Abstract
Epithelial ovarian cancer is one of the most lethal of gynecological malignancies. Due to its lack of early symptoms, detection usually occurs when the tumor is no longer confined to the ovary. We previously identified Fbxw15, a gene encoding an F-box protein in the mouse ovary, and showed that its expression is developmentally regulated. Here we report the molecular analysis of its human homologue, FBXW12 in epithelial ovarian tumors. To search for FBXW12 gene mutations, we PCR-amplified and sequenced the coding region of FBXW12, the gene’s 5-untranslated region and the proximal promoter in each of 30 EOC tumors. Promoter methylation was determined by DNA bisulfite conversion, followed by methylation specific PCR. FBXW12 intracellular localization was identified by means of immunohistochemistry. A complete deletion of the gene’s coding region, the 5’-UTR and the proximal promoter, was observed in 3 EOC samples. Eight of the remaining 27, had a deletion of the 5’-UTR, and the proximal promoter. FBXW12 mRNA was detected in 2 of the 19 samples without deletions. The methylation specific PCR results demonstrated CpGs methylation in the FBXW12 proximal promoter. Immunohistochemistry assay revealed that within the normal ovary, FBXW12 has an oocyte specific expression, whereas in EOC samples it is present in the ovarian surface epithelium. Our results indicate that the FBXW12 gene is deleted in approximately ten percent of the EOC cases studied; such deletions comprised either the FBXW12 promoter or the mRNA-encoding region. Moreover, FBXW12 could be epigenetically silenced by CpGs methylation in some of these EOC cases.
Keywords: F-box proteins, epithelial ovarian cancer
Introduction
Ovarian cancer represents a leading cause of death in women, and one of the most lethal of gynecological malignancies [1]. Unfortunately, due to the lack of specific symptoms at early stages of development, it is generally diagnosed when metastasis has already occurred, which leads to a poor prognosis and less than 20% five-year survival rate after the initial diagnosis [2]. Epithelial ovarian cancer (EOC) accounts for 90% of all ovarian malignancies [3]. It usually occurs sporadically, which results in a low (2-5%) life-time risk for this type of cancer. Women bearing a genetic predisposition to EOC represent only 5 to 10% of the affected individuals. Most of them carry germ line mutations in tumor suppressor genes like BRCA1, BRCA2 and/or p53, which are involved in DNA damage response and the regulation of cell cycle and cell proliferation [4,5].
Like most human cancers, EOC involves disruption of various molecular components including those participating in the G1-S phase transition of the cell cycle [6]. The SCF ubiquitin ligase complex may be especially vulnerable to such disruption, because of its central involvement in the degradation of proteins regulating G1-S cell-cycle progression [7,8]. E3 ubiquitin protein ligase complexes are termed SCFs, because their basic components include Scp1, Cullin and an F-box protein [9]. SCFs mediate the phosphorylation-dependent ubiquitination of proteins, and appear to be essential components of proteolytic events regulating not only cycle progression, but also signal transduction [9,10]. Within the SCF complex, F-box proteins provide substrate specificity by recognizing and recruiting proteins to the complex. The recruited substrates are then targeted by ubiquitin and degraded by the 26 s proteasome [9,11].
An involvement of the SCF complex in cancer was first suggested by its role in the stabilization of p27, a tumor suppressor gene. Li and colleagues, demonstrated that 1, 25-dihydroxyvitamin D3 (1, 25(OH)2D3) arrests in G1 an ovarian cancer cell line (OVCAR3), by increasing the abundance of p27. 1, 25(OH)2D3 decreases the amounts of cyclin E and cyclin dependent kinase 2. This results in reduced p27 phosphorylation, which in turn reduces p27 interaction with the F box protein Skp2 leading to diminished p27 proteosome-mediated degradation [12]. Another F-box encoding gene implicated in cancer is FBXW7. This gene encodes an F-box protein responsible for regulating levels of Cyclin E, NOTCH and other oncoproteins like c-MYC and c-JUN. Mutations of FBXW7 have been found in different types of cancer, including ovarian carcinomas [13-16]. The latter suggests that inactivating mutations in FBXW7 may lead to the abnormal expression of pro-oncogenic genes [17,18]. An additional example was provided by Gütgemann and colleagues, who reported significant overexpression of Emi1 in ovarian clear cell carcinoma. Emi1 is an F-box protein implicated in the inactivation of the anaphase promoting complex/cyclosome (APC/C) ubiquitin ligase pathway. It blocks the association of this ubiquitin ligase with its substrates, suggesting that Emi1 dependent APC/C misregulation leads to mitotic alterations and genomic instability, which contributes to tumorigenesis [19]. It has also been postulated that F-box proteins act as tumor suppressors, which are transcriptionally silenced by epigenetic modifications leading to a reduced degradation of downstream targets and subsequently to ovarian cancer [20,21].
In an earlier study we employed DNA microarrays to interrogate the mouse genome, and identified an oocyte-specific gene whose expression increases biphasically during feto-neonatal development. This gene, termed Fbxw15, is a novel member of the F-box proteins family, which regulates cell proliferation by interacting with the histone acetyltransferase binding to origin recognition complex (HBO1), in order to mediate its ubiquitination and further degradation in the cytoplasm [22,23]. Here we report that in eleven of thirty EOC cases, the coding region, the 5’UTR or the proximal promoter region of the human homologue, FBXW12 are deleted and that in some of these thirty EOC cases, the proximal promoter could be epigenetically silenced by CpGs methylation. Moreover, FBXW12 is identified as oocyte-specific within the normal ovary, while its localization changes in EOC, where it is detected in the neoplastic ovarian surface epithelium.
Materials and methods
Clinical specimens
Thirty primary epithelial ovarian tumors collected at the Women’s Hospital in Mexico City were snap-frozen in liquid nitrogen at the time of surgery. All patients were of Mexican-Mestizo ethnic origin with no family history of ovarian cancer, aged 17 to 69 years (mean = 44.4±18.6). Written informed consent was obtained from all patients before enrollment in the study. In addition, 9 formalin-fixed, paraffin-embedded samples (2 healthy ovarian tissues, 4 benign tumours and 3 EOCs), obtained also from the Women’s Hospital of the Secretaría de Salud were processed for immunohistochemistry analysis. The study was performed after obtaining authorization from the National Committee of Scientific Research and in accordance with the respective international guidelines. Histological information on the type of cancer as well as grade of malignancy according to the International Federation of Gynecology and Obstetrics (FIGO) are shown in Table 1.
Table 1.
