Abstract
Gliomas are the most common and aggressive form of primary brain tumor. Although EGF-containing fibulin-like extracellular matrix protein 2 (EFEMP2), an extracellular matrix (ECM) glycoprotein, is regarded as a candidate oncogene, little is known about the association of EFEMP2 and gliomas. Here, the expression of EFEMP2 was significantly increased in glioma tissues (n=60) compared to non-tumorous brain tissues (n=25). Silencing of EFEMP2 expression through RNA interference in two glioma cell lines (U87 and U373) remarkably inhibited cell proliferation and G1/S transition. More importantly, EFEMP2 silencing significantly induced cell apoptosis via increasing the ratio of Bax and Bcl-2. Additionally, knockdown of EFEMP2 significantly inhibited the invasive ability of both glioma cells, which was associated with the downregulated expression of metalloproteinase-2 (MMP-2) and MMP-9. In conclusion, expression of EFEMP2 was associated with the oncogenic potential of gliomas and silencing of its expression can suppress cancer cell growth and metastasis. Inhibition of EFEMP2 may be a therapeutic strategy for gliomas.
Keywords: Apoptosis, EFEMP2, G1/S transition, gliomas, metalloproteinase
Introduction
Gliomas, arising from glia cells, are the most common primary brain tumor [1]. According to the World Health Organization (WHO) classification, gliomas are classified into four grades (I, II, III and IV), of which glioblastoma multiforme (GBM, WHO grade IV ) is the most frequent and aggressive malignant type [2]. Regardless of recent advances in the diagnostic and therapeutic strategies, glioma remains one of the most lethal cancer worldwide [3]. The median survival of patients with GBM is still less than one year [4]. The advances in glioma therapy have been restricted because the pathophysiological mechanisms underlying the development of this disease are not fully known. Therefore, elucidating the molecular mechanism for the glioma development will help the identification of novel therapeutic targets and the development of strategies for glioma treatment.
Fibulins are a family of extracellular matrix (ECM) glycoproteins defined by epidermal growth factor (EGF)-like domains, calcium-binding-EGF-like domains and a unique fibulin C-terminal domain [5]. Gene-targeting experiments in animal models [6-10] and the identification of heritable diseases in humans [11-13] have recently highlighted the importance of fibulins in development and disease. EGF-containing fibulin-like extracellular matrix protein 2 (EFEMP2, also known as MBP1 orfibulin-4), a member of fibulins, has been reported as a candidate oncogene [14]. The ectopic over expression of EFEMP2 increased both neoplastic transformation and tumor-cell growth in a human lung carcinoma cell line. Moreover, EFEMP2 was found to be elevated in human colorectal tumors at the mRNA level [15]. However, it remains to be seen whether EFEMP2 expression is also altered in gliomas and whether it exerts biological functions in gliomas.
In the present study, we found that EFEMP2 mRNA expression was significantly increased in glioma tissues compared with non-neoplastic brain tissues. Then, we investigated the biological function of EFEMP2 in glioma cell lines by RNA interference. We found that EFEMP2 was involved in multiple cellular progress including cell proliferation, cell cycle progression, apoptosis and invasion. These data suggest that EFEMP2 is a potent oncogene in glioma and a possible target for the treatment of glioma.
Materials and methods
Tissue collection
Fresh frozen human tissue samples of 60 glioma and 25 non-neoplastic brain tissues from surgical procedures for epilepsy were obtained from Department of Neurosurgery, Renmin Hospital of Wuhan University (Wuhan, China) between January 2008 and December 2009. The research has been carried out in accordance with the Declaration of Helsinki (2008) of the World Medical Association. All investigations described in this study were performed after written informed consent was obtained and in accordance with the guidelines of the local independent ethics committee at Department of Neurosurgery, Renmin Hospital of Wuhan University (Wuhan, China).
