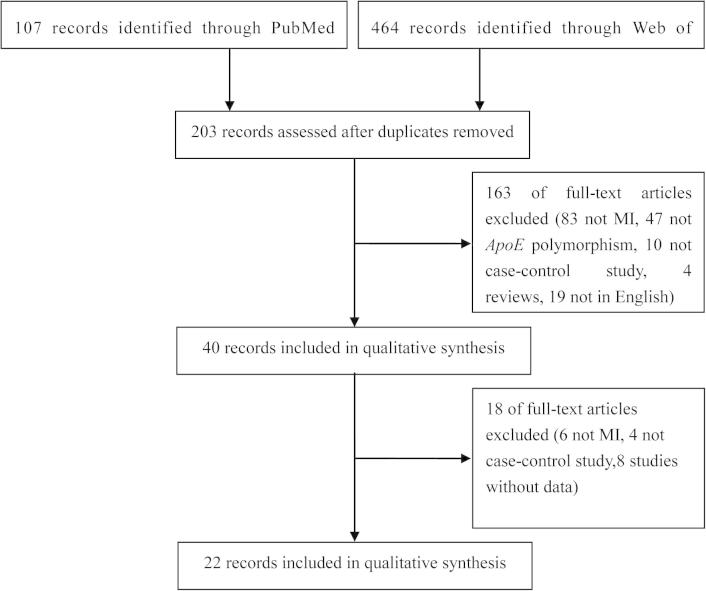
Based on the search strategy, 571 potentially eligible studies were identified in the initial search. Among these, 22 studies were enrolled in this meta-analysis based on the inclusion criteria [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32] (Fig. 1). The study by Luc et al. [30] investigated in four countries and was divided into four studies. The main characteristics of the enrolled 22 studies are summarized in Table 1.
Fig. 1.
Flow diagram of the study selection process.
Table 1.
The characteristics of the enrolled studies in this meta-analysis.
| First author | Year | Country | Ethnicity | Source of control | Genotyping method | Sample size (cases/controls) | HWE |
|---|---|---|---|---|---|---|---|
| Tanguturi | 2013 | India | Asian | PB | PCR-RFLP | 202/210 | 0.097 |
| Anand | 2009 | Mixed | Mixed | Mixed | IlluminaGoldenGate technology | 4017/4017 | 0.091 |
| Al-Bustan | 2009 | Kuwaiti | Asian | HB | PCR-RFLP | 88/122 | <0.050 |
| Koch | 2008 | Germany | Caucasian | PB | TaqMan | 3657/1211 | 0.558 |
| Ranjith | 2004 | Indian | African | PB | PCR-RFLP | 195/300 | <0.050 |
| Keavney | 2004 | UK | Caucasian | PB | PCR-RFLP | 4685/3460 | – |
| Keavney | 2003 | UK | Caucasian | PB | PCR-RFLP | 4484/5757 | 0.463 |
| Mamotte | 2003 | Australia | Caucasian | PB | PCR-RFLP | 359/639 | 0.732 |
| Wang | 2001 | Xinjiang | Asian | PB | PCR-RFLP | 54/71 | 0.479 |
| Raslova | 2001 | Bratislava | Caucasian | PB | PCR-RFLP | 71/71 | 0.183 |
| Batalla | 2000 | Asturias | Caucasian | PB | PCR-RFLP | 220/200 | 0.776 |
| Joven | 1998 | Spanish | Caucasian | PB | PCR-RFLP | 250/250 | 0.109 |
| Luc | 1994 | Belfast | Caucasian | PB | NA | 183/176 | 0.405 |
| Luc | 1994 | Lille | Caucasian | PB | NA | 64/150 | 0.932 |
| Luc | 1994 | Strasbourg | Caucasian | PB | NA | 187/172 | 0.35 |
| Luc | 1994 | Toulouse | Caucasian | PB | NA | 140/182 | 0.698 |
| Lenzen | 1986 | NA | Caucasian | PB | NA | 570/624 | 0.081 |
| Kolovou | 2002 | Greek | Caucasian | PB | PCR-RFLP | 124/240 | 0.552 |
| Kumar | 2003 | North India | Asian | PB | PCR-RFLP | 35/45 | <0.050 |
| Baum | 2006 | Hong Kong | Asian | HB | PCR-RFLP | 234/336 | 0.659 |
| Nakai | 1998 | Japan | Asian | PB | PCR-RFLP | 254/422 | 0.175 |
| Hergenc | 1995 | Turkish | Caucasian | PB | PCR-RFLP | 50/60 | 0.117 |
| Utermann | 1984 | Germany | Caucasian | PB | NA | 523/1031 | <0.050 |
NA: not available; PB: population based; HB: hospital based; PCR-RFLP: restriction fragment length polymorphism; HWE: Hardy–Weinberg equilibrium. –: the data of the study are not enough.
