Abstract
Membrane trafficking in male germ cells contributes to their development via cell morphological changes and acrosome formation. TBC family proteins work as Rab GTPase accelerating proteins (GAPs), which negatively regulate Rab proteins, to mediate membrane trafficking. In this study, we analyzed the expression of a Rab GAP, TBC1D9, in mouse organs and the intracellular localization of the gene products. Tbc1d9 showed abundant expression in adult mice testis. We found that the Tbc1d9 mRNA was expressed in primary and secondary spermatocytes, and that the TBC1D9 protein was expressed in spermatocytes and round spermatids. In 293T cells, TBC1D9-GFP proteins were localized in the endosome and Golgi apparatus. Compartments that were positive for the constitutive active mutants of Rab7 and Rab9 were also positive for TBC1D9 isoform 1. In addition, TBC1D9 proteins were associated with Rab7 and Rab9, respectively. These results indicate that TBC1D9 is expressed mainly in spermatocytes, and suggest that TBC1D9 regulates membrane trafficking pathways related to Rab9- or Rab7-positive vesicles.
Keywords: Rab7, Rab9, Spermatogenesis, TBC1D9
Introduction
Spermatogenesis is a complex process involving male germ cell division and differentiation, starting from primordial germ cells. The primordial germ cell differentiates into spermatogonium, followed by primary spermatocytes, secondary spermatocytes, spermatids and sperm. This process involves a number of important events such as stem cell differentiation, spermatogonium division, meiosis, and haploid spermatids differentiation and maturation into the sperm accompanying with morphogenesis [9].
Membrane trafficking has a number of important and diverse roles in cell physiology. Rab GTPases are known to control the process of membrane trafficking, such as sorting, formation, motility, docking and fusion of transport-vesicles [35]. These Rab proteins are regulated by three factors; guanine nucleotide exchange factor (GEF), GTPase accelerating proteins (GAPs) and GDP dissociation inhibitor. Rabs are converted to a GTP-bound “active” state by specific GEF [10, 25]. In contrast, GAPs bind to the Rab to catalyze the hydrolysis of bound GTP to GDP and, thereby, convert the Rab back to its GDP-bound “inactive” state [10, 25]. In addition, Rab GAP proteins contain conserved TBC (Tre2/Bub2/Cdc16) domains that confer GAP activity [10]. The TBC domain contains two highly conserved amino acid residues, arginine and glutamine, which promote GTP hydrolysis [3, 24]. Rab GAPs are essentially implicated in the spatial and temporal dynamics of the cellular endomembrane system by regulating Rab proteins [3]. Thus, Rab proteins work as molecular switches that oscillate between GTP- and GDP-bound conformations, and this on/off regulatory function is restricted to the membrane compartments in which they are localized [37].
Membrane trafficking via Rab protein is widely observed in spermatogenesis: for example, cytokinesis [19, 27, 28], acrosome biogenesis [20, 30] and junction dynamics [14, 21]. However, although some TBC domain family proteins, such as TBC1D7 and TBC1D21 (also known as MgcRabGAP), were abundantly observed in the testis [15, 33], levels of Rab GAPs in spermatogenesis were reported to be much lower than their target proteins Rabs. Here we focused on a novel Rab GAP gene, Tbc1d9, which is abundantly expressed in the mouse testis and localized in the Rab9- or Rab7-positive late endosome. These observations suggest that TBC1D9 is associated in spermatogenesis via regulation of the late-endosome, particularly the Rab9- and/or Rab7-positive vesicle, trafficking pathway.
Materials and Methods
Animals
C57BL/6J mice were maintained in the animal breeding rooms in the School of Veterinary Medicine, Tottori University. Animal breeding rooms were kept at 23 ± 2°C and 50 ± 10% relative humidity with a 12 h light-dark cycle. Research was conducted according to the Guidelines for the Care and Use of Laboratory Animals of Tottori University. The experimental protocols were approved by the Institutional Animal Care and Use Committee of Tottori University.
Reverse transcription-PCR (RT-PCR)
Total RNA from various organs in 8-week-old mice was isolated using Trizol reagent (Life Technologies, Carlsbad, CA, USA). Reverse transcription was performed with 1 µg total RNA, Super-Script III reverse transcriptase (Life Technologies), and an oligo dT primer, according to the manufacturer’s recommended protocol. ExTaq (TaKaRa Bio Inc., Shiga, Japan) was used for RT-PCR. Primer sequences are shown in Table 1.
Table 1. Primer sequences.
