Abstract
Trichomonadid protozoa have been found in the intestinal tracts of common marmosets (Callithrix jacchus). However, there is little information available on species identification and the pathogenicity of these trichomonads. In this study, we conducted a fecal survey of a common marmoset colony maintained as laboratory animals in Japan and identified the trichomonad species. Screening using a fecal smear examination revealed that 66% (58/88) of the marmosets had trichomonadid trophozoites in their feces. The trichomonads were found in both normal feces (31/49, 63%) and diarrhea (27/39, 69%), with no significant difference in frequency. The protozoa were identified as Pentatrichomonas hominis using morphological characters and the 100% identity of the nucleotide sequence of the partial 18S rRNA gene (297 bp). The intraspecific genetic variability between P. hominis from the marmosets in this study and P. hominis from other reported mammal hosts was ≤1% in the nucleotide sequence, including the internal transcribed spacer (ITS)-1, 5.8S rRNA gene, and ITS-2 (293 bp). P. hominis inhabits the large intestine of various mammalian hosts, including primates, and is considered nonpathogenic. These results suggest that P. hominis is transmitted among marmosets and other mammals but is not a primary cause of bowel disease in marmosets.
Keywords: marmoset, Pentatrichomonas hominis, protozoa, Trichomonas
Introduction
The common marmoset (Callithrix jacchus), a New World primate, has been increasingly used for biomedical and preclinical research in recent decades. This laboratory nonhuman primate has the advantages of small body size, easy handling, high fertility, and low zoonotic risk compared with other nonhuman primates and has become popular in various research fields including neuroscience, regenerative medicine, infectious disease, and drug testing [24, 26, 35]. Recent progress in transgenic and developmental engineering technology has expanded the research use of this laboratory animal [31, 36]. In this situation, improved management of marmoset health is necessary, and more information on spontaneous diseases and pathogens is needed. Diarrhea and gastrointestinal diseases are important health problems in marmoset colonies worldwide, but the exact cause of the diarrhea remains unknown; chronic diarrhea is associated with wasting marmoset syndrome, which is characterized by impaired weight gain, weight loss, muscle atrophy, alopecia, and diarrhea, and the etiology of this syndrome is poorly understood [3, 23, 29].
Trichomonad protozoa have been found in the large intestines of some callitrichid species in several colonies [8, 27]. However, there is little information available on species identification and the pathogenicity of these trichomonads. Intestinal trichomonad species include Pentatrichomonas hominis, which has been found and is considered nonpathogenic in a wide variety of mammalian hosts, including cats, dogs, rodents, and primates [18, 33, 38]. Nevertheless, several studies have indicated that some trichomonad species are associated with bowel diseases. Tritrichomonas suis is a causative agent of chronic large-bowel diarrhea in cats [11, 21]. Reports of trichomonad-induced lesions in nonhuman primates are rare, but invasive trichomoniasis involving the gastric mucosa and the colonic mucosa suspected of being caused by Tritrichomonas mobilensis has been reported in rhesus macaques (Macaca mulatta) infected with simian immunodeficiency virus (SIV) [20] and a titi monkey (Callicebus sp.) [4], respectively. There have been no detailed reports of intestinal trichomonads in common marmosets. Therefore, this study conducted fecal screening and identified the trichomonad species in a facility-bred common marmoset colony to investigate the relationship between parasitism and bowel disease.
Materials and Methods
Animals
This survey was performed at the Central Institute for Experimental Animals (CIEA) in Kawasaki, Japan. The animal experimental procedures were reviewed by the Institutional Animal Care and Use Committee and approved according to the Regulations for Animal Experiments in CIEA established based on the Basic Policies on Animal Experiments conducted in Research Institutions (Notice No. 71 of the Ministry of Education, Culture, Sports, Science and Technology, June 2006) and the Guidelines for the Proper Conduct of Animal Experiments (Science Council of Japan, 2006).
The common marmosets used in this survey were from a commercial breeder, CLEA Japan (Tokyo, Japan), or were born in the CIEA facility. Ultimately, all of the animals trace back to a breeding colony that has been maintained for more than 20 years in the indoor facilities of CLEA Japan. The animals were kept in stainless steel wire cages on a 12 h:12 h light:dark cycle at 25–28°C and 40–60% humidity. They were given a New World primate diet (CMS-1M; CLEA Japan) with added vitamins and tap water ad libitum.
