Abstract
Multidrug resistance is a prevailing phenomenon leading to chemotherapy treatment failure in cancer patients. In the current study two known cytotoxic pseudoguaianolide sesquiterpene lactones; neoambrosin (1) and damsin (2) that circumvent MDR were identified. The two cytotoxic compounds were isolated using column chromatography, characterized using 1D and 2D NMR, MS, and compared with literature values. The isolated compounds were investigated for their cytotoxic potential using resazurin assays and thereafter confirmed with immunoblotting and in silico studies. MDR cells overexpressing ABC transporters (P-glycoprotein, BCRP, ABCB5) did not confer cross-resistance toward (1) and (2), indicating that these compounds are not appropriate substrates for any of the three ABC transporters analyzed. Resistance mechanisms investigated also included; the loss of the functions of the TP53 and the mutated EGFR. The HCT116 p53-/- cells were sensitive to 1 but resistant to 2. It was interesting to note that resistant cells transfected with oncogenic ΔEGFR exhibited hypersensitivity CS toward (1) and (2) (degrees of resistances were 0.18 and 0.15 for (1) and (2), respectively). Immunoblotting and in silico analyses revealed that 1 and 2 silenced c-Src kinase activity. It was hypothesized that inhibition of c-Src kinase activity may explain CS in EGFR-transfected cells. In conclusion, the significant cytotoxicity of 1 and 2 against different drug-resistant tumor cell lines indicate that they may be promising candidates to treat refractory tumors.
Keywords: ABC-transporters, Asteraceae, EGFR, p53, Src inhibition, targeted chemotherapy
Introduction
Chemotherapy still represents a major treatment option for cancer, despite its limitations in specificity and toxicity. Resistance is an everlasting obstacle in cancer therapy eventually leading to MDR that result to therapy failure. Various factors account for inherent (primary) and acquired (secondary) resistance including: point mutations in target and effector genes, genetic, and epigenetic heterogeneity of cancer cells, increased activity of drug efflux pump such as ABC transporters, activation of EGFR leading to increased proliferation by auto-activation of downstream signaling cascades, activation of drug metabolizing enzymes such as CYP450, impairment of DNA repair mechanisms and suppression of apoptosis by mutated p53 (Gottesman, 2002; Holohan et al., 2013).
Resistance to chemotherapy remains a key challenge; therefore, targeted chemotherapy emerged as new concept in the treatment regime. In this treatment, a drug specifically inhibit tumor growth by targeting cellular molecules that are pivotal for growth, progression, and metastasis, for example using monoclonal antibodies or natural or synthetic small molecule inhibitors. This approach increased the specificity of treatment for different types of malignancies, including breast, colorectal, lung, and pancreatic cancers, as well as lymphoma, leukemia, and multiple myeloma (Gerber, 2008).
Natural products considerably impact drug discovery due to their wide structural diversity. More than 60% of clinically used anticancer agents are derived from natural sources (plants, marine organisms, and microorganisms; Newman and Cragg, 2012).
Ambrosia maritima L. (Asteraceae) is gray-hairy herb, richly branched with finely dissected, fragrant leaves. This plant is widely distributed in the central and northern Sudan, near the banks of Nile River and locally known as Damsisa. It is used in folk medicine to treat diabetes, hypertension, urinary tract infections, gastrointestinal disturbances, kidney stones, and also cancer (Khalid et al., 2012; Dirar et al., 2014). A. maritima has been planted in Egypt, on canals and banks of the Nile Delta as a molluscocide against the schistosomiasis intermediate host (El-Sawy et al., 1981). Recently, it has been reported that A. maritima exhibited considerable cytotoxic activity against various MDR tumor cells (Saeed et al., 2015). Several genera of Asteraceae are rich of sesquiterpene lactones (SLs), which are considered as promising class of compounds because of their diverse biological activity with interesting therapeutic mechanisms, including anti-inflammatory, antitumor, antimicrobial, and antiviral properties (Zhang et al., 2005; Fraga, 2011; Merfort, 2011).
In the present work, two pseudoguaianolide SLs, neoambrosin (1) and damsin (2), were characterized from A. maritima L. Their anticancer activities against drug-resistant tumor cell lines were determined, in which either MDR-conferring ABC-transporters [P-glycoprotein (P-gp), BCRP, ABCB5] or mutation-activated EGFR were overexpressed. Furthermore, drug-resistant TP53-knockout cells were analyzed. The focus of this study was on EGFR-downstream signals due to the striking hypersensitivity CS of EGFR-transfected cells toward 1 and 2. The two compounds which are structurally related, specifically inhibited c-Src kinase activity, indicating that this oncogenic kinase is a suitable target of 1 and 2, explaining the CS exhibited by EGFR-transfected cells to these compounds.
Materials and Methods
General Experimental Procedures
Pure DCM was used to prepare crude extract from whole plant of A. maritima. Crude extract was analyzed by high pressure liquid chromatography (HPLC, Agilent 1100 Series, equipped with a LiChrospher RP 18 column (3 mm × 125 mm; 5 μm, Merck, Darmstadt, Germany) at 40°C and a flow rate of 1 mL/min with an elution gradient composed of H2O + 0.1% (v/v) trifluoroacetic acid and acetonitrile. The molecular weight of the selected peaks was determined by HPLC-MS (Agilent 1260 Series LC and 6130 Series Quadrupole MS System). The mass spectra were recorded using atmospheric pressure chemical ionization (APCI) with positive and negative polarization. A Superspher RP 18 (125 mm × 2 mm; 4 μm, Merck) column was used at 40°C. For every run, 1 μL of a sample (1 mg/mL) was injected. Chromabond column (C18 ec-column, Macherey–Nagel, Düren, Germany) was used to obtain the pure compounds. 1D and 2D NMR data were recorded with a Bruker AVANCE III-600 MHz spectrometer equipped with a 5 mm inverse TCI cryoprobe using standard pulse sequences to elucidate the structures of neoambrosin (1) and damsin (2).
Plant Material
The whole plant of A. maritima was identified and authenticated by Prof. Hassan Khalid at medicinal and aromatic plants research institute (MAPRI) in Khartoum. A voucher specimen (A23) represents the plant was deposited in the herbarium of National Research Institute, Khartoum, Sudan (Saeed et al., 2015).
Extraction and Isolation of Neoambrosin (1) and Damsin (2) from the Whole Plant of A. maritima
Plant materials (∼100 g) was dried and ground before being subjected to cold maceration with DCM. The plant powder was macerated with overnight gentle shaking in DCM into stoppered flask at room temperature. The extracted solvent was separated and filtered through Whatman no. 1 filter paper. The DCM was evaporated under reduced pressure using rotatory evaporator, resulting in A. maritima crude extract.
One gram of the whole plant of A. maritima extract was subjected to separation using a silica gel-60 matrix eluting with cyclohexane (C6H10): Ethyl acetate (EtOAc) gradient in increasing polarities. This resulted into 10 fractions each including: 1 (2 mg), 2 (103 mg), 3 (66 mg), 4 (107 mg), 5 (67 mg), 6 (71 mg), 7 (37 mg), 8 (45 mg), 9 (37 mg), 10 (415 mg). The 10 samples were fractionated via HPLC in micro-titer plates in order to determine their cytotoxic potential. The samples were fractionated using the analytical HPLC method given above. A subsequent located fraction collector (Agilent 1100 Series) was used equipped with 96-well plates for collecting the flow through resulting in 92 different fractions of 250 μl each. These fractions could be used in biological assays. Fractions 3 and 4 of the main column revealed interesting cytotoxic activities (IC50 values of 4.5 and 4.8 μM for fractions 3 and 4) against CCRF-CEM cells, respectively. These fractions were subsequently subjected to solid phase extraction using C18- Chromabond column as the stationary phase eluting with neat methanol (MeOH). The resultant fractions were further purified by preparative HPLC using 65% acetonitrile with 35% H2O + 0.1% (v/v) trifluoracetic acid for fraction 3, and 60% acetonitrile with 40% H2O + 0.1% (v/v) trifluoracetic acid, with 17 mL/min isocratic flow-rate, yielding compounds 1 (20.0 mg) and 2 (50.5 mg), respectively.