Tumor classification of the 30 samples analyzed based on histology and grade of malignancy. For labeling purposes, each tumor sample was numbered upon receipt in a consecutive order
| Tumor type* | Number of samples | Stage (grade of malignancy) | Number assigned |
|---|---|---|---|
| Serous | 21 | 1c, n = 5 | 3, 4, 23, 24, 26 |
| 2a, n = 9 | 5, 8, 19-21, 27-30 | ||
| 3a, n = 6 | 1, 6, 7, 9,16,18 | ||
| 4, n = 1 | 2 | ||
| Endometroid | 3 | 1c | 10, 11, 25 |
| Mucinous | 4 | 1b | 12, 13, 17, 22 |
| Mucinous borderline | 1 | 14 | |
| Mixt serous and endometroid | 1 | 1b | 15 |
According to the International Federation of Gynecology and Obstetrics (FIGO).
DNA isolation
Genomic DNA isolated by standard techniques [24] from leukocytes of healthy women was employed as positive control. Genomic DNA from the 30 tumor samples was isolated following Sambrook’s protocol for rapid isolation of mammalian DNA [25]. Briefly, all frozen samples were grinded to a powder using a mortar and pestle, both prechilled in liquid nitrogen. Twenty mg of each sample were placed in a 1.5 ml microfuge tube containing 600 µl of ice cold cell lysis buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA pH 8.0, 0.1% SDS) and quickly homogenized with 30-50 strokes of a microfuge pestle. The homogenates were incubated overnight at 37°C with 3 µl of proteinase K (30 U/mg) (Promega Corp., Madison, WI). All digestions were allowed to cool down to room temperature before incubation with 3 µl of DNAse-free RNAse (10 U/µl) (Promega Corp.), for 1 hour at 37°C. Thereafter, 200 µl of potassium acetate solution (60 ml of 5 M CH3CO2K, 11.5 ml of glacial acetic acid, 28.5 ml of sH2O) were added to each sample and the contents were mixed by vortexing vigorously for 20 seconds. To pellet the protein/SDS complexes, the samples were centrifuged for 3 minutes at 4°C. To precipitate the DNA, supernatants were transferred to a new microfuge tube containing 600 µl of isopropanol. DNA was recovered by centrifugation again at maximum speed for 1 minute at room temperature. The supernatant of each sample was removed by aspiration, following the addition of 600 µl of 70% ethanol to each DNA pellet and then centrifugation, the supernatants were removed by aspiration and each DNA pellet was allowed to air-dry for approximately 10 minutes. DNA was dissolved in 40 µl of Tris-EDTA (pH 7.6) and quantitated by measuring absorbance at 260 nm, followed by storage at -85°C until used.
DNA sequence analysis
The 10 coding exons of the FBXW12 transcript variant 1 (GeneBank: NM_207102.2), the 5’-untranslated region encoded by exons 1 and 2, and the proximal promoter (360 bp upstream from the transcriptional start site, TSS), were amplified by PCR. All reactions were carried out in a final volume of 20 µl including: 0.5 µg of DNA template, 10 µl of PyroStartTM Fast PCR Master Mix (2×) (Fermentas International, Inc., Glen Burnie, MD, USA) and 15 pmol of each primer set (Table 2). The primer sequences derived from GenBank Accession No. NT_022517. The PCR conditions used for amplification of all exons were 1 min at 95°C for an initial denaturation step, followed by 30 cycles of denaturation for 5 sec at 94°C, 10 sec of annealing at specific temperatures for each pair of primers (Table 2), and 25 sec of extension at 72°C, ending with a 10 sec extension at 72°C. The PCR products were electrophoresed on a 2% agarose gel stained with ethidium bromide and the corresponding bands were purified with the QIAEX II Gel Extraction Kit (QIAGEN, Inc., Valencia, CA). Purified samples were then sequenced on an Applied Biosystems DNA Sequencer model 377, using the Big DyeTM Terminator Sequencing Ready Reaction kit version 3.1 (Applied Biosystems, Foster City, CA). Sequencing was performed following the protocol supplied by the manufacturer. Sequencing results were compared against the GenBank sequence database by means of the Basic Local Alignment Search Tool algorithm of the National Center for Biotechnology Information (NCBI).
Table 2.
Primers complementary to Fbxw12 genomic sequence
| Primer Number | Primer Sequence | Amplicon Size (Base Pairs) | Annealing Temperature (°C) Per Primer Pair | Primer Location Within Genomic Contig Nt022517.17 (Base Pair Number) |
|---|---|---|---|---|
| 5’ flanking region primers | ||||
| 1AF | 5’-TCCTCGCATGATATGAACAGGGCT-3’ | 790 | 60 | 2468-2491 |
| 1AR | 5’-ATGCCCATCTGTGTGCGGTAGAAA-3’ | 3257-3234 | ||
| Coding region primers | ||||
| 2F | 5’-AGAAAGGAAAGTGGATGTGGGT-3’ | 193 | 54 | 3325-3346 |
| 2R | 5’-AGTGGTGCTCAAACTGGT-3’ | 3517-3500 | ||
| 3F | 5’-CCCTTGCTCTCTGATGTAAC-3’ | 192 | 55 | 3845-3864 |
| 3R | 5’-ATCTCACAACACAGCCACAC-3’ | 4036-4017 | ||
| 4F | 5’-GCAGCAACTTCACCAATCAA-3’ | 167 | 55 | 4186-4167 |
| 4R | 5’-CTGCTCATGTTCCTCTAGGG-3’ | 4314-4333 | ||
| 5F | 5’-TTCTAGCATTTGAGACGGAG-3’ | 179 | 56 | 5960-5941 |
| 5R | 5’-GAACAGGATGGATGCAAATATG-3’ | 6098-6119 6119 | ||
| 6F | 5’-GTGTTACTCTCGTTTTCTAGG-3’ | 241 | 51 | 8110-8890 |
| 6R | 5’-GGCTCACTTACCATCAGGAA-3’ | 9111-9130 | ||
| 7F | 5’-GATATGGTGGGGATGCTTTG-3’ | 183 | 57 | 9985-9966 |
| 7R | 5’-TGGCAAGGTACGACTGTATG-3’ | 10129-10148 | ||
| 8F | 5’-GATGCTCTCTTTAGGTAT-3’ | 225 | 51 | 11298-11281 |
| 8R | 5’-CTTGGATGACTGTTTGGCCT-3’ | 11505-11486 | ||
| 9F | 5’-AAAACAGCATATGAGATCG-3’ | 192 | 51 | 12286-12304 |
| 9R | 5’-TACATCCACCCAGAAGTTG-3’ | 12478-12459 | ||
| 10F | 5’-ATCCTTGCTATGTGCTCACC-3’ | 149 | 51 | 12549-12568 |
| 10R | 5’-CATCACAAAACAGAGCTTAG-3’ | 12698-12679 | ||
| 11F | 5’-CTCCAGTGTGATGTGTGATAATG-3’ | 163 | 57 | 25134-25156 |