Fecal examination and species identification of trichomonads
Fresh feces of common marmosets were collected from 88 cages. These cages housed 123 animals (53 females, 70 males) individually (53 cages) or in pairs (35 cages). The ages of the animals ranged from 6 months to 9 years old.
The feces were examined using a direct smear method; feces mixed with a drop of saline on a slide glass were covered with a cover slip and examined under a microscope. Four samples of trichomonad-positive feces were examined morphologically using Giemsa-stained fecal smears and were cultured on Trichomonas medium (CM0161, Oxoid, Basingstoke, UK ). After a 5-day culture, medium containing the protozoa was used for DNA extraction with a MagExtractor Genome kit (Toyobo, Tokyo, Japan). Extracted DNA samples were used for the following analyses.
A species-specific PCR assay to identify P. hominis was performed using a primer pair, 5′-TGTAAACGATGCCGACAGAG-3′ (Th3) and 5′-CAACACTGAAGCCAATGCGAGG-3′ (Th5) designed to amplify a 339-bp sequence of the 18S ribosomal RNA (rRNA) gene [7]. PCR was performed in a 20-µl reaction volume containing 0.5 U of PrimeSTAR HS DNA Polymerase (Takara, Tokyo, Japan), 4 µl of 5× PrimeSTAR Buffer, 200 µM of each dNTP, 0.3 µM of each primer, and 1 µl of DNA extract. DNA amplification consisted of 30 cycles of 98°C for 10 s, 55°C for 5 s, and 72°C for 20 s. The PCR products were confirmed by electrophoresis on an agarose gel with ethidium bromide staining. After purifying the PCR products with DNA Clean & Concentrator-5 (Zymo Research, Irvine, CA, USA), the nucleotide sequences of the reaction products were determined using a BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) and ABI Prism 310 Genetic Analyzer (Applied Biosystems).
To investigate intraspecies variation between P. hominis from these common marmosets and P. hominis from other mammals, a 338-bp fragment consisting of the internal transcribed spacer region (ITS)-1, 5.8S rRNA gene, and ITS-2 of the obtained DNA samples was also amplified by PCR using a primer pair, 5′-TGCTTCAGTTCAGCGGGTCTTCC-3′ (TFR1) and 5′-CGGTAGGTGAACCTGCCGTTGG-3′ (TFR2) [13] and the nucleotide sequences were determined by sequencing. Homology between the obtained nucleotide sequences and known sequences from the GenBank database was analyzed using a BLAST search [2] on the website of the DNA Data Bank of Japan (DDBJ). The phylogenetic relationships among P. hominis from marmosets and P. hominis from other hosts were inferred using the neighbor-joining method [30]. The evolutionary distances were computed using the Kimura two-parameter method [19], and they are given as the number of base substitutions per site. The evolutionary analyses were conducted in MEGA5 [37].
Statistical analysis
The difference in positive rates of trichomonad trophozoites was analyzed using Fisher’s exact probability test.
Results
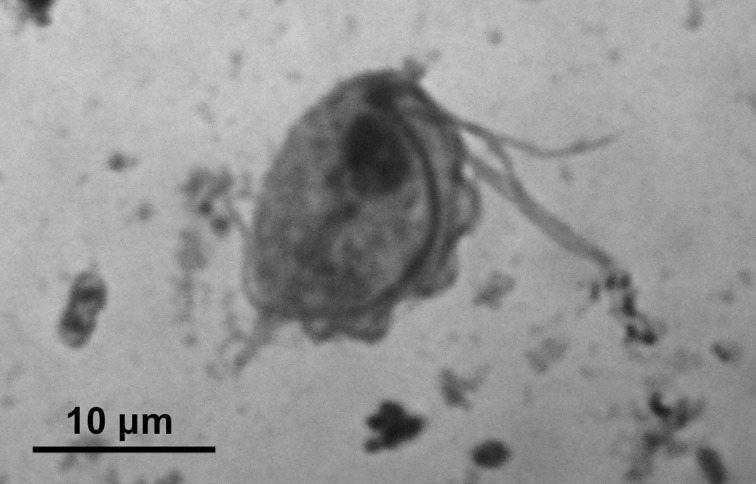
Trichomonadid trophozoites were detected in 58 (66%) of the 88 samples. There was no significant difference in positive rate between normal feces (31/49, 63%) and diarrheal stools (27/39, 69%) (Table 1). According to age, the positive rate in samples from young (0–1 years old) animals was high (8/10, 80%), but it was not significantly different from that in adults (2–9 years old) (50/78, 64%). There was also no significant difference in the positive rate between females (16/25, 64%) and males (27/43, 63%), excluding the 20 samples from male-female pairs. The trichomonadid trophozoites were similar morphologically in all samples, and no other protozoans were detected. The detected trophozoites were piliform with multiple flagella and an undulating membrane. In Giemsa-stained specimens, the trophozoites had a nucleus, axostyle, four or five anterior flagella, and a posterior flagellum that aligned along the undulating membrane and extended freely beyond the body (Fig. 1). Their bodies were 10–15 µm long by 7–12 µm wide according to measurements of ten trophozoites from each of the two specimens. These morphological characteristics of the detected trichomonadid trophozoites conformed to those of P. hominis.
