Abstract
Recent studies have highlighted the role of long non-coding RNAs (lncRNAs) in carcinogenesis and have suggested that genes of this class might be used as biomarkers in cancer. However, whether lncRNAs are involved in ovarian cancer (OC) remains largely unknown. In the present study, we focused on lncRNAH19 and investigated the expression and functional role of H19 in OC. H19 expression was measured in 70 pairs of ovarian cancer tissue samplescompared with normal controls by real-time quantitative RT-PCR. The effects of H19 on ovarian cancer cells were studied by RNA interference approach. Apoptosis and cell cycle were analyzed by flow cytometry. Cells viability was evaluated using cell counting Kit-8. Our results demonstrated that that H19 silencing inhibited OV90 and SKOV3 OC cell proliferation in vitro. Further investigation into the mechanisms responsible for the growth inhibitory effects by H19 silencing revealed that its knockdown resulted in the induction of cell cycle arrest and apoptosis through certain cell cycle-related and apoptosis-related proteins. Together, our data suggest that LncRNAH19 plays an important role in OC cell proliferation and contributes to a better understanding of the importance of dysregulated lncRNAs in OC progression.
Keywords: Ovarian cancer, long non-coding RNA H19, proliferation, apoptosis, cell cycle
Introduction
Ovarian cancer is the fifth leading cause of cancer-related death worldwide and is still the most lethal gynecologic malignancy among women [1]. One of the main reasons for such a high mortality is that the majority of cases are diagnosed at an advanced stage. Therefore, looking for new biomarkers and therapeutic targets has been a major research focus for many years.
Recently, various lncRNAs have been implicated in many human diseases including cancers, and emerging studies are beginning to unravel the molecular mechanisms underlying lncRNA function in these pathological processes [2-5]. H19 gene is known as a 2.3 kb of long noncoding RNA molecules, the gene expression of matrilineal and patrilineal imprinting, present in mammals evolved on the conservative is one of the earliest identified imprinted genes [6-9]. Increasing evidence showed that H19 expression was upregulated in breast, liver, endometrial, lung, cervical, esophageal, and bladder tumors and promoted cancer cell proliferation, suggesting an oncogenic function for H19 [10-18]. A recent study has demonstratedthat the transcription factor c-Myc causes H19 upregulation that plays a crucial role in cellular transformation [11]. Despite the importance of H19 in various cancers, the role of H19 in ovarian cancer remains unknown.
In the present study, we sought to examine the expression and functional role of H19 in human ovarian cancer. We confirmed that H19 expression is markedly increased in ovarian cancer tissues compared with adjacent normal tissues. Ectopic expression of H19 increased cell proliferation, whereas H19 siRNA treatment contributed to cellapoptosis through certain cell cycle-related and apoptosis-related proteins in human ovarian cancer cell lines. To our knowledge, our findings for the first time revealed the involvement of H19 in human ovarian cancer.
Materials and methods
Tissue samples
All specimens were handled and made anonymous according to the ethical and legal standards. Paired tissue specimens (tumor and adjacent normal tissues) from 70 patients with ovarian cancer were obtained and histologically confirmed by a pathologist at Changhai Hospital, Second Military Universityfrom January 2010 to December 2010. All samples were derived from patients who had not received adjuvant treatment including chemotherapy or radiotherapy prior to the surgery in order to eliminate potential treatment-induced changes to gene expression profiles. After excision, tissue specimens were immediately frozen in liquid nitrogen for subsequent analysis. Patient descriptions are detailed in Table 1.
Table 1.
