Abstract
MicroRNA (miR-126) was reported to be downregulated and to act as a tumor suppressor in cancers of the lung, cervix, bladder, breast, liver and prostate. However, the precise roles and underling mechanisms of miR-126 in glioma remain largely unknown. This study is aimed to study the role of miR-126 in the progression of glioma and to elucidate underlying miR-126-mediated mechanisms in glioma. Our results revealed that miR-126 was downregulated in the collected glioma specimen, compared with non-cancerous brain tissues. Restored miR-126 expression inhibited cell proliferation, colony formation, migration and invasion, and induced cell cycle arrest at G0/G1 phase and cell apoptosis of U-87 MG glioma cells. Overexpression of miR-126 was also able to suppress the growth of U-87 MG glioma xenografts in mice. Furthermore, insulin receptor substrate 1 (IRS-1) were identified as a target of miR-126, and showed that it was negatively regulated by miR-126 in glioma cells. We also demonstrated that overexpression of miR-126 suppressed PI3K and AKT activation, which contribute to suppress tumor growth of glioma. Taken together, these findings showed that miR-126 functions as a tumor suppressor in glioma cells by targeting IRS-1 expression via the PI3K/AKT signaling pathways, suggesting that miR-126 might be a novel target for therapeutic strategies in glioma.
Keywords: miR-126, glioma, insulin receptor substrate 1, PI3K/AKT
Introduction
Malignant gliomas are the most common primary tumors of the central nervous system with high morbidity and mortality [1,2]. The high lethality of the disease is mainly because of the highly invasive/migratory nature of glioma cells, which are capable of diffusely infiltrating and widely migrating in the surrounding brain tissue [3,4]. Although treating with combinations of surgery, radiotherapy and chemotherapy, the prognosis and tumor recurrence in glioma patients have not been significantly improved [5,6], especially, the most malignant glioma, glioblastoma multiform (GBM), is only 15 months average life expectancy due to difficulties in complete resection and the low sensitivity to radio-/chemo- therapeutic agents [7]. Thus, it is quite urgent to reveal the molecular mechanisms by which glioma initiates, progresses, invades, and recurs to develop novel and effective therapeutic strategies.
MicroRNAs (miRNAs) are a large family of endogenous small non-coding RNAs (19-24 net) that regulate gene expression by antisense complementarity to specific mRNA [8]. It has been firmly confirmed that miRNAs played critical role in the regulation of diverse physiological and pathological processes [9], especially in tumorigenesis [10,11]. Growing evidences show that miRNAs modulate cell proliferation, cell cycle, apoptosis, migration, invasion, and metastasis in human cancers [12,13]. Among them, miR-126 has been demonstrated to function as a tumor suppressor, and loss of miR-126 expression has been reported in many cancer types, such as renal cell carcinoma [14], hepatocellular carcinoma [15], osteosarcoma [16], gastric cancer [17], non-small cell lung cancer [18], colorectal tumors [19], cervix [20] and colon cancer [21], restoration of miRNA-126 reduced cell proliferation, decreased cell invasive capacity and increased apoptosis in several human cancers [22]. However, to our knowledge, the precise roles and mechanisms of miR-126 in glioma have not been determined until now.
In the present study, we investigated miR-126 expression level in glioma tissue and adjacent noncancerous brain tissues, and found that the expression of miR-126 was significantly downregulated in glioma tissues than those in the adjacent normal tissues (P < 0.01). We also investigated the functional role of miR-126 in glioma, both in vitro and in vivo. We found that that in glioma cell lines miR-126 functioned as a tumor suppressor and overexpression of miR-126 reduced cell proliferation, decreased cell migration and invasive capacity and increased apoptosis and cell arrest at G0/G1 phase in vitro, and suppressed tumor growth in vivo. Furthermore, we showed that miR-126 directly targets the 3’UTR IRS-1 of to regulate its expression and downstream signaling proteins. Our findings provide the first evidence of a role for miR-126 as a tumor suppressor in glioma mediated by targeting IRS-1, suggesting that miR-126 might be a novel target for therapeutic strategies in glioma.
Materials and methods
Clinical glioma samples
Human glioma samples were collected from the Department of Neurosurgery, the first hospital of Jilin University (Changchun, China) from 30 adult patients, freshly resected during surgery. All samples were immediately frozen in liquid nitrogen and stored at -80°C until use. There were 12 low-grade (5 from WHO grades I and 7 from WHO grades II) and 18 high-grade tumors (8 from WHO grades III and 10 from WHO grades IV). Normal brain tissues adjacent to the tumor were taken from 3 cm away from the tumor cells. None of patients had received chemotherapy, immunotherapy and radiotherapy prior to surgery. All patients gave written informed consent according to a study protocol were approved by the ethics committee of the first the first hospital of Jilin University (Changchun, China).