| 11R | 5’-GGGCTTGCAAATAGAATTTC-3’ | 25297-25278 | ||
| FBXW12 met F | 5’-GTTAGAATTTTAACGTAGGAATTTTGA-3’ | 341 | 51 | 2599-2625 |
| FBXW12 met R | 5’-TCTACTTACCCTATATCTATACCTACC-3’ | 2939-2913 |
RNA isolation
Total RNA (1 µg/µl) from normal human ovary was purchased from Clontech (Clontech Laboratories, Inc. Mountain View, CA). Total RNA from the 30 primary tumors was isolated using the RNeasy Mini Kit (QIAGEN, Valencia, CA) following the manufacturer’s instructions. Briefly, tissue from the 30 EOC samples was homogenized in TRI Reagent (Molecular Research Center, Cincinnati, OH), and the aqueous and organic phases were separated by addition of 1 vol bromo-3-chloropropane (Sigma Chemicals, St. Louis, MO), followed by centrifugation at 13,000 rpm for 15 min at 4°C. Thereafter, 350 µl of 70% ethanol (Sigma) were added to all samples, and each sample was applied to an RNeasy mini-column; all mini-columns were washed by centrifugation with buffers containing guanidine and ethanol. To elute the RNA, 30 µl of RNAse-free water were added directly onto the silica-gel membrane of the columns, which were then centrifuged for 1 min at 13,000 rpm. RNA was quantitated by measuring absorbance at 260 nm and stored at -85°C until used. The quality of each RNA sample was assessed on 2% formaldehyde denaturing agarose gels.
Reverse transcription
Reverse transcription was performed employing the Superscript-First Strand kit (Invitrogen Life Technologies, Inc., Carlsbad, CA) in accordance with the manufacturer’s instructions. All reactions were carried out in a total volume of 20 µl. Initially, 1 µg of total RNA was annealed at 65°C for 5 min to 0.5 µg of random primer (0.5 µg/µl) and 1 µl of a 10 mM dNTPs cocktail. The annealed RNA-primer samples were incubated for 1 hour at 42°C with RT-buffer (10×), MgCl2 (25 mM), RNAseOUT (40 U/µl) and 50 units of the Superscript II reverse transcriptase (50 U/µl). The reactions were terminated by incubation at 70°C for 15 min, following incubation at 37°C for 20 min with 2 units of E. coli RNAse H (2 U/µl).
These reverse transcribed products served as templates for the PCR amplification of a portion (831 base pairs) of the FBXW12 coding region and a 654 base pair (bp) segment of a constitutive gene (β-actin). All PCR reactions were carried out in a 20 µl reaction containing: 10 µl of PyroStartTM Fast PCR Master Mix (2×) (Fermentas), 15 pmoles of the corresponding primer pair and 1µl of the cDNA template. The PCR conditions used were as described above. Equal volumes of the PCR reactions were electrophoresed on 2% agarose gels stained with ethidium bromide.
Identification of CpG islands
To determine if FBXW12 is epigenetically silenced in EOC by hypermethylation of CpG islands we searched the entire gene’s sequence for CpG islands using the on-line tool http://www.ebi.ac.uk/Tools/emboss/cpgplot/index.html.
Bisulfite conversion and methylation specific PCR of the FBXW12 proximal promoter region
Bisulfite conversion of 2 µg of genomic DNA isolated from one stage IV serous ovarian carcinoma sample was performed with the EZ DNA Methylation kit (Zymo Research, Orange, CA) following the manufacturer’s protocol. Bisulfite-modified DNA was then amplified by PCR employing specific primers for the proximal promoter of FBXW12 (Table 2). The PCR was carried out in a final volume of 20 µl containing GoTaq Flexi Buffer 1X; 1.5 mM MgCl2; 0.2 mM dNTP’s; 5 pmol of each corresponding primer and 1.25 U of GoTaq Flexi DNA polymerase (Promega Corp). Conditions for the PCR were an initial denaturation step of 94°C for 5 min followed by 40 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 45 sec; and a final extension step at 72°C for 10 min. PCR products were electrophoresed on a 2% agarose gel stained with ethidium bromide and the corresponding bands were purified with the QIAEX II Gel Extraction Kit (QIAGEN, Inc., Valencia, CA). The purified products were ligated into pGEMT-Easy kit (Promega Corp.). After bacterial transformation, 18 recombinant plasmids expected to contain a 341 bp fragment of the proximal promoter were sequenced as described previously. In order to determine the methylation degree, sequencing results were compared against the in silico bisulfite converted DNA sequence by means of the MultiAlin 5.4.1 program [26].
Immunohistochemistry
Four μm sections from the nine formalin-fixed, paraffin-embedded samples obtained from the Women’s Hospital, were mounted on glass slides previously coated with poly-L-lysine, deparaffinized and rehydrated in serial alcohols (100, 90, 70 and 30%) until water. Sections were then microwave heated with antigen retrieval solution (Vector Laboratories, Burlingame, CA, USA), rinsed in 1x PBS pH 7.4 and incubated for 30 min on 3% H2O2 in methanol to inactivate endogenous peroxidase, and subsequently blocked with 10% BSA in 1x PBS for 30 min. Tissues were then incubated with primary anti-FBXW12 (LS-C166094) antibody, from LifeSpan BioSciences, Inc. (Seattle WA, USA) at 4°C overnight. Sections were washed in PBS, incubated at room temperature for 2 hours with the Mouse/Rabbit Immunodetector HRP/DAB (Bio SB Inc. CA, USA) and washed with 1x PBS. The peroxidase reaction was developed with diaminobenzidine and H2O2 generating a brown precipitate. Finally, slides were counterstained with hematoxylin, dehydrated and mounted with synthetic resin. Breast cancer tissues were used as positive control and negative controls consisted of BSA in PBS instead of primary antibody.