Association between the expression of H19 and clinic opathological parameters of patients
| Clinical pathological parameters | Total | H19 | P value | |
|---|---|---|---|---|
|
| ||||
| n=70 | High No. cases | Low No. cases | ||
| Age (years) | ||||
| ≤50 | 28 | 24 | 4 | |
| >50 | 42 | 33 | 9 | 0.549 |
| FIGO disease stage | ||||
| I~II | 21 | 13 | 8 | |
| III~IV | 49 | 44 | 5 | 0.041 |
| Grade | ||||
| Well (G1) | 28 | 23 | 5 | |
| Moderately (G2) | 11 | 8 | 3 | |
| Poorly (G3) | 31 | 26 | 5 | 0.052 |
| Lymph node | ||||
| Negative | 33 | 27 | 6 | |
| Positive | 37 | 30 | 7 | 0.729 |
| Residual disease | ||||
| ≤2 cm | 49 | 41 | 8 | |
| >2 cm | 21 | 16 | 5 | 0.285 |
| Ascites | ||||
| Absent | 32 | 14 | 6 | |
| Present | 38 | 16 | 7 | 0.789 |
FIGO=International Federation of Gynecology and Obstetrics.
Cell culture
Ovarian cancer cell lines SKOV3, OV90, TOV112D and ES2 were purchased from American Type Culture Collection. Cells were maintained in DMEM/F12 (Invitrogen, Carlsbad, CA) supplemented with 1% sodium pyruvate (Invitrogen), 0.2% non-essential amino acids (Invitrogen), and 5% FBS in a humidified atmosphere containing 5% CO2 at 37°C.
Immortalized normal ovarian surface epithelial cell line IOSE80 was obtained as a generous gift from the laboratory of Dr Nelly Auersperg (The University of British Columbia, Vancouver, Canada). The cells were grown in a 1:1 combination of two media, Medium 199 (Invitrogen) and MCDB 105 (Cell Applications Inc., San Diego, CA) with 10% FBS in a humidifiedatmosphere containing 5% CO2 at 37°C. Ovarian cancercelllines and normal ovarian cell lines were cultured in monolayerin the above stated conditions.
Quantitative real-time RT-PCR
Total RNA was extracted using Trizol reagents (Invitrogen) according to the manufacturer’s instructions and diluted to 200 ng/mL. Then, quantitative real-time RT-PCR (qRT-PCR) was performed using One Step SYBR® Prime Script™ RT-PCR Kit II (TaKaRa, China) according to standard protocol. GAPDH gene was used as an internal control. The relative gene expression was calculated using 2-ΔΔCt method. Primers used in qRT-PCR were as follows:
H19: 5’-GCGGGTCTGTTTCTTTACTTC-3’ (forward), 5’-TTTCATGTTGTGGGTTCTGG-3’ (reverse); GAPDH: 5’-GGACCAATACGACCAAATCCG-3’ (forward), 5’-AGCCACATCGCTCAGACAC-3’ (reverse).
Western blotting analysis
Cells were harvested and homogenized with cell lysis buffer (Beyotime, China). Then, the homogenates were centrifuged for 30 min at 4°C, 12000 rpm, and the supernatants were collected as protein samples. Protein amounts were measured using BCA Protein Assay Kit (Beyotime, China). Equal amounts of protein samples were separated by denaturing 10% SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes. Membranes were incubated in a 5% skim milk TBST blocking solution at room temperature (RT) for 1 h. And, membranes were incubated with agitation at 4°C overnight with specific primary antibodies against Caspase-3 (1:1000), Caspase-8 (1:2000), Caspase-9 (1:1000), Bax (1:1500), Bcl-2 (1:1000), Cyclin A2 (1:2000), Cyclin B1 (1:1000), Cyclin D1 (1:5000), Cyclin E1 (1:1500), CDC2 (1:2000) and p-CDC2 (1:1000). Then, membranes incubated by secondary antibodies (1:1000) conjugated with horseradish peroxidase (HRP) at RT for 50 min. Finally, protein bands were visualized using an enhanced chemiluminescence (ECL) Western blotting detection system (GE Healthcare, Amersham, UK).
Small interfering RNA (siRNA)
Cells were seeded (2×105 cells/well) in six-well plates. After incubation for 24 hours, cells were transfected with siRNA targeting H19, or negative control (si-Scramble) using Lipofectamine 2000 transfection reagent. The sequences of siRNAs were as follows:
si-H19-1: 5’-GGAGAGTTAGCAAAGGTGACATCTT-3’, si-H19-2: 5’-AGAGTTAGCAAAGGTGACATCTTCT-3’, si-H19-3: 5’-CGGCAAGAAGCGGGTCTGTTTCTTT-3’, si-Scramble: 5’-GGATGATCGAAGATGAGACTAGCTT-3’.