Cell line and culture
Four human glioma cell lines, including U251, U87, U118 and LN18 and primary normal human astrocytes (NHA) were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China), and were cultured at 37°C with 5% CO2 in DMEM (Gibson, Gaithersburg, USA) supplemented with 10% fetal bovine serum (FBS, Gibson BRL, Gaithersburg, MD, USA), 100 units/ml penicillin and 100 mg/ml streptomycin.
Real time quantitative RT-PCR
RNA was isolated from harvested cells or human tissues with Tirol reagent according to the manufacturer’s instruction (Invitrogen, CA, USA). Total RNA (3 μg) was reverse transcribed into cDNA using M-MLV reverse transcriptase (Promega, USA) with the specific primers according to the manufacturer’s instructions. Then miR-126 expression level was quantified as described by Liu et al [23]. Expression of U6 was used as an endogenous control. To determine the mRNA levels of IRS-1, total RNAs were reversely transcribed by oligo dT primer using PrimeScript RT Reagent Kit (Takara, Dalian, China). Housekeeping gene β-actin was used as internal control, primers of IRS-1 and β-actin were used as described previously [24]. The cDNAs were amplified by qRT-PCR using SYBR Premix DimerEraser (Takara, Dalian, China) on an ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). The comparative 2-∆∆Ct method was used for relative quantification and statistical analysis.
Transfection of cells with miR-126
miR-126 mimic or corresponding negative control (miR-NC) were purchased from GenePharma (Shanghai, China). U87 cells were transfected with either miR-126 mimic or miR-NC at final concentration 50 nM using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s protocol. At 48 h post-transfection, Transfection efficiencies were evaluated in every experiment by qRT-PCR.
Cell proliferation
To determine the effects of miR-126 on growth of glioma cells, 2 × 103 transfected cells/well were seeded in 96-well plates. The absorptions of the cells were measured at a wavelength of 450 nm by an enzyme-linked immunosorbent assay reader (Thermo Labsystems, Finland) using a CCK8 kit (Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s instruction at different indicated time points. Data were from at least three separate experiments with three replications each time.
Cell cycle and apoptosis analysis
Cells cycle and apoptosis analysis was performed on U87 cells 48 h after transfection. Transfected cells were harvested, washed twice with cold PBS, fixed in ice-cold 70% ethanol, incubated with RNase A (Sigma, USA) at 37°C for 30 min and then stained with propidium iodide (PI, Sigma, USA) at 4°C for 30 min in the dark, and were analyzed by fluorescence-activated cell sorting (FACS) (BD Biosciences, Mansfield, MA, USA).
For analysis of apoptosis, transfected cells were incubated with PE Annexin-V and 7AAD following the PE Annexin-V Apoptosis Detection Kit I (BD Pharmingen, CA, USA) protocol, and then analyzed by FACS. The apoptotic rate and cycle distribution and were analyzed using CellQuest software (BD Biosciences San Jose, CA, USA). All experiments were run in triplicate.
Migration and invasion assays
For the transwell migration assay, 5 × 104 transfected cells were placed in the upper chamber of each insert (Corning, Cambridge, USA). For the invasion assay, 5 × 104 transfected cells were placed on the upper chamber of each insert coated with 150 mg Matrigel (BD Biosciences, Bedford, MD). The lower chamber of the transwell was filled with the DMEM medium supplemented with 20% FBS. After 24 h or 48 h of incubation (24 h for migration assays; 48 h for invasion assays), the upper surface of the membrane was wiped with a cotton tip and cells attached to the lower surface were stained with crystal violet for 20 min. The invaded or migrated cells were photographed and were counted in five random fields of view at 100 × magnification and expressed as the average number of cells per field of view.
Vector construction and luciferase reporter assay
The human IRS-1 3’UTR oligonucleotides harboring the wild-type (wt) or mutant (mt) miR-126 binding site of were cloned into the psiCHECK2 vector (Promega, USA) in the XhoI/NotI sites. For luciferase assay, U87 cells (3 × 104/well) in 24-well plates were co- transfected with 100 ng psiCHECK2-UTR vectors (wt/mt) and the miR-126 mimics or miR-NC at a final concentration of 50 nM. 48 h after transfection, cells were lysed and reporter gene expression was determined using the Dual-luciferase reporter assay system (Promega) according to the manufacturer’s instructions. The specific activity is expressed as the fold changes of the experimental group versus the control group. Experiments were performed three times in triplicates.