3.2. Quantitative synthesis
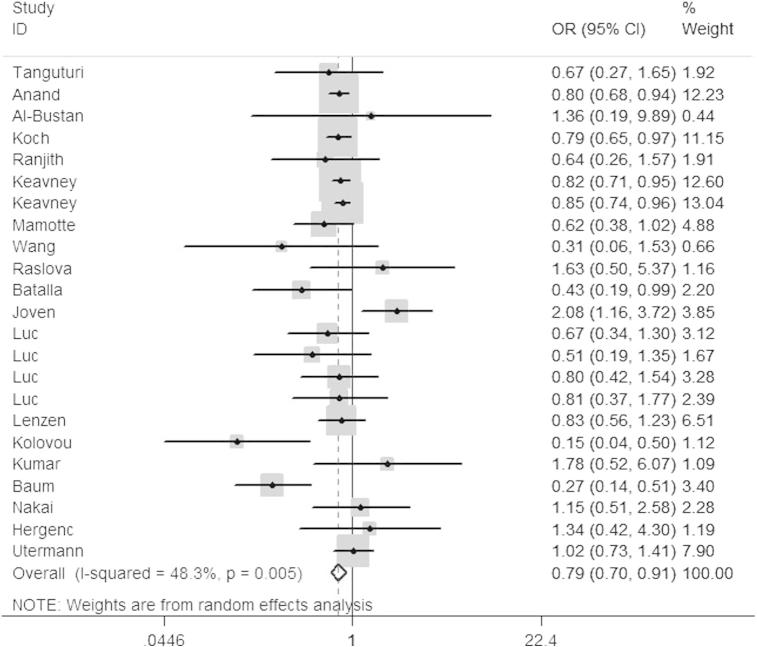
In the pooled analysis, significant association was observed between ApoE polymorphism and risk of myocardial infarction. The main results were presented in Table 2, and the result of ε2/ε3 vs. ε3/ε3 was also shown in Fig. 2.
Table 2.
Stratified analyses of the ApoE polymorphism and MI risk.
| Variables | na | ε2/ε2 vs. ε3/ε3 |
ε2/ε3 vs. ε3/ε3 |
ε2/ε4 vs. ε3/ε3 |
ε3/ε4 vs. ε3/ε3 |
ε4/ε4 vs. ε3/ε3 |
ε2 allele vs. ε3 allele |
ε4 allele vs. ε3 allele |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR(95%CI) | Pb | I2 | OR(95%CI) | Pb | I2 | OR(95%CI) | Pb | I2 | OR(95%CI) | Pb | I2 | OR(95%CI) | Pb | I2 | OR(95%CI) | Pb | I2 | OR(95%CI) | Pb | I2 | ||
| Total | 23 | 0.56(0.31,1.02)c | 0.001 | 63.4 | 0.79(0.70,0.91)c | 0.005 | 48.3 | 0.96(0.81,1.14) | 0.750 | 0.0 | 1.20(1.08,1.34)c | 0.000 | 58.0 | 1.39(1.19,1.64) | 0.194 | 20.6 | 0.74(0.65,0.85)c | 0.000 | 64.3 | 1.22(1.11,1.35)c | 0.000 | 65.5 |
| Ethnicity | ||||||||||||||||||||||
| Caucasian | 15 | 0.64(0.27,1.51)c | 0.000 | 76.5 | 0.83(0.72,0.96)c | 0.026 | 46.1 | 1.05(0.99,1.12) | 0.390 | 5.6 | 1.12(1.00,1.26)c | 0.007 | 54.0 | 1.26(1.05,1.52) | 0.914 | 0.0 | 0.77(0.65,0.91)c | 0.000 | 70.6 | 1.11(1.01,1.21)c | 0.041 | 42.6 |
| Asian | 6 | 0.52(0.18,1.49) | 0.866 | 0.0 | 0.68(0.34,1.37)c | 0.022 | 61.9 | 1.16(0.57,2.35) | 0.864 | 0.0 | 1.52(1.19,1.93) | 0.333 | 12.8 | 5.67(2.68,12.02) | 0.553 | 0.0 | 0.58(0.43,0.79) | 0.071 | 50.7 | 1.92(1.26,2.92)c | 0.004 | 70.9 |
| Other | 2 | 0.40(0.20,0.83) | 0.704 | 0.0 | 0.79(0.68,0.93) | 0.629 | 0.0 | 0.94(0.77,1.14) | 0.555 | 0.0 | 1.72(0.69,4.27)c | 0.001 | 91.3 | 1.34(0.91,1.98) | 0.186 | 42.7 | 0.77(0.67,0.89) | 0.388 | 0.0 | 1.42(0.85,2.38)c | 0.022 | 81.0 |
Statistically significant results were in bold.