Cell Lines
Drug sensitive CCRF-CEM and multidrug-resistant P-gp-overexpressing CEM/ADR5000 leukemic cells were cultured in RPMI-1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin (Invitrogen, Darmstadt, Germany). Doxorubicin (5000 ng/mL) was added to maintain overexpression of P-gp (MDR1, ABCB1) in resistant cells (Kimmig et al., 1990). Breast cancer cells transfected with a control vector (MDA-MB-231-pcDNA3) or with cDNA for the breast cancer resistance protein BCRP/ABCG2 (MDA-MB-231-BCRP clone 23) were cultured and maintained as reported (Doyle et al., 1998). Expression of ABCG2 in resistant cells was maintained by geneticin (800 ng/mL) (Efferth et al., 2003). Human HEK293-ABCB5 embryonic kidney cells transfected with another ABC-transporter, ABCB5, were propagated in DMEM medium supplemented with 10% FBS and 1% penicillin/streptomycin (Invitrogen) (Kawanobe et al., 2012). Non-transfected HEK293 cells served as control. Human wild-type HCT116 colon cancer cells (p53+/+) and knockout clones (p53-/-) derived by homologous recombination (Waldman et al., 1995; Bunz et al., 1998) were generously provided by Dr. B. Vogelstein and H. Hermeking (Howard Hughes Medical Institute, Baltimore, MD, USA). Both colon cancer cells were cultured in DMEM medium supplemented with 10% FBS and 1% penicillin/streptomycin (Invitrogen). Wild-type human GBM U87MG cells and cells transfected with control mock vector or an expression vector harboring EGFR cDNA with a deletion in exons 2–7 (U87MGΔEGFR), were kindly provided by Dr. W. K. Cavenee (Ludwig Institute for Cancer Research, San Diego, CA, USA; Huang et al., 1997). Six non-transfected human GBM cell lines were used, which overexpress c-Src including: A172, T98G, and U251MG cells, obtained from the German Cancer Research Center (DKFZ, Heidelberg, Germany). DK-MG and SNB-19 cells were kindly provided by Dr. Tcholpon Djuzenova (Department of Radiation Oncology, University Hospital, Würzburg, Germany). BS153 cells were obtained from Dr. Alexander Schulte (Department of Neurosurgery, University Medical Center Hamburg-Eppendorf, Hamburg, Germany) and originally generated by Dr. Adrian Merlo (Jones et al., 2001). All GBM cells were cultured in DMEM medium supplemented with 10% FBS and 1% penicillin/streptomycin (Invitrogen). All cell lines were incubated in a humidified 5% CO2 atmosphere at 37°C and passaged twice weekly.
Cytotoxicity Assays
The resazurin (Promega, Mannheim, Germany) reduction assay was performed to assess cytotoxicity of neoambrosin (1) and damsin (2) in a concentration range of 0.001–1000 μM as previously described (Kuete and Efferth, 2013). The IC50 values have been calculated from dose response curves and resistance ratios were determined by dividing the IC50 of resistant cells by the IC50 of the corresponding parental cells. Each assay has been done thrice with six replicates for each concentration.
Epidermal Growth Factor Receptor Signaling Proteins Analyses by SDS-PAGE and Immunoblotting
Tumor cells (500,000 cells/well) were treated with neoambrosin (1) and damsin (2), harvested after 24 h, and washed with PBS. Total proteins were extracted by the Mammalian Protein Extraction Reagent (M-PER) (Thermo Scientific, Rockford, IL, USA) containing protease inhibitor and phosphatase inhibitor (Roche, Mannheim, Germany). Cells were shaken at 4°C for 30 min, cell lysates were centrifuged at 14,000 ×g for 15 min and supernatants were transferred to new tubes. Protein concentrations in lysates were detected by NanoDrop1000 (PEQLAB, Erlangen, Germany). Thirty micrograms of protein were electrophoresed on 10% SDS-polyacrylamide gels and blotted onto polyvinylidene difluoride (Ruti®-PVDF) membrane (Millipore Corporation, Billerica, MA, USA) using wet sandwich blotting. Membranes were washed using Tris-buffered saline containing 0.5% Tween-20 (TBST), then blocked with 5% (w/v) bovine serum albumin in TBST for 1 h at room temperature. Membranes were incubated at 4°C overnight with primary antibodies (Cell Signaling Technology, Frankfurt, Germany) including: phosphorylated EGFR (Tyr1068) (1:1000), phosphorylated STAT5 (Tyr694) (1:1000), phosphorylated Akt (Ser473) (1:1000), phosphorylated MAPK (Thr202/Tyr204) (1:1000), phosphorylated Src (Tyr416) (1:1000), Src (1:1000), and β-actin (1:2000). After washing membranes three times with TBST, the blots were probed with horseradish peroxidase-linked anti-rabbit IgG secondary antibodies (1:2000) for 2 h at room temperature. Finally, LuminataTM Classico Western HRP substrate (Merck Millipore, Schwalbach, Germany) was added for 5 min in the dark. Alpha Innotech FluorChem Q system (Biozym, Oldendorf, Germany) was used for documentation and band analysis (Zhao et al., 2015).
Molecular Docking
Molecular docking predicts in silico the interaction, affinity, and geometry of ligands with target proteins. The affinity of neoambrosin (1) and damsin (2) to c-Src kinase domain compared with the known c-Src inhibitor dasatinib (3) (Buettner et al., 2008; Ceppi et al., 2009) was studied using Autodock4, Lamarckian algorithm (Morris et al., 2009). The compounds were prepared as 3D-structures and converted to PDBQT format. The c-Src kinase domain was obtained from the Protein Data Bank (PDB ID: 3QLG). Non-standard residues of protein (HETATM) were excluded, hydrogens added, non-polar hydrogen atoms merged, missing atoms supplemented and attached to solvation parameters. Afterward, a grid box was assigned to cover the c-Src kinase domain (PDBQT format). Docking parameters were set to 250 runs and 2,500,000 energy evaluations for each cycle. Docking was performed three times independently to calculate mean values and standard deviations of lowest binding energies and predicted inhibition constants (Pki), which were obtained from the docking log files (dlg). Binding poses between ligands (neoambrosin (1), damsin (2) and dasatinib (3)) and the macromolecule (c-Src kinase domain) were visualized by Visual Molecular Dynamics software (University of Illinois, Champaign, IL, USA).
Results
Identification and Structure Elucidation of Neoambrosin (1) and Damsin (2)
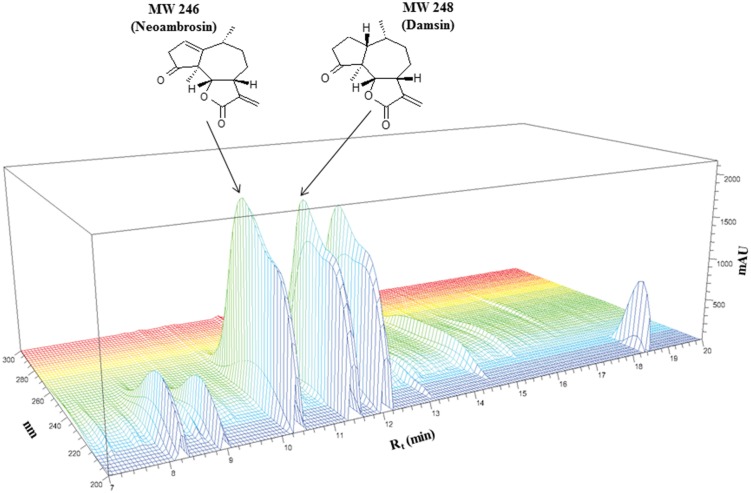
Two compounds 1 and 2 were identified from pure DCM extract of the whole plant of A. maritima L. (Figure 1). Neoambrosin (1) was obtained as colorless oil. Its structure was established by NMR. The 13C NMR spectrum revealed 15 signals. According to HSQC, they originated from CH3 groups (δ 1.14/15.2, δ 1.18/21.5), CH2 groups including one olefinic [δ (1.77, 1.82)/31.4, δ (2.06, 2.15)/24.7, δ (2.74, 3.22)/40.7, (δ 5.64, 6.19)/120.5], CH groups including one olefinic (δ 2.93/40.2, δ 3.49/44.8, δ 4.55/81.6, δ 6.03/125.7), and quaternary carbons (δ 59.9, 140.8, 150.4, 172.2, 216.2). These spectra data were suggestive of SLs. The skeleton and the relative configuration of the pseudoguaianolide (1) was confirmed from 2D NMR analysis (including COSY, HMBC, and NOESY) and comparison with literature values from previous studies. The 1D and 2D spectral data of this compound completely matched those of neombrosin (1), previously isolated from the aerial parts of A. maritima (Salam et al., 1984) and the vegetative parts of a related species, A. hispida by Herz et al. (1981).
FIGURE 1.
Chemical structures of neoambrosin (1) and damsin (2) and HPLC profiling of Ambrosia maritima L. dichloromethane (DCM) extract.