Western blot analysis
Protein was extracted from tissues and cells using RIPA lysis buffer containing proteinase inhibitor (Sigma). Protein concentration was determined using the BCA Protein Assay Kit (Vigorous Biotechnology Beijing Co. Ltd, Beijing, China). Equal amounts of protein lysates (20 μg each lane) was separated using 10% SDS-PAGE gel and then electroblotted onto nitrocellulose membranes (Millipore, Wisconsin, USA). Membranes were blocked for 2 h with 5% non-fat dry milk in Tris-buffered saline containing 0.1% Tween-20, and incubated at 4°C overnight with following primary antibody: mouse monoclonal anti-human IRS-1 (1:1000, Santa Cruz, USA), mouse monoclonal anti-human phospho-Akt (p-Akt) (Ser473) (1:100, Cell Signaling, USA), mouse monoclonal anti-human anti-Akt (1:1000, Cell Signaling, USA), mouse monoclonal anti-human PI3K (1:500, Santa Cruz, USA), mouse monoclonal anti-human phosphorylated (p)-PI3K(Tyr458, 1:1000, Cell Signaling Technology) and mouse monoclonal anti-human β-actin (1:5000, Santa Cruz, USA). β-actin was used as an internal control for protein loading. The membrane was further incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (1:5000, Santa Cruz, USA) for 1 h at room temperature. The immune complexes were detected by enhanced chemiluminescence (ECL, Cell Signaling Technology). The integrated density of the band was quantified by Quantity One software (Bio-Rad, USA).
Tumorigenesis in nude mice
Thirty male BALB/c nude mice (6-weeks-old) were purchased from were obtained from Experiments Animal Center of Changchun Biological Institute (Changchun, China), and maintained in special pathogen-free (SPF) condition for one week. Animal handling and experimental procedures were in accordance with the Guide for the Care and Use of Laboratory Animals, and approved by the Animal Experimental Ethics Committee of Jilin University.
U87 cells and U87 cells stably expressing miR-126 or miR-NC were injected subcutaneously into both flanks of nude mice (2 × 106 cells in 100 μl). Tumor sizes were measured using vernier caliper every five days and tumor volume was calculated by formula: volume = 1/2 × Length × Width2. 30 days after implantation, mice were sacrificed and tumors were dissected, and tumor weight was measured. Total RNAs of tumor tissues were extracted to determine miR-126 expression level by qRT-PCR.
Statistical analysis
All experiments were repeated at least three times independently. Data were expressed as the mean ± SD (standard deviation) of repeated experiments. Statistical analysis between two samples was performed using Student t- test, and more than two groups was performed using one-way ANOVA followed by a Tukey post hoc test. The software Graphpad Prism 5.0 software (GraphPad Software, San Diego, CA, USA) was used for statistical analyses. P values < 0.05 were considered significant.
Results
miR-126 expression was downregulated in glioma tissue and cell lines
To determine whether miR-126 was involved in the tumorigenesis or development of glioma, we first evaluated the expression levels of miR-126 glioma tissues and corresponding normal brain tissues by qRT-PCR (Figure 1A). The results showed that the expression levels of miR-126 were consistently lower in the glioma tissues than corresponding normal brain tissues. Meanwhile, the miR-126 expression was lower in grade III and IV gliomas compared to grades I and II tumors (Figure 1A). In addition, qRT-PCR was performed on a panel of 4 human glioma cell lines and primary normal human astrocytes (NHA). It was found that the miR-126 expression in four glioma cell lines was remarkable downregulated compared to NHA (Figure 1B). The U87 cell line, which possessed the lowest levels of miR-126 expression among four cell lines, was selected for next studies. These results suggested that miR-126 could be closely related to human glioma and loss of miR-126 might be related to glioma oncogenesis and progression.
Figure 1.