Number of comparisons.
P value of Q-test for heterogeneity test.
Random-effect model was applied when P value for heterogeneity <0.05; otherwise, fixed-effect model was applied.
Fig. 2.
Forest plot for ApoE polymorphism and MI risk in the genetic model of ε2/ε3 vs. ε3/ε3.
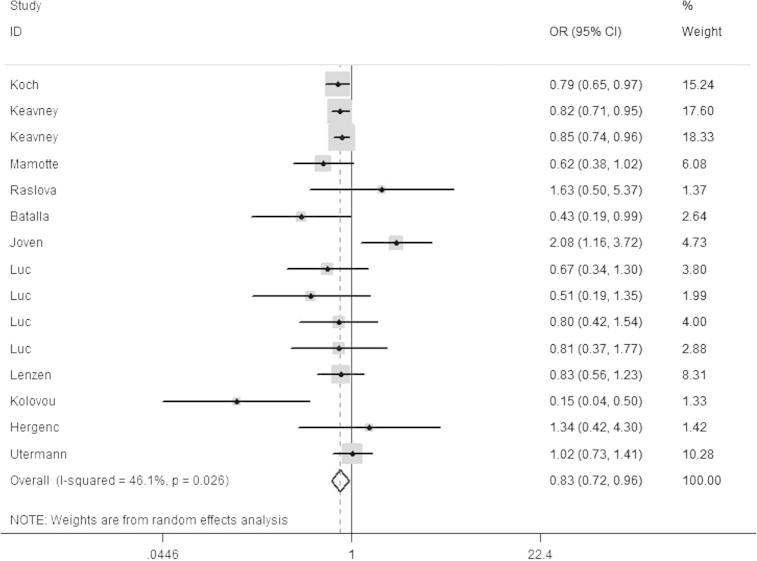
Additionally, the subgroup analysis by ethnicity was also conducted, and significant associations with myocardial infarction were observed in Caucasian (Fig.3) and Asian population. The main results were presented in Table 2.
Fig. 3.
Forest plot for ApoE polymorphism and MI risk among the Caucasian population in the genetic model of ε2/ε3 vs. ε3/ε3.