Table 1. Detection of trichomonad trophozoites in the feces of common marmosets.
| No. positive / no. examined (%) | |||
|---|---|---|---|
| Normal feces | Diarrheal feces | Total | |
| Young (< 2 years) | 6/7 (86) | 2/3 (67) | 8/10 (80) |
| Adult (2–9 years) | 25/42 (60) | 25/36 (69) | 50/78 (64) |
| Total | 31/49 (63) | 27/39 (69) | 58/88 (66) |
Fig. 1.
A trichomonad trophozoite in a Giemsa-stained fecal smear from a common marmoset.
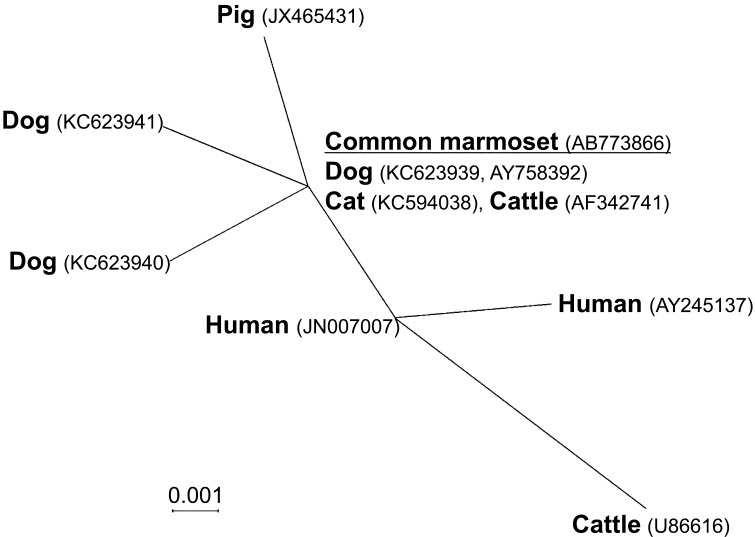
All four cultured samples from trichomonadid trophozoite-positive feces were positive for P. hominis species-specific PCR of the partial 18S rRNA gene (339 bp) (Fig. 2). The nucleotide sequence of a 297-bp product without primers was determined for two cultured samples; these two sequences were identical (GenBank no. AB773865). The sequence showed 100% identity to known P. hominis sequences from dogs [10], cats [6, 16], cattle [12], and nonhuman primates in zoos, including common marmosets [34], and 99.7% identity to those from dogs [15] and humans [9]. The nucleotide sequence of a 293-bp fragment containing ITS-1, the 5.8S rRNA gene, and ITS-2 without primers was determined for two cultured samples; these two sequences were identical (GenBank no. AB773866). The sequence showed 100% identity to known sequences of P. hominis from dogs [15, 17], cats [6], and cattle [39] and 99.0–99.7% identity to those from dogs [17], pigs [22], cattle [13], and humans [5, 28]. Fig. 3 shows the phylogenetic relationships among these sequences.
Fig. 2.