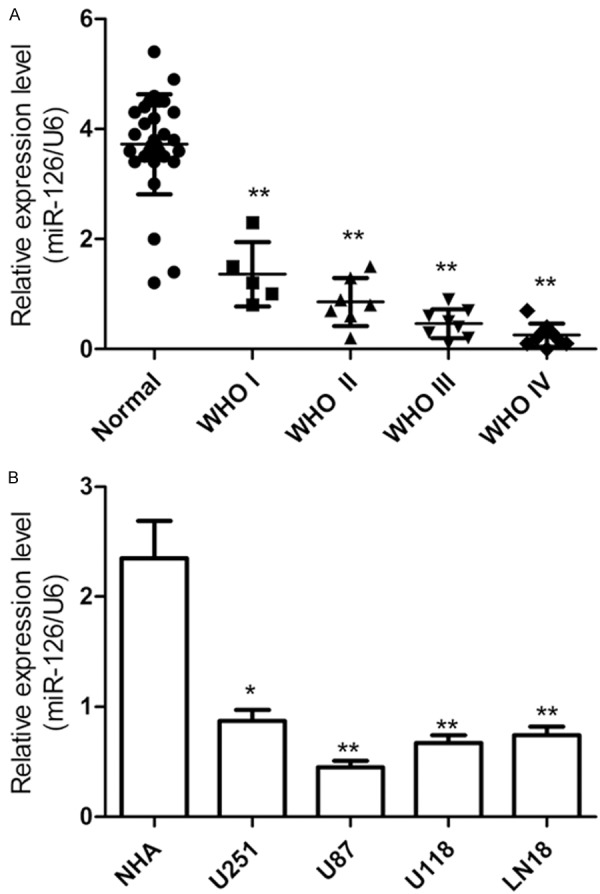
miR-126 was down-regulated in human glioma tissues and cell lines. A. Real-time quantitative RT-PCR (qRT-PCR) analysis of miR-126 expression in glioma tissues from different grades and adjacent normal brain tissues. U6 was used as loading control. B. Real-time quantitative RT-PCR (qRT-PCR) analysis of miR-126 expression in four glioma cell lines and primary normal human astrocytes (NHA). *P < 0.05 and **P < 0.01.
Ectopic expression of miR-126 inhibits cell proliferation, migration and invasion and induces cell apoptosis of glioma cells
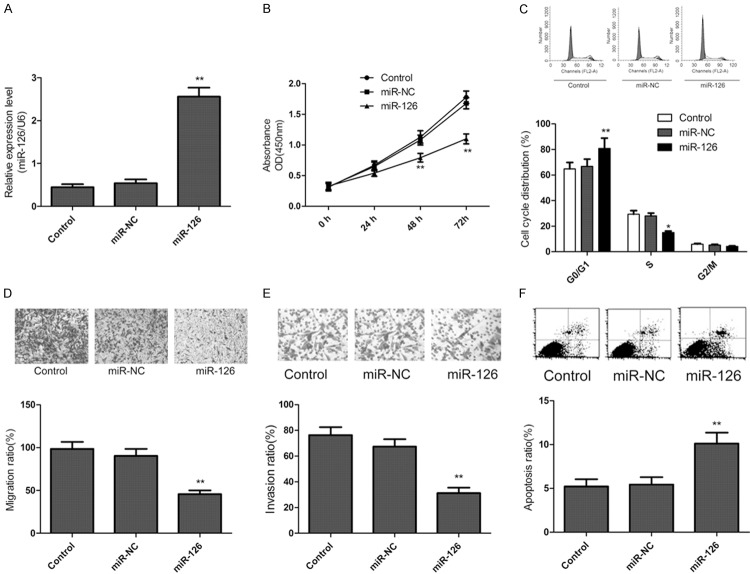
To explore the relevance of miR-126 and glioma cell growth, miR-126 mimic or miR-NC was transfected into U87 cells. The intracellular level of miR-126 was significantly higher in U87 cells after transfecting with miR-126 mimic compared with the miR-NC group (Figure 2A). Then, cell proliferation rate was measured using CCK-8 assays. It was found that restored expression of miR-126 led to significantly decrease of proliferation in U87 cells (Figure 2B). As proliferation directly linked to cell cycle distribution, we investigate the effect of miR-126 on cell cycle progression. As expected, the percentage of S phase cells decreased, and the percentage of G1 phase cells increased in U87 cells transfected with miR-126 mimic compared to the miR-NC group (Figure 2C).Furthermore, to reveal the biological role of miR-126 on migration and invasion, transwell assay were performed. We find that restored expression of miR-126 could significantly inhibit migration (Figure 2D) and invasion (Figure 2E) in U87 cells. In addition, to reveal miR-126 on apoptosis in U87 cells, PE Annexin-V staining was used. As shown in Figure 2F, transfection with miR-126 increased cells apoptosis compared to miR-NC and control group. These findings indicated that ectopic expression of miR-126 inhibited cell proliferation, migration and invasion and induced cell apoptosis of glioma cells.
Figure 2.
Ectopic expression of miR-126 inhibits cell proliferation, migration and invasion and induces cell apoptosis of glioma cells. (A) The expression of miR-126 was detected in U87 cells by qRT-PCR after transfected with miR-126 mimic or miR-NC. (B) Cell viability was measured at posttransfection by CCK-8 assay. (C) Cell cycle was measured in glioma cells at posttransfection by flow cytometry. (D, E) Cell migration (D) and invasion (E) were determined in glioma cells by transwell assay after transfected with miR-126 mimic or miR-NC. (F) Cell apoptosis were measured in glioma cells by flow cytometry after transfected with miR-126 mimic or miR-NC. *P < 0.05, **P < 0.01 versus control.