3.3. Sensitivity analysis
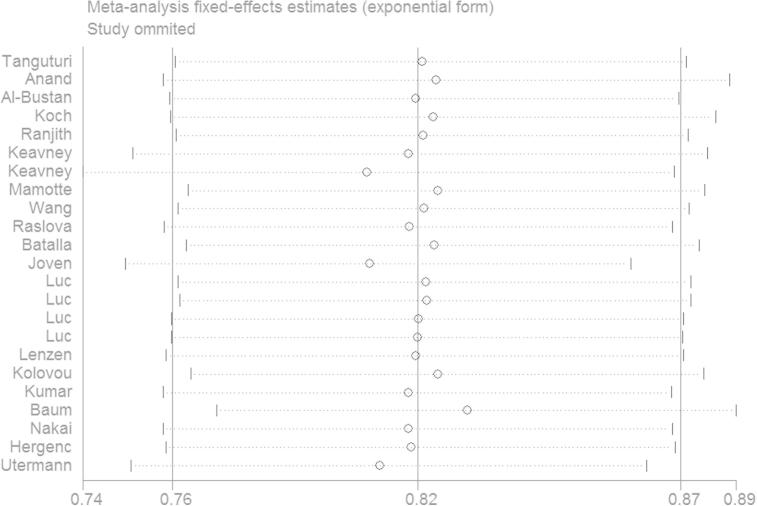
To assess the stability of the results and assess the source of the heterogeneity, the sensitivity analysis was performed by omitting individual eligible study to reflect the influence of the individual data on the summary ORs. The pooled ORs were not altered for all comparison models. Among all the enrolled studies, four studies did not follow HWE, the corresponding summary ORs were not materially altered with or without these studies. Therefore, the results of current study were statistically robust. The result of ε2/ε2 vs. ε3/ε3 was shown in Fig. 4.
Fig. 4.
The sensitivity analysis in the genetic model of ε2/ε2 vs. ε3/ε3. The omitted study is indicated by the first author’s last name.
3.4. Heterogeneity analysis
There was significant between-study heterogeneity in ε2/ε2 vs. ε3/ε3 (P = 0.001, I2 = 63.4), ε2/ε3 vs. ε3/ε3 (P = 0.005, I2 = 48.3), ε3/ε4 vs. ε3/ε3 (P = 0.000, I2 = 58.0), ε2 allele vs. ε3 allele (P = 0.000, I2 = 64.3) and ε4 allele vs. ε3 allele (P = 0.000, I2 = 65.5). In contract, no significant heterogeneity was observed in other two genetic models (ε2/ε4 vs. ε3/ε3: P = 0.750, I2 = 0.0; ε4/ε4 vs. ε3/ε3: P = 0.194, I2 = 20.6). In order to detect the sources of heterogeneity, the sensitivity analysis was performed based on HWE and ethnicity. However, the heterogeneity was not materially altered. As a consequence, we conducted a Galbraith plot to graphically assess the source of heterogeneity. The results indicated that a total of eight studies contributed to the heterogeneity. Two studies were the main sources for ε2/ε2 vs. ε3/ε3 [20], [32] (Fig. 5), three studies for ε2/ε3 vs. ε3/ε3 [15], [21], [27], four studies for ε3/ε4 vs. ε3/ε3 [16], [19], [27], [32], four studies for ε2 allele vs. ε3 allele [15], [21], [27], [32] and five studies for ε4 allele vs. ε3 allele [11], [16], [19], [27], [32], and after removal of these outlier studies, the heterogeneity was effectively removed (ε2/ε2 vs. ε3/ε3: P = 0.640, I2 = 0.0; ε2/ε3 vs. ε3/ε3: P = 0.828, I2 = 0.0; ε3/ε4 vs. ε3/ε3: P = 0.800, I2 = 0.0; ε2 allele vs. ε3 allele: P = 0.651, I2 = 0.0; ε4 allele vs. ε3 allele: P = 0.566, I2 = 0.0). Meanwhile, the corresponding pooled ORs were not materially altered in all comparisons. As a consequence, the results of heterogeneity analysis indicated that our results were statistically robust and credible.
Fig. 5.
Galbraith plot for ApoE gene polymorphism and MI risk in the genetic model of ε2/ε2 vs. ε3/ε3.
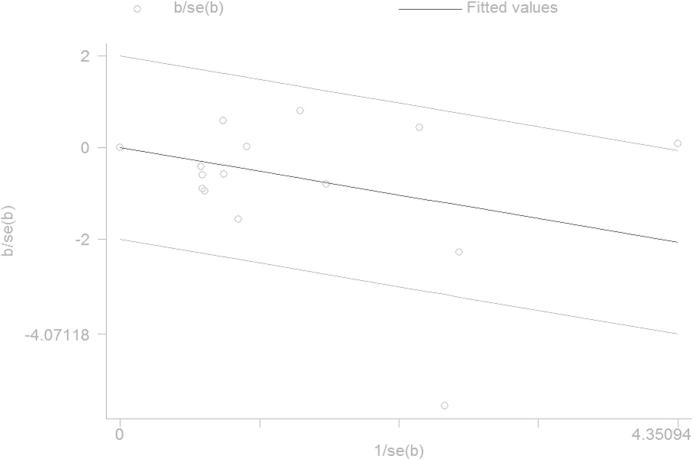
3.5. Publication bias
To evaluate the publication bias of enrolled studies, the Begg’s funnel plot and Egger’s test were performed. The shapes of funnel plots did not show any obvious asymmetry in all genetic models. Therefore, the Egger’s test was performed to provide statistical evidence of funnel plot symmetry, and the results confirmed the absence of publication bias (Table 3).