| Gene and type of use | Primer sequences (5’–3’) | Product size (bp) | Position | Reference | |
|---|---|---|---|---|---|
| Forward | Reverse | ||||
| Tbc1d9 isoform 1 and 2, RT-PCR | TCAAGATGAGCCTGACTCCGCC | TACGCACCAGGACTGTGTCCTC | 743 | 3549-4291 (isoform 1) 2850-3592 (isoform 2) |
NM_001111304 (isoform 1) NM_027758 (isoform 2) |
| Gapdh, RT-PCR | AGTCGGAGTGAACGGATTTGG | AGTTGTCATGGATGACCTTGG | 488 | 249-736 | NM_008084 |
| Rab5, cloning for RFP-fusion protein | AAGCTTCGATGGCTAATCGAGGAGC | TCAGTTACTACAACACTGGCTTC | 656 | 468-1115 | NM_025887 |
| Rab7, cloning for RFP-fusion protein | CTCGAGCTATGACCTCTAGGAAGAAAGTG | GTCGACTCAACAACTGCAGCTTTCTGCGG | 638 | 1-624 | NM_009005 |
| Rab9, cloning for RFP-fusion protein | CTCGAGCTATGGCAGGAAAATCGTCTCT | TCAACAGCAAGATGAGTTTG | 614 | 387-992 | NM_019773 |
| Rab11, cloning for RFP-fusion protein | AAGCTTCGATGGGCACCCGCGACGACG | TTAGATGTTCTGACAGCACTGC | 659 | 134-784 | NM_017382 |
| Rab7, site-directed mutagenesis | TGGGACACAGCCGGTCTGGAACGG | CCGTTCCAGACCGGCTGTGTCCCA | (184-207) | NM_009005 | |
| Rab9, site-directed mutagenesis | TGGGACACAGCTGGCCTGGAACGC | GCGTTCCAGGCCAGCTGTGTCCCA | (567-590) | NM_019773 | |
| Rab7, cloning for FLAG-fusion protein | AAGCTTATGACCTCTAGGAAGAAA | GGTACCTCAACAACTGCAGCTTTC | 630 | 1-624 | NM_009005 |
| Rab9, cloning for FLAG-fusion protein | GCGGCCGCGATGGCAGGAAAATCGTCT | GGATCCTCAACAGCAAGATGAGTT | 621 | 387-992 | NM_019773 |
| Rutbc2, cloning for GFP-fusion protein | CTCGAGGCCACCATGGCTTCGGTCCCTGCGGAGGCCGA | AAGCTTCTTGTTCTCAATTAGAATCTGCACC | 3297 | 95-3373 | NM_172718 |
In situ hybridization
The detailed procedure for in situ hybridization was previously described [22]. Two non-overlapping 45-mer antisense oligonucleotide DNA probes for each mRNA were designed to be complementary to the following sequences: 5′-TCTGCTCTGAAACGTGATCTGGATGCCAGAGCGAAGAGCGAGCGA-3′ [1561–1605 of mouse Tbc1d9, isoform 1 (NM_001111304)], and 5′-TCCTGTGATCAAGATGAGCCTGACTCCGCCTTTGAAGCCA-3′ [3541–3580 of mouse Tbc1d9, isoform 1 and 2842–2880 of mouse Tbc1d9, isoform 2 (NM_027758)]. Frozen sections from adult mice testes, 14-µm-thick, were fixed and hybridized with a hybridization buffer containing 33P-labeled probes (10,000 cpm/ml). The hybridized sections were rinsed and dipped in an autoradiographic emulsion (NTB-2; Kodak, Rochester, NY, USA) at 4°C for 8 weeks. The hybridized sections were counterstained with hematoxylin after development. An in situ hybridization technique using the two non-overlapping antisense probes exhibited identical labeling in all the tissues examined. The specificity of the hybridization was also confirmed by the disappearance of signals upon the addition of an excess of an unlabeled antisense probe.
Immunohistochemistry
Testes from 8-week-old mice were fixed in Bouin’s solution, dehydrated, and embedded in paraffin blocks. Sections were then deparaffinized, rehydrated, and heated at 95°C in 1 mM EDTA (pH 6.0) for 45 min, and washed 3 times with PBS containing 0.1% Tween 20 (PBS-T). The sections were then treated with 0.45% hydrogen peroxide in absolute methanol at 4°C for 30 min. The sections were washed with PBS-T and incubated with anti-human TBC1D9 antibody (1:100 in dilution; Sigma-Aldrich, St. Louis, MO, USA), which recognizes both isoforms 1 (amino acid 902–1007) and 2 (amino acid 669–774), or anti-mouse TBC1D9 antiserum (1:500 in dilution; Kazusa DNA Research Institute, Chiba, Japan), which only recognizes isoform 1 (amino acid 193–361), for 60 min at room temperature. After washing with PBS-T, the sections were immunostained using an DAKO ENVISION kit HRP (DAKO, Glostrup, Denmark) according to the manufacturer’s recommended protocol. Sections were counterstained with haematoxylin, dehydrated, cleaned and mounted. Preimmune rabbit IgG (Vector Laboratories, Inc., Burlingame, CA, USA) or rabbit serum were used as the negative control of the primary antibody.
Plasmid construction
Mouse cDNA clones of Tbc1d9 isoform 1 (FANTOM clone ID: 6330501J07) and isoform 2 (FANTOM clone ID: C230095G18) were purchased from Dnaform (Kanagawa, Japan). These cDNAs were recloned into a GFP-fused expression plasmid, pEGFP-C1 (Clontech, Mountain View, CA, USA). Organelle marker plasmids were cloned as follows. As a lysosome marker, mouse Lamp1 cDNA (Accession no. NM_010684) [5]; and as a Golgi apparatus marker, cDNA corresponding with amino acid 1–88 of human β1,4-galactosyltransferase (NM_001497) [16] were cloned into a RFP-fused expression plasmid, pTagRFP-N (Enrogen, Moscow, Russia). As an endosome marker, mouse Rhob cDNA (NM_007483) [1] was cloned into a plasmid, pTagRFP-C (Enrogen). As an endoplasmic reticulum (ER) maker, cDNA of the signal sequence from human Calreticulin (NM_007591), RFP and the carboxyl terminal KDEL sequence [32] were cloned into an expression plasmid, pCl-neo (Promega, Madison, WI, US). The cDNA fragments of wild-type Rab5, Rab7, Rab9, Rab11, and Rutbc2 of the mouse were amplified by RT-PCR using the primers shown in Table 1. The cDNA fragments of constitutively active mutants of Rab7 [Rab7 (Q67L)] and Rab9 [Rab9 (Q66L)] [17, 23] were amplified by overlapping PCR using the primers shown in Table 1. These cDNA fragments were cloned into pTagRFP-C, and Rutbc2 cDNA was cloned into pEGFP-C1. The cDNAs of wild-type Rab7, Rab9 and Rab9 (Q66L) were also cloned into a FLAG-tagged protein expression vector, p3×FLAG-Myc-CMV (Sigma-Aldrich).
Cell culture, transfection and confocal fluorescence microscopy analysis
The 293T cells were cultured in DMEM (Sigma-Aldrich) containing 10% fetal bovine serum (Life Technologies) and 1% penicillin/streptomycin solution (Nacalai Tesque, Kyoto, Japan). Transfection using expression plasmids was performed with Lipofectamine 2000 reagent (Life Technologies).