IRS-1 is a direct target of miR-126
IRS-1 has been identified as a direct target of miR-126 in breast [24], colorectal [25], and colon cancer cells [26]. However, the link between miR-126 and IRS-1 is still unclear in glioma. To verify whether IRS-1 is a direct target of miR-126 in glioma, a human IRS 3’ UTR fragment containing the binding sites of miR-126 or the mutant sites (Figure 3A) was cloned to were cloned into the psiCHECK2 vector (Promega, USA), then together with miR-126 or miR-NC were cotransfected into U87 cells, and cultured for 48 h, and luciferase activities in those cells were measured. We found that luciferase activities were significantly reduced in those cells transfected with the wild sequence IRS-1 and miR-126, but not in the cells with the mutant sequence IRS-1 and miR-126 (Figure 3B). Then, qRT-PCR and western blotting analysis was performed to measure IRS-1 on mRNA and protein level, we found that the expression of IRS-1 was downregulated both mRNA level (Figure 3C) and protein level (Figure 3D) in miR-126 treated cells. These results suggest that miR-126 directly targets IRS-1 by binding its seed region to their 3’-UTRs in glioma cells.
Figure 3.
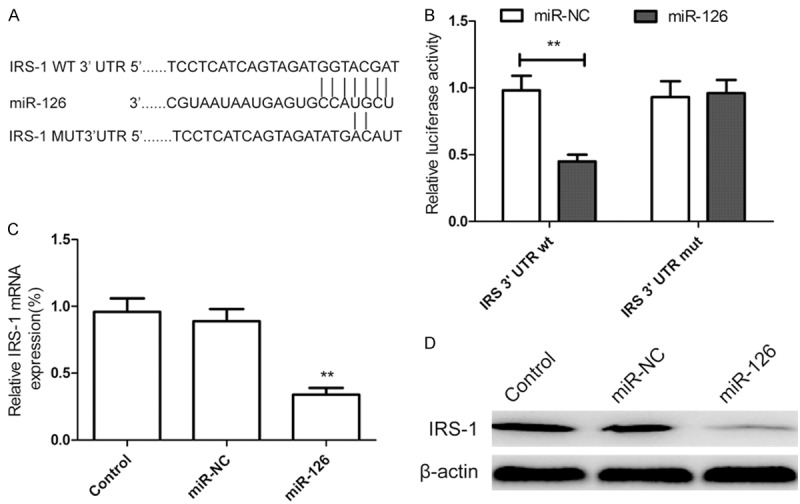
miR-126 targets the 3’-UTR of IRS-1 and down-regulates its expression. A. The predicted binding sites for miR-126 in the 3’UTR of IRS-1 and the mutations in the binding sites are shown. B. U87 cells were transfected with the wild type or mutated version of the luciferase-IRS 3’-UTR reporter vector as well as the miR-126 mimics or miR-NC for 48 h, then luciferase reporter assay were performed. C. Measurement of IRS-1 mRNA expression levels in U87 cells by qRT-PCR after transfected with the miR-126 mimics or miR-NC. The endogenous expression levels of the β-actin mRNA were used for normalization. D. Measurement of IRS-1 expression levels in U87 cells by western blot analysis after transfected with the miR-126 mimics or miR-NC. The endogenous expression levels of the β-actin protein were used for normalization. *P < 0.05, **P < 0.01 versus control.
Effects of miR-126 on PI3K/AKT signaling pathways
The PI3K/AKT signaling pathway plays an important role in carcinogenesis for several types of cancer including glioma [27]. In addition, it has been confirmed that IRS-1 transduces extracellular signals into cells through AKT signaling pathways via miR-126 in colorectal cancer cells [25]. To determine whether there were some changes in this signaling pathway in miR-126-transfected cells, we examined the phosphorylation status of PI3K and AKT by western blot. It was found that miR-126 overexpression led to decreased levels of phosphorylated PI3K and AKT (Figure 4), while there was no difference in the levels of total PI3K and AKT in each group (Figure 4). These data suggest that miR-126 may be an important regulator in the PI3K/AKT signaling pathways.
Figure 4.
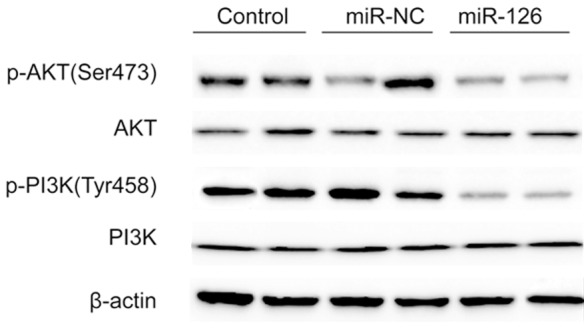
Effects of miR-126 on activation of PI3K/AKT signaling pathways in glioma cells. PI3K, p-PI3K, AKT, p-AKT protein expression levels in U87 cells were determined by western blot after transfected with miR-126 mimic or miR-NC. The endogenous expression levels of the β-actin protein were used for normalization.