Table 3.
Egger’s test for ApoEpolymorphism.
| Egger’s test | ε2/ε2 vs. ε3/ε3 | ε2/ε3 vs. ε3/ε3 | ε2/ε4 vs. ε3/ε3 | ε3/ε4 vs. ε3/ε3 | ε4/ε4 vs. ε3/ε3 |
|---|---|---|---|---|---|
| t | −0.53 | −0.73 | 0.6 | 1.46 | 0.48 |
| p | 0.607 | 0.474 | 0.554 | 0.159 | 0.638 |
4. Discussion
A total of 22 studies were included in this meta-analysis to investigate the association between ApoE polymorphisms and MI. The results of the overall studies showed that the ε2/ε3 genotype was associated with a decreased risk of MI, while the ε3/ε4 and ε4/ε4 genotypes were associated with an increased risk of MI. The current results were in accord with the previous observers, which support the ε4 allele as a risk factor of MI [16], [33], [34]. For the ε2 allele, it was found significantly protective against MI [12], [21]. The discrepancy of the effect on MI risk between different ApoE genotypes might be supported by earlier studies [35], [36]. It provided evidence that the ApoE E4 coding by ε4 allele exhibits enhanced transfer from HDL to TG-rich lipoproteins, promoting hepatic remnant clearance by apoE receptors and decreasing LDLR, thereby increasing cholesterol levels. On the contrary, the ApoE E2 coding by ε2 allele binds LDLR poorly, which can increase the LDLR numbers, thereby lowering cholesterol level. A previous meta-analysis showed no significant association between ε2 carriers and MI risk (ε2 carriers vs. ε3/ε3: OR = 0.90, 95%CI = 0.76–1.06, P = 0.120), whereas an increased MI risk with ε4 carriers (ε4 carriers vs. ε3/ε3: OR = 1.18, 95%CI = 1.05–1.33, P = 0.003) [37]. By comparison, our results were not completely consistent with previous meta-analysis. The discrepancy may partly result from the genetic diversity among ethnicities.
Furthermore, the subgroup analysis by ethnicity showed a decreased MI risk in ε2 carriers, while an increased MI risk in ε4 carriers compared with ε3 carriers in both Asian and Caucasian population. These results were consistent with the studies enrolled in our meta-analysis [11], [12], [15], [19], [26], [30]. Furthermore, sensitivity analysis was also performed to make sure whether modification of the inclusion criteria of the meta-analysis affected the final results. The results showed that corresponding pooled ORs were not materially altered in all genetic models which indicated that the results were statistically robust.
Heterogeneity is a potentially important factor to influence the interpretation of the current results. In this meta-analysis, significant heterogeneity existed in ε2/ε2 vs. ε3/ε3, ε2/ε3 vs. ε3/ε3, ε3/ε4 vs. ε3/ε3, ε2 allele vs. ε3 allele and ε4 allele vs. ε3 allele. Common reasons of heterogeneity may attribute to the diversity in design, study quality, sample-sizes, genotyping methods, inclusion criteria and some studies without HWE. To explore the sources of heterogeneity, we first performed the sensitivity analyses based on HWE and ethnicity. However, the heterogeneity was not effectively removed. Therefore, a Galbraith plot was performed to further evaluate the source of heterogeneity. After excluding eight outlier studies, the heterogeneity was effectively removed. Moreover, the corresponding pooled ORs were not materially altered in all comparisons, which also suggested that our results were statistically robust.