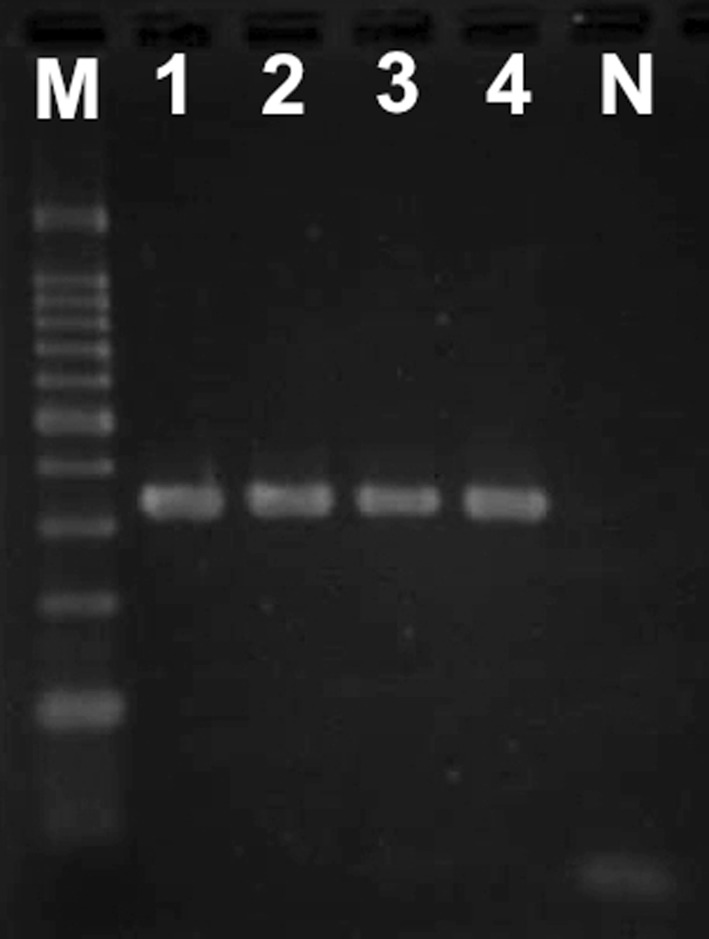
Agarose gel electrophoresis of the products (339 bp) of Pentatrichomonas hominis species-specific PCR. M, 100-bp ladder; 1-4, DNA samples of cultures from trichomonad trophozoites-positive feces; N, water (negative control).
Fig. 3.
Phylogenetic relationship of Pentatrichomonas hominis from common marmosets and other mammal hosts using the nucleotide sequence of the internal transcribed spacer region (ITS)-1, the 5.8S rRNA gene, and ITS-2 (297 bp) and the neighbor-joining method. The information shown for each taxon includes the host names and GenBank accession numbers. The sequence data for P. hominis from common marmosets obtained in this study are underlined. Data for other hosts were from previous reports [5, 6, 13, 15, 17, 22, 28, 39]. The genetic distances were computed using the Kimura two-parameter method.
Discussion
This fecal screening and genetic species identification revealed that P. hominis was prevalent in a colony of facility-bred common marmosets in Japan. P. hominis is regarded as a nonpathogenic opportunist in the large intestines of various mammalian hosts, including nonhuman primates [18, 38]. Our survey found similar positive rates for the trophozoites in normal and diarrheal feces, implying that P. hominis infection was not the primary cause of the diarrhea or colitis in marmosets. In histopathological examinations, no invasive or inflammatory lesions involving trichomonads have been found in the intestinal mucosa of marmosets (data not shown). This supports the assertion that P. hominis is nonpathogenic in marmosets. Nevertheless, there is no evidence that refutes any relationship between P. hominis infection and disease in mammalian hosts. Our data were not sufficient to make conclusions regarding whether or not P. hominis is nonpathogenic in marmosets and further investigations, such as experimental infection and anti-parasite treatment experiments, are needed to clarify this.