Overexpression of miR-126 inhibits tumor growth in vivo. As we showed overexpression of miR-126 played important role in inhibiting glioma cells growth in vitro, we further investigated whether miR-126 would have similar antitumor effect in vivo. U87 cells and U87 cells stable expression miR-NC or miR-126 were subcutaneously inoculated in nude mice (n = 10 for each group). The sizes of U-87 MG tumors established in mice were measured with caliper every five today. The results revealed that tumor volumes were significantly smaller in the group treated with miR-126 mimic than controls group and miR-NC group (Figure 5A). The tumors were extracted 30 days after implantation, tumor weight were measured and found that tumors weight were significantly lower in miR-126 mimic group compared to control group and miR-NC group (Figure 5B, 5C). In addition, we also determined miR-126 expression level in xenograft tumors by qRT-PCR. The results showed that miR-126 expression was upregulated in the xenograft tumors of miR-126 mimic group compared to the xenograft tumors of miR-NC group and control group (P < 0.05, Figure 5D). These results might imply that upregulation of miR-126 could inhibit glioma growth in vivo.
Figure 5.
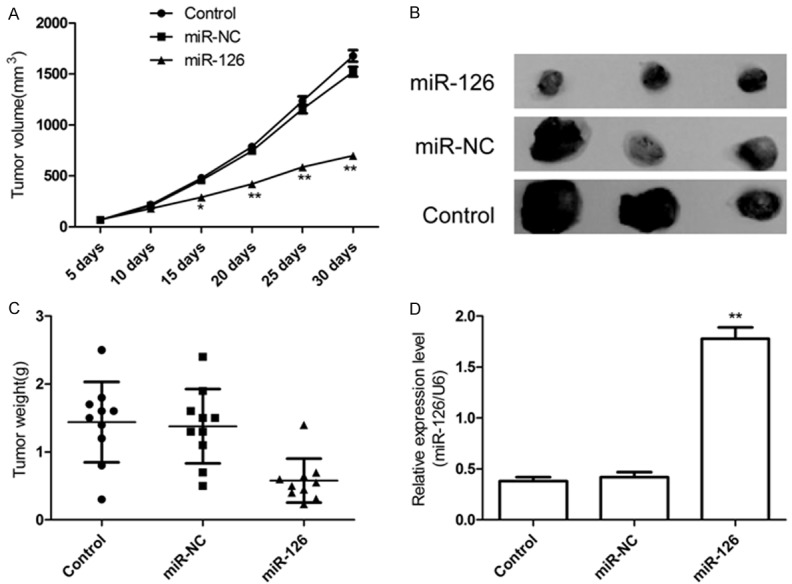
miR-126 suppressed tumor growth of glioma in a xenograft model. A. Growth curves for tumor volumes in xenografts of nude mice were established based on the tumor volume measured every five days until 30 days. B. Photographs of tumor tissue with different group at day 30. C. Tumor weights of different group at day 30. D. miR-126 level in tumor tissues of different group was determined by qRT-PCR. *P < 0.05, **P < 0.01 versus control group.
Discussion
MicroRNAs (miRNA), a novel class of regulatory molecules, have been showed to play important role in regulation of tumorigenesis, differentiation, proliferation, and survival through the inhibition of major cellular pathways in diverse human cancers [28]. miRNAs have been documented to function both as tumor suppressor genes and oncogenes, regulating many cellular events, such as proliferation, apoptosis, migration, differentiation and invasion [28]. Recent studies have been reported that miR-126 plays a potential role as a tumor suppressor in many kinds of cancers [14-25]. However, there are no results referring to the role of miR-126 in glioma so far. In this study, to our knowledge, we first showed that the expression of miR-126 is downregulated in glioma samples and glioma cell lines. Moreover, overexpression of miR-126 significantly inhibited glioma cell proliferation, migration, invasion, and induced cell apoptosis in vitro and suppressed tumor growth in vivo.
IRS-1, a docking protein, is highly expressed in many kinds of cancers such as pancreatic cancer [29], breast [24], colorectal [25], and colon cancer cells [26]. Consistent with these results, the present study showed that IRS-1 protein expression is up-regulated in glioma tissue compared with normal brain tissue (Figure 3D). Dearth et al found that transgenic mice over-expressing IRS-1 or IRS-2 in the mammary gland could result in progressive mammary hyperplasia, tumorigenesis, and metastasis [30].