Some limitations of the meta-analysis should be addressed. Firstly, the potential factors such as gender, age, smoking, drinking, living habits were not considered in this meta-analysis. Secondly, between-study heterogeneity should be paid attention, which may affect the results. Thirdly, only studies in English were enrolled in this meta-analysis, which may lose some studies in other languages consistent with inclusion criteria. Regardless of such limitations, this meta-analysis still had some advantages. Firstly, all enrolled studies were consistent with inclusion criteria well. Secondly, no publication bias was observed indicating that the whole pooled results might be unbiased.
In conclusion, the current meta-analysis of 22 studies indicated that ApoE ε2 allele was a protective factor of MI, while ε4 allele was a dangerous factor of MI, especially in Asian and Caucasian population. However, the results should be further conformed in well-designed, unbiased, powered studies.
Author contributions
YLW conceived and designed the project. LMS, LZ and HTX acquired the data. ZD and LQ analyzed and interpreted the data. YLW and MLW wrote the paper.
Conflict of interest
The authors declare that they have no competing interests.
Acknowledgment
None.
References
- 1.Luo J.Q., Wen J.G., Zhou H.H., Chen X.P., Zhang W. Endothelial nitric oxide synthase gene g894t polymorphism and myocardial infarction: a meta-analysis of 34 studies involving 21068 subjects. PLoS One. 2014;9(1):e87196. doi: 10.1371/journal.pone.0087196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mozas P., Castillo S., Reyes G. Apolipoprotein E genotype is not associated with cardiovascular disease in heterozygous subjects with familial hypercholesterolemia. Am. Heart J. 2003;145(6):999–1005. doi: 10.1016/S0002-8703(02)94788-5. [DOI] [PubMed] [Google Scholar]
- 3.Mendes-Lana A., Pena G.G., Freitas S.N. Apolipoprotein E polymorphism in Brazilian dyslipidemic individuals: ouro preto study. Braz. J. Med. Biol. Res. 2007;40(1):49–56. doi: 10.1590/s0100-879x2007000100007. [DOI] [PubMed] [Google Scholar]
- 4.Stampfer M.J., Sacks F.M., Salvini S., Willett W.C., Hennekens C.H. A prospective study of cholesterol, apolipoproteins, and the risk of myocardial infarction. N. Engl. J. Med. 1991;325(6):373–381. doi: 10.1056/NEJM199108083250601. [DOI] [PubMed] [Google Scholar]
- 5.Lahiri D.K., Sambamurti K., Bennett D.A. Apolipoprotein gene and its interaction with the environmentally driven risk factors: molecular, genetic and epidemiological studies of Alzheimer’s disease. Neurobiol. Aging. 2004;25(5):651–660. doi: 10.1016/j.neurobiolaging.2003.12.024. [DOI] [PubMed] [Google Scholar]
- 6.Pollin T.I., Hsueh W.C., Steinle N.I., Snitker S., Shuldiner A.R., Mitchell B.D. A genome-wide scan of serum lipid levels in the old order amish. Atherosclerosis. 2004;173(1):89–96. doi: 10.1016/j.atherosclerosis.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 7.Pilia G., Chen W.M., Scuteri A. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2006;2(8):e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Handoll H.H. Systematic reviews on rehabilitation interventions. Arch. Phys. Med. Rehabil. 2006;87(6):875. doi: 10.1016/j.apmr.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Midgette A.S., Wong J.B., Beshansky J.R., Porath A., Fleming C., Pauker S.G. Cost-effectiveness of streptokinase for acute myocardial infarction: a combined meta-analysis and decision analysis of the effects of infarct location and of likelihood of infarction. Med. Decis. Making. 1994;14(2):108–117. doi: 10.1177/0272989X9401400203. [DOI] [PubMed] [Google Scholar]