Fluorescence in the cells were observed and quantified by confocal fluorescence microscopy (FLUOVIEW FV10i, OLYMPUS, Tokyo, Japan) according to manufacturer’s instructions. Co-localization analysis was performed as follows. Fluorescent images of the cells were obtained from confocal fluorescence microscopy, and the intensity of the fluorescence from GFP and RFP was measured from each pixel in the image. The pixels in which the fluorescent intensity was higher than the background were counted, and then categorized as GFP-positive/RFP-positive, GFP-positive/RFP-negative, or GFP-negative/RFP-positive. The percentage of co-localized (GFP-positive/RFP-positive) pixels and non co-localized (GFP-positive/RFP-negative and GFP-negative/RFP-positive) pixels to the total fluorescent pixels was the calculated. Values are means ± SEM calculated from 5 images from independent visual fields.
Immunoprecipitation
The TBC1D9-GFP expression vector, RUTBC2-GFP expression vector, 3×FLAG-Rab7, 3×FLAG-Rab9 and 3×FLAG-Rab9 (Q67L) expression vectors were transfected in 293T cells. At 1 d post-transfection, the cells were lysed by cell lysis buffer as previously described [23]. Three hundred µg of cellular protein was incubated with anti-FLAG antibody (Sigma-Aldrich) for 1 h at 4°C, and precipitated for 1 h at 4°C with pre-cleared Protein A Sepharose (GE-Healthcare). Pellets were washed in Tris-buffered saline and used for SDS-PAGE. Immunoblotting was performed using anti-GFP antibody (Nacalai Tesque). Twenty µg of cellular protein was also immunoblotted using antibodies against GFP and FLAG.
Statistics
Statistical analyses were performed by unpaired t-test with Welch’s correction using GraphPad Prism 6 software (GraphPad software, La Jolla, CA, USA).
Results
Tbc1d9 expression in mouse organs
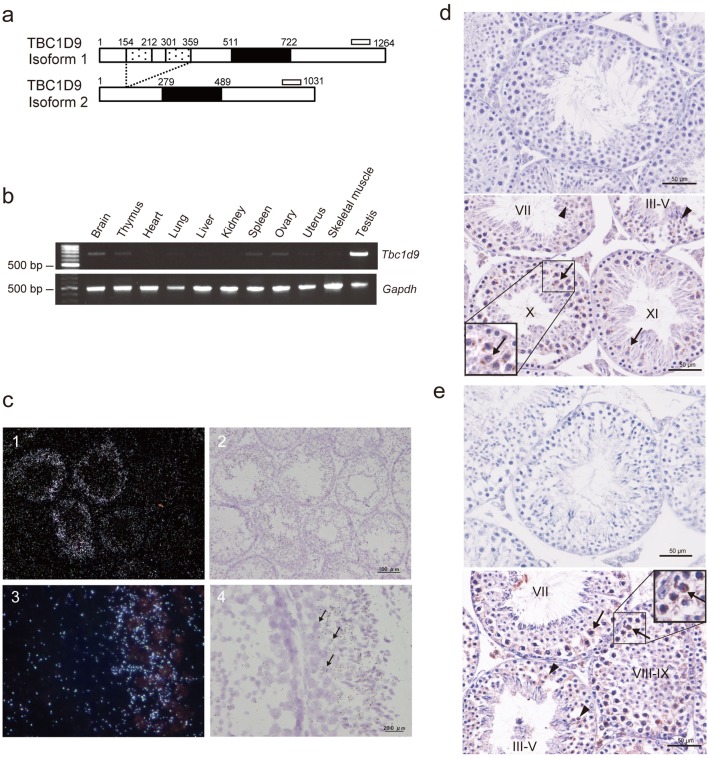
The expression of Rab GAP genes in mice spermatocytes or spermatids was surveyed by screening spermatogenesis-related genes in the Gene Expression Omnibus (GEO) database (GDS2390). This survey identified one Rab GAP gene, TBC family member number 9 (Tbc1d9), which was expected to be expressed abundantly in spermatocytes and spermatids. The murine Tbc1d9 gene has two alternative splicing variants: Tbc1d9 isoform 1 (NM_001111304) contains two GRAM (glucosyltransferases, Rab-like GTPase activators and myotubularins) domains and one TBC domain, whereas Tbc1d9 isoform 2 (NM_027758) is a short form that contains only the TBC domain (Fig. 1a).
Fig. 1.
Analyses of TBC1D9 expression in 8-week-old mouse tissues. (a) Scheme of mouse TBC1D9 isoforms. Isoform 1 protein (1264 amino acids) possesses two GRAM domains (dotted box). A TBC domain (solid box) exists in both isoform 1 and isoform 2 (1031 amino acids). The open bars represent regions amplified by RT-PCR. (b) RT-PCR analysis of Tbc1d9 and Gapdh mRNA in mouse tissues. Top, amplification of both isoforms of Tbc1d9; bottom, amplification of Gapdh. (c) In situ hybridization of Tbc1d9 mRNA in the mouse testis. Light microscopic dark (1, 3) and light field (2, 4) images of Tbc1d9 mRNA in sections of the testis. Signal is seen in the cytoplasm of spermatocytes (arrows). (d) and (e) Immunohistochemical analysis of TBC1D9 proteins in the mouse testis using anti-mouse TBC1D9 antiserum (d) and anti-human TBC1D9 antibody (e). Top, negative control; bottom, immunoreactivity is seen in the cytoplasm of spermatocytes (arrows) and round spermatids (arrowheads). Roman numerals, the stages in the seminiferous tubules.
We examined the mRNA expression level of Tbc1d9 in mouse organs by RT-PCR. Fig. 1b shows mRNA expression in mouse organs analyzed by RT-PCR. When the set of primers for both isoforms (see Fig. 1a) was used, cDNA was more abundantly amplified in the testis than in other organs.
TBC1D9 expression in the mouse testis
We next examined the mRNA expression of Tbc1d9 in the mice testis by in situ hybridization using probes for both isoforms. The signals for Tbc1d9 were detected in some seminiferous tubules, while other tubules are free from the signals (Fig. 1c, panels 1 and 2). At a higher magnification, the signals for Tbc1d9 are restricted to primary and secondary spermatocytes (Fig. 1c, panels 3 and 4).