Results
Search for FBXW12 mutations in EOC
The coding sequence of the FBXW12 gene (NM_207102.2) was analyzed in the 30 ovarian tumors by PCR and direct sequencing of genomic DNA. No inversions, indels or single nucleotide polymorphisms (SNPs) were found in any of the samples studied. Despite this lack of common genetic variation, we identified a complete deletion of the FBXW12 coding region in 10% of the EOC samples (three out of thirty) (samples 1, 2 and 18; Figure 1A-C). In contrast, amplification of a 654 bp fragment of a constitutive gene (β-actin) was observed in all cases including these three samples (Figure 1D). In addition, all FBXW12 exons were PCR-amplified from DNA isolated from white cells of 30 healthy women (positive control) (PC) (Figure 1A-C). No genomic deletions within the coding region were detected in the DNA isolated from the remaining 27 tumors (Figure 1A and 1B). Most of these tumors were classified as low-grade EOCs (Table 1). Of note, the three samples showing deletion of the FBXW12 coding region were from patients with tumors of advanced stage (stage III and IV serous carcinoma, respectively, according to FIGO) (Table 1). Information on family history of ovarian cancer indicated that in all patients the disease was sporadic. This was corroborated by PCR and direct sequencing of the 10 coding exons of FBXW12 from DNA isolated from white cells of all patients (data not shown). We could not determine the length of the deletions, because information on conventional cytogenetic karyotyping was not available.
Figure 1.
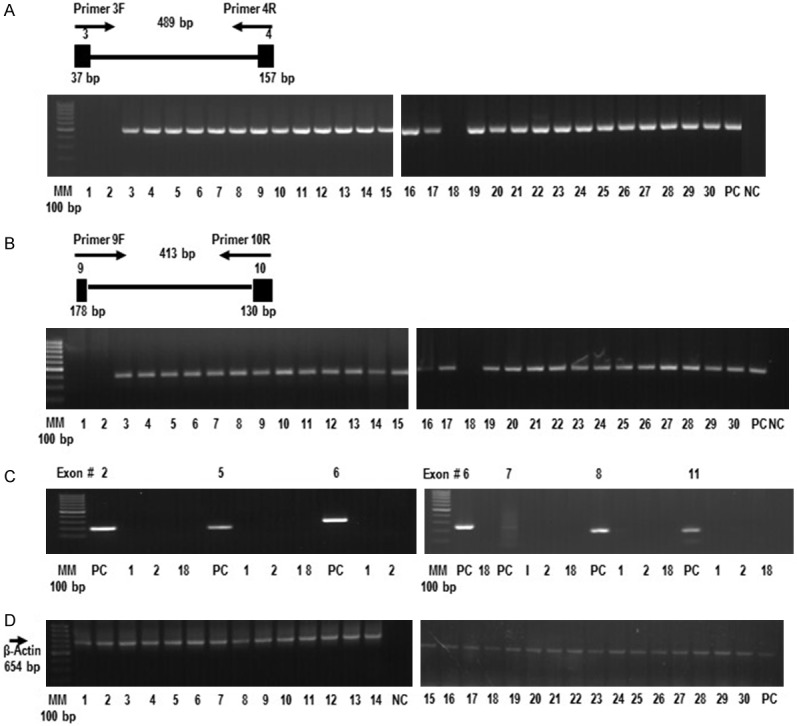
The FBXW12 gene is deleted in some EOCs. (A) Gel image showing the absence of a 489 bp PCR product in three tumor samples (1, 2 and 18), after amplification of ovarian genomic DNA, using primers targeting exons 3 and 4 and the corresponding intronic sequence of the FBXW12 gene. (B) Gel image showing the absence of PCR products following amplification of ovarian genomic DNA from tumors 1, 2 and 18, using primers targeting a 413 bp fragment comprising exons 9 and 10 and the corresponding intronic sequence of FBXW12. (C) Gel image showing the absence of PCR products derived from tumors 1, 2 and 18, using primers targeting the remaining 6 exons (2, 5, 6, 7, 8 and 11) of the FBXW12 coding sequence. (D) The lack of PCR products in tumors 1, 2 and 18 is in contrast to the amplification of a 654 bp fragment of a constitutive gene (β-actin) from the same tumor samples. Genomic DNA isolated from white cells of healthy women was used as positive control (PC). Also in all PCR experiments, a reaction with all PCR components with the exception of DNA was used as negative control (NC). MM = molecular marker, PC = positive control, NC = negative control, 1 = stage III serous EOC with complete deletion of FBXW12 coding region, 2 = stage IV serous EOC with complete deletion of FBXW12 coding region, 18 = stage III serous EOC with complete deletion of FBXW12 coding region, 3-30 = EOCs of different histotypes with no deletion of FBXW12 coding region. The diagrams on top of panels (A) and (B) depict the approximate positions of the primers used for amplification.
Next, we considered the possibility of genomic mutations in the 5’-UTR and proximal promoter of FBXW12, and analyzed all 30 tumors by PCR-amplifying a 790 bp fragment comprising exon 1, part of exon 2 (where the translational start site is located), the corresponding intronic sequence between them and 360 bp of the proximal promoter. The reactions intended to amplify this 790 bp segment, did not yield a PCR product in eleven of the thirty tumors (Figure 2A), indicating that the 5’-untranslated region and proximal promoter are deleted in these eleven samples. In contrast, PCR products of the correct size were obtained from the amplification of genomic DNA derived from white cells of a healthy woman (PC) (Figure 2A). Again, amplification of the constitutive gene (β-actin) was observed in all samples (Figure 2B).
Figure 2.
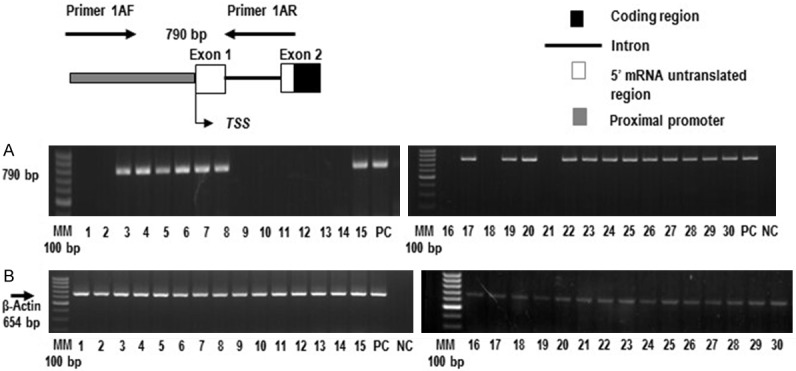
The 5’ untranslated region (UTR) and proximal promoter of FBXW12 are deleted in some of the 30 EOCs. (A) Absence of the FBXW12 5’ UTR region and proximal promoter in eleven of 30 EOCs, as assessed by the lack of amplification of a PCR product targeting a 790 bp genomic fragment upstream of the FBXW12 start codon. (B) Amplification of a 654 bp fragment of the constitutive gene (β-actin) from all tumor samples. Genomic DNA isolated from white cells of healthy women was used as positive control (PC). In all PCR experiments, a reaction with all PCR components with the exception of DNA was used as negative control (NC). PC = positive control, NC = negative control, 1-30 = tumor EOC samples. The diagram on top of panel A depicts the approximate positions of the primers used for amplification.