Damsin (2) was isolated as colorless oil. Its 13C NMR spectra also contained 15 signals. The lack of two olefinic resonances suggested 2 as a reduced form of 1. Based on HSQC correlations, two CH3 groups (δ 1.08/14.0, δ 1.08/16.0), five CH2 groups including one olefinic exo-methylene (δ 1.75,1.80/33.6, δ 1.80, 2.01/24.1, δ 1.80, 2.01/25.9, δ 2.24, 2.48/36.2, δ5.54, 6.28/121.1), four CH groups (δ 2.20/34.5, δ 2.01/46.3, δ 3.30/44.6, δ 4.53/81.9) and four quaternary carbons (δ 55.1, 139.7, 170.3, 219.1) were identified. The HMBC correlations indicated another pseudoguaianolide. The relative stereochemistry and the structure matched that of damsin, as previously reported by Herz et al. (1981).
Cytotoxicity of Neoambrosin (1) and Damsin (2)
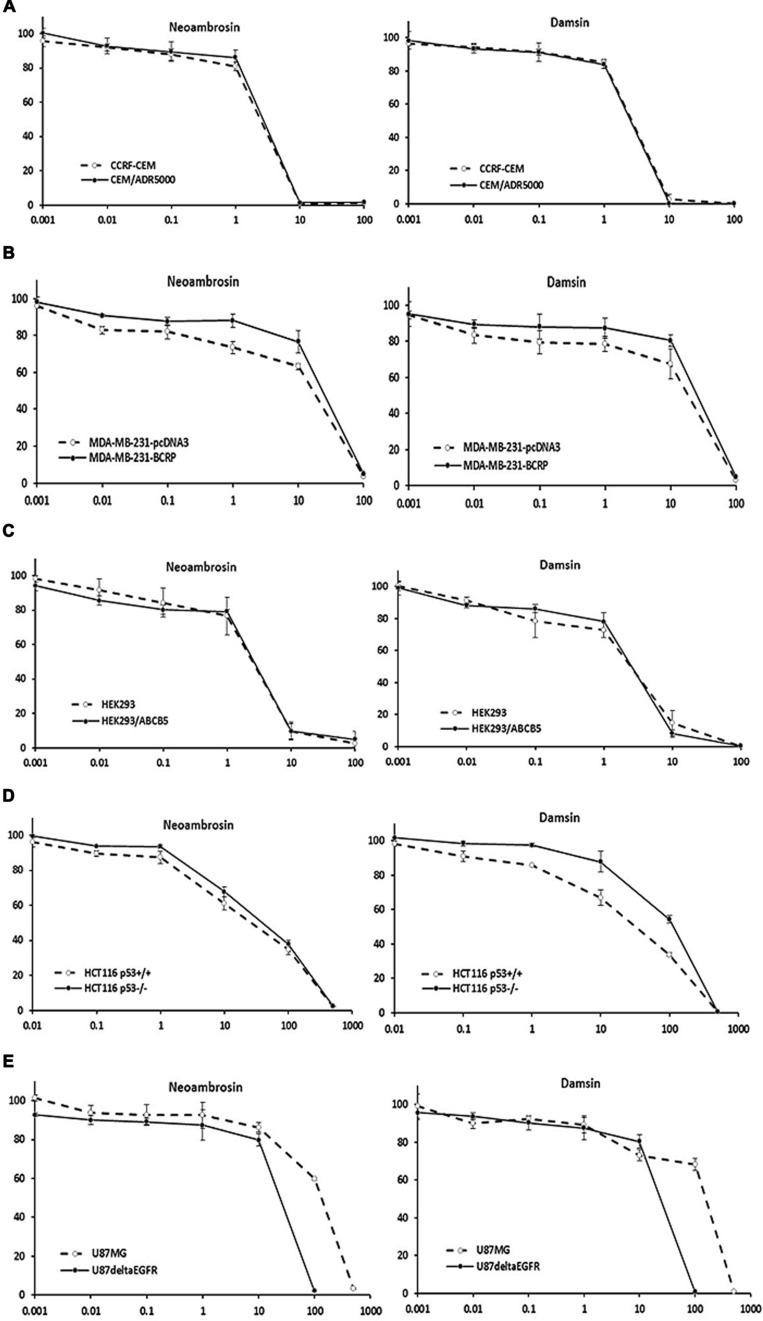
Neoambrosin (1) and damsin (2) were tested in a range from 10-3 to 103 μM in resazurin assays. Firstly, the effects of 1 and 2 on drug sensitive CCRF-CEM and resistant CEM/ADR5000 cells were analyzed. No cross-resistance was observed (Figure 2A; Table 1). The IC50 values were similar in both cell lines (degrees of resistance were 1.07 and 1 for 1 and 2, respectively). Similarly, BCRP-transfected MDA-MB-231 cells were not cross-resistant to these compounds (Figure 2B; Table 1). Multidrug-resistant ABCB5-transfectants did also not show resistance toward these SLs (Figure 2C; Table 1).
FIGURE 2.
Dose response curves of neoambrosin (1) and damsin (2) as determined by resazurin assays. (A) Cytotoxicity against drug-sensitive parental CCRF-CEM tumor cells and their P-glycoprotein (P-gp; MDR1/ABCB1)-expressing, multidrug-resistant subline, CEM/ADR5000. (B) Cytotoxicity against MDA-MB-231-pc DNA cells and their BCRP-transduced subline, MDA-MB-231-BCRP. (C) Cytotoxicity against HEK293 cells and their ABCB5-transfectant subline, HEK293/ABCB5. (D) Cytotoxicity against HCT116 cells (p53+/+) and their knockout subline (p53-/-). (E) Cytotoxicity against U87MG cells and their EGFR- transduced subline, U87MGΔEGFR. The dose response curves show mean values ± SD of three independent experiments with each six parallel measurements.
Table 1.
IC50 values of different drug-sensitive and -resistant cell lines of neoambrosin (1) and damsin (2).
| Compound | Cell lines | Resistance ratio (RR) | |
|---|---|---|---|
| CCRF-CEM | CEM/ADR5000 | ||
| Neoambrosin | 4.5 ± 0.2 μM | 4.8 ± 0.2 μM | 1.07 |
| Damsin | 4.8 ± 0.3 μM | 4.7 ± 0.2 μM | 1 |
| MDA-MB-231-pcDNA3 | MDA-MB-231-BCRP | ||
| Neoambrosin | 29.9 ± 1.7 μM | 43.3 ± 4.9 μM | 1.4 |
| Damsin | 33.8 ± 8.4 μM | 46.4 ± 2.6 μM | 1.4 |
| HEK293 | HEK293/ABCB5 | ||
| Neoambrosin | 4.5 ± 0.6 μM | 4.8 ± 0.2 μM | 1.07 |
| Damsin | 4.5 ± 0.1 μM | 4.6 ± 0.6 μM | 1 |
| HCT116 P53+/+ | HCT116 P53-/- | ||
| Neoambrosin | 26.5 ± 6.9 μM | 39.4 ± 6.4 μM | 1.5 |
| Damsin | 32.5 ± 6 μM | 133.6 ± 18.4 μM | 4.1 |
| U87MG | U87ΔEGFR | ||
| Neoambrosin | 132.2 ± 2.4 μM | 24.1 ± 1.4 μM | 0.18 |
| Damsin | 154.4 ± 8.9 μM | 24.1 ± 1.5 μM | 0.15 |
Then, neoambrosin (1) and damsin (2) were tested on wild-type HCT116 p53+/+ and HCT116 p53-/- knockout cells (Figure 2D; Table 1). Both cell lines showed sensitivity to neoambrosin (1) (degree of resistance was 1.5), whereas knockout cells exhibited 4.1-fold cross-resistance to damsin (2) compared to wild-type cells.
Surprisingly, U87.MGΔEGFR cells were hypersensitive toward both compounds as compared to U87MG wild-type cells. The degrees of resistances were 0.18 and 0.15 for neoambrosin (1) and damsin (2), respectively (Figure 2E; Table 1). This phenomenon is known as collateral sensitivity (CS) (Hall et al., 2009) and suggests that both SLs may target proteins involved in EGFR signaling.