Our analysis showed that the genetic variation of P. hominis was low, even in the ITS regions, which are highly variable sites. There was no difference in the analyzed nucleotide sequences between P. hominis from marmosets in this survey and those reported from other nonhuman primates, dogs, cats, and cattle. This suggests that isolated protozoa have been transmitted among various mammal hosts. From the perspective of animal facility management, managers should consider the possibility that P. hominis spreads from marmosets to other animals. The protozoa have been found in most laboratory animal species, including dogs, mice, rats, and hamsters [17, 32]. P. hominis from a beagle dog was transmitted experimentally to mice and rats by oral administeration of the trophozoites, and they were detected in the rodents 2 months later [14]. The protozoa are transmitted by oral ingestion of trophozoites or pseudocysts excreted in feces. To prevent transmission, good hygiene is important. There was a slight difference between the protozoa isolated from marmosets and those reported from humans. This genetic difference suggests that there is a low risk of P. hominis transmission from marmosets to humans. The zoonotic potential of P. hominis is controversial [25], but P. hominis occurs infrequently in humans [1, 33], and there is no clear evidence of its pathogenicity. Therefore, the zoonotic risk of P. hominis from marmosets to humans is quite low.
In conclusion, this survey found that laboratory-bred common marmosets harbored P. hominis but that was not a primary cause of diarrhea and had low intraspecific genetic variation among various mammal hosts.
Acknowledgments
We thank Dr. Oku Yuzaburo of Tottori University for kindly telling us about the Trichomonas culture methods and all of the members of the marmoset research group at CIEA for their warm support.
References
- 1.Adu-Sarkodie Y., Opoku B.K., Crucitti T., Weiss H.A., Mabey D.2007. Lack of evidence for the involvement of rectal and oral trichomonads in the aetiology of vaginal trichomoniasis in Ghana. Sex. Transm. Infect. 83: 130–132. doi: 10.1136/sti.2006.020941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J.1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. doi: 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baxter V.K., Shaw G.C., Sotuyo N.P., Carlson C.S., Olson E.J., Zink M.C., Mankowski J.L., Adams R.J., Hutchinson E.K., Metcalf Pate K.A.2013. Serum albumin and body weight as biomarkers for the antemortem identification of bone and gastrointestinal disease in the common marmoset. PLoS ONE 8: e82747. doi: 10.1371/journal.pone.0082747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bunton T.E., Lowenstine L.J., Leininger R.1983. Invasive trichomoniasis in a Callicebus moloch. Vet. Pathol. 20: 491–494. doi: 10.1177/030098588302000412 [DOI] [PubMed] [Google Scholar]
- 5.Cepicka I., Kutisová K., Tachezy J., Kulda J., Flegr J.2005. Cryptic species within the Tetratrichomonas gallinarum species complex revealed by molecular polymorphism. Vet. Parasitol. 128: 11–21. doi: 10.1016/j.vetpar.2004.11.003 [DOI] [PubMed] [Google Scholar]
- 6.Ceplecha V., Svoboda M., Cepička I., Husník R., Horáčková K., Svobodová V.2013. InPouch™ TF-Feline medium is not specific for Tritrichomonas foetus. Vet. Parasitol. 196: 503–505. doi: 10.1016/j.vetpar.2013.04.015 [DOI] [PubMed] [Google Scholar]
- 7.Crucitti T., Abdellati S., Ross D.A., Changalucha J., Dyck E., Buve A.2004. Detection of Pentatrichomonas hominis DNA in biological specimens by PCR. Lett. Appl. Microbiol. 38: 510–516. doi: 10.1111/j.1472-765X.2004.01528.x [DOI] [PubMed] [Google Scholar]
- 8.Deinhardt J.B., Devine J., Passovoy M., Pohlman R., Deinhardt F.1967. Marmosets as laboratory animals. I. Care of marmosets in the laboratory, pathology and outline of statistical evaluation of data. Lab. Anim. Care 17: 11–29. [Google Scholar]