FBXW12 proximal promoter is epigenetically silenced in some cases of EOC
Because a portion (831 bp) of FBXW12 mRNA was only observed in 2 samples without deletions of either the coding region or the 5’-UTR and proximal promoter (Figure 3A and 3B), we considered the possibility that CpGs in FBXW12 are methylated in EOC, resulting in FBXW12 silencing. The sequencing results derived from the methylation specific PCR of the FBXW12 proximal promoter amplified from a stage IV ovarian serous adenocarcinoma which cDNA was not detected by PCR, demonstrated CpGs methylation in 6 portions of the FBXW12 proximal promoter. Moreover, 3 of these sites with methylated cytosines, are within recognition sequences for three transcription factors: the CCAAT/Enhancer Binding Protein (C/EBP), known to regulate cellular differentiation and proliferation in a wide array of tissues [27], Lyf-1, initially described as a lymphoid-specific transcription factor which regulates gene expression and Oct-1, whose high expression in cervical cancer has been recently reported [28,29] (Figure 4).
Figure 3.
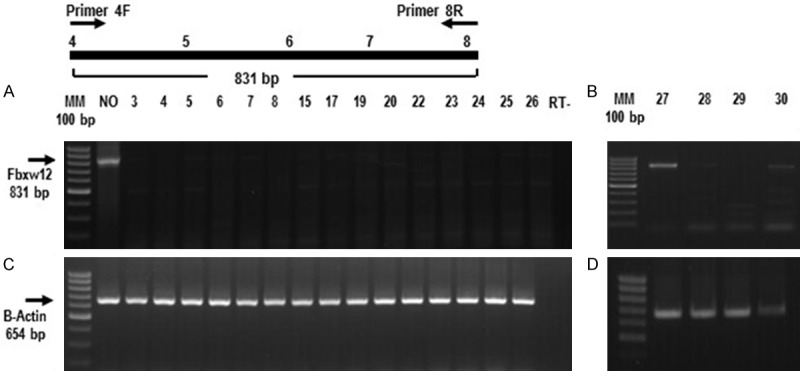
FBXW12 mRNA is heterogeneously expressed in EOC. (A) and (B) Representative images showing either the lack or low expression of FBXW12 mRNA in nineteen tumors with no deletion of FBXW12 coding region, 5’UTR or proximal promoter, in contrast to the detection of FBXW12 mRNA in total RNA from a normal ovary. The arrow to the left indicates the size of the mRNA fragment amplified by the primers used (targeting the sequences encoded by exons 4 to 8). (C) and (D) Amplification of a 654 bp fragment of the constitutive gene actin. NO = normal ovary, 3-30 = EOC samples of three different histotypes (serous, mucinous and endometroid) with no deletion of FBXW12 coding region, 5’UTR or proximal promoter, RT- = reverse transcription negative control. The diagram on top of panel A depicts the approximate positions of the primers used for amplification.
Figure 4.
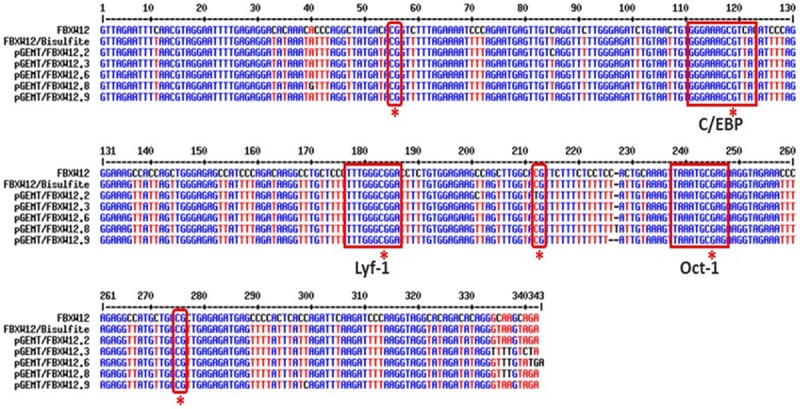
FBXW12 proximal promoter is methylated in some cases of EOC. Representative image of the sequencing results from 5 clones containing a 345 bp fragment of the FBXW12 proximal promoter. Sequences were compared against the in silico bisulfite converted fragment of the promoter (second top sequence) by means of the MultiAlin 5.4.1 program. The six CpG methylation sites are indicated in red squares. Three of these sites were identified within recognition sequences for three different transcription factors (C/EBP, Lyf-1 and Oct-1, respectively).
FBXW12 is differentially expressed in the normal human ovary and in EOC
FBXW12 localization within normal human ovary and epithelial ovarian carcinoma samples was determined by means of immunohistochemistry. In sections of two paraffin embedded normal ovaries, FBXW12 immunostaining was detected specifically in oocytes of growing follicles which is similar to what was reported for the murine orthologue Fbxw15 [22]. FBXW12 was not detected neither in follicular cells, nor in the ovarian surface epithelium (OSE) of the normal ovary (Figure 5A, 5I, 5II). In spite of not being present in normal cells of epithelial origin, FBXW12 signal was observed in both the benign EOC samples analyzed and in malignant tissue (Figure 5A, 5III, 5IV, 5B). The latter suggests that within the normal ovary, FBXW12 is somehow silenced in cells of epithelial origin, while in the EOC samples, its transcription is active allowing the presence of the corresponding protein. Breast cancer tissue was used as positive control to FBXW12 antibody (Figure 5A, 5V) No signal was observed in ovarian samples without addition of primary antibody (Figure 5A, 5V).
Figure 5.