Inhibition of EGFR Signaling Cascades by Neoambrosin (1) and Damsin (2)
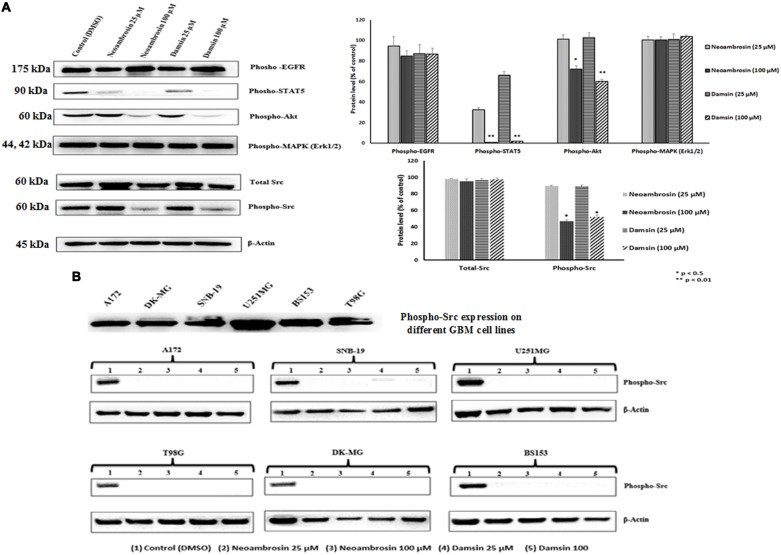
In order to have a closer insight into the phenomenon of CS of U87.MG cells transfected with deletion-activated EGFR toward neoambrosin (1) and damsin (2), the phosphorylation status of EGFR signaling proteins as parameter of their activation during signal transduction was investigated. Two concentrations of each compound were selected: 25 μM (∼IC50) and 100 μM (4 × IC50). Both compounds inhibited STAT5 and AKT phosphorylation at 100 μM, whereas other signaling proteins were not affected, including EGFR (Figure 3A). It was surmised that STAT5 and AKT activation works independently from EGFR activation (Okutani et al., 2001; Haynes et al., 2003; Summy and Gallick, 2006). Then the two compounds 1 and 2 were hypothesized to target c-Src. Remarkably, they inhibited c-Src autophosphorylation at Tyr416, whereas non-phosphorylated (total) c-Src was not affected. These results indicated that both compounds targeted c-Src and silenced its downstream signaling.
FIGURE 3.
Western blot analysis of the effect of neoambrosin (1) and damsin (2) on EGFR and its downstream signaling cascade proteins. (A) Effect on phosphorylation of ΔEGFR, STAT5, Akt, MAPK (Erk1/2), c-Src, and total c-Src. Bands were normalized to β-actin in order to obtain numerical values (Mean ± SEM). Bars of the average and error bars of three independent experiments are shown. Statistical analysis was done by paired student’s t-test ∗∗p < 0.01,∗p < 0.05; paired t-test. (B) Effect on phosphorylation of c-Src in different GBM cell lines that highly overexpressed c-Src (A172, DK-MG, SNB-19, U251.MG, BS153, and T98G). β-actin was used as loading control.
For confirmation, six other glioblastoma cell lines with known c-Src overexpression were investigated. Indeed, neoambrosin (1) and damsin (2) inhibited c-Src phosphorylation in all concentrations tested (Figure 3B).
In silico Binding of Neoambrosin and Damsin to c-Src
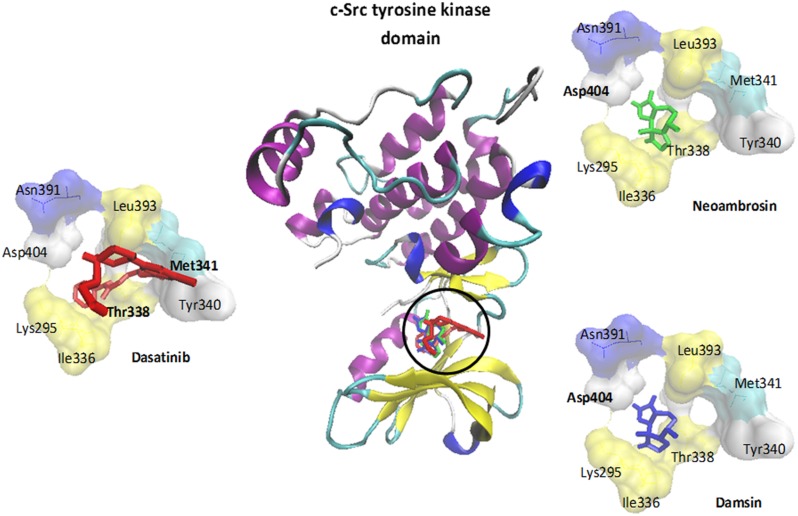
In silico molecular docking of the c-Src kinase domain to both SLs and dasatinib (3) as positive control, were performed. The binding energy of dasatinib (3) to c-Src was slightly lower than those of neoambrosin (1) and damsin (2) (Table 2). Interestingly, all three compounds bound to the same site on the c-Src kinase domain. The c-Src kinase domain consists of characteristic small amino-terminal and large carboxyl-terminal lobes. The catalytic ATP binding site lies in a cleft between the two lobes. These lobes move relative to each other and open or close the cleft. The open form is necessary to allow access of ATP to phosphorylate c-Src. The residues, which are essential for c-Src phosphorylation are Lys295 and Asp404. Any molecule able to interrupt the open/closed c-Src form is considered to have an inhibitory effect (Hanks et al., 1988; Roskoski, 2004). Importantly, neoambrosin (1), damsin (2), and dasatinib (3) have the ability to bind to the cleft between the lobes. Although dasatinib revealed better affinity, both SLs bind nevertheless to the same binding site as dasatinib (3) (Table 2; Figure 4). This result explains the inhibition of c-Src phosphorylation by neoambrosin (1) and damsin (2).
Table 2.
Molecular docking of dasatinib (3), neoambrosin (1), and damsin (2) to the c-Src kinase domain.
| Compound | Lowest binding energy (kcal/mol) | Pki (μM) | AA involved in H-bonds |
|---|---|---|---|
| Dasatinib | –9.85 ± 0.03 | 0.06 , <0.00 | Thr338, Met341 |
| Neoambrosin | –7.23 ± <0.00 | 4.99 , <0.00 | Asp404 |
| Damsin | –7.25 ± 0.01 | 4.8 , 0.01 | Asp404 |
Shown are lowest binding energy, predicted inhibition constant (Pki), and amino acids (AA) involved in hydrogen bonding. Each docking experiment has been repeated three times.
FIGURE 4.
Molecular docking of dasatinib (3), neoambrosin (1), and damsin (2) to the c-Src kinase domain. The protein was represented by its secondary structure in a new carton format, whereas ligands were represented in dynamic bond format with different colors: dasatinib (3) (red), neoambrosin (1) (green) and damsin (2) (blue). Amino acids (AA) involved in H-bonds were shown in a bold font Met341 and Thr338 (dasatinib); Asp404 (neoambrosin and damsin).
Discussion
Sesquiterpene lactones gathered attention for cancer treatment (Lopez-Anton et al., 2007; Staudt, 2010). The cytotoxicity of pseudoguaianolide SLs, neoambrosin (1) and damsin (2) from A. maritima L. was tested. Neoambrosin (1) and damsin (2) inhibited all cell lines tested in the micromolar ranges. While the cytotoxicity of damsin (2) against cancer cells has been reported elsewhere (Villagomez et al., 2013), neoambrosin’s cytotoxic activity is reported here for the first time to the best of our knowledge.
ABC-transporter-mediated MDR leads to reduced chemotherapeutic drugs accumulation inside the cells, including anthracyclines, Vinca alkaloids, colchicine, epipodophyllotoxins, taxanes, camptothecins, imatinib, methotrexate, and mitoxantrone (Raaijmakers et al., 2006; Efferth et al., 2008; Fletcher et al., 2010). Among the 48 members of the ABC transporter family, P-gp and BCRP have been extensively characterized (Gros et al., 1986; Ueda et al., 1986; Doyle et al., 1998). Frank et al. cloned a novel ABC transporter named ABCB5, which controls progenitor cell fusion by altering the membrane potential in normal melanocytes expressing the stem cell marker CD133 (Frank et al., 2003). ABCB5 is closely related to P-gp and confers doxorubicin resistance in malignant melanoma (Frank et al., 2005). Furthermore, it mediates resistance to paclitaxel and docetaxel (Kawanobe et al., 2012). Cell models, which overexpressed these three ABC transporters were used. Importantly, they did not confer cross-resistance to neoambrosin (1) and damsin (2), indicating that they are not substrates for any of these three transporters. This implies that these SLs may be useful to treat MDR- tumors.