To examine the localization of TBC1D9 proteins in the mice testis, immunohistochemical analysis was performed using an anti-mouse TBC1D9 antiserum that recognized the GRAM domain in isoform 1. As shown in Fig. 1d, strong immunoreactivity for TBC1D9 isoform 1 protein was detected in the cytoplasm of germ cells, particularly spermatocytes. The weak signal was seen in round spermatids. In addition, we also detected immunoreactivity for TBC1D9 isoform 1 and 2 proteins in spermatocytes and round spermatids using an anti-human TBC1D9 antibody that recognized the C-terminal region in both isoforms (Fig. 1e).
Intracellular localization of TBC1D9 proteins
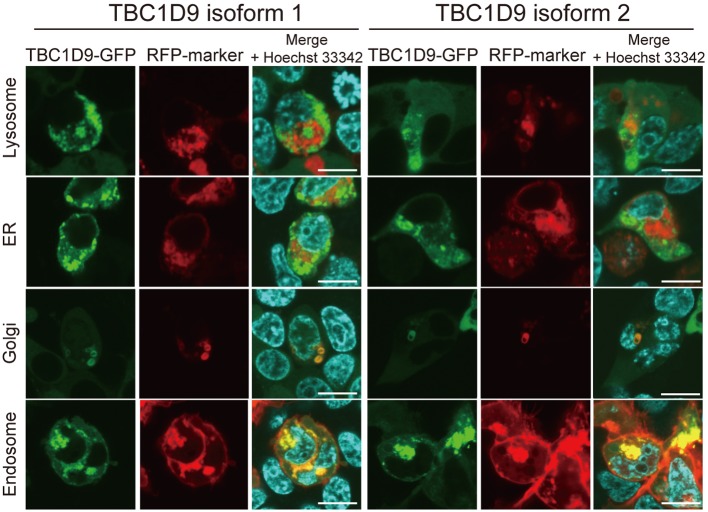
The intracellular localization of TBC1D9 proteins was examined. Expression vectors of each TBC1D9 protein fused to GFP (TBC1D9-GFP) and organelle marker proteins fused to RFP that localize in the lysosome, ER, Golgi apparatus and endosome, respectively, were co-transfected in 293T cells, and the co-localization of fluorescence was analyzed by confocal microscopy. As shown in Fig. 2, TBC1D9 was localized in the endosomes and partially in the Golgi apparatus of 293T cells. No differences were observed in the localization of organelles between the two isoforms. These results suggested that TBC1D9 was mainly localized in the endosome and/or Golgi apparatus and regulates membrane trafficking therein.
Fig. 2.
The intracellular localization analysis of TBC1D9 in 293T cell. Fluorescence of TBC1D9-GFP (left: isoform 1, right: isoform 2) and RFP-fused organelle markers (lysosome, ER, Golgi apparatus and endosome) are shown. Scale bar, 10 µm.
Co-localization of TBC1D9 protein with Rab7- and Rab9-positive vesicles
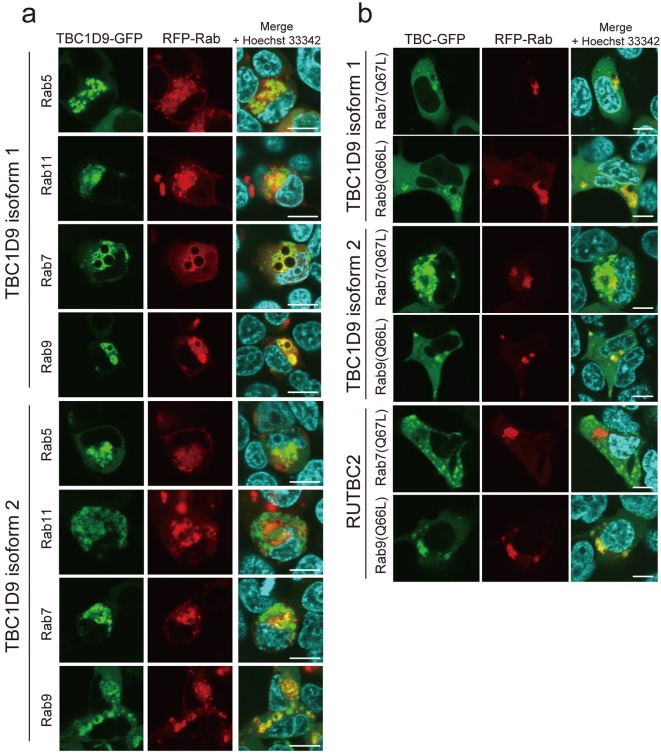
The intracellular localization of several Rab proteins has been demonstrated previously [35]. To determine whether Rab proteins were co-localized with TBC1D9 in cells, the following expression vectors for RFP-Rab fusion proteins localized in the endosome were constructed; Rab5, localized in the early-endosome; Rab11, localized in the recycling-endosome; Rab7, localized in the late-endosome and related to the late-endosome/lysosome pathway, and Rab9, also localized in the late-endosome and transferred to the trans-Golgi network [34, 35]. Each RFP-Rab expression vector was co-transfected with TBC1D9-GFP in 293T cells, and the intracellular localization of each protein was compared (Fig. 3a). We quantified the co-localization of TBC proteins and Rab5/Rab7/Rab9/Rab11 by measuring the percentage of the fluorescence-positive pixels in 4 visual fields (Table 2). As shown in Fig. 3a and Table 2, RFP-Rab5, RFP-Rab7 and RFP-Rab9 showed tendency to co-localize with TBC1D9-GFP, indicating that TBC1D9 is largely localized in the early- and late-endosome. In contrast, RFP-Rab11 tended not to co-localize with TBC1D9-GFP. Interestingly, RFP-Rab7 showed a tendency of fewer co-localization to TBC1D9 isoform 2-GFP than RFP-Rab5 and RFP-Rab9.
Fig. 3.