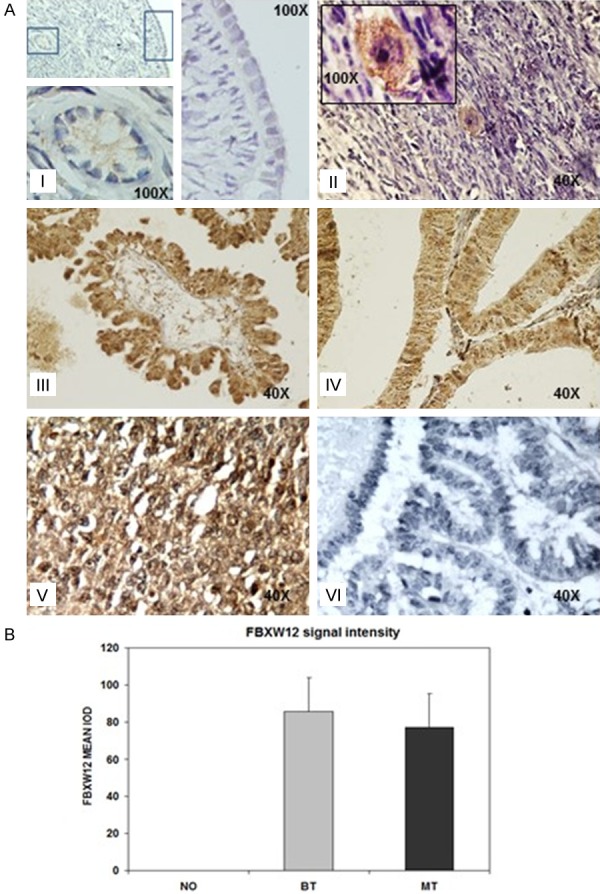
FBXW12 expression in normal human ovary and in EOC. (A) Immunoreaction of FBXW12 in normal ovary. (a) No FBXW12 reaction is present in the ovarian surface epithelium, however in this sample a weak cytoplasmic reaction is observed in oocytes (100X). (b) Strong immunoreaction of FBXW12 in the cytoplasm of oocytes in the second sample of a normal ovary (100X). Stromal cells did not show immunoreaction to FBXW12 (40X). Difference in signal intensity between (a) and (b) is due to intrinsic experimental variability. (c) Immunopositivity for FBXW12 in the benign ovarian tumor where a reaction in the cytoplasm of epithelial cells is mainly observed (40X). (d) Cytoplasmic immunopositivity for FBXW12 is detected in epithelial cells of a malignant ovarian epithelial tumor (40X). (e) Breast cancer tissue was used as positive control to FBXW12 antibody according to the supplier instructions (40X). (f) A negative control in benign ovarian tumor consisted in the omission of the primary antibody (40X). (B) Densitometry analysis of FBXW12 expression in the epithelium of normal ovary (NO), benign epithelial ovarian tumor (BT) and malignant epithelial ovarian tumor (MT).
Discussion
In this study, we performed the molecular analysis of FBXW12 in 30 EOC samples, identifying a complete deletion of its coding region in three of them, and deletion of the proximal promoter and 5’UTR in 11 cases, including the three samples lacking the coding region.
The association between chromosomal alterations and cancer development is well established [30]; a large number of studies employing different technical approaches, have focused on the identification of chromosomal amplifications or deletions. As a result, recurrent regions of copy number gain, loss, or allelic imbalance have been identified [31,32]. Among such regions, gains in copy number within 3q are reported as frequent, but deletions within this chromosomal segment, or within 3p are not considered as markers with prognostic significance [32-34]. The deletion of FBXW12, which is located on 3p21.31 is, to our knowledge, one of the few deletions on this locus reported in ovarian cancer.
It is known that genes involved in cell cycle regulation are frequently damaged in tumor cells. EOC is not an exception, as some cell cycle genes-including genes encoding F-box proteins - have been shown to be affected in these cells [6-8]. Because Skp2, an F-box protein necessary for DNA replication, is overexpressed in ovarian adenocarcinoma [35], we initially hypothesized that FBXW12 mRNA abundance would be increased in primary EOC over the low levels reported by Zeng et al., in healthy ovaries [36].
Contrary to this expectation, we only observed mRNA expression in 2 EOC samples without deletion of the coding region, the 5’-UTR or the proximal promoter, as opposed to the expression observed in the normal human ovary. Even though FBXW12 has no discernible CpG islands, which suggested that epigenetic silencing by DNA hypermethylation was not responsible for the lack of mRNA expression seen in these EOC cases, we decided to determine if the proximal promoter was indeed not silenced by CpG methylation. Therefore to analyze the FBXW12 proximal promoter region, we employed the DNA from one of the EOC samples lacking mRNA expression. To our surprise, the sequencing results of a 341 bp fragment of the FBXW12 proximal promoter, identified six sites of CpG methylation within this region, this could account for the absence of FBXW12 mRNA in the majority of the EOC samples without deletions. In this context, the promoter of FBX032, another F-box encoding gene is silenced by hypermethylation in ovarian cancer cell lines [20].
Because the murine orthologue of FBXW12 is selectively expressed in oocytes [22], another explanation for the absence of FBXW12 mRNA in the EOC samples is that this tissue is devoid of oocytes, and thus it does not contain FBXW12-expressing cells. The results derived from the immunohistochemistry experiments demonstrated that like its murine orthologue, FBXW12 in the normal human ovary is oocyte specific and surprisingly, its localization changes to the OSE neoplastic cells of the EOC samples analyzed. Our results are in accordance with a study in which a high heterogeneity of the D-glucuronyl C5 epimerase (GLCE) expression was observed. The authors reported a significant decrease in GLCE mRNA expression in 53% of the prostate tumors but also normal or elevated levels of GLCE mRNA expression in the remaining 47%, they also demonstrated changes in the localization pattern of the corresponding protein between normal and benign samples versus the malignant ones; due to possible alterations in epigenetic mechanisms that regulate the expression of the gene encoding GLCE [37]. Regarding FBXW12, we can speculate that in the healthy human ovary, this gene is normally silenced in the epithelial cells comprising the ovarian surface epithelium. In some EOC cases, it can also remain silenced, while in others this epigenetic silencing is deregulated in the neoplastic cells of the OSE. Besides silencing by CpGs methylation, at least three possibilities may be entertained to explain the absence of FBXW12 mRNA in EOC: 1) that enhancer sequences located further upstream of the proximal promoter region, as shown for the GnRH gene [38], are deleted., b) that mRNA degradation is enhanced by aberrant microRNA processing. Growing evidence indicates that de-regulation of microRNAs is closely associated with ovarian tumorigenesis [39,40]. Or c) that the life-span of FBXW12 mRNA is too short to be easily amplified by PCR.