One of the most frequently mutated genes is the TP53. It is impaired in approximately 50% of all tumors (Nigro et al., 1989; Olivier et al., 2002). As transcription factor, p53 maintains the genomic integrity after genotoxic stress by inducing cell cycle arrest, DNA repair, and apoptosis (Vogelstein et al., 2000). Activated p53 spurs p21/WAF1, an inhibitor for G2/M-specific cell division control protein 2 kinase and cyclin-dependent G1 kinase subsequently leading to G2/M and G1 checkpoint control. Failure in arresting cells at both checkpoints due to mutated p53 confers drug resistance (Agarwal et al., 1995; Piovesan et al., 1998). HCT116 p53-/- cells, which have been reported for their resistance to 5-FU, irinotecan, and oxaliplatin (Boyer et al., 2004) were tested. Interestingly, wild-type and knockout HCT116 cells were sensitive to neoambrosin (1), while knockout cells were cross-resistant to damsin (2). This indicates that both SLs may activate wild-type p53 triggering their programmed death. This result is in agreement with a previous study on another SL, parthenolide, which activated p53 by enhancing ubiquitylation and degradation of the p53-negative regulator Mdm2 (Gopal et al., 2009). The sensitivity of p53-knockout cells to neoambrosin (1) could be due to targeting other molecular determinants, whereas damsin (2) failed to address these targets. This result warrants more detailed investigation in the future.
Epidermal growth factor receptor and its downstream signaling proteins are associated with MDR and up-regulated co-expression of ABC-transporters (Hirsch-Ernst et al., 1995; Garcia et al., 2006; Katayama et al., 2007; Hoffmann et al., 2011). Oncogenic ΔEFGR confers resistance by inducing expression of the anti-apoptotic protein Bcl-xL (Nagane et al., 1998). Several EGFR mutations have been described. A common mutant is ΔEFGR, which causes poor prognosis and survival times, especially in glioblastoma patients (Shinojima et al., 2003). An 801 bp in-frame deletion of the extracellular EGFR domain results in ligand-independent receptor activation, consequent auto-phosphorylation of tyrosine residues in the kinase domain and induction of downstream signaling (Nishikawa et al., 1994). Surprisingly, neoambrosin (1) and damsin (2) revealed CS in U87MGΔEGFR cells compared to U87MG wild-type cells. CS represents a strategy to preferentially kill resistant cells (Hall et al., 2009). CS is well known from MDR phenotypes with P-gp overexpression. Several hypotheses attempted to explain CS, e.g. increased production of reactive oxygen species (ROS) in expense of ATP hydrolysis, expelling endogenous vital substrates, exploitation of energetic sensitivities and plasma membrane disruption (Pluchino et al., 2012). CS in EGFR-expressing cells has been recently described for the first time by us (Saeed et al., 2014). SLs induce oxidative stress and disturb the redox status by increased ROS production through interaction with the sulfhydryl moiety in the cysteine of the antioxidant glutathione (Sun et al., 2010). This is in accordance with the assumption that CS occurs because of ROS accumulation. However, this explanation may be insufficient, because in this case all other resistant cell lines tested by us should also exert CS because of glutathione abundance in all these cell lines.
To further explore CS, we performed phosphorylation-specific immunoblotting analyses of EGFR and its downstream signal transducers. Interestingly, only AKT and STAT5 phosphorylation was downregulated, whereas EGFR and MAPK remained unaffected by the two compounds. This result raises the question about other targets of these SLs. Another key player of EGFR signaling is the oncogenic tyrosine kinase, c-Src. Worth to mention, inhibition of c-Src kinase activity diminished tyrosine phosphorylation of STAT5 in K562 cells, indicating that Src activates STAT5 (Okutani et al., 2001; Silva, 2004; Figure 5). Furthermore, c-Src activated AKT through the formation of a complex with regulatory subunit p85 of PI3K with subsequent activation of the PI3K/Akt pathway (Haynes et al., 2003; Summy and Gallick, 2006; Figure 5). Thus, c-Src inhibition could silence STAT5 and AKT phosphorylation.
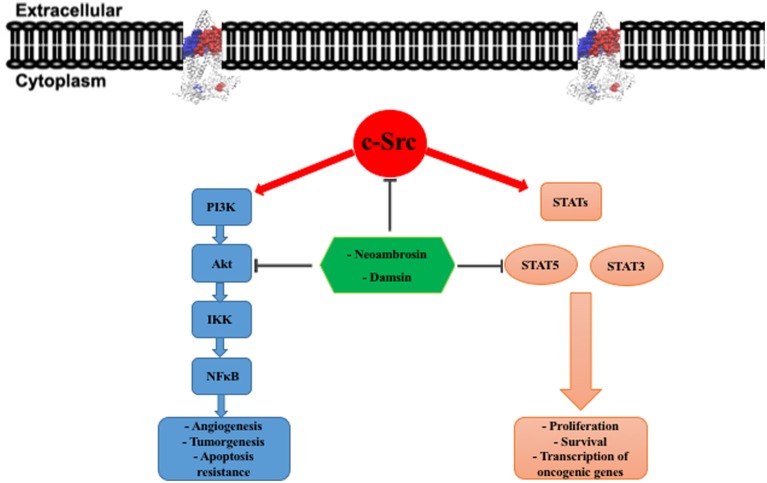
FIGURE 5.
c-Src downstream signaling pathways. In tumor cells, overexpression of c-Src leads to constitutive activation of PI3K/Akt and STATs (STAT3 and STAT5) pathways. This activation governing cancer development and progression. PI3K, phosphatidylinositol-3-kinase; IKK, IκB kinase; NFκB, nuclear factor kappa-light-chain-enhancer of activated B cells; STAT, signal transducers and activators of transcription.
Src was discovered in Rous sarcoma virus that causes transmissible tumors in chicken (Rous, 1911; Martin, 1970; Courtneidge, 2002). The viral gene product, v-Src, is a tyrosine kinase with a cellular homolog, c-Src (Czernilofsky et al., 1980). Upon c-Src activation, it becomes an oncogene involved in adhesion, growth, angiogenesis, and progression (Irby and Yeatman, 2000). Several studies reported elevated expression and kinase activity of c-Src in many tumor cells as compared with normal cells, suggesting that c-Src is crucial for cancer development and progression (Cartwright et al., 1989; Budde et al., 1994; Muthuswamy et al., 1994; Irby and Yeatman, 2000). c-Src has a great metastatic potential (Yeatman, 2004). The activity of c-Src tyrosine kinase interacts with receptor of tyrosine kinases such as EGFR in promoting tumor growth. The combination of EGFR and c-Src expressions in fibroblasts leads to synergistic tumorigenicity, if compared with EGFR or c-Src alone (Maa et al., 1995; Mao et al., 1997; Tice et al., 1999; Irby and Yeatman, 2000). c-Src activation increased growth rates and invasion characteristics of tumors. It also decreased apoptosis in tumor cells (Irby and Yeatman, 2000). Therefore, it is generally assumed that inhibition of c-Src function represents an attractive target to stop tumor growth. The recent use of c-Src inhibitors or antisense therapy in nude mouse studies, pancreatic cell growth, and leukemia cells supports the validity of this concept (Staley et al., 1997; Lutz et al., 1998; Roginskaya et al., 1999).
It has been postulated that c-Src could be a target for neoambrosin (1) and damsin (2) and that CS emerged in EGFR-expressing cells due to c-Src inhibition. We further investigated, how c-Src expression is affected in different other brain tumor models with c-Src overexpression upon treatment with these two SLs. Interestingly, both compounds inhibited c-Src phosphorylation not only in ΔEGFR-transfected cells, but also in six other c-Src-overexpressing cell lines. Further confirmation came from in silico molecular docking studies. Neoambrosin (1), damsin (2), and dasatinib (3) bound to the cleft between the two c-Src’s lobes. These small amino-terminal and large carboxyl lobes are of crucial importance for ATP binding to activate c-Src, thus, blocking of this cleft leads to impaired c-Src activation. These results indicate that neoambrosin (1) and damsin (2) are interesting c-Src inhibitors. Recent studies have shown that targeted inhibition of c-Src by dasatinib leads to decreased growth, and induced cell-cycle arrest and apoptosis in a subset of thyroid cancer and breast cancer cells. This therapeutic approach has been verified in vitro by immunoblotting analysis (Nautiyal et al., 2009; Chan et al., 2012). In this respect, it is remarkable that neoambrosin (1) and damsin (2) reveal similar c-Src-inhibiting features as dasatinib. Hence, it is reasonable to conclude that the inhibition of c-Src function (i.e., phosphorylation) led to the inhibition of tumor cell growth, which we observed in resazurin assays.