Confocal microscopic analysis of co-localization of TBC1D9-GFP and RFP-Rab proteins. (a) Fluorescence images of TBC1D9-GFP and RFP-Rab proteins (Rab5, Rab11, Rab7 and Rab9) localized in the endosome are shown. (b) Fluorescence images of TBC1D9-GFP, RUTBC2-GFP, RFP-Rab7 (Q67L) and RFP-Rab9 (Q66L) are shown. Scale bar, 10 µm.
Table 2. Co-localization analysis of fluorescent pixels in images from 293T cells expressing TBC-GFP and RFP-Rab proteins.
| Transfected genes | Co-localization (%)* | Non co-localization (%)** |
|---|---|---|
| TBC1D9 isoform 1 | ||
| + Rab5 | 26.3 ± 2.0 | 73.7 ± 2.0 |
| + Rab7 | 24.3 ± 8.1 | 75.7 ± 8.1 |
| + Rab9 | 24.9 ± 4.1 | 75.1 ± 4.1 |
| + Rab11 | 13.2 ± 4.4 | 86.8 ± 4.4 |
| TBC1D9 isoform 2 | ||
| + Rab5 | 24.5 ± 1.8 | 75.5 ± 1.8 |
| + Rab7 | 13.3 ± 1.6 | 86.7 ± 1.6 |
| + Rab9 | 34.4 ± 3.7 | 65.6 ± 3.7 |
| + Rab11 | 10.7 ± 6.8 | 89.3 ± 6.8 |
*Percentage of pixels that were both GFP-positive and RFP-positive. **Percentage of pixels that were GFP-positive/RFP-negative or GFP-negative/RFP-positive.
Previous studies also demonstrated that constitutively active mutants of Rab proteins deficient in GTP hydrolytic activity showed more apparent co-localization with several TBC proteins, suggesting that TBC proteins prefer to bind to GTP-binding Rab proteins [2, 11, 23]. We, therefore, co-transfected constitutively active mutants of Rab7 and Rab9 [Rab7 (Q67L) and Rab9 (Q66L)] fused to RFP with the TBC1D9-GFP fusion protein in the 293T cells (Fig. 3b). Almost all of the RFP-Rab9 (Q66L)-positive structures were TBC1D9-GFP-positive. In addition, RFP-Rab7 (Q67L)-positive structures were also positive for TBC1D9-GFP. In contrast, RUTBC2-GFP, which was previously reported to bind specifically with Rab9 [23], was not co-localized with RFP-Rab7 (Q67L) (Fig. 3b).
As shown in Table 3, we also quantified the co-localization of TBC proteins and Rab7 (Q67L)/Rab9 (Q66L) by measuring the percentage of the fluorescence-positive pixels in 5 visual fields. The percentage of pixels in which both TBC1D9 isoform 1-GFP and RFP-Rab9 (Q66L) were positive was found to be approximately 40%. This value was similar to that in which both TBC1D9 isoform 2-GFP and RFP-Rab9 (Q66L) were positive. The percentage of pixels in which both RUTBC2-GFP and RFP-Rab9 (Q67L) were positive was 48.3 ± 6.4%, which was also comparable to the values for the TBC1D9 proteins. The percentage of pixels in which both TBC1D9-GFP and RFP-Rab7 (Q67L) were positive was lower than that in which both TBC1D9-GFP and RFP-Rab9 (Q66L) were positive. In particular, the value of TBC1D9-GFP isoform 2 and RFP-Rab7 (Q67L) was significantly lower than that of TBC1D9-GFP isoform 2 and RFP-Rab9 (Q66L). In contrast, the percentage in which both RUTBC2-GFP and RFP-Rab7 (Q67L) were positive was 1.3 ± 1.0%, a value much lower than observed for the other proteins. These results indicate that TBC1D9 strongly co-localized with Rab9 (Q66L), whereas the co-localization with Rab7 (Q67L) was relatively weak.
Table 3. Co-localization analysis of fluorescent pixels in images from 293T cells expressing TBC-GFP and RFP-Rab mutants.
| Transfected genes | Co-localization (%)* | Non co-localization (%)** |
|---|---|---|
| TBC1D9 isoform 1 | ||
| + Rab9(Q66L) | 39.0 ± 7.3 | 61.0 ± 7.3 |
| + Rab7(Q67L) | 31.6 ± 5.4 | 68.4 ± 5.4 |
| TBC1D9 isoform 2 | ||
| + Rab9(Q66L) | 46.7 ± 7.0 | 53.3 ± 7.0 |
| + Rab7(Q67L) | 16.4 ± 4.9† | 83.6 ± 4.9 |
| RUTBC2 | ||
| + Rab9(Q66L) | 48.3 ± 6.4 | 51.7 ± 6.4 |
| + Rab7(Q67L) | 1.3 ± 1.0† | 98.7 ± 1.0 |
*Percentage of pixels that were both GFP-positive and RFP-positive. **Percentage of pixels that were GFP-positive/RFP-negative or GFP-negative/RFP-positive. †P<0.05 vs. respective + Rab9 (Q66L) by unpaired t test with Welch’s correction.