Cell proliferation assay
The proliferation of ovarian cancer cells was evaluated using cell counting Kit-8 (Beyotime, China) according to the manufacturer’s instruction. Briefly, cells were transferred into a 96-well cell culture plate, with 200 μL suspension per well, and grown overnight. After 24 h, the cells were transfected with siRNAs or si-Scramble. All groups were performed in triplicate. At 1 d, 2 d, 3 d, 4 d and 5 d, 20 Μl CCK-8 was added to each well respectively, and then the plates were incubated for 2 h. Finally, absorbance was measured at 490 nm with a microplate reader (BioRad).
Determination of apoptosis
The extent of apoptosis was determined by the flow cytometric measurement through Annexin V-FITC apoptosis detection kit (Beyotime, China). Cells treated as above described. After 4 d, cells were harvested and washed twice with cold PBS. Then, cells were stained in 1 mL Annexin V binding buffer with 10 μL of PI solution and 5 μL of Annexin V -FITC for 10 min at RT and analyzed by flow cytometry.
Cell cycle analysis
Ovarian cancer cells were seeded at a density of approximately 5×105 in 100 mm plates and transfection with siRNAs for 48 h. Cell cycle was analyzed by flow cytometry with propidium iodide (PI) staining using Cell Cycle Analysis Kit (Beyotime, China). Briefly, cells were harvested and washed with PBS and fixed with 70% ice-cold ethanol at 4°C overnight. Then cells were incubated in a PBS solution containing 10 mg/mL RNase and 1 mg/mL PI for 1 h at room temperature. Finally, the percentage of cells in different phases of the cell cycle was measured by flow cytometry (FACSCalibur, BD Biosciences, NY, US). All samples were examined in triplicate.
Statistical analysis
Data are reported as mean ± standard deviation (SD). Statistical significance was determined using Double-sided Student’s t test. Multiple groups were analyzed using ANOVA. A P value of less than 0.05 was considered to be significant.
Results
H19 was highly expressed in ovarian cancer tissues and cell lines
To assess the role of H19 in ovarian cancer progression, we firstexamined the H19 expression levels in ovarian cancer tissues andovarian cancer cell using quantitative real-time PCR. We verified that H19 expression levels are significantly upregulated in most ovarian cancer tissues (Figure 1A). H19 mRNA level in ovarian cancer tissues were nearly 2-fold enhanced compare to those in adjacent non-tumor samples. Subsequently, we also detected the expression of H19 in 4 human ovarian cancer cell lines including SKOV3, OV90, TOV112D and ES2 using qRT-PCR. Immortalized normal ovarian surface epithelial cell line IOSE80 used as a negative control. High expression of H19 in ovarian cancer cell lines was also observed, especially in OV90 and SKOV3 cells (Figure 1B). Also, a direct correlation between clinical cancer stages and H19 was noted. There was asignificantly positive correlation between H19 expression andtumor stages (P<0.05) and tumor size (P<0.05), while no correlation was observed between H19 expression and age, Grade, Lymph node or Ascites (Table 1). These data suggest that upregulated expression of H19 is related to the progression of ovarian cancer.
Figure 1.