- 10.Egger M., Davey Smith G., Schneider M., Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanguturi P., Pullareddy B., Kumar P.S., Murthy D.K. Association between apolipoprotein E gene polymorphism and myocardial infarction. Biochem. Genet. 2013;51(5–6):398–405. doi: 10.1007/s10528-013-9572-2. [DOI] [PubMed] [Google Scholar]
- 12.Anand S.S., Xie C., Pare G. Genetic variants associated with myocardial infarction risk factors in over 8000 individuals from five ethnic groups: the INTERHEART Genetics Study. Circ. Cardiovasc. Genet. 2009;2(1):16–25. doi: 10.1161/CIRCGENETICS.108.813709. [DOI] [PubMed] [Google Scholar]
- 13.Al-Bustan S.A., Alkhalaf M., Al-Rashdan I. Apolipoprotein E, CI and B gene polymorphisms in a sample of patients with coronary heart disease in the Kuwaiti population. Med. Princ. Pract. 2009;18(4):294–299. doi: 10.1159/000215727. [DOI] [PubMed] [Google Scholar]
- 14.Koch W., Hoppmann P., Schomig A., Kastrati A. Apolipoprotein E gene epsilon2/epsilon3/epsilon4 polymorphism and myocardial infarction: case-control study in a large population sample. Int. J. Cardiol. 2008;125(1):116–117. doi: 10.1016/j.ijcard.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Baum L., Ng H.K., Wong K.S. Associations of apolipoprotein E exon 4 and lipoprotein lipase S447X polymorphisms with acute ischemic stroke and myocardial infarction. Clin. Chem. Lab. Med. 2006;44(3):274–281. doi: 10.1515/CCLM.2006.047. [DOI] [PubMed] [Google Scholar]
- 16.Ranjith N., Pegoraro R.J., Rom L., Rajput M.C., Naidoo D.P. Lp(a) and apoE polymorphisms in young South African Indians with myocardial infarction. Cardiovasc. J. S. Afr. 2004;15(3):111–117. [PubMed] [Google Scholar]
- 17.Keavney B., Palmer A., Parish S. Lipid-related genes and myocardial infarction in 4685 cases and 3460 controls: discrepancies between genotype, blood lipid concentrations, and coronary disease risk. Int. J. Epidemiol. 2004;33(5):1002–1013. doi: 10.1093/ije/dyh275. [DOI] [PubMed] [Google Scholar]
- 18.Marques-Vidal P., Bongard V., Ruidavets J.B., Fauvel J., Perret B., Ferrieres J. Effect of apolipoprotein E alleles and angiotensin-converting enzyme insertion/deletion polymorphisms on lipid and lipoprotein markers in middle-aged men and in patients with stable angina pectoris or healed myocardial infarction. Am. J. Cardiol. 2003;92(9):1102–1105. doi: 10.1016/j.amjcard.2003.06.008. [DOI] [PubMed] [Google Scholar]
- 19.Kumar P., Luthra K., Dwivedi M., Behl V.K., Pandey R.M., Misra A. Apolipoprotein E gene polymorphisms in patients with premature myocardial infarction: a case-controlled study in Asian Indians in North India. Ann. Clin. Biochem. 2003;40(Pt 4):382–387. doi: 10.1258/000456303766477020. [DOI] [PubMed] [Google Scholar]
- 20.Keavney B., Parish S., Palmer A. Large-scale evidence that the cardiotoxicity of smoking is not significantly modified by the apolipoprotein E epsilon2/epsilon3/epsilon4 genotype. Lancet. 2003;361(9355):396–398. doi: 10.1016/S0140-6736(03)12386-0. [DOI] [PubMed] [Google Scholar]
- 21.Kolovou G., Yiannakouris N., Hatzivassiliou M. Association of apolipoprotein E polymorphism with myocardial infarction in Greek patients with coronary artery disease. Curr. Med. Res. Opin. 2002;18(3):118–124. doi: 10.1185/030079902125000444. [DOI] [PubMed] [Google Scholar]
- 22.Mamotte C.D., Burke V., Taylor R.R., van Bockxmeer F.M. Evidence of reduced coronary artery disease risk for apolipoprotein epsilon2/3 heterozygotes. Eur. J. Intern. Med. 2002;13(4):250–255. doi: 10.1016/s0953-6205(02)00030-4. [DOI] [PubMed] [Google Scholar]