Interaction of TBC1D9 with Rab7 and Rab9
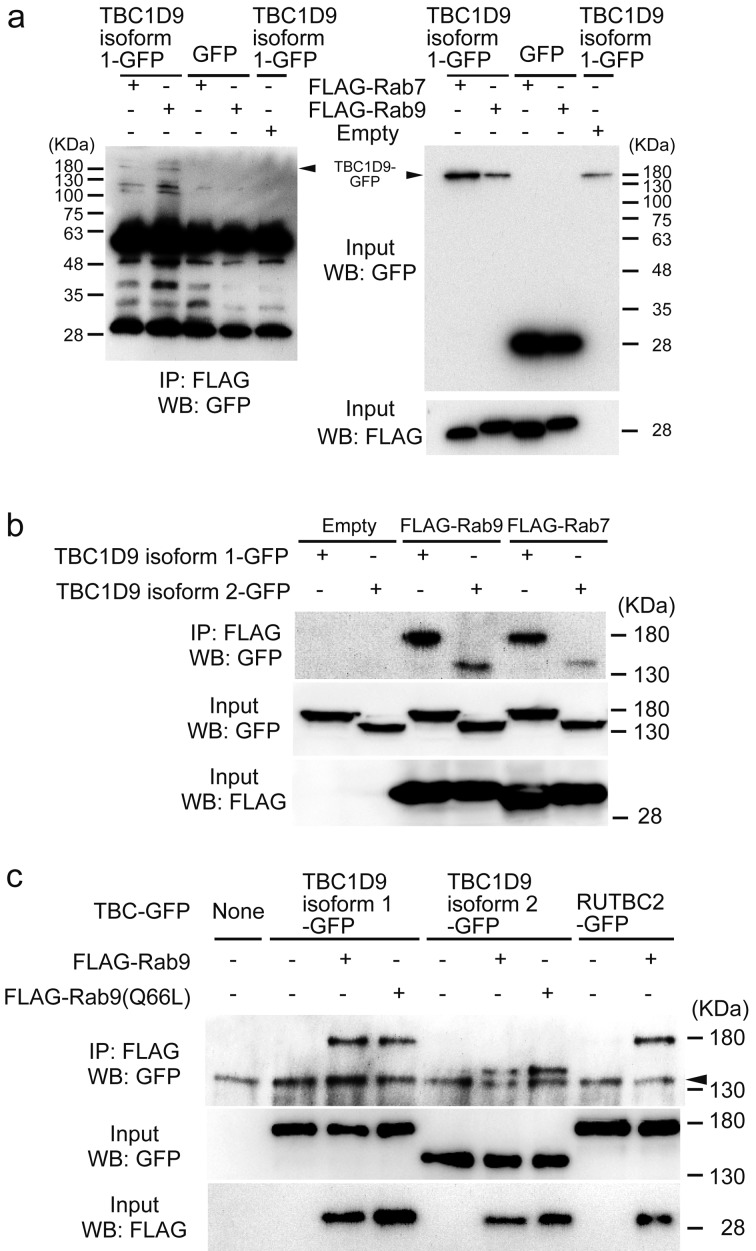
A previous study reported that RUTBC2 was not a Rab9 GAP but bound to Rab9, suggesting that RUTBC2 acts as a Rab9 effector [23]. We, therefore, undertook an immunoprecipitation study to confirm the association of TBC1D9 with Rab7/Rab9. We co-transfected FLAG-Rab7 and FLAG-Rab9 with TBC1D9-GFP or GFP in 293T cells (Fig. 4). We found that both FLAG-Rab7 and FLAG-Rab9 co-immunoprecipitated with TBC1D9 isoform 1-GFP, but not with GFP (Fig. 4a). In addition, TBC1D9 isoform 2 also co-immunoprecipitated with Rab7 and Rab9, respectively (Fig. 4b). Furthermore, the two isoforms of TBC1D9 could interact with wild-type and constitutively active Rab9 in the 293T cells, respectively (Fig. 4c). A similar result was observed when using RUTBC2 and Rab9.
Fig. 4.
Interaction of TBC1D9 with Rab7 and Rab9 in cells. Immunoprecipitation (IP) was performed with anti-FLAG antibody and immunoblotted (WB) with anti-GFP antibody.
(a) IP of FLAG-Rab9 with TBC1D9-GFP or GFP. (b) IP of TBC1D9-GFP with FLAG-Rab7 and FLAG-Rab9. (c) IP of TBC1D9-GFP with FLAG-Rab9 and FLAG-Rab9 (Q66L). Arrowhead; non-specific bands, overlapping with TBC1D9.
Discussion
In the present study, we demonstrated that Tbc1d9 mRNA was expressed at particularly high levels in the spermatocytes of the mice. Our results indicate that at least TBC1D9 isoform 1 protein was detected in late-stage spermatocytes and round spermatids. As mRNA and protein of isoform 2 were overlapped with those of isoform 1, TBC1D9 isoform 2 alone was not able to detect. However, the protein may also be expressed in these male germ cells because the immunoreactivity for TBC1D9 proteins was detected in spermatocytes and round spermatids, not in other types of cells when using antibody that recognized the C-terminal region in both isoforms. Another TBC family gene, MgcRabGAP, was also reported to be expressed in the testis of the mice and its protein was observed in elongated spermatids [15]. These differences suggest the existence of step-wise regulation of membrane trafficking mediated by switching of the expression of each TBC protein in developing male germ cells.
In the 293T cells, both isoforms of TBC1D9 proteins were localized in the late-endosomes and Golgi apparatus. This result indicates that TBC1D9 may regulate the membrane trafficking pathway to the late-endosome and/or Golgi apparatus. Although there are only a few studies on membrane trafficking events via Rab proteins in the late-endosomes and Golgi apparatus in spermatocytes, several membrane trafficking events, such as proacrosome formation [13, 20], morphological changes in the Golgi apparatus [20, 36] and endocytosis of the sex-steroid binding protein and transferrin receptor [7, 8, 26], have been reported in spermatocytes. We suggest that these events may be regulated by TBC1D9 proteins.
In this study, it was demonstrated that TBC1D9 proteins tended to co-localize with Rab5, Rab7 and Rab9, compared with Rab11. We also showed that co-localization of TBC1D9 with Rab7 (Q67L) was weaker than that with Rab9 (Q66L), although RUTBC2 was not co-localized with Rab7 (Q67L). These results indicate the possibility that TBC1D9 regulates the trafficking of Rab7- or Rab9-positive vesicles. Rab7 is known to localize in the acrosome [29], although the mechanisms of Rab9 or Rab9-positive vesicles in spermatogenesis remain unclear. Rab9 mRNA, on the other hand, was detected in the mouse testis (data not shown). We also showed that TBC1D9 was able to interact with Rab7/Rab9. Therefore, TBC1D9 may function as an effector of GTP-bound Rab7/Rab9 in spermatocytes. However, the co-localization of and binding between TBC proteins and Rab protein does not only indicate that the TBC proteins have Rab GAP activity against the Rab protein binding them [12]. For example in yeast, a Rab GAP, Gyp1, binds to GTP-bound Ypt32 Rab, and is recruited to the compartment containing Ypt1, which is inactivated by Gyp1 [31]. A recent report showed that TBC1D9B, which is a homolog of TBC1D9 and contains two GRAM domains, can bind to Rab11a via TBC domain and activate a GTPase activity of Rab11a. In addition, TBC1D9B is also able to bind Rab4a [6]. Further studies, such as Rab GAP assays, are necessary to clarify the relationship between Rab9 or Rab7 and TBC1D9.