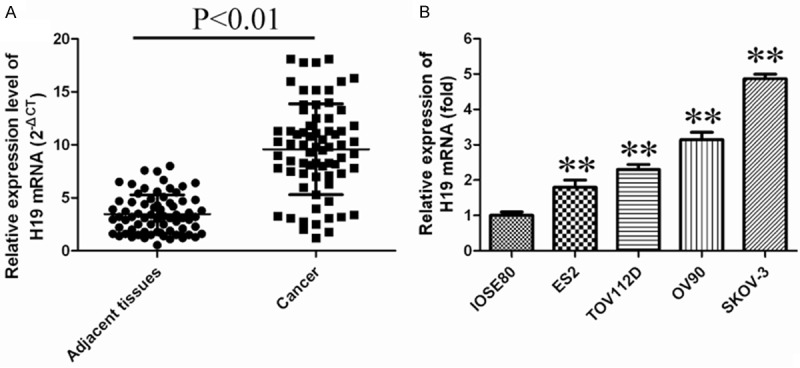
LncRNAH19 levels were up-regulated in ovarian cancer cells and tissues. A. Relative expression of lncRNAH19 in ovarian cancer tissues (n=70) compared with adjacent non-tumor normal tissues (n=70). LncRNAH19 expression was examined by qRT-PCR and normalized to GAPDH expression. B. H19 expression wasassessed by real-time PCR using SYBRGreen in ovarian cancer cell lines. H19 levelswere normalized to β-actin levels in IOSE80cells. Asterisks indicate values that aresignificantly different from that in IOSE80 (P<0.05).
Silencing of H19 inhibits cell growth and induces apoptosis in human ovarian cancer cell lines
To study the biological role of H19 in cell growth, siRNAexperiment was used to down-regulate H19 expression in OV90. As shown in Figure 2A, the mRNA level of H19 in siRNAs transfected cells down to 0.8-fold, 0.4-fold and 0.6-flod respectively, comparing with scramble group, and down regulation of H19 significantly inhibited OV90 cell growth (Figure 2B). Similarly, down-regulated H19 expression in SKOV3 cells suppressed cell growth (Figure 2C, 2D). These data suggest that H19 positively regulates growth of ovarian cancer cells.
Figure 2.
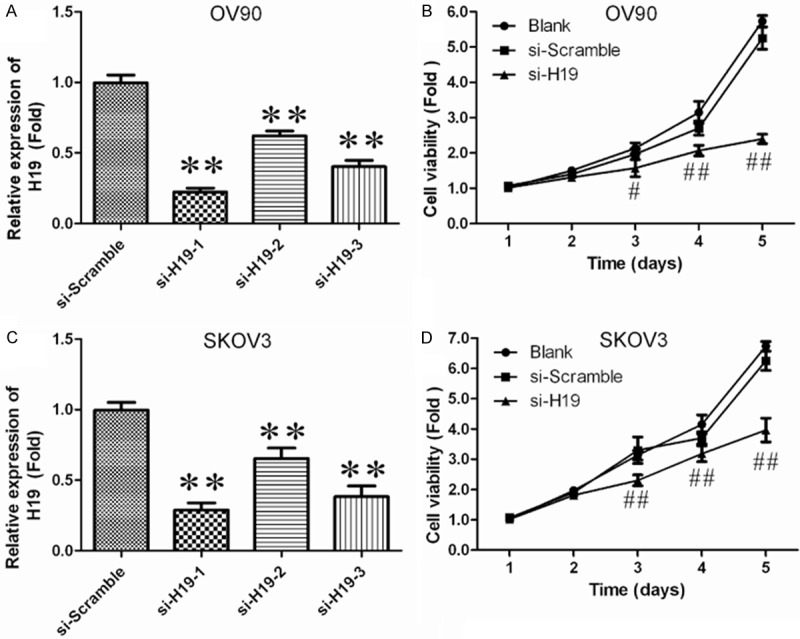
Effects of lncRNAH19 knockdown on cell growth in ovarian cancer cells. A. Silencing efficiency of si-H19-1, si-H19-2 and si-H19-3 was verified in OV90by qRT-PCR. *P<0.05, **P<0.01 vs. si-Scramble group. B. OV90 cells were plated in a 24-well plate and transfected with si-H19-2, and cell proliferation was determined at the indicated time points using CCK-8 kit. #P<0.05, ##P<0.01 vs. Blank group. C. Silencing efficiency of si-H19-1, si-H19-2 and si-H19-3 were verified in SKOV3 by qRT-PCR. *P<0.05, **P<0.01 vs. si-Scramble group. *P<0.05, **P<0.01 vs. si-Scramble group. D. SKOV3 cells were plated in a 24-well plate and transfected with si-H19-2, and cell proliferation was determined at the indicated time points using CCK-8 kit. #P<0.05, ##P<0.01 vs. Blank group.