Searching for deletions of either the 5’ UTR or the proximal promoter of FBXW12 revealed the absence of these regions in eleven of the thirty samples studied. Because these eleven samples include those three lacking the FBXW12 coding region, it is apparent that mutations of FBXW12 in EOC involve varying portions of the gene, compromising either the mRNA encoding sequence or the proximal promoter. Such genomic deletions are consistent with recent evidence, gathered with the help of high resolution arrays, showing the existence of complex chromosomal rearrangements or copy number alterations of small regions of DNA in EOC [41]. Still we did not observe deletions of either the 5’ UTR, proximal promoter or mRNA encoding region in nineteen of the 30 tumors, suggesting that deletion of the FBXW12 is not a universal feature of EOC pathology.
Conclusion
Because FBXW12 is an F-box protein, it is possible that deletions of different portions of its gene or a de-regulation of its silencing mechanism may compromise ubiquitin-mediated proteosomal degradation resulting in an altered expression of downstream oncogenic targets directly involved in ovarian cancer development.
Acknowledgements
We thank M. Sc. Laura Márquez from the Laboratorio de Biología Molecular del Instituto de Biología, UNAM, for her kind assistance with the sequencing reactions. This work was supported by the Coordinación de Investigación en Salud, Instituto Mexicano del Seguro Social, México, Grant: 2005/1/I/032 (ED), and by NIH grants HD24870 and 8P51OD011092-53 for the operation of the Oregon National Primate Research Center (SRO).
Disclosure of conflict of interest
None.
References
- 1.Gubbels J AA, Claussen N, Kapur AK, Connor JP, Patankar MS. The detection, treatment, and biology of epithelial ovarian cancer. J Ov Res. 2010;3:8–18. doi: 10.1186/1757-2215-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nossov V, Amneus M, Su F, Lang J, Janco JM, Reddy ST, Farias-Eisner R. The early detection of ovarian cancer: from tradicional methods to proteomics. Can we really do better than serum CA-125? Am J Obstet Gynecol. 2008;199:21–23. doi: 10.1016/j.ajog.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Holschneider C, Berek J. Ovarian cancer: epidemiology, biology, and prognostic factors. Semin Surg Oncol. 2000;19:3–10. doi: 10.1002/1098-2388(200007/08)19:1<3::aid-ssu2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 4.Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tulinius H, Thorlacius S, Eerola H, Nevanlinna H, Syrjäkoski K, Kallioniemi OP, Thompson D, Evans C, Peto J, Lalloo F, Evans DG, Easton DF. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiyotsugu Y, Yoshio M. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Science. 2004;95:866–871. doi: 10.1111/j.1349-7006.2004.tb02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D’Andrilli G, Giordano A, Bovicelli A. Epithelial Ovarian Cancer: The Role of Cell Cycle Genes in the Different Histotypes. Open Clin Cancer J. 2008;2:7–12. doi: 10.2174/1874189400802010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhong W, Feng H, Santiago FE, Kipreos ET. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature. 2003;423:885–890. doi: 10.1038/nature01747. [DOI] [PubMed] [Google Scholar]
- 8.Li A, Blow JJ. Degradation ensures integrity. Nature. 2003;423:818–819. doi: 10.1038/423818b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craig KL, Tyers M. The F-box: A new motif for ubiquitin dependent proteolysis in cell cycle regulation and signal transduction. Progress in Biophys Mol Biol. 1999;72:299–328. doi: 10.1016/s0079-6107(99)00010-3. [DOI] [PubMed] [Google Scholar]
- 10.Kipreos ET, Pagano M. The F-box protein family. Genome Biol. 2000;1:REVIEWS3002. doi: 10.1186/gb-2000-1-5-reviews3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winston JT, Koepp DM, Zhu C, Elledge SJ, Harper JW. A family of mammalian F-box proteins. Curr Biology. 1999;9:1180–1182. doi: 10.1016/S0960-9822(00)80021-4. [DOI] [PubMed] [Google Scholar]
- 12.Li P, Li C, Zhao X, Zhang X, Nicosia SV, Bai W. P27kip1 stabilization and G1 arrest by 1, 25-dihydroxyvitamin D3 in ovarian cancer cells mediated through down-regulation of cyclin E/cyclin-dependent kinase 2 and Skp1-Cullin-F-box protein/Skp2 ubiquitin ligase. J Biol Chem. 2004;279:25260–7. doi: 10.1074/jbc.M311052200. [DOI] [PubMed] [Google Scholar]
- 13.Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- 14.Kwak EL, Moberg KH, Wahrer DC, Quinn JE, Gilmore PM, Graham CA, Hariharan IK, Harkin DP, Haber DA, Bell DW. Infrequent mutations of Archipelago (hAGO, hCDC4, Fbw7) in primary ovarian cancer. Gynecol Oncol. 2005;98:124–128. doi: 10.1016/j.ygyno.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Sgambato A, Cittadini A, Masciullo V, Di Salvatore M, Graziani C, Rettino A, Valdivieso P, Scambia G, Bianchino G, Zupa A, Improta G, Cifarelli RA. Low frequency of hCDC4 mutations in human primary ovarian cancer. Gyencol Oncol. 2007;105:553–5. doi: 10.1016/j.ygyno.2007.01.026. [DOI] [PubMed] [Google Scholar]
- 16.Tan Y, Sangfelt O, Spruck C. The Fbxw7/hCdc4 tumor suppressor in human cancer. Cancer Lett. 2008;271:1–12. doi: 10.1016/j.canlet.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 17.Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, Cepeda D, Fiegl H, Dafou D, Marth C, Mueller-Holzner E, Corcoran M, Dagnell M, Nejad SZ, Nayer BN, Zali MR, Hansson J, Egyhazi S, Petersson F, Sangfelt P, Nordgren H, Grander D, Reed SI, Widschwendter M, Sangfelt O, Spruck C. FBXW7/hCDC4 is a general tumour suppressor in human cancer. Cancer Res. 2007;67:9006–9012. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Ye X, Liu Y, Wei W, Wang Z. Aberrant regulation of FBW7 in cancer. Oncotarget. 2014;5:1–16. doi: 10.18632/oncotarget.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gütgemann I, Lehman NL, Jackson PK, Longacre TA. Emi1 protein accumulation implicates misregulation of the anaphase promoting complex/cyclosome pathway in ovarian clear cell carcinoma. Modern Pathology. 2008;21:445–454. doi: 10.1038/modpathol.3801022. [DOI] [PubMed] [Google Scholar]