In this study, no differences in intracellular localization were observed between isoforms, indicating that the GRAM domain is not involved in intracellular localization. The GRAM domain was found in putative membrane-associated proteins such as TBC domain-containing proteins, and it was speculated that the GRAM domain was an intracellular protein-binding or lipid-binding signaling domain [4]. For example, a phosphatidylinositol 3-phosphatase, myotubularin-related protein 6 (MTMR6), binds to Rab1B via its GRAM domain [18]. The absence of two GRAM domains in TBC1D9 isoform 2 in comparison to the TBC1D9 isoform 1 may abolish its binding ability to a certain type of proteins expressed in spermatocytes. Indeed, at least in our results, there was no difference in the ability of both isoforms to associate with Rab9, indicating that TBC1D9 associates with Rab9 via a region outside the GRAM domain. In contrast, TBC1D9 isoform 2, compared with isoform 1, have weaker interaction with Rab7 (Fig. 4b and Tables 2 and 3). To clarify the differences in the roles of the two isoforms of TBC1D9 proteins in germ cell development, further studies are required to identify the proteins in the male germ cells that interact with the GRAM domain in TBC1D9 isoform 1.
References
- 1.Adamson P., Marshall C.J., Hall A., Tilbrook P.A.1992. Post-translational modifications of p21rho proteins. J. Biol. Chem. 267: 20033–20038. [PubMed] [Google Scholar]
- 2.Bains M., Zaegel V., Mize-Berge J., Heidenreich K.A.2011. IGF-I stimulates Rab7-RILP interaction during neuronal autophagy. Neurosci. Lett. 488: 112–117. doi: 10.1016/j.neulet.2010.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr F., Lambright D.G.2010. Rab GEFs and GAPs. Curr. Opin. Cell Biol. 22: 461–470. doi: 10.1016/j.ceb.2010.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doerks T., Strauss M., Brendel M., Bork P.2000. GRAM, a novel domain in glucosyltransferases, myotubularins and other putative membrane-associated proteins. Trends Biochem. Sci. 25: 483–485. doi: 10.1016/S0968-0004(00)01664-9 [DOI] [PubMed] [Google Scholar]
- 5.Falcón-Pérez J.M., Nazarian R., Sabatti C., Dell’Angelica E.C.2005. Distribution and dynamics of Lamp1-containing endocytic organelles in fibroblasts deficient in BLOC-3. J. Cell Sci. 118: 5243–5255. doi: 10.1242/jcs.02633 [DOI] [PubMed] [Google Scholar]
- 6.Gallo L.I., Liao Y., Ruiz W.G., Clayton D.R., Li M., Liu Y.J., Jiang Y., Fukuda M., Apodaca G., Yin X.M.2014. TBC1D9B functions as a GTPase-activating protein for Rab11a in polarized MDCK cells. Mol. Biol. Cell 25: 3779–3797. doi: 10.1091/mbc.E13-10-0604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerard A.1995. Endocytosis of androgen-binding protein (ABP) by spermatogenic cells. J. Steroid Biochem. Mol. Biol. 53: 533–542. doi: 10.1016/0960-0760(95)00109-D [DOI] [PubMed] [Google Scholar]
- 8.Gerard A., Nya A.E., Egloff M., Domingo M., Degrelle H., Gerard H.1991. Endocytosis of human sex steroid-binding protein in monkey germ cells. Ann. N. Y. Acad. Sci. 637: 258–276. doi: 10.1111/j.1749-6632.1991.tb27314.x [DOI] [PubMed] [Google Scholar]
- 9.Hermo L., Pelletier R.M., Cyr D.G., Smith C.E.2010. Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 1: background to spermatogenesis, spermatogonia, and spermatocytes. Microsc. Res. Tech. 73: 241–278. doi: 10.1002/jemt.20783 [DOI] [PubMed] [Google Scholar]
- 10.Hutagalung A.H., Novick P.J.2011. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 91: 119–149. doi: 10.1152/physrev.00059.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Itoh T., Kanno E., Uemura T., Waguri S., Fukuda M.2011. OATL1, a novel autophagosome-resident Rab33B-GAP, regulates autophagosomal maturation. J. Cell Biol. 192: 839–853. doi: 10.1083/jcb.201008107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jean S., Kiger A.A.2012. Coordination between RAB GTPase and phosphoinositide regulation and functions. Nat. Rev. Mol. Cell Biol. 13: 463–470. doi: 10.1038/nrm3379 [DOI] [PubMed] [Google Scholar]
- 13.Kierszenbaum A.L., Tres L.L., Rivkin E., Kang-Decker N., van Deursen J.M.A.2004. The acroplaxome is the docking site of Golgi-derived myosin Va/Rab27a/b- containing proacrosomal vesicles in wild-type and Hrb mutant mouse spermatids. Biol. Reprod. 70: 1400–1410. doi: 10.1095/biolreprod.103.025346 [DOI] [PubMed] [Google Scholar]
- 14.Lau A.S.N., Mruk D.D.2003. Rab8B GTPase and junction dynamics in the testis. Endocrinology 144: 1549–1563. doi: 10.1210/en.2002-220893 [DOI] [PubMed] [Google Scholar]
- 15.Lin Y.H., Lin Y.M., Kuo Y.C., Wang Y.Y., Kuo P.L.2011. Identification and characterization of a novel Rab GTPase-activating protein in spermatids. Int. J. Androl. 34: e358–e367. doi: 10.1111/j.1365-2605.2010.01126.x [DOI] [PubMed] [Google Scholar]