Real-time reverse transcription PCR
Total RNA was extracted from tissue samples or from the cell lines using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. cDNA was generated by using a first-strand cDNA synthesis kit (Fermentas, Hanover, MD, USA), amplified by using a standard SYBR Green PCR kit (Thermo Fisher Scientific, Rockford, IL, USA) and loaded on the 7300 Real-time PCR Detection System (Applied Biosystems, Foster City, CA, USA). The cycling parameters were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 45 s. PCR primers used were as follows: EFEMP2 (NM_016938.4), forward primer: 5’-CCCAGAGATTTGGACTTG-3’; reverse primer: 5’-GTTTGAGGCAGAGTTAGG-3’; GAPDH (NM_001256799.1), forward primer: 5’-CACCCACTCCTCCACCTTTG-3’; reverse primer: 5’-CCACCACCCTGTTGCTGTAG-3’.
Cell culture and gene silencing
The human glioblastoma cell lines, U251, SHG44, U373, U87 and U138 were from cell bank of Shanghai Biology Institute, Chinese Academy of Science (Shanghai, China) and maintained in a humidified atmosphere with 5% CO2 at 37°C. U373, U87 and U138 were cultured in Eagle’s Minimum Essential Medium (MEM, Hyclone, Logan, UT, USA), while U251 and SHG44 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Hyclone). All culture media were supplemented with 10% fetal bovine serum (FBS, Hyclone) and 100 U/mL penicillin/streptomycin (Gibco, Carlsbad, CA, USA).
Small interfering (si) RNA oligonucleotides targeting EFEMP2 (siEFEMP2) and non-silencing siRNA (siNC) were purchased from GenePharma (Shanghai, China). Two glioma cell lines, U87 and U373 were transfected with siEFEMP2 or siNC using Lipofectamine 2000 (Invitrogen). All the following assays were performed 48 h after transfection.
Western blot analysis
Cell lysates were extracted with RIPA cell lysis buffer with protease inhibitor cocktail (Sigma, St. Louis, MO, USA). Protein samples were subjected to SDS gel electrophoresis and immunoblotting using antibodies against EFEMP2, Bcl-2, Bax, matrix metalloproteinase-2 (MMP-2), MMP-9 (Abcam, Cambridge, MA, USA), cleaved caspase 3, GAPDH (Cell Signaling Technology, Danvers, MA, USA). Detection of specific proteins was performed with enhanced chemiluminescence (ECL, Millipore, Bredford, USA). Band density was measured (ImageJ software) and normalized to GAPDH.
Cell viability assay
Cell viability was assayed using the Cell Counting Kit-8 (CCK-8, Dojindo Laboratories, Tokyo, Japan) as described by the manufacturer’s protocol. Briefly, cells were transfected with siEFEMP2 or siNC. After 2 days, transfected and WT cells were seeded onto 96-well plates at a density of 5×103 cells per well and incubated for 1, 2 or 3 days. Cells were then incubated with CCK-8 solution for 1 h. The absorbance value (optical density) of each well at 450 nm was measured. For each experimental condition, three wells were used. All experiments were repeated at least three times.
Analysis of cell cycle distribution
Treated and untreated cells were harvested, fixed in 70% ethanol at -20°C overnight, washed in PBS, and stained with 0.1 mg/ml propidium iodide (PI, Sigma) containing 1 mg/ml RNase A for 30 min at 37°C. DNA content was then analyzed on a FACScan flow cytometry.
Evaluation of cell apoptosis by flow cytometry
The percentage of cells actively undergoing apoptosis was determined. Treated and untreated cells were harvested, double-labeled with Annexin V-fluorescein isothiocyanate (FITC) and PI apoptosis detection kits (KeyGEN Biotech., Nanjing, China), and analyzed using a FACS can flow cytometry. The experiments were performed in triplicate.
Cell invasion assays
In vitro cell invasion assays were performed using 24-well transwell chambers (8-μm pores, Coring Incorporated, NY, USA) with Matrigel-precoated membrane. The treated and untreated cells were cultured in the top chamber with serum-free media at a density of 5×104 cells per well. In the lower chamber, media with 10% FBS was added as chemoattractant. After 24 h, the chamber was gently wiped to remove cells on the top of the membrane. Invaded cells were fixed in 4% paraformaldehyde, stained with 0.5A% crystal violet solution and counted in five fields (×200) under a microscope.
Statistical analysis
Data are presented as mean ± S.D of at least three independent experiments performed in triplicate. Two-tailed Student’s t-test was used for comparisons between groups. Differences were considered statistically significant when P<0.05.