A previous study demonstrated that damsin (2) inhibited STAT3 and NF-κB expression in Jurkat and Hela cells by interfering with their signaling pathways (Villagomez et al., 2013). This result is in accordance with our finding that damsin (2) inhibited a master regulator upstream of STAT3 and NF-κB. Interestingly, c-Src directly activates STAT3, and NF-κB is activated by the PI3K/Akt pathway (Silva, 2004; Summy and Gallick, 2006). AKT enhances the transcriptional activity of NF-κB by phosphorylation and subsequent degradation of inhibitor of κB (IκB) (Bai et al., 2009). Moreover, several studies stated that PI3K/Akt pathway regulates the expression of P-gp (ABCB1) and BCRP (ABCG2) (Figure 5) (Barancík et al., 2006; Chiarini et al., 2008; Pick and Wiese, 2012).
Noteworthy, there is a cross talk between c-Src and p53. c-Src suppressed the apoptotic p53 pathway (Courtneidge, 2002; Polonio-Vallon et al., 2014). This may explain the sensitivity of wild-type 53 cells to neoambrosin (1) and damsin (2) and that they probably exert their cytotoxicity toward wild-type 53 cells by c-Src inhibition.
Conclusion
Neoambrosin (1) and damsin (2) isolated from the whole plant of A. maritima L. possess remarkable cytotoxicity against different drug-resistant tumor cell lines. Both SLs silenced c-Src phosphorylation, which represents an interesting approach for argeted therapy, because c-Src is activated in many human cancers (Yeatman, 2004). The remarkable CS of neoambrosin (1) and damsin (2) toward EGFR-expressing cells opened prospects for synergistic tumor killing if combined with established anticancer drugs. Clinically, tumors develop MDR and activate alternative pathways to survive and disseminate. Thus, combination therapies with these two SLs might improve cure rates of cancer patients (Shapiro, 1955; Devita et al., 1970; Rougier and Lepère, 2005; Li et al., 2014).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Financial support by the Carl Zeiss foundation (project ChemBioMed) is gratefully acknowledged.
ABBREVIATIONS
- ABC
ATP-binding cassette
- CS
collateral sensitivity
- DCM
dichloromethane
- EGFR
epidermal growth factor receptor
- GBM
glioblastoma multiforme
- MDR
multidrug resistance
- TP53
tumor suppressor p53.
References
- Agarwal M. L., Agarwal A., Taylor W. R., Stark G. R. (1995). p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 92 8493–8497. 10.1073/pnas.92.18.8493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai D., Ueno L., Vogt P. K. (2009). Akt-mediated regulation of NFkappaB and the essentialness of NFkappaB for the oncogenicity of PI3K and Akt. Int. J. Cancer 125 2863–2870. 10.1002/ijc.24748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barancík M., Bohácová V., Sedlák J., Sulová Z., Breier A. (2006). LY294,002, a specific inhibitor of PI3K/Akt kinase pathway, antagonizes P-glycoprotein-mediated multidrug resistance. Eur. J. Pharm. Sci. 29 426–434. 10.1016/j.ejps.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Boyer J., Mclean E. G., Aroori S., Wilson P., Mcculla A., Carey P. D., et al. (2004). Characterization of p53 wild-type and null isogenic colorectal cancer cell lines resistant to 5-fluorouracil, oxaliplatin, and irinotecan. Clin. Cancer Res. 10 2158–2167. 10.1158/1078-0432.CCR-03-0362 [DOI] [PubMed] [Google Scholar]
- Budde R. J., Ke S., Levin V. A. (1994). Activity of pp60c-src in 60 different cell lines derived from human tumors. Cancer Biochem. Biophys. 14 171–175. [PubMed] [Google Scholar]
- Buettner R., Mesa T., Vultur A., Lee F., Jove R. (2008). Inhibition of Src family kinases with dasatinib blocks migration and invasion of human melanoma cells. Mol. Cancer Res. 6 1766–1774. 10.1158/1541-7786.MCR-08-0169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J. P., et al. (1998). Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282 1497–1501. 10.1126/science.282.5393.1497 [DOI] [PubMed] [Google Scholar]
- Cartwright C. A., Kamps M. P., Meisler A. I., Pipas J. M., Eckhart W. (1989). pp60c-src activation in human colon carcinoma. J. Clin. Invest. 83 2025–2033. 10.1172/JCI114113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceppi P., Papotti M., Monica V., Lo Iacono M., Saviozzi S., Pautasso M., et al. (2009). Effects of Src kinase inhibition induced by dasatinib in non-small cell lung cancer cell lines treated with cisplatin. Mol. Cancer Ther. 8 3066–3074. 10.1158/1535-7163.MCT-09-0151 [DOI] [PubMed] [Google Scholar]
- Chan C. M., Jing X., Pike L. A., Zhou Q., Lim D. J., Sams S. B., et al. (2012). Targeted inhibition of Src kinase with dasatinib blocks thyroid cancer growth and metastasis. Clin. Cancer Res. 18 3580–3591. 10.1158/1078-0432.CCR-11-3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarini F., Del Sole M., Mongiorgi S., Gaboardi G. C., Cappellini A., Mantovani I., et al. (2008). The novel Akt inhibitor, perifosine, induces caspase-dependent apoptosis and downregulates P-glycoprotein expression in multidrug-resistant human T-acute leukemia cells by a JNK-dependent mechanism. Leukemia 22 1106–1116. 10.1038/leu.2008.79 [DOI] [PubMed] [Google Scholar]
- Courtneidge S. A. (2002). Role of Src in signal transduction pathways. The Jubilee Lecture. Biochem. Soc. Trans. 30 11–17. 10.1042/bst0300011 [DOI] [PubMed] [Google Scholar]
- Czernilofsky A. P., Levinson A. D., Varmus H. E., Bishop J. M., Tischer E., Goodman H. M. (1980). Nucleotide sequence of an avian sarcoma virus oncogene (src) and proposed amino acid sequence for gene product. Nature 287 198–203. 10.1038/287198a0 [DOI] [PubMed] [Google Scholar]
- Devita V. T., Serpick A. A., Carbone P. P. (1970). Combination chemotherapy in the treatment of advanced Hodgkin’s disease. Ann. Int. Med. 73 881–895. 10.7326/0003-4819-73-6-881 [DOI] [PubMed] [Google Scholar]
- Dirar A. I., Mohamed M. A., Osman W. J. A., Abdalgadir H., Khalid H. S. (2014). A Phytopharmacological review on four antitumor medicinal plants grown in sudan. Am. J. Pharm. Technol. Res. 4 28–41. [Google Scholar]
- Doyle L. A., Yang W., Abruzzo L. V., Krogmann T., Gao Y., Rishi A. K., et al. (1998). A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 95 15665–15670. 10.1073/pnas.95.26.15665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efferth T., Konkimalla V. B., Wang Y. F., Sauerbrey A., Meinhardt S., Zintl F., et al. (2008). Prediction of broad spectrum resistance of tumors towards anticancer drugs. Clin. Cancer Res. 14 2405–2412. 10.1158/1078-0432.CCR-07-4525 [DOI] [PubMed] [Google Scholar]
- Efferth T., Sauerbrey A., Olbrich A., Gebhart E., Rauch P., Weber H. O., et al. (2003). Molecular modes of action of artesunate in tumor cell lines. Mol. Pharmacol. 64 382–394. 10.1124/mol.64.2.382 [DOI] [PubMed] [Google Scholar]
- El-Sawy M. F., Bassiouny H. K., El-Magdoub A. I. (1981). Biological combat of schistosomiasis Ambrosia maritima (Damsissa) for snail control. J. Egypt Soc. Parasitol. 11 99–117. [PubMed] [Google Scholar]
- Fletcher J. I., Haber M., Henderson M. J., Norris M. D. (2010). ABC transporters in cancer: more than just drug efflux pumps. Nat. Rev. Cancer 10 147–156. 10.1038/nrc2789 [DOI] [PubMed] [Google Scholar]
- Fraga B. M. (2011). Natural sesquiterpenoids. Nat. Prod. Rep. 28 1580–1610. 10.1039/c1np00046b [DOI] [PubMed] [Google Scholar]
- Frank N. Y., Margaryan A., Huang Y., Schatton T., Waaga-Gasser A. M., Gasser M., et al. (2005). ABCB5-mediated doxorubicin transport and chemoresistance in human malignant melanoma. Cancer Res. 65 4320–4333. 10.1158/0008-5472.CAN-04-3327 [DOI] [PubMed] [Google Scholar]