- 9.Delgado-Viscogliosi P., Viscogliosi E., Gerbod D., Kulda J., Sogin M.L., Edgcomb V.P.2000. Molecular phylogeny of parabasalids based on small subunit rRNA sequences, with emphasis on the Trichomonadinae subfamily. J. Eukaryot. Microbiol. 47: 70–75. doi: 10.1111/j.1550-7408.2000.tb00013.x [DOI] [PubMed] [Google Scholar]
- 10.Dimasuay K.G.B., Lavilla O.J.Y., Rivera W.L.2013. New hosts of Simplicimonas similis and Trichomitus batrachorum identified by 18S ribosomal RNA gene sequences. J. Parasitol. Res. 2013: 831947. doi: 10.1155/2013/831947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doi J., Hirota J., Morita A., Fukushima K., Kamijyo H., Ohta H., Yamasaki M., Takahashi T., Katakura K., Oku Y.2012. Intestinal Tritrichomonas suis (=T. foetus) infection in Japanese cats. J. Vet. Med. Sci. 74: 413–417. doi: 10.1292/jvms.11-0171 [DOI] [PubMed] [Google Scholar]
- 12.Dufernez F., Walker R.L., Noël C., Caby S., Mantini C., Delgado-Viscogliosi P., Ohkuma M., Kudo T., Capron M., Pierce R.J., Villanueva M.R., Viscogliosi E.2007. Morphological and molecular identification of non-Tritrichomonas foetus trichomonad protozoa from the bovine preputial cavity. J. Eukaryot. Microbiol. 54: 161–168. doi: 10.1111/j.1550-7408.2007.00247.x [DOI] [PubMed] [Google Scholar]
- 13.Felleisen R.S.1997. Comparative sequence analysis of 5.8S rRNA genes and internal transcribed spacer (ITS) regions of trichomonadid protozoa. Parasitology 115: 111–119. doi: 10.1017/S0031182097001212 [DOI] [PubMed] [Google Scholar]
- 14.Fukushima T., Mochizuki K., Yamazaki H., Watanabe Y., Yamada S., Aoyama T., Sakurai Y., Mori H., Nakazawa M.1990. [Pentatrichomonas hominis from beagle dogs—detection method, characteristics and route of infection]. Jikken Dobutsu 39: 187–192(in Japanese). [PubMed] [Google Scholar]
- 15.Gookin J.L., Birkenheuer A.J., St John V., Spector M., Levy M.G.2005. Molecular characterization of trichomonads from feces of dogs with diarrhea. J. Parasitol. 91: 939–943. doi: 10.1645/GE-474R.1 [DOI] [PubMed] [Google Scholar]
- 16.Gookin J.L., Stauffer S.H., Levy M.G.2007. Identification of Pentatrichomonas hominis in feline fecal samples by polymerase chain reaction assay. Vet. Parasitol. 145: 11–15. doi: 10.1016/j.vetpar.2006.10.020 [DOI] [PubMed] [Google Scholar]
- 17.Grellet A., Brunopolack, Feugier A., Boucraut-Baralon C., Grandjean D., Vandewynckel L., Cian A., Meloni D., Viscogliosi E.2013. Prevalence, risk factors of infection and molecular characterization of trichomonads in puppies from French breeding kennels. Vet. Parasitol. 197: 418–426. doi: 10.1016/j.vetpar.2013.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honigberg B.1978. Trichomonads of importance in human medicine. pp. 275–279. In: Parasitic protozoa Vol 2 (Kreier, J. ed.), Academic Press, New York. [Google Scholar]
- 19.Kimura M.1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16: 111–120. doi: 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- 20.Kondova I., Simon M.A., Klumpp S.A., MacKey J., Widmer G., Domingues H.G., Persengiev S.P., O’Neil S.P.2005. Trichomonad gastritis in rhesus macaques (Macaca mulatta) infected with simian immunodeficiency virus. Vet. Pathol. 42: 19–29. doi: 10.1354/vp.42-1-19 [DOI] [PubMed] [Google Scholar]
- 21.Levy M.G., Gookin J.L., Poore M., Birkenheuer A.J., Dykstra M.J., Litaker R.W.2003. Tritrichomonas foetus and not Pentatrichomonas hominis is the etiologic agent of feline trichomonal diarrhea. J. Parasitol. 89: 99–104. doi: 10.1645/0022-3395(2003)089[0099:TFANPH]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 22.Li W., Li W., Gong P., Meng Y., Li W., Zhang C., Li S., Yang J., Li H., Zhang X., Li J.2014. Molecular and morphologic identification of Pentatrichomonas hominis in swine. Vet. Parasitol. 202: 241–247. doi: 10.1016/j.vetpar.2014.01.028 [DOI] [PubMed] [Google Scholar]