Growing evidence have been showed that that IRS-1 could act as oncogene, and play major roles on in cell growth, proliferation, migration, invasion and differentiation [24,26,29,31]. Of note, IRS-1 has been identified as a direct target of miR-126 in several human cancer cells [24-26]. Here, we also found that insertion of the wild type of miR-126 binding site in the IRS-1 3’UTR in a dual-luciferase reporter construct led to a reduction of the reporter gene expression in glioma cells; and that overexpression of miR-126 induced the inhibition of IRS-1, specifically, suggesting that miR-126 directly targets IRS-1 by binding its seed region to their 3’-UTRs in glioma cells.
IRS-1, as an adaptor of IGF1R, has been showed to play an important role in cell growth and proliferation mainly through the PI3K/Akt pathway [24,31]. In addition, recently a study showed that miR-126 may act as a tumor suppressor in colon cancer by downregulating the p58b subunit through the PI3K signaling pathway [32]. Another study indicated found that up-regulation of miR-126 could down-regulate IRS-1, inhibit colorectal cancer cells proliferation, migration, invasion, and induce cell cycle arrest partly through AKT and ERK1/2 signaling pathways. In the present study, our results showed that up-regulation of miR-126 could inhibit IRS-1 expression and suppress activation PI3K/AKT signal pathway in glioma, suggesting that miR-126 functions as a tumor suppressor in glioma block glioma cell growth by regulating IRS-1 expression via the PI3K/AKT signaling pathways.
In summary, the present study provides evidence that miR-126 are down-regulated in glioma tissues and glioma cell line, its downregulation negative correlated with advanced WHO grades, that miR-126 functions as a tumor suppressor to inhibit cell proliferation, migration and invasion, and to induce cell apoptosis and cell cycle arrest at G0/G1 phase in glioma cells, as well as to suppress tumor growth of glioma in nude mice model. This study suggested that IRS-1 is one of target genes of miR-126. In addition, miR-126 decreased IRS-1 expression and inhibited activation of PI3K/AKT signaling pathways. These results suggested that miR-126 may be a novel candidate for glioma therapeutics.
Disclosure of conflict of interest
None.
References
- 1.Reardon DA, Rich JN, Friedman HS, Bigner DD. Recent advances in the treatment of malignant astrocytoma. J. Clin. Oncol. 2006;24:1253–1265. doi: 10.1200/JCO.2005.04.5302. [DOI] [PubMed] [Google Scholar]
- 2.Holland EC. Gliomagenesis: genetic alterations and mouse models. Nat Rev Genet. 2001;2:120–129. doi: 10.1038/35052535. [DOI] [PubMed] [Google Scholar]
- 3.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Malignant astrocytic glioma: genetics, biology and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 4.Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN, Cavenee WK, DePinho RA. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- 5.Clarke J, Butowski N, Chang S. Recent advances in therapy for glioblastoma. Arch Neurol. 2010;67:279–283. doi: 10.1001/archneurol.2010.5. [DOI] [PubMed] [Google Scholar]
- 6.Babu R, Kranz PG, Agarwal V, McLendon RE, Thomas S, Friedman AH, Bigner DD, Adamson C. Malignant brainstem gliomas in adults: clinicopathological characteristics and prognostic factors. J Neurooncol. 2014;119:177–185. doi: 10.1007/s11060-014-1471-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grauer OM, Wesseling P, Adema GJ. Immunotherapy of diffuse gliomas: biological background, current status and future developments. Brain Pathol. 2009;19:674–693. doi: 10.1111/j.1750-3639.2009.00315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. Int J Genomics. 2014;2014:970607. doi: 10.1155/2014/970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 10.Akaza H, Kawahara N, Masui T, Takeyama K, Nogimori M, Roh JK. Union for International Cancer Control International Session: healthcare economics: the significance of the UN Summit non-communicable diseases political declaration in Asia. Cancer Sci. 2013;104:773–778. doi: 10.1111/cas.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 13.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khella HW, Scorilas A, Mozes R, Mirham L, Lianidou E, Krylov SN, Lee JY, Ordon M, Stewart R, Jewett MA, Yousef GM. Low Expression of miR-126 Is a Prognostic Marker for Metastatic Clear Cell Renal Cell Carcinoma. Am J Pathol. 2015;185:693–703. doi: 10.1016/j.ajpath.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 15.Zhao C, Li Y, Zhang M, Yang Y, Chang L. miR-126 inhibits cell proliferation and induces cell apoptosis of hepatocellular carcinoma cells partially by targeting Sox2. Hum Cell. 2015;28:91–9. doi: 10.1007/s13577-014-0105-z. [DOI] [PubMed] [Google Scholar]