Results
EFEMP2 mRNA was overexpressed in human glioma tissues
We first analyzed EFEMP2 mRNA levels in 60 snap-frozen glioma tissues and 25 non-neoplastic brain tissues by real-time PCR. EFEMP2 expression was up-regulated in glioma tissues (1.39±0.07), compared with non-neoplastic brain tissues (0.64±0.06, Figure 1, P<0.0001).
Figure 1.
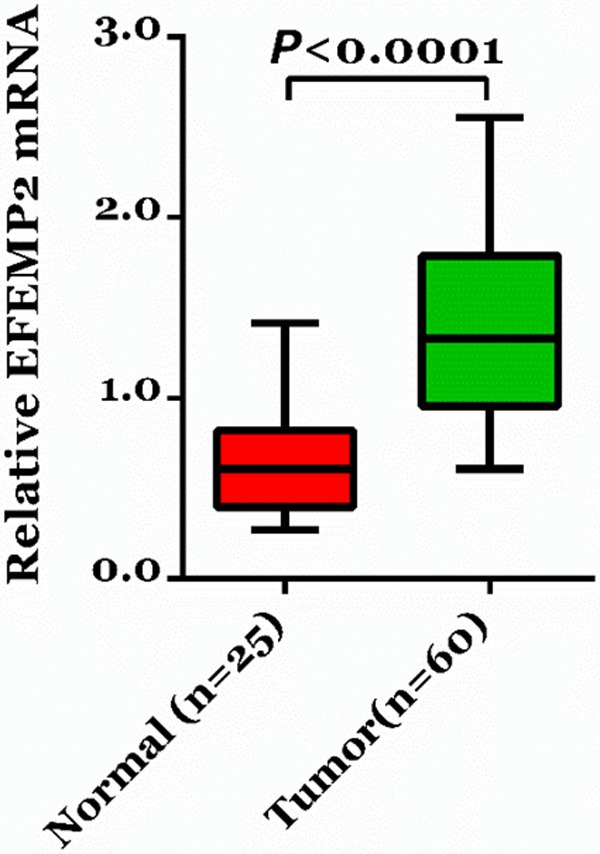
Over expression of EFEMP2 in glioma tissues. The mRNA level of EFEMP2 in glioma and non-neoplastic brain tissues collected from patients admitted to Department of Neurosurgery, Renmin Hospital of Wuhan University (Wuhan, China) between January 2008 and December 2009 was detected by real-time PCR. EFEMP2 mRNA was significantly higher in glioma tissues as compared with non-neoplastic tissues (P<0.0001).
Knockdown of EFEMP2 expression by siRNA transfection
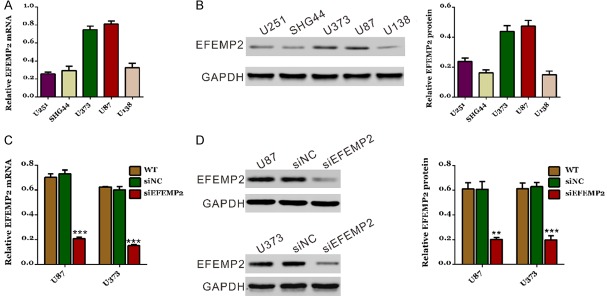
To investigate the functional role of EFEMP2 in glioma cells, its expression was detected in diverse glioma cell lines. mRNA (Figure 2A) and protein levels (Figure 2B) of EFEMP2 were higher in U373 and U87 cells, which were chosen for the following assays. As shown in Figure 2C and 2D, EFEMP2-siRNA efficiently suppressed endogenous EFEMP2 expression in both glioma cells, whereas EFEMP2 expression remained unaffected in siNC-transfected cells.
Figure 2.
Knockdown of EFEMP2 expression by siRNA transfection. EFEMP2 expression level in five glioma cell lines was analyzed by real-time PCR (A) and Western blot (B). (C, D) RNAi efficiency of EFEMP2 in U87 and U373 cells was analyzed by real-time PCR (C) and Western blot (D). GAPDH was also detected as the control of sample loading. Representative western blots (left panel) and quantitative results (right panel) were shown. siNC: scrambled siRNA transfected cells; siEFEMP2: EFEMP2-siRNA transfected cells (***P<0.001 V.S. siNC).