H19 promotes G1 arrest and causes apoptosis in ovarian cancer cells
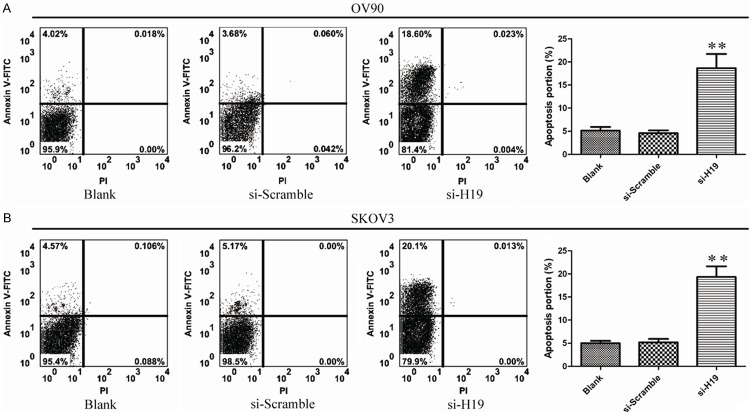
To investigate the influence of apoptosis caused by silencing of H19. Ovarian cancer cells apoptosis was measured by Annexin-V FITC/PI double staining assay. As shown, the percentages of apoptotic cells were significantly increased in the treated group compared to control group (P<0.05; Figure 3A and 3B). These results showed that H19 knockdown increased apoptotic rate of ovarian cancer cells. To further determine the mechanism of H19 on apoptosis, the expression of Caspase-3, Caspase-8, Caspase-9, Bax and Bcl-2 was analyzed by Western Blot and qPCR. As shown in Figure 4, the expression of Caspase-3, Caspase-9 and Bax were significantly increased and Bcl-2 was significantly decreased with treatment of si-H19-2. But the expression of Caspase-8 has no obvious change. These dates provided evidence that silencing H19 induced apoptosis through mitochondrial and caspases dependent pathway.
Figure 3.
The effect of lncRNAH19 knockdown on ovarian cancer apoptosis in vitro. A, B. Flow cytometry analyze apoptosis of H19knockdown cells stained with Annexin V-FITC and PI. The data represent the mean ± SD from three independent experiments. **P<0.01.
Figure 4.
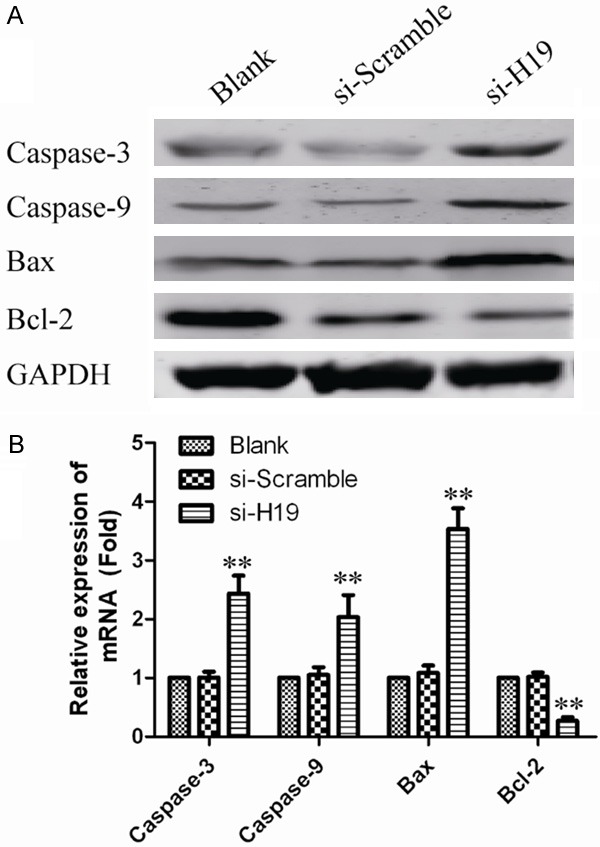
The effect of lncRNAH19 knockdown on the expression of apoptosis relevant protein. A. Caspase3, Caspase8, Caspase9, Bax and Bcl-2 were analyzed through Western blot. GAPDH was used as an internal control. B. Quantifying Western blot using BandScan 5.0 software. Values represent mean ± SD of three independent experiments. **P<0.01 vs. GAPDH group.