- 16.Jiang L, Tao C, He A, He X. Overexpression of miR-126 sensitizes osteosarcoma cells to apoptosis induced by epigallocatechin-3-gallate. World J Surg Oncol. 2014;12:383. doi: 10.1186/1477-7819-12-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H, Li L, Wang S, Lei Y, Ge Q, Lv N, Zhou X, Chen C. Reduced miR-126 expression facilitates angiogenesis of gastric cancer through its regulation on VEGF-A. Oncotarget. 2014;5:11873–11885. doi: 10.18632/oncotarget.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim MK, Jung SB, Kim JS, Roh MS, Lee JH, Lee EH, Lee HW. Expression of microRNA miR-126 and miR-200c is associated with prognosis in patients with non-small cell lung cancer. Virchows Arch. 2014;465:463–471. doi: 10.1007/s00428-014-1640-4. [DOI] [PubMed] [Google Scholar]
- 19.Yamaguchi T, Iijima T, Wakaume R, Takahashi K, Matsumoto H, Nakano D, Nakayama Y, Mori T, Horiguchi S, Miyaki M. Underexpression of miR-126 and miR-20b in hereditary and nonhereditary colorectal tumors. Oncology. 2014;87:58–66. doi: 10.1159/000363303. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, Zheng ZM. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li N, Li X, Huang S, Shen S, Wang X. [miR-126 inhibits colon cancer proliferation and invasion through targeting IRS1, SLC7A5 and TOM1 gene] . Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2013;38:809–817. doi: 10.3969/j.issn.1672-7347.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 22.Ebrahimi F, Gopalan V, Smith RA, Lam AK. miR-126 in human cancers: clinical roles and current perspectives. Exp Mol Pathol. 2014;96:98–107. doi: 10.1016/j.yexmp.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Liu LY, Wang W, Zhao LY, Guo B, Yang J, Zhao XG, Hou N, Ni L, Wang AY, Song TS, Huang C, Xu JR. Mir-126 inhibits growth of SGC-7901 cells by synergistically targeting the oncogenes PI3KR2 and Crk and the tumor suppressor PLK2. Int J Oncol. 2014;45:1257–1265. doi: 10.3892/ijo.2014.2516. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Du YY, Lin YF, Chen YT, Yang L, Wang HJ, Ma D. The cell growth suppressor, mir-126, targets IRS-1. Biochem Biophys Res Commun. 2008;377:136–140. doi: 10.1016/j.bbrc.2008.09.089. [DOI] [PubMed] [Google Scholar]
- 25.Zhou Y, Feng X, Liu YL, Ye SC, Wang H, Tan WK, Tian T, Qiu YM, Luo HS. Down-regulation of miR-126 is associated with colorectal cancer cells proliferation, migration and invasion by targeting IRS-1 via the AKT and ERK1/2 signaling pathways. PLoS One. 2013;8:e81203. doi: 10.1371/journal.pone.0081203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li N, Li X, Huang S, Shen S, Wang X. [miR-126 inhibits colon cancer proliferation and invasion through targeting IRS1, SLC7A5 and TOM1 gene] . Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2013;38:809–817. doi: 10.3969/j.issn.1672-7347.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 27.Sami A, Karsy M. Targeting the PI3K/AKT/mTOR signaling pathway in glioblastoma: novel therapeutic agents and advances in understanding. Tumour Biol. 2013;34:1991–2002. doi: 10.1007/s13277-013-0800-5. [DOI] [PubMed] [Google Scholar]
- 28.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 29.Bergmann U, Funatomi H, Kornmann M, Beger HG, Korc M. Increased expression of insulin receptor substrate-1 in human pancreatic cancer. Biochem Biophys Res Commun. 1996;220:886–890. doi: 10.1006/bbrc.1996.0500. [DOI] [PubMed] [Google Scholar]
- 30.Dearth RK, Cui X, Kim HJ, Kuiatse I, Lawrence NA, Zhang X, Divisova J, Britton OL, Mohsin S, Allred DC, Hadsell DL, Lee AV. Mammary tumorigenesis and metastasis caused by overexpression of insulin receptor substrate 1 (IRS-1) or IRS-2. Mol Cell Biol. 2006;26:9302–9314. doi: 10.1128/MCB.00260-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baserga R. The contradictions of the insulin-like growth factor 1 receptor. Oncogene. 2000;19:5574–5581. doi: 10.1038/sj.onc.1203854. [DOI] [PubMed] [Google Scholar]
- 32.Guo C, Sah JF, Beard L, Willson JK, Markowitz SD, Guda K. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer. 2008;47:939–946. doi: 10.1002/gcc.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]