Silencing of EFEMP2 inhibited growth and cell cycle progression of glioma cells
Subsequently, cell proliferation was detected by CCK-8 assay (Figure 3A and 3B). Silencing of EFEMP2 led to a significant reduction at 24 h, 48 h and 72 h in viability compared with siNC group both in U87 and U373 cells (P<0.01). These results indicated the inhibitory role of EFEMP2-siRNA in the proliferation of glioma cells.
Figure 3.
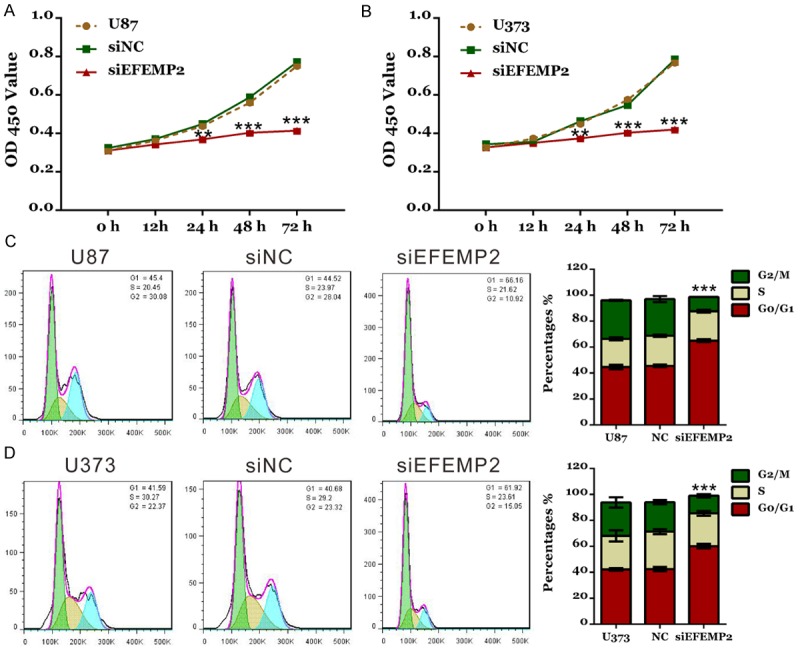
Silencing of EFEMP2 suppressed cell growth and G1/S phase transition in glioma cells. A, B: Cell proliferation was detected by CCK-8 assay in U87 and U373 cells. C, D: Cell cycle profile was analyzed using PI staining and flow cytometry. siNC: scrambled siRNA transfected cells; siEFEMP2: EFEMP2-siRNA transfected cells (**P<0.01, ***P<0.001 V.S. siNC).
To explore whether EFEMP2 affected the cell cycle of glioma cells, cell cycle distribution was then evaluated by PI staining and flow cytometry analysis (Figure 3C and 3D). Our data revealed that EFEMP2-siRNA transfection induced a remarkable G1 cell cycle arrest. Compared with the corresponding control cells, the population of G0/G1 phase cells in U87 and U373 with EFEMP2 silenced was significantly increased by 42.5% and 41.6%, respectively (P<0.001).
Knockdown of EFEMP2 induced apoptosis of glioma cells
We then assessed the apoptotic function of EFEMP2 in glioma cells by Annexin V-FITC/PI staining assay. As shown in Figure 4A and 4B, knockdown of EFEMP2 in both glioma cells resulted in a notable increase in cell apoptosis compared with corresponding control cells.
Figure 4.