- Frank N. Y., Pendse S. S., Lapchak P. H., Margaryan A., Shlain D., Doeing C., et al. (2003). Regulation of progenitor cell fusion by ABCB5 P-glycoprotein, a novel human ATP-binding cassette transporter. J. Biol. Chem. 278 47156–47165. 10.1074/jbc.M308700200 [DOI] [PubMed] [Google Scholar]
- Garcia R., Franklin R. A., Mccubrey J. A. (2006). EGF induces cell motility and multi-drug resistance gene expression in breast cancer cells. Cell Cycle 5 2820–2826. 10.4161/cc.5.23.3535 [DOI] [PubMed] [Google Scholar]
- Gerber D. E. (2008). Targeted therapies: a new generation of cancer treatments. Am. Fam. Physician 77 311–319. [PubMed] [Google Scholar]
- Gopal Y. N., Chanchorn E., Van Dyke M. W. (2009). Parthenolide promotes the ubiquitination of MDM2 and activates p53 cellular functions. Mol. Cancer Ther. 8 552–562. 10.1158/1535-7163.MCT-08-0661 [DOI] [PubMed] [Google Scholar]
- Gottesman M. M. (2002). Mechanisms of cancer drug resistance. Annu. Rev. Med. 53 615–627. 10.1146/annurev.med.53.082901.103929 [DOI] [PubMed] [Google Scholar]
- Gros P., Ben-Neriah Y., Croop J. M., Housman D. E. (1986). Isolation and expression of a complementary-DNA that confers multidrug resistance. Nature 323 728–731. 10.1038/323728a0 [DOI] [PubMed] [Google Scholar]
- Hall M. D., Handley M. D., Gottesman M. M. (2009). Is resistance useless? Multidrug resistance and collateral sensitivity. Trends Pharmacol. Sci. 30 546–556. 10.1016/j.tips.2009.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks S. K., Quinn A. M., Hunter T. (1988). The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241 42–52. 10.1126/science.3291115 [DOI] [PubMed] [Google Scholar]
- Haynes M. P., Li L., Sinha D., Russell K. S., Hisamoto K., Baron R., et al. (2003). Src kinase mediates phosphatidylinositol 3-kinase/Akt-dependent rapid endothelial nitric-oxide synthase activation by estrogen. J. Biol. Chem. 278 2118–2123. 10.1074/jbc.M210828200 [DOI] [PubMed] [Google Scholar]
- Herz W., Gage D., Kumar N. (1981). Damsinic acid and ambrosanolides from vegetative Ambrosia hispida. Phytochemistry 20 1601–1604. 10.1016/S0031-9422(00)98540-6 [DOI] [Google Scholar]
- Hirsch-Ernst K. I., Ziemann C., Schmitz-Salue C., Foth H., Kahl G. F. (1995). Modulation of P-glycoprotein and mdr1b mRNA expression by growth factors in primary rat hepatocyte culture. Biochem. Biophys. Res. Commun. 215 179–185. 10.1006/bbrc.1995.2450 [DOI] [PubMed] [Google Scholar]
- Hoffmann K., Xiao Z., Franz C., Mohr E., Serba S., Buchler M. W., et al. (2011). Involvement of the epidermal growth factor receptor in the modulation of multidrug resistance in human hepatocellular carcinoma cells in vitro. Cancer Cell Int. 11 40 10.1186/1475-2867-11-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan C., Van Schaeybroeck S., Longley D. B., Johnston P. G. (2013). Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer 13 714–726. 10.1038/nrc3599 [DOI] [PubMed] [Google Scholar]
- Huang H. S., Nagane M., Klingbeil C. K., Lin H., Nishikawa R., Ji X. D., et al. (1997). The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J. Biol. Chem. 272 2927–2935. 10.1074/jbc.272.5.2927 [DOI] [PubMed] [Google Scholar]
- Irby R. B., Yeatman T. J. (2000). Role of Src expression and activation in human cancer. Oncogene 19 5636–5642. 10.1038/sj.onc.1203912 [DOI] [PubMed] [Google Scholar]
- Jones G., Machado J., Merlo A. (2001). Loss of focal adhesion kinase (FAK) inhibits epidermal growth factor receptor-dependent migration and induces aggregation of nh(2)-terminal FAK in the nuclei of apoptotic glioblastoma cells. Cancer Res. 61 4978–4981. [PubMed] [Google Scholar]
- Katayama K., Yoshioka S., Tsukahara S., Mitsuhashi J., Sugimoto Y. (2007). Inhibition of the mitogen-activated protein kinase pathway results in the down-regulation of P-glycoprotein. Mol. Cancer Ther. 6 2092–2102. 10.1158/1535-7163.MCT-07-0148 [DOI] [PubMed] [Google Scholar]
- Kawanobe T., Kogure S., Nakamura S., Sato M., Katayama K., Mitsuhashi J., et al. (2012). Expression of human ABCB5 confers resistance to taxanes and anthracyclines. Biochem. Biophys. Res. Commun. 418 736–741. 10.1016/j.bbrc.2012.01.090 [DOI] [PubMed] [Google Scholar]
- Khalid H., Abdalla W. E., Abdelgadir H., Opatz T., Efferth T. (2012). Gems from traditional north-African medicine: medicinal and aromatic plants from Sudan. Nat. Prod. Bioprospect. 2 92–103. 10.1007/s13659-012-0015-2 [DOI] [Google Scholar]
- Kimmig A., Gekeler V., Neumann M., Frese G., Handgretinger R., Kardos G., et al. (1990). Susceptibility of multidrug-resistant human leukemia cell lines to human interleukin 2-activated killer cells. Cancer Res. 50 6793–6799. [PubMed] [Google Scholar]
- Kuete V., Efferth T. (2013). Molecular determinants of cancer cell sensitivity and resistance towards the sesquiterpene farnesol. Pharmazie 68 608–615. [PubMed] [Google Scholar]
- Li F., Zhao C., Wang L. (2014). Molecular-targeted agents combination therapy for cancer: developments and potentials. Int. J. Cancer 134 1257–1269. 10.1002/ijc.28261 [DOI] [PubMed] [Google Scholar]
- Lopez-Anton N., Hermann C., Murillo R., Merfort I., Wanner G., Vollmar A. M., et al. (2007). Sesquiterpene lactones induce distinct forms of cell death that modulate human monocyte-derived macrophage responses. Apoptosis 12 141–153. 10.1007/s10495-006-0331-2 [DOI] [PubMed] [Google Scholar]
- Lutz M. P., Esser I. B., Flossmann-Kast B. B., Vogelmann R., Lührs H., Friess H., et al. (1998). Overexpression and activation of the tyrosine kinase Src in human pancreatic carcinoma. Biochem. Biophys. Res. Commun. 243 503–508. 10.1006/bbrc.1997.8043 [DOI] [PubMed] [Google Scholar]
- Maa M. C., Leu T. H., Mccarley D. J., Schatzman R. C., Parsons S. J. (1995). Potentiation of epidermal growth factor receptor-mediated oncogenesis by c-Src: implications for the etiology of multiple human cancers. Proc. Natl. Acad. Sci. U.S.A. 92 6981–6985. 10.1073/pnas.92.15.6981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao W., Irby R., Coppola D., Fu L., Wloch M., Turner J., et al. (1997). Activation of c-Src by receptor tyrosine kinases in human colon cancer cells with high metastatic potential. Oncogene 15 3083–3090. 10.1038/sj.onc.1201496 [DOI] [PubMed] [Google Scholar]
- Martin G. S. (1970). Rous sarcoma virus: a function required for the maintenance of the transformed state. Nature 227 1021–1023. 10.1038/2271021a0 [DOI] [PubMed] [Google Scholar]
- Merfort I. (2011). Perspectives on sesquiterpene lactones in inflammation and cancer. Curr. Drug Targets 12 1560–1573. 10.2174/138945011798109437 [DOI] [PubMed] [Google Scholar]
- Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., et al. (2009). AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuswamy S. K., Siegel P. M., Dankort D. L., Webster M. A., Muller W. J. (1994). Mammary tumors expressing the neu proto-oncogene possess elevated c-Src tyrosine kinase activity. Mol. Cell Biol. 14 735–743. 10.1128/MCB.14.1.735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagane M., Levitzki A., Gazit A., Cavenee W. K., Huang H. J. (1998). Drug resistance of human glioblastoma cells conferred by a tumor-specific mutant epidermal growth factor receptor through modulation of Bcl-XL and caspase-3-like proteases. Proc. Natl. Acad. Sci. U.S.A. 95 5724–5729. 10.1073/pnas.95.10.5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nautiyal J., Majumder P., Patel B. B., Lee F. Y., Majumdar A. P. (2009). Src inhibitor dasatinib inhibits growth of breast cancer cells by modulating EGFR signaling. Cancer Lett. 283 143–151. 10.1016/j.canlet.2009.03.035 [DOI] [PubMed] [Google Scholar]