- 16.Llopis J., McCaffery J.M., Miyawaki A., Farquhar M.G., Tsien R.Y.1998. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc. Natl. Acad. Sci. USA 95: 6803–6808. doi: 10.1073/pnas.95.12.6803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Méresse S., Gorvel J.P., Chavrier P.1995. The rab7 GTPase resides on a vesicular compartment connected to lysosomes. J. Cell Sci. 108: 3349–3358. [DOI] [PubMed] [Google Scholar]
- 18.Mochizuki Y., Ohashi R., Kawamura T., Iwanari H., Kodama T., Naito M., Hamakubo T.2013. Phosphatidylinositol 3-phosphatase myotubularin-related protein 6 (MTMR6) is regulated by small GTPase Rab1B in the early secretory and autophagic pathways. J. Biol. Chem. 288: 1009–1021. doi: 10.1074/jbc.M112.395087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montagnac G., Echard A., Chavrier P.2008. Endocytic traffic in animal cell cytokinesis. Curr. Opin. Cell Biol. 20: 454–461. doi: 10.1016/j.ceb.2008.03.011 [DOI] [PubMed] [Google Scholar]
- 20.Moreno R.D., Ramalho-Santos J., Sutovsky P., Chan E.K., Schatten G.2000. Vesicular traffic and golgi apparatus dynamics during mammalian spermatogenesis: implications for acrosome architecture. Biol. Reprod. 63: 89–98. doi: 10.1095/biolreprod63.1.89 [DOI] [PubMed] [Google Scholar]
- 21.Mruk D.D., Lau A.S.N.2009. RAB13 participates in ectoplasmic specialization dynamics in the rat testis. Biol. Reprod. 80: 590–601. doi: 10.1095/biolreprod.108.071647 [DOI] [PubMed] [Google Scholar]
- 22.Nio-Kobayashi J., Iwanaga T.2012. Galectin-1 and galectin-3 in the corpus luteum of mice are differentially regulated by prolactin and prostaglandin F2α. Reproduction 144: 617–624. doi: 10.1530/REP-11-0495 [DOI] [PubMed] [Google Scholar]
- 23.Nottingham R.M., Pusapati G.V., Ganley I.G., Barr F.A., Lambright D.G., Pfeffer S.R.2012. RUTBC2 protein, a Rab9A effector and GTPase-activating protein for Rab36. J. Biol. Chem. 287: 22740–22748. doi: 10.1074/jbc.M112.362558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan X., Eathiraj S., Munson M., Lambright D.G.2006. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature 442: 303–306. doi: 10.1038/nature04847 [DOI] [PubMed] [Google Scholar]
- 25.Park H.H.2013. Structural basis of membrane trafficking by rab family small g protein. Int. J. Mol. Sci. 14: 8912–8923. doi: 10.3390/ijms14058912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrie R.G., Jr, Morales C.R.1992. Receptor-mediated endocytosis of testicular transferrin by germinal cells of the rat testis. Cell Tissue Res. 267: 45–55. doi: 10.1007/BF00318690 [DOI] [PubMed] [Google Scholar]
- 27.Polevoy G., Wei H.C., Wong R., Szentpetery Z., Kim Y.J., Goldbach P., Steinbach S.K., Balla T., Brill J.A.2009. Dual roles for the Drosophila PI 4-kinase four wheel drive in localizing Rab11 during cytokinesis. J. Cell Biol. 187: 847–858. doi: 10.1083/jcb.200908107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prekeris R., Gould G.W.2008. Breaking up is hard to do - membrane traffic in cytokinesis. J. Cell Sci. 121: 1569–1576. doi: 10.1242/jcs.018770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramalho-Santos J., Moreno R.D.2001. Targeting and fusion proteins during mammalian spermiogenesis. Biol. Res. 34: 147–152. doi: 10.4067/S0716-97602001000200021 [DOI] [PubMed] [Google Scholar]
- 30.Ramalho-Santos J., Moreno R.D., Wessel G.M., Chan E.K., Schatten G.2001. Membrane trafficking machinery components associated with the mammalian acrosome during spermiogenesis. Exp. Cell Res. 267: 45–60. doi: 10.1006/excr.2000.5119 [DOI] [PubMed] [Google Scholar]
- 31.Rivera-Molina F.E., Novick P.J.2009. A Rab GAP cascade defines the boundary between two Rab GTPases on the secretory pathway. Proc. Natl. Acad. Sci. USA 106: 14408–14413. doi: 10.1073/pnas.0906536106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roderick H.L., Campbell A.K., Llewellyn D.H.1997. Nuclear localisation of calreticulin in vivo is enhanced by its interaction with glucocorticoid receptors. FEBS Lett. 405: 181–185. doi: 10.1016/S0014-5793(97)00183-X [DOI] [PubMed] [Google Scholar]
- 33.Sato N., Koinuma J., Ito T., Tsuchiya E., Kondo S., Nakamura Y., Daigo Y.2010. Activation of an oncogenic TBC1D7 (TBC1 domain family, member 7) protein in pulmonary carcinogenesis. Genes Chromosomes Cancer 49: 353–367. [DOI] [PubMed] [Google Scholar]
- 34.Seabra M.C., Mules E.H., Hume A.N.2002. Rab GTPases, intracellular traffic and disease. Trends Mol. Med. 8: 23–30. doi: 10.1016/S1471-4914(01)02227-4 [DOI] [PubMed] [Google Scholar]
- 35.Stenmark H.2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10: 513–525. doi: 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- 36.Suarez-Quian C.A., An Q., Jelesoff N., Dym M.1991. The Golgi apparatus of rat pachytene spermatocytes during spermatogenesis. Anat. Rec. 229: 16–26. doi: 10.1002/ar.1092290104 [DOI] [PubMed] [Google Scholar]
- 37.Zerial M., McBride H.2001. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2: 107–117. doi: 10.1038/35052055 [DOI] [PubMed] [Google Scholar]