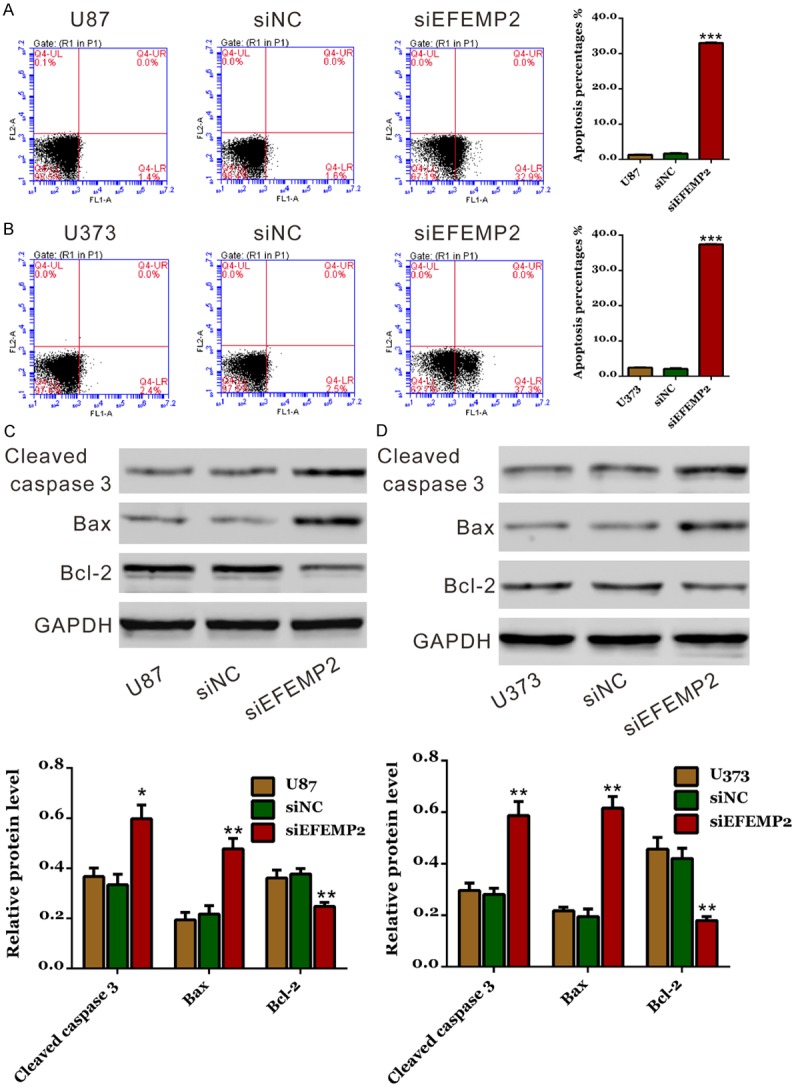
EFEMP2 knockdown induced cell apoptosis in glioma cells. A, B: Cells were stained with Annexin V-FITC/PI and apoptotic rates were analyzed using flow cytometry. C, D: Expression of apoptotic factors, cleaved caspase 3, Bcl-2 and Bax was evaluated by western blot. siNC: scrambled siRNA transfected cells; siEFEMP2: EFEMP2-siRNA transfected cells (*P<0.05, **P<0.01, ***P<0.001 V.S. siNC).
The effects of EFEMP2 siRNA on the protein levels of apoptosis regulating proteins, cleaved caspase 3, Bax and Bcl-2 were investigated. EFEMP2 knockdown in U87 and U373 cells significantly increased the expression of the apoptosis marker protein (cleaved caspase 3) and apoptosis promoting protein (Bax), but notably decrease the expression of the anti-apoptosis protein (Bcl-2) (Figure 4C and 4D). The ratio of Bax/Bcl2 was significantly increased when EFEMP2 expression was suppressed. These results indicated the anti-apoptosis role of EFEMP2 in glioma cells.
Silencing of EFEMP2 suppressed the invasive ability of glioma cells
To investigate the function of EFEMP2 in the metastasis, the invasive ability of U87 and U373 cells were evaluated by transwell assay (Figure 5A and 5B). Silencing of EFEMP2 expression significantly reduced the invasive ability of both glioma cells compared with control cells. The number of invaded knockdown cells was about 60.2% and 56.1% of that of the control cells in U87 and U373 cells, respectively (U87: WT, 83±9; siNC, 83±5; siEFEMP2, 50±6; U373: WT, 80±9; siNC, 82±13; siEFEMP2, 46±5). These data suggested a promotion role of EFEMP2 in glioma cell invasion.
Figure 5.