- 23.Wang X., Wang G., Yang C., Li X. Apolipoprotein E gene polymorphism and its association with human longevity in the Uygur nationality in Xinjiang. Chin. Med. J. (Engl.) 2001;114(8):817–820. [PubMed] [Google Scholar]
- 24.Raslova K., Smolkova B., Vohnout B., Gasparovic J., Frohlich J.J. Risk factors for atherosclerosis in survivors of myocardial infarction and their spouses: comparison to controls without personal and family history of atherosclerosis. Metabolism. 2001;50(1):24–29. doi: 10.1053/meta.2001.19499. [DOI] [PubMed] [Google Scholar]
- 25.Batalla A., Alvarez R., Reguero J.R. Synergistic effect between apolipoprotein E and angiotensinogen gene polymorphisms in the risk for early myocardial infarction. Clin. Chem. 2000;46(12):1910–1915. [PubMed] [Google Scholar]
- 26.Nakai K., Fusazaki T., Zhang T. Polymorphism of the apolipoprotein E and angiotensin I converting enzyme genes in Japanese patients with myocardial infarction. Coron. Artery Dis. 1998;9(6):329–334. doi: 10.1097/00019501-199809060-00002. [DOI] [PubMed] [Google Scholar]
- 27.Joven J., Simo J.M., Vilella E. Lipoprotein(a) and the significance of the association between platelet glycoprotein IIIa polymorphisms and the risk of premature myocardial infarction. Atherosclerosis. 1998;140(1):155–159. doi: 10.1016/s0021-9150(98)00076-8. [DOI] [PubMed] [Google Scholar]
- 28.O’Malley J.P., Maslen C.L., Illingworth D.R. Angiotensin-converting enzyme DD genotype and cardiovascular disease in heterozygous familial hypercholesterolemia. Circulation. 1998;97(18):1780–1783. doi: 10.1161/01.cir.97.18.1780. [DOI] [PubMed] [Google Scholar]
- 29.Hergenc G., Taga Y., Emerk K., Cirakoglu B. Apolipoprotein E genotyping in Turkish myocardial infarction survivors and healthy controls. J. Biomed. Sci. 1995;2(1):46–49. doi: 10.1007/BF02257924. [DOI] [PubMed] [Google Scholar]
- 30.Luc G., Bard J.M., Arveiler D. Impact of apolipoprotein E polymorphism on lipoproteins and risk of myocardial infarction. The ECTIM Study. Arterioscler. Thromb. 1994;14(9):1412–1419. doi: 10.1161/01.atv.14.9.1412. [DOI] [PubMed] [Google Scholar]
- 31.Lenzen H.J., Assmann G., Buchwalsky R., Schulte H. Association of apolipoprotein E polymorphism, low-density lipoprotein cholesterol, and coronary artery disease. Clin. Chem. 1986;32(5):778–781. [PubMed] [Google Scholar]
- 32.Utermann G., Hardewig A., Zimmer F. Apolipoprotein E phenotypes in patients with myocardial infarction. Hum. Genet. 1984;65(3):237–241. doi: 10.1007/BF00286509. [DOI] [PubMed] [Google Scholar]
- 33.Lehtinen S., Lehtimaki T., Sisto T. Apolipoprotein E polymorphism, serum lipids, myocardial infarction and severity of angiographically verified coronary artery disease in men and women. Atherosclerosis. 1995;114(1):83–91. doi: 10.1016/0021-9150(94)05469-y. [DOI] [PubMed] [Google Scholar]
- 34.Singh P.P., Singh M., Bhatnagar D.P., Kaur T.P., Gaur S.K. Apolipoprotein E polymorphism and its relation to plasma lipids in coronary heart disease. Indian J. Med. Sci. 2008;62(3):105–112. [PubMed] [Google Scholar]
- 35.Davignon J., Gregg R.E., Sing C.F. Apolipoprotein E polymorphism and atherosclerosis. Arteriosclerosis. 1988;8(1):1–21. doi: 10.1161/01.atv.8.1.1. [DOI] [PubMed] [Google Scholar]
- 36.Lahoz C., Schaefer E.J., Cupples L.A. Apolipoprotein E genotype and cardiovascular disease in the framingham heart study. Atherosclerosis. 2001;154(3):529–537. doi: 10.1016/s0021-9150(00)00570-0. [DOI] [PubMed] [Google Scholar]
- 37.Song Y., Stampfer M.J., Liu S. Meta-analysis: apolipoprotein E genotypes and risk for coronary heart disease. Ann. Intern. Med. 2004;141(2):137–147. doi: 10.7326/0003-4819-141-2-200407200-00013. [DOI] [PubMed] [Google Scholar]