- Newman D. J., Cragg G. M. (2012). Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 75 311–335. 10.1021/np200906s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro J. M., Baker S. J., Preisinger A. C., Jessup J. M., Hostetter R., Cleary K., et al. (1989). Mutations in the p53 gene occur in diverse human tumour types. Nature 342 705–708. 10.1038/342705a0 [DOI] [PubMed] [Google Scholar]
- Nishikawa R., Ji X. D., Harmon R. C., Lazar C. S., Gill G. N., Cavenee W. K., et al. (1994). A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc. Natl. Acad. Sci. U.S.A. 91 7727–7731. 10.1073/pnas.91.16.7727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okutani Y., Kitanaka A., Tanaka T., Kamano H., Ohnishi H., Kubota Y., et al. (2001). Src directly tyrosine-phosphorylates STAT5 on its activation site and is involved in erythropoietin-induced signaling pathway. Oncogene 20 6643–6650. 10.1038/sj.onc.1204807 [DOI] [PubMed] [Google Scholar]
- Olivier M., Eeles R., Hollstein M., Khan M. A., Harris C. C., Hainaut P. (2002). The IARC TP53 database: new online mutation analysis and recommendations to users. Hum. Mutat. 19 607–614. 10.1002/humu.10081 [DOI] [PubMed] [Google Scholar]
- Pick A., Wiese M. (2012). Tyrosine kinase inhibitors influence ABCG2 expression in EGFR-positive MDCK BCRP cells via the PI3K/Akt signaling pathway. ChemMedChem 7 650–662. 10.1002/cmdc.201100543 [DOI] [PubMed] [Google Scholar]
- Piovesan B., Pennell N., Berinstein N. L. (1998). Human lymphoblastoid cell lines expressing mutant p53 exhibit decreased sensitivity to cisplatin-induced cytotoxicity. Oncogene 17 2339–2350. 10.1038/sj.onc.1202147 [DOI] [PubMed] [Google Scholar]
- Pluchino K. M., Hall M. D., Goldsborough A. S., Callaghan R., Gottesman M. M. (2012). Collateral sensitivity as a strategy against cancer multidrug resistance. Drug Resist. Updat. 15 98–105. 10.1016/j.drup.2012.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polonio-Vallon T., Kirkpatrick J., Krijgsveld J., Hofmann T. G. (2014). Src kinase modulates the apoptotic p53 pathway by altering HIPK2 localization. Cell Cycle 13 115–125. 10.4161/cc.26857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers M. H., De Grouw E. P., Van Der Reijden B. A., De Witte T. J., Jansen J. H., Raymakers R. A. (2006). ABCB1 modulation does not circumvent drug extrusion from primitive leukemic progenitor cells and may preferentially target residual normal cells in acute myelogenous leukemia. Clin. Cancer Res. 12 3452–3458. 10.1158/1078-0432.CCR-05-1945 [DOI] [PubMed] [Google Scholar]
- Roginskaya V., Zuo S., Caudell E., Nambudiri G., Kraker A. J., Corey S. J. (1999). Therapeutic targeting of Src-kinase Lyn in myeloid leukemic cell growth. Leukemia 13 855–861. 10.1038/sj.leu.2401429 [DOI] [PubMed] [Google Scholar]
- Roskoski R. (2004). Src protein-tyrosine kinase structure and regulation. Biochem. Biophys. Res. Commun. 324 1155–1164. 10.1016/j.bbrc.2004.09.171 [DOI] [PubMed] [Google Scholar]
- Rougier P., Lepère C. (2005). Metastatic colorectal cancer: first- and second-line treatment in 2005. Semin. Oncol. 32 15–20. 10.1053/j.seminoncol.2005.07.020 [DOI] [PubMed] [Google Scholar]
- Rous P. (1911). A sarcoma of the fowl transmissible by an agent separable from the tumor cells. J. Exp. Med. 13 397–411. 10.1084/jem.13.4.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed M., Kuete V., Kadioglu O., Börtzler J., Khalid H., Greten H. J., et al. (2014). Cytotoxicity of the bisphenolic honokiol from Magnolia officinalis against multiple drug-resistant tumor cells as determined by pharmacogenomics and molecular docking. Phytomedicine 21 1525–1533. 10.1016/j.phymed.2014.07.011 [DOI] [PubMed] [Google Scholar]
- Saeed M. E., Abdelgadir H., Sugimoto Y., Khalid H. E., Efferth T. (2015). Cytotoxicity of 35 medicinal plants from Sudan towards sensitive and multidrug-resistant cancer cells. J. Ethnopharmacol. 10.1016/j.jep.2015.07.005 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Salam N. A. A., Mahmoud Z. F., Ziesche J., Jakupovic J. (1984). Sesquiterpene lactones from Ambrosia maritima (Damssissa). Phytochemistry 23 2851–2853. 10.1016/0031-9422(84)83028-9 [DOI] [Google Scholar]
- Shapiro D. M. (1955). Combination chemotherapy of cancer based upon quantitative biochemical differences. Surg. Forum. 5 646–650. [PubMed] [Google Scholar]
- Shinojima N., Tada K., Shiraishi S., Kamiryo T., Kochi M., Nakamura H., et al. (2003). Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 63 6962–6970. [PubMed] [Google Scholar]
- Silva C. M. (2004). Role of STATs as downstream signal transducers in Src family kinase-mediated tumorigenesis. Oncogene 23 8017–8023. 10.1038/sj.onc.1208159 [DOI] [PubMed] [Google Scholar]
- Staley C. A., Parikh N. U., Gallick G. E. (1997). Decreased tumorigenicity of a human colon adenocarcinoma cell line by an antisense expression vector specific for c-Src. Cell Growth Differ. 8 269–274. [PubMed] [Google Scholar]
- Staudt L. M. (2010). Oncogenic activation of NF-kappaB. Cold Spring Harb. Perspect. Biol. 2:a000109 10.1101/cshperspect.a000109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summy J. M., Gallick G. E. (2006). Treatment for advanced tumors: SRC reclaims center stage. Clin. Cancer Res. 12 1398–1401. 10.1158/1078-0432.CCR-05-2692 [DOI] [PubMed] [Google Scholar]
- Sun Y., St Clair D. K., Xu Y., Crooks P. A., St Clair W. H. (2010). A NADPH oxidase-dependent redox signaling pathway mediates the selective radiosensitization effect of parthenolide in prostate cancer cells. Cancer Res. 70 2880–2890. 10.1158/0008-5472.CAN-09-4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice D. A., Biscardi J. S., Nickles A. L., Parsons S. J. (1999). Mechanism of biological synergy between cellular Src and epidermal growth factor receptor. Proc. Natl. Acad. Sci. U.S.A. 96 1415–1420. 10.1073/pnas.96.4.1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K., Cornwell M. M., Gottesman M. M., Pastan I., Roninson I. B., Ling V., et al. (1986). The mdr1 gene, responsible for multidrug-resistance, codes for P-glycoprotein. Biochem. Biophys. Res. Commun. 141 956–962. 10.1016/S0006-291X(86)80136-X [DOI] [PubMed] [Google Scholar]
- Villagomez R., Rodrigo G. C., Collado I. G., Calzado M. A., Munoz E., Akesson B., et al. (2013). Multiple anticancer effects of damsin and coronopilin isolated from Ambrosia arborescens on cell cultures. Anticancer. Res. 33 3799–3805. [PubMed] [Google Scholar]
- Vogelstein B., Lane D., Levine A. J. (2000). Surfing the p53 network. Nature 408 307–310. 10.1038/35042675 [DOI] [PubMed] [Google Scholar]
- Waldman T., Kinzler K. W., Vogelstein B. (1995). p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 55 5187–5190. [PubMed] [Google Scholar]
- Yeatman T. J. (2004). A renaissance for SRC. Nat. Rev. Cancer 4 470–480. 10.1038/nrc1366 [DOI] [PubMed] [Google Scholar]
- Zhang S., Won Y. K., Ong C. N., Shen H. M. (2005). Anti-cancer potential of sesquiterpene lactones: bioactivity and molecular mechanisms. Curr. Med. Chem. Anticancer Agents 5 239–249. 10.2174/1568011053765976 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Kretschmer N., Bauer R., Efferth T. (2015). Shikonin and its derivatives inhibit the epidermal growth factor receptor signaling and synergistically kill glioblastoma cells in combination with erlotinib. Int. J. Cancer 137 1446–1456. 10.1002/ijc.29483 [DOI] [PubMed] [Google Scholar]