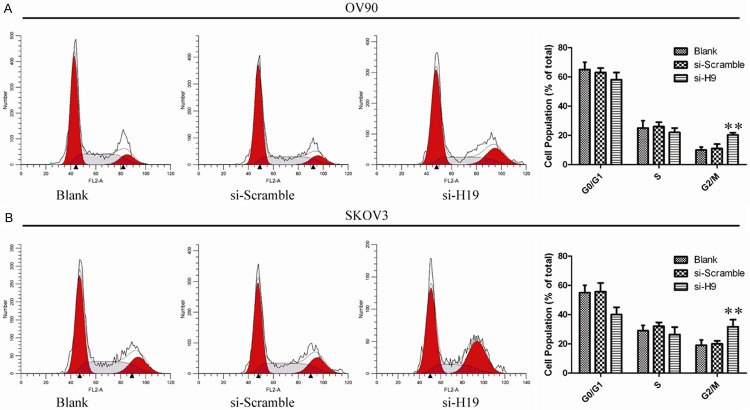
To determine whether the effects of H19 on the proliferation of ovarian cancer cells were mediated by inhibition of cell cycle progression, we followed cell cycle progression in OV90 and SKOV3 cells with flow cytometry. Compared with untreated cells and si-scramble transfected cells, H19 silenced cells were arrested in G2/M phase (Figure 5). In order to further illuminate the G2/M arrest observed, we examined cyclins A2, B1, D1, E1 as well as Cdc2 (cyclin dependent kinases), p-Cdc2 (phosphorylated Cdc2). As shown in Figure 6, H19 silencing led to a decrease in cyclins A2, B1 as well as a decrease in protein expression of p-Cdc2. However, the expression levels of cyclin D1, cyclin E1 and total Cdc2 remained unchanged. These data suggest that G2/M arrest is associated with activation of the cyclin B1/Cdc2.
Figure 5.
The effect of lncRNAH19 knockdown on ovarian cancer cell cycle. A, B. The bar chart represents the percentage of cells in G0/G1, S, or G2/M phase, as indicated. The data represent the mean ± SD from three independent experiments. **P<0.01.
Figure 6.
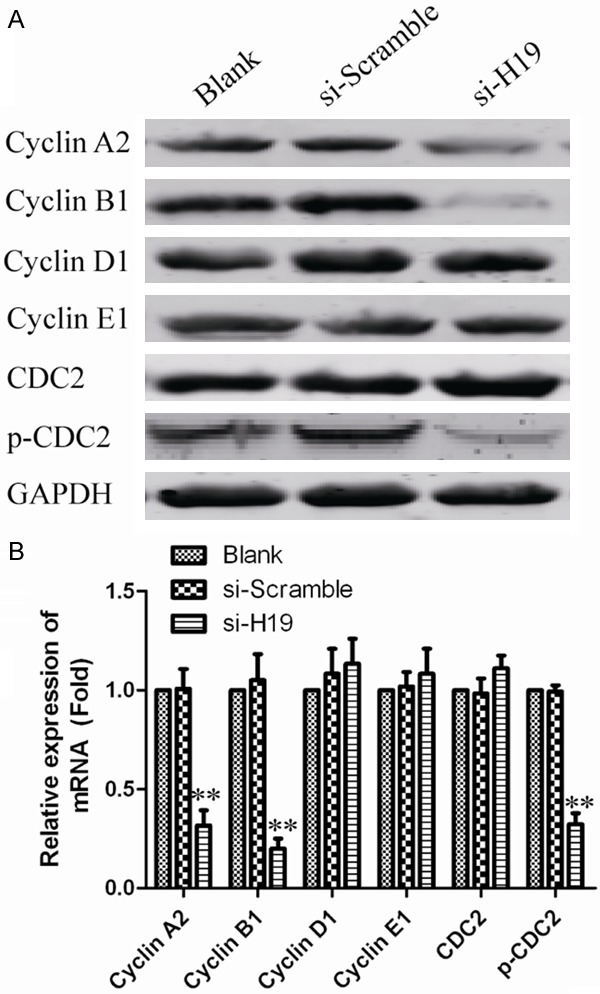
The effect of lncRNAH19 knockdown on the expression of cell cycle relevant protein. LncRNAH19 silenced ovarian cancer cells and controls were subjected to western blot for determining the expression levels of cell cycle related proteins. GAPDH was used as an internal control. B. Quantifying Western blot using BandScan 5.0 software. Values represent mean ± SD of three independent experiments. **P<0.01 vs. GAPDH group.