- 20.Chou JL, Su HY, Chen LY, Liao YP, Hartman-Frey C, Lai YH, Yang HW, Deatherage DE, Kuo CT, Huang YW, Yan PS, Hsiao SH, Tai CK, Lin HJ, Davuluri RV, Chao TK, Nephew KP, Huang TH, Lai HC, Chan MW. Promoter hypermethylation of FBXO32, a novel TGF-β/SMAD4 target gene and tumor suppressor, is associated with poor prognosis in human ovarian cancer. Lab Invest. 2010;90:414–425. doi: 10.1038/labinvest.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Liu P, Inuzuka H, Wei W. Roles of F-box proteins in cancer. Nat Rev Cancer. 2014;14:233–247. doi: 10.1038/nrc3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De La Chesnaye E, Kerr B, Paredes A, Merchant-Larios H, Méndez JP, Ojeda SR. Fbxw15/Fbxo12J Is an F-Box Protein-Encoding Gene Selectively Expressed in Oocytes of the Mouse Ovary. Biol Reprod. 2008;78:714–25. doi: 10.1095/biolreprod.107.063826. [DOI] [PubMed] [Google Scholar]
- 23.Zou C, Chen Y, Smith RM, Snavely C, Li J, Coon TA, Chen BB, Zhao Y, Mallampalli RK. SCFFbxw15 mediates histone acetyltransferase binding to origin recognition complex (HBO1) ubiquitin-proteosomal degradation to regulate cell proliferation. J Biol Chem. 2013;288:6306–16. doi: 10.1074/jbc.M112.426882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T. Preparation and analysis of eukaryotic genomic DNA. In: Nolan C, editor. Molecular Cloning: A Laboratory Manual. 2nd edition. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. pp. 9.16–9.23. [Google Scholar]
- 25.Sambrook J, Russell DW. Rapid Isolation of mammalian DNA. In: Nolan C, editor. Molecular Cloning: A Laboratory Manual. 3th edition. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. pp. 6.28–6.30. [Google Scholar]
- 26.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucl Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ko CY, Chang WC, Wang YM. Biological roles of CCAAT/Enhancer binding protein delta during inflammation. J Biomed Sci. 2015;22:6. doi: 10.1186/s12929-014-0110-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Francis OL, Payne JL, Su RJ, Payne KJ. Regulator of myeloid differentiation and function: The secret life of Ikaros. World J Biol Chem. 2011;2:119–125. doi: 10.4331/wjbc.v2.i6.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao S, Liao S, Zhou Y, Jiang B, Li Y, Xue M. High expression of octamer transcription factor 1 in cervical cancer. Oncology letters. 2014;7:1889–1894. doi: 10.3892/ol.2014.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Höglund M, Gisselsson D, Hansen GB, Säll T, Mitelman F. Ovarian carcinoma develops through multiple modes of chromosomal evolution. Cancer Res. 2003;63:3378–3385. [PubMed] [Google Scholar]
- 31.Kiechle M, Jacobsen A, Schwarz-Boeger U, Hedderich J, Pfisterer J, Arnold N. Comparative genomic hybridization detects genetic imbalances in primary ovarian carcinomas as correlated with grade of differentiation. Cancer. 2001;91:534–540. [PubMed] [Google Scholar]
- 32.Hauptmann S, Denkert C, Koch I, Petersen S, Schlüns K, Reles A, Dietel M, Petersen I. Genetic alterations in epithelial ovarian tumors analyzed by comparative genomic hybridization. Hum Pathol. 2002;33:632–641. doi: 10.1053/hupa.2002.124913. [DOI] [PubMed] [Google Scholar]
- 33.Sonoda G, Palazzo J, du Manoir S, Godwin AK, Feder M, Yakushiji M, Testa JR. Comparative genomic hybridization detects frequent overrepresentation of chromosomal material from 3q26, 8q24 and 20q13 in human ovarian carcinomas. Genes Chromosomes Cancer. 1997;20:320–328. [PubMed] [Google Scholar]
- 34.Jancﬞarková N, Krkavcová M, Janashia M, Freitag P, Dusková J, Cibula D. Chromosomal rearrangements and their relations to histopathological and clinical parameters in epithelial ovarian cancer. Folia Biologica (Praha) 2008;54:58–64. doi: 10.14712/fb2008054020058. [DOI] [PubMed] [Google Scholar]
- 35.Shigemasa K, Gu L, O’Brien TJ, Ohama K. Skp2 overexpression is a prognostic factor in patients with ovarian adenocarcinoma. Clinical Cancer Res. 2003;9:1756–1763. [PubMed] [Google Scholar]
- 36.Zeng L, Gu S, Li Y, Wang W, Huang Y, Ye X, Xu J, Zhao E, Ji C, Ying K, Xie Y, Mao Y. cDNA cloning and expression analysis of a novel human F-box domain containing gene. Mol Biol Rep. 2004;31:51–57. doi: 10.1023/b:mole.0000013502.72069.19. [DOI] [PubMed] [Google Scholar]
- 37.Prudnikova TY, Soulitzis N, Kutsenko OS, Mostovich LA, Haraldson K, Ernberg I, Kashuba VI, Spandidos DA, Zabarovsky ER, Grigorieva EV. Heterogeneity of D-glucuronyl C5-epimerase expression and epigenetic regulation in prostate cancer. Cancer Med. 2013;2:654–661. doi: 10.1002/cam4.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Novaira HJ, Yates M, Diaczok D, Kim H, Wolfe A, Radovick S. The gonadotropin-releasing hormone cell-specific element is required for normal puberty and estrous ciclicity. J Neurosci. 2011;31:3336–3343. doi: 10.1523/JNEUROSCI.5419-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang N, Kaur S, Volinia S, Greshock J, Lassus H, Hasegawa K, Liang S, Leminen A, Deng S, Smith L, Johnstone CN, Chen XM, Liu CG, Huang Q, Katsaros D, Calin GA, Weber BL, Bützow R, Croce CM, Coukos G, Zhang L. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res. 2008;68:10307–10314. doi: 10.1158/0008-5472.CAN-08-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balch C, Fang F, Matei DF, Huang TH, Nephew KP. Minireview: Epigenetic changes in ovarian cancer. Endocrinology. 2009;150:4003–4011. doi: 10.1210/en.2009-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gorringe KL, Jacobs Sh, Thompson ER, Sridhar A, Qiu W, Choong DY, Campbell IG. High-resolution single nucleotide polymorphism array analysis of epithelial ovarian cancer reveals numerous microdeletions and amplifications. Clin Cancer Res. 2007;13:4731–4739. doi: 10.1158/1078-0432.CCR-07-0502. [DOI] [PubMed] [Google Scholar]