- 23.Ludlage E., Mansfield K.2003. Clinical care and diseases of the common marmoset (Callithrix jacchus). Comp. Med. 53: 369–382. [PubMed] [Google Scholar]
- 24.Mansfield K.2003. Marmoset models commonly used in biomedical research. Comp. Med. 53: 383–392. [PubMed] [Google Scholar]
- 25.Maritz J.M., Land K.M., Carlton J.M., Hirt R.P.2014. What is the importance of zoonotic trichomonads for human health? Trends Parasitol. 30: 333–341. doi: 10.1016/j.pt.2014.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okano H., Hikishima K., Iriki A., Sasaki E.2012. The common marmoset as a novel animal model system for biomedical and neuroscience research applications. Semin. Fetal Neonatal Med. 17: 336–340. doi: 10.1016/j.siny.2012.07.002 [DOI] [PubMed] [Google Scholar]
- 27.Potkay S.1992. Diseases of the Callitrichidae: a review. J. Med. Primatol. 21: 189–236. [PubMed] [Google Scholar]
- 28.Reinmann K., Müller N., Kuhnert P., Campero C.M., Leitsch D., Hess M., Henning K., Fort M., Müller J., Gottstein B., Frey C.F.2012. Tritrichomonas foetus isolates from cats and cattle show minor genetic differences in unrelated loci ITS-2 and EF-1α. Vet. Parasitol. 185: 138–144. doi: 10.1016/j.vetpar.2011.09.032 [DOI] [PubMed] [Google Scholar]
- 29.Rensing S., Oerke A.K.2005. Husbandry and Management of New World Species: Marmosets and Tamarins. pp. 145–162. In: The Laboratory Primate (Wolfe-Coote, S. ed.), Elsevier, Amsterdam. [Google Scholar]
- 30.Saitou N., Nei M.1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 31.Sasaki E., Suemizu H., Shimada A., Hanazawa K., Oiwa R., Kamioka M., Tomioka I., Sotomaru Y., Hirakawa R., Eto T., Shiozawa S., Maeda T., Ito M., Ito R., Kito C., Yagihashi C., Kawai K., Miyoshi H., Tanioka Y., Tamaoki N., Habu S., Okano H., Nomura T.2009. Generation of transgenic non-human primates with germline transmission. Nature 459: 523–527. doi: 10.1038/nature08090 [DOI] [PubMed] [Google Scholar]
- 32.Saxe L.H.1954. Transfaunation studies on the host specificity of the enteric protozoa of rodents. J. Protozool. 1: 220–230. doi: 10.1111/j.1550-7408.1954.tb00821.x [DOI] [Google Scholar]
- 33.Schwebke J.R., Burgess D.2004. Trichomoniasis. Clin. Microbiol. Rev. 17: 794–803. doi: 10.1128/CMR.17.4.794-803.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smejkalová P., Petrželková K.J., Pomajbíková K., Modrý D., Čepička I.2012. Extensive diversity of intestinal trichomonads of non-human primates. Parasitology 139: 92–102. doi: 10.1017/S0031182011001624 [DOI] [PubMed] [Google Scholar]
- 35.'t Hart B.A., Abbott D.H., Nakamura K., Fuchs E.2012. The marmoset monkey: a multi-purpose preclinical and translational model of human biology and disease. Drug Discov. Today 17: 1160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi T., Hanazawa K., Inoue T., Sato K., Sedohara A., Okahara J., Suemizu H., Yagihashi C., Yamamoto M., Eto T., Konno Y., Okano H., Suematsu M., Sasaki E.2014. Birth of healthy offspring following ICSI in in vitro-matured common marmoset (Callithrix jacchus) oocytes. PLoS ONE 9: e95560. doi: 10.1371/journal.pone.0095560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S.2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toft J.D., 2nd1982. The pathoparasitology of the alimentary tract and pancreas of nonhuman primates: a review. Vet. Pathol. Suppl. 19:(Suppl 7): 44–92. doi: 10.1177/030098588201907s06 [DOI] [PubMed] [Google Scholar]
- 39.Walker R.L., Hayes D.C., Sawyer S.J., Nordhausen R.W., Van Hoosear K.A., BonDurant R.H.2003. Comparison of the 5.8S rRNA gene and internal transcribed spacer regions of trichomonadid protozoa recovered from the bovine preputial cavity. J. Vet. Diagn. Invest. 15: 14–20. doi: 10.1177/104063870301500104 [DOI] [PubMed] [Google Scholar]