Taken together, these data indicated that H19 plays a critical role inovarian cancer and silencing of H19 inhibits cells growth and induces apoptosis in ovarian cancer cells.
Discussion
In the present study, our experiments demonstrate that lncRNAH19 plays a critical role inovarian cancer. Remarkably, H19 was found to be upregulated ovarian cancer tissues and cell lines, which was consistent with previous findings that H19 is reduced in several human tumors and cancer cell lines compared with normal adjacent lung tissues and normal cell lines [19]. In addition, the effects of this lncRNA on cell proliferation suggest that it promotes tumorigenesis in ovarian cancer.
Long non-coding RNAs (lncRNAs) represent a new class of non-protein-coding RNAs which are longer than 200 bases, and do not function as templates for protein synthesis [20]. Previous studies have proved that lncRNAs play a critical role in the development and progression of cancer, such as HOTAIR, MALAT-1 and H19 have been implicated as playing a functional rolein carcinogenesis or cancer growth [21-26]. Additionally, multiple studies have indicated that the expression levels of lncRNAs are dysregulated in different kinds of tumors, including ovarian cancer. For example, Endo et al. found HOTAIR expression is increased in primary breasttumors and metastases, and the HOTAIR expressionlevel in primary tumors is a powerful predictor of eventual metastasis and death [27]. Zheng et al. suggested that expression of lncRNA MALAT1 was increased in colorectal cancer compared to that in the adjacent normal tissues, patients with higher expression of MALAT1 showed a significantly worse overall survival and disease free survival than that patients with lower expression of MALAT1 [28].
Recently, emerging studies have shown that the dysregulation of H19 is associated with diverse human genetic disorders and cancers, and the dual roles for H19 acting either as a tumor suppressoror an oncogene have been suggested [11,16]. Ying et al. found that ectopic expression of H19 increases gastric cancer cell proliferation. Feng et al. also demonstrated that ectopic expression of H19 increases cell proliferation, whereas H19 siRNA treatment contributes to cell apoptosis in AGS human gastric cancer cell lines [29]. These reports indicated that H19 may serve as a therapeutic target for tumors. Despite these promising findings, investigations related to H19 function inhematologic malignancies appear to be fragmentary, and are notas extensively depicted as those in solid tumors.
In the study, we found that H19 expression levels are remarkably increased in ovarian cancer cells and ovarian cancer tissues compared with normal control. This implies that H19 may also play a role in the development and progression of human ovarian cancer. Hence, we knockdown H19 in human ovarian cancer cell lines by siRNA. We found silencing H19 dramatically suppressed human ovarian cancer cell growth, induced cell apoptosis and arrested cell division at the G2/M phase. Thus, we assumed H19 is critical in maintaining ovarian cancer cells growth.
According to the results of our study, lncRNAH19 could serve as a potential therapeutic target for the following reasons. First, lncRNAH19 expression is significant increased in ovarian cancer tissues and ovarian cancer cell lines. The second reason is lncRNAH19 could predict the prognosis of patients with ovarian cancer. Third, Down-regulated expression of lncRNAH19 could inhibit ovarian cancer cells proliferation, and induces apoptosis.