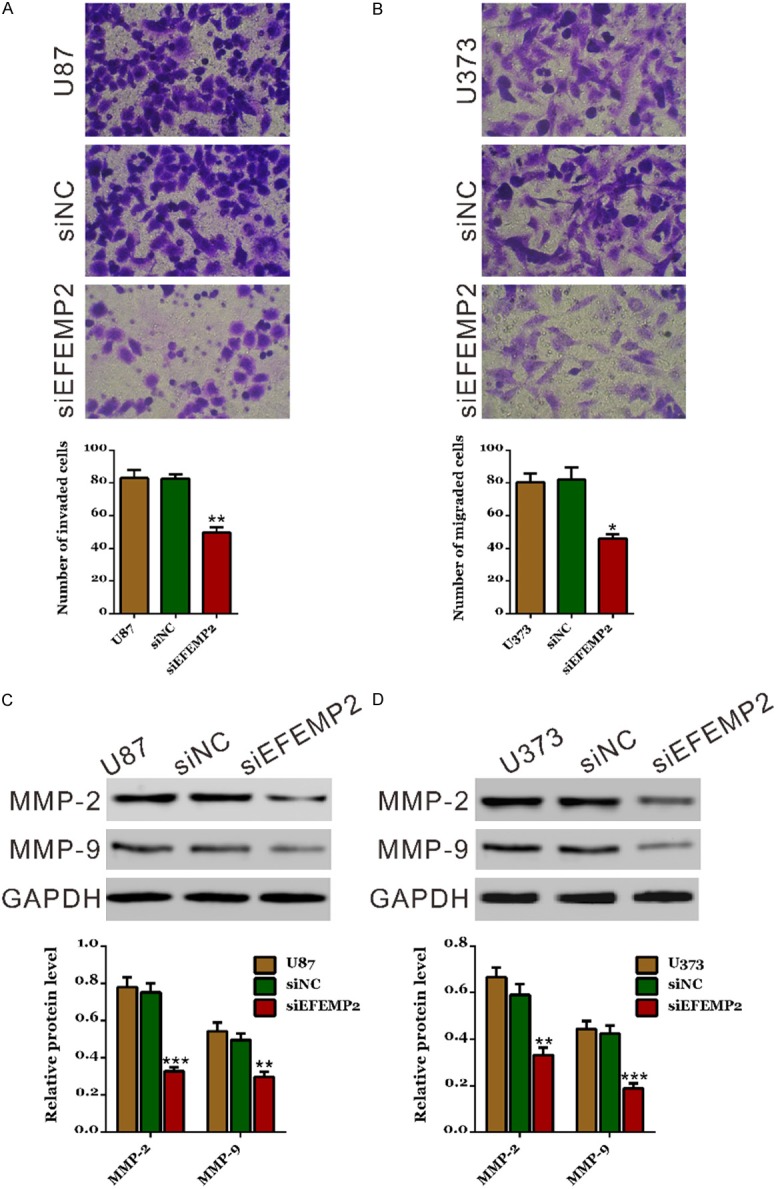
Suppressing EFEMP2 expression repressed cell invasion in glioma cells. A, B: Invasion assay of control and EFEMP2 knockdown cells in Matrigel-coated transwell chambers. Cells that invaded into the lower well were stained, photographed and counted under a microscope in five fields (×200). Representative images and quantitative results of cell invasion assay were shown. C, D: Expression of MMP-2 and MMP-9 was evaluated by Western blot. siNC: scrambled siRNA transfected cells; siEFEMP2: EFEMP2-siRNA transfected cells (*P<0.05, **P<0.01, ***P<0.001 V.S. siNC).
Moreover, the expression levels of important factors to regulate invasion were then estimated by Western blot. The mRNA and protein levels of MMP-2 and MMP-9 were significantly down-regulated by EFEMP2 knockdown in both glioma cells. These results further indicated that the EFEMP2 played an important role in the invasion of glioma cells.
Discussion
A previous study has proposed that EFEMP2 is an oncogene by in vitro experiments [14]. High expression of EFEMP2 is observed in colorectal cancer [15,16] although its functions in tumors are not known. Hu et al. identified fibulin-3, another member of fibulins, as proinvasive molecule specific to the glioma ECM [17]. In the present study, we found that EFEMP2 mRNA levels were raised in glioma tissues, which suggested a role of EFEMP2 in gliomas (Figure 1). Then we investigated the biological functions of EFEMP2 in two glioma cell lines (U87 and U373) by suppressing its expression.