In summary, our findings demonstrated that lncRNAH19 plays a vital role in the development and progression of ovarian cancer. The development of H19-based therapeutic strategies for the downregulation of such oncogenic lncRNAs may provide a novel and promising alternative therapeutic approach for future ovarian cancer treatment.
Disclosure of conflict of interest
None.
References
- 1.Lengyel E. Ovarian cancer development and metastasis. Am J Pathol. 2010;177:1053–64. doi: 10.2353/ajpath.2010.100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32:473–480. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- 8.Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forne T, Jammes H, Ainscough JF, Surani MA, Journot L, Dandolo L. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136:3413–3421. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- 9.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H, Min W, Bennett AM, Gregory RI, Ding Y, Huang Y. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ariel I, Sughayer M, Fellig Y, Pizov G, Ayesh S, Podeh D, Libdeh BA, Levy C, Birman T, Tykocinski ML, de Groot N, Hochberg A. The imprinted H19 gene is a marker of early recurrence in human bladder carcinoma. Mol Pathol. 2000;53:320–323. doi: 10.1136/mp.53.6.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, Tsao MS, Penn LZ. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66:5330–5337. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- 12.Berteaux N, Lottin S, Monte D, Pinte S, Quatannens B, Coll J, Hondermarck H, Curgy JJ, Dugimont T, Adriaenssens E. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem. 2005;280:29625–29636. doi: 10.1074/jbc.M504033200. [DOI] [PubMed] [Google Scholar]
- 13.Byun HM, Wong HL, Birnstein EA, Wolff EM, Liang G, Yang AS. Examination of IGF2 and H19 loss of imprinting in bladder cancer. Cancer Res. 2007;67:10753–10758. doi: 10.1158/0008-5472.CAN-07-0329. [DOI] [PubMed] [Google Scholar]
- 14.Hibi K, Nakamura H, Hirai A, Fujikake Y, Kasai Y, Akiyama S, Ito K, Takagi H. Loss of H19 imprinting in esophageal cancer. Cancer Res. 1996;56:480–482. [PubMed] [Google Scholar]
- 15.Kondo M, Suzuki H, Ueda R, Osada H, Takagi K, Takahashi T, Takahashi T. Frequent loss of imprinting of the H19 gene is often associated with its overexpression in human lung cancers. Oncogene. 1995;10:1193–1198. [PubMed] [Google Scholar]
- 16.Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, Hochberg A, Galun E. The H19 non-coding RNA is essential for human tumor growth. PLoS One. 2007;2:e845. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanos V, Ariel I, Prus D, De-Groot N, Hochberg A. H19 and IGF2 gene expression in human normal, hyperplastic, and malignant endometrium. Int J Gynecol Cancer. 2004;14:521–525. doi: 10.1111/j.1048-891x.2004.014314.x. [DOI] [PubMed] [Google Scholar]
- 18.Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ, Kwok TT. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350–358. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]
- 19.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333:213–221. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 20.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 21.Benetatos L, Vartholomatos G, Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int J Cancer. 2011;129:773–779. doi: 10.1002/ijc.26052. [DOI] [PubMed] [Google Scholar]
- 22.Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S, Negrini M, Miotto E, Croce CM, Patel T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750–4756. doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 24.Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 25.Matouk IJ, Mezan S, Mizrahi A, Ohana P, Abu-Lail R, Fellig Y, Degroot N, Galun E, Hochberg A. The oncofetal H19 RNA connection: hypoxia, p53 and cancer. Biochim Biophys Acta. 2010;1803:443–451. doi: 10.1016/j.bbamcr.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Gejman R, Mahta A, Zhong Y, Rice KA, Zhou Y, Cheunsuchon P, Louis DN, Klibanski A. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 2010;70:2350–2358. doi: 10.1158/0008-5472.CAN-09-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woo CJ, Kingston RE. HOTAIR lifts noncoding RNAs to new levels. Cell. 2007;129:1257–1259. doi: 10.1016/j.cell.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 28.Zheng HT, Shi DB, Wang YW, Li XX, Xu Y, Tripathi P, Gu WL, Cai GX, Cai SJ. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7:3174–3181. [PMC free article] [PubMed] [Google Scholar]
- 29.Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]