Uncontrolled proliferation is a hallmark of cancer cells. Here, we found that knockdown of EFEMP2 in glioma cells significantly suppressed cell growth (Figure 3A and 3B). Deregulation of cell cycle occurs frequently in most common malignancies, resulting in aberrant cell proliferation [18]. Suppressing of EFEMP2expression by siRNA significantly induced G1-phase arrest of cell cycle progression (Figure 3C and 3D), which indicated that the repression of cell proliferation is owing to cell cycle inhibition. Furthermore, G1-phase arrest cells either undergo repairing or apoptosis process. Apoptosis is considered as a protective mechanism against cancer progression [18]. Here, our data demonstrated that silencing of EFEMP2 resulted in a significant induction of apoptosis by Annixin V-PI staining in glioma cells (Figure 4A and 4B), which was further confirmed by the increased expressed of apoptosis marker [19], cleaved caspase 3 (Figure 4C and 4D). The proteins of the Bcl-2 family either promote (e.g., Bax) or inhibit programmed cell death (e.g., Bcl-2) [20]. EFEMP2 knockdown notably increased Bax expression, but decreased Bcl-2 expression (Figure 4C and 4D), which suggested that EFEMP2 siRNA promoted cell apoptosis via up-regulating the ratio of Bax/Bcl-2. Because of its anti-apoptosis role in gliomas, EFEMP2 may be a potential therapy target worth further investigation.
Glioma cells have highly invasive ability to invade adjacent areas by degrading ECM components and diffusing into normal brain tissue [21]. Matrix metalloproteinases, including MMP-2 and MMP-9, play a critical role in tumor invasion and metastasis [22]. Here, our data indicated that EFEMP2 siRNA notably inhibited the invasion of glioma cells, and decrease the expression of MMP-2 and MMP-9 (Figure 5). Taken together, we speculated that EFEMP2 may perform its invasion-promoting function through regulating MMP-2 and MMP-9.
In summary, our study provides for the first time that EFEMP2 was overexpressed in glioma tissues. In addition, our data indicated that EFEMP2 played an essential role in the proliferation, apoptosis and invasion of glioma cells. EFEMP2 is a potentially important, tumor-specific, highly accessible ECM target for therapy strategies of gliomas.
Disclosure of conflict of interest
None.
References
- 1.Avgeropoulos NG, Batchelor TT. New treatment strategies for malignant gliomas. Oncologist. 1999;4:209–224. [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Penas-Prado M, Gilbert MR. Molecularly targeted therapies for malignant gliomas: advances and challenges. Expert Rev Anticancer Ther. 2007;7:641–61. doi: 10.1586/14737140.7.5.641. [DOI] [PubMed] [Google Scholar]
- 4.Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109:93–108. doi: 10.1007/s00401-005-0991-y. [DOI] [PubMed] [Google Scholar]
- 5.Argraves WS, Greene LM, Cooley MA, Gallagher WM. Fibulins: physiological and disease perspectives. EMBO Rep. 2003;4:1127–1131. doi: 10.1038/sj.embor.7400033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McLaughlin PJ, Chen Q, Horiguchi M, Starcher BC, Stanton JB, Broekelmann TJ, Marmorstein AD, McKay B, Mecham R, Nakamura T, Marmorstein LY. Targeted disruption of fibulin-4 abolishes elastogenesis and causes perinatal lethality in mice. Mol Cell Biol. 2006;26:1700–1709. doi: 10.1128/MCB.26.5.1700-1709.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kostka G, Giltay R, Bloch W, Addicks K, Timpl R, Fässler R, Chu ML. Perinatal lethality and endothelial cell abnormalities in several vessel compartments of fibulin-1-deficient mice. Mol Cell Biol. 2001;21:7025–7034. doi: 10.1128/MCB.21.20.7025-7034.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamura T, Lozano PR, Ikeda Y, Iwanaga Y, Hinek A, Minamisawa S, Cheng CF, Kobuke K, Dalton N, Takada Y, Tashiro K, Ross J Jr, Honjo T, Chien KR. Fibulin-5/DANCE is essential for elastogenesis in vivo. Nature. 2002;415:171–175. doi: 10.1038/415171a. [DOI] [PubMed] [Google Scholar]
- 9.Sicot FX, Tsuda T, Markova D, Klement JF, Arita M, Zhang RZ, Pan TC, Mecham RP, Birk DE, Chu ML. Fibulin-2 is dispensable for mouse development and elastic fiber formation. Mol Cell Biol. 2008;28:1061–1067. doi: 10.1128/MCB.01876-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooley MA, Kern CB, Fresco VM, Wessels A, Thompson RP, McQuinn TC, Twal WO, Mjaatvedt CH, Drake CJ, Argraves WS. Fibulin-1 is required for morphogenesis of neural crest-derived structures. Dev Biol. 2008;319:336–345. doi: 10.1016/j.ydbio.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stone EM, Lotery AJ, Munier FL, Héon E, Piguet B, Guymer RH, Vandenburgh K, Cousin P, Nishimura D, Swiderski RE, Silvestri G, Mackey DA, Hageman GS, Bird AC, Sheffield VC, Schorderet DF. A single EFEMP1 mutation associated with both Malattia Leventinese and Doyne honeycomb retinal dystrophy. Nat Genet. 1999;22:199–202. doi: 10.1038/9722. [DOI] [PubMed] [Google Scholar]
- 12.Debeer P, Schoenmakers EF, Twal WO, Argraves WS, De Smet L, Fryns JP, Van De Ven WJ. The fibulin-1 gene (FBLN1) is disrupted in a t(12;22) associated with a complex type of synpolydactyly. J Med Genet. 2002;39:98–104. doi: 10.1136/jmg.39.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loeys B, Van Maldergem L, Mortier G, Coucke P, Gerniers S, Naeyaert JM, De Paepe A. Homozygosity for a missense mutation in fibulin-5 (FBLN5) results in a severe form of cutis laxa. Hum Mol Genet. 2002;11:2113–2118. doi: 10.1093/hmg/11.18.2113. [DOI] [PubMed] [Google Scholar]
- 14.Gallagher WM, Argentini M, Sierra V, Bracco L, Debussche L, Conseiller E. MBP1: a novel mutant p53-specific protein partner with oncogenic properties. Oncogene. 1999;18:3608–3616. doi: 10.1038/sj.onc.1202937. [DOI] [PubMed] [Google Scholar]
- 15.Gallagher WM, Greene LM, Ryan MP, Sierra V, Berger A, Laurent-Puig P, Conseiller E. Human fibulin-4: analysis of its biosynthetic processing and mRNA expression in normal and tumour tissues. FEBS Lett. 2001;489:59–66. doi: 10.1016/s0014-5793(00)02389-9. [DOI] [PubMed] [Google Scholar]
- 16.Yao L, Lao W, Zhang Y, Tang X, Hu X, He C, Hu X, Xu LX. Identification of EFEMP2 as a serum biomarker for the early detection of colorectal cancer with lectin affinity capture assisted secretome analysis of cultured fresh tissues. J Proteome Res. 2012;11:3281–3294. doi: 10.1021/pr300020p. [DOI] [PubMed] [Google Scholar]
- 17.Hu B, Thirtamara-Rajamani KK, Sim H, Viapiano MS. Fibulin-3 is uniquely upregulated in malignant gliomas and promotes tumor cell motility and invasion. Mol Cancer Res. 2009;7:1756–1770. doi: 10.1158/1541-7786.MCR-09-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 19.Riemenschneider MJ, Betensky RA, Pasedag SM, Louis DN. AKT activation in human glioblastomas enhances proliferation via TSC2 and S6 kinase signaling. Cancer Res. 2006;66:5618–5623. doi: 10.1158/0008-5472.CAN-06-0364. [DOI] [PubMed] [Google Scholar]
- 20.Adams JM, Cory S. The Bcl-2 protein family: arbiters of cell survival. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 21.Giese A, Westphal M. Glioma invasion in the central nervous system. Neurosurgery. 1996;39:235–252. doi: 10.1097/00006123-199608000-00001. [DOI] [PubMed] [Google Scholar]
- 22.Curran S, Murray GI. Matrix metalloproteinases in tumour invasion and metastasis. J Pathol. 1999;189:300–308. doi: 10.1002/(SICI)1096-9896(199911)189:3<300::AID-PATH456>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]