Abstract
Background: Dilated cardiomyopathy (DCM) is one type of primary myocardial disease, partly caused by immunity dysfunctions. BTNL2 (butyrophilin-like 2) has already been confirmed to be involved in the etiology of autoimmune disorders and GWAS (genome wide association study) has also identified mutants of a SNP (single nucleotide polymorphism) near BTNL2 could modulate risk of coronary heart disease (also cardiomyopathy). The current study, therefore, was aimed to investigate whether polymorphisms within or around BTNL2 would be correlated with susceptibility to and prognosis of DCM. Material and methods: Peripheral blood samples were gathered from 82 DCM patients and 75 healthy controls. Nine tag-SNPs within or near BTNL2 were obtained from HapMap Database and previously published studies. Eligible haplotypes were gained on the basis of SHesis software. Genotyping of SNPs was implemented with aid of Sequenom MassArray iPLEX platform and subsequently analyzed via MALDI-TOF mass spectrometry. The odd ratios and their 95% confidence interval (95% CI) were utilized to evaluate the correlations between SNPs/haplotypes and DCM risks. Finally, Cox proportional hazard models and Kaplan-Meier curves were performed to assess association of SNPs/haplotypes with prognosis of DCM patients. The statistical analyses were conducted with SPSS 19.0 software. Results: Under the allelic model, rs3763313 (A > C), rs9268494 (C > A), rs9268492 (C > G) and rs9268402 (A > G) were remarkably associated with susceptibility to grade IV of DCM classified by NYHA (New York heart association) (OR = 0.43, 95% CI: 0.22-0.84; P = 0.018; OR = 0.49, 95% CI: 0.27-0.91; P = 0.024; OR = 0.50, 95% CI: 0.27-0.94; P = 0.035; OR = 0.53, 95% CI: 0.28-0.97; P = 0.048). Haplotype C-C-A-T (rs9268492, rs9268494, rs3763313 and rs3763317 synthesized) was also regarded as a protective factor for DCM patients compared with carriers of other haplotypes (OR = 0.50, 95% CI: 0.26-0.97, P = 0.038). Moreover, the univariate survival analysis and multivariate Cox regression analysis both indicated noticeable correlations between rs9268402 and haplotype C-C-A-T and prognosis of DCM patients (NYHA IV), respectively (Long-Rank P = 0.029, HR: 0.241, 95% CI: 0.089-0.650, P = 0.005; Long-Rank P = 0.036; HR = 0.126, 95% CI: 0.035-0.457, P = 0.002). Nonetheless, rs3763313 was found only associated with prognosis of DCM patients (NYHA IV) expressed in the Kaplan-Meier curve (P = 0.009). Conclusion: The genetic mutations within or around BTNL2 (rs3763313, rs9268494, rs9268492 and rs9268402) could alter susceptibility to grade IV of DCM in a Chinese population, and the 2 SNPs (rs3763313 and rs9268402) therein added with haplotype C-C-A-T might separately predict the prognosis of DCM patients. However, additional studies regarding diverse ethnicities need to be furthered to validate our results.
Keywords: Dilated cardiomyopathy, butyrophilin-like 2, single nucleotide polymorphism, haplotype, susceptibility, prognosis
Introduction
Dilated cardiomyopathy (DCM) has been defined as a primary myocardial disorder, primarily embodied in augmentation of left or/right ventricles accompanied by failure of ventricle systole [1]. It has been demonstrated that DCM appears to be prevalent with an incidence of nearly 0.365‰ population per year, merely a half of whom would ultimately gain a survival in excess of 5 years since their initial diagnosis [2,3]. Despite estimations that the occurrence of DCM would be exacerbated commonly due to myocardial ischemia, virus infection, metabolic and immunologic dysfunctions and so on, approximately a third of DCM sufferers could be attributed to inherited causes, among which mutations of autosomal dominants with decreased penetrance could not be ignored [4-6]. The identified genes (e.g. ACTC1, TPM1, TNNT2, TNNC1, TNNI3, MYH7, TTN) were chiefly concerned with encoding of sarcomeric proteins, such as filamentous actin, tropomyosin, troponin subunits, β-myosin heavy chain (β-MHC), myosin binding protein C and giant protein titin [7-10]. Nonetheless, genes encoding nonsarcomeric proteins should also be underscored with respect to their fundamental role in the pathogenesis of DCM. For instance, autoimmune-related genetic variants found in lymphocytes or macrophages could weaken resistance against environmental elements (e.g. viral agents) and further cause myocardial lesion, although the polymorphisms might not be directly causative for DCM development [11]. Specifically, it has been reported that a SNP (single-nucleotide polymorphism) involved in encoding cytotoxic T-lymphocyte antigen 4 (CTLA4) could elevate susceptibility to DCM [12]. Albeit with the above achievements, the underlying pathology regarding autoimmune-mediated DCM remains ambiguous [13,14].
BTNL2 (butyrophilin-like 2) belongs to the immunoglobulin gene superfamily situated close to the HLA class 2-3 regions, a gene-dense part that embraces immunomodulating genes [15]. It has been shown that a protein that BTNL2 encodes manifests notable amino acid and domain structure homology with the B7.1 receptor, which is of vital significance in the cross-talk between B and T lymphocytes [16,17]. Additionally, binding of BTNL2 to a putative receptor could lead to activation of T cells and inhibition of T cell amplification in a mouse model [18]. The above phenomena revealed that BTNL2 could function as a hazard for autoimmune disorders, such as sarcoidosis [19] and rheumatoid arthritis [20], suggesting a potential role of BTNL2 in DCM immune-pathogenesis. Previous GWAS (genome-wide association study) has also identified rs9268402 within C6orf10-BTNL2 to be associated with coronary artery disease (CAD) and it was explained that rs9268402 exhibited strong linkage disequilibrium (LD) with rs2076530 in BTNL2 [21], which seemed to be correlated with risk of Kawasaki disease (KD), a disorder that would increase the likelihood of suffering from ischemic heart disease [22]. Conclusively, certain SNPs on and near BTNL2 (including rs9405098, rs3763313, rs3763317, rs9268494, rs9268492, rs9268402, rs2076523, rs2076530 and rs1555115) were selected in the current study to explore their potential associations with DCM allowing for the fact that BTNL2 pertained to autoimmune genes and it was linked with certain myocardial dysfunctions.
As an individual SNP might impose relatively low influence on the occurrence of DCM, haplotype analysis incorporating multi-SNPs could aid in more accurate prediction of the disorder. Thus, the current study was aimed to provide solid foundations for estimating the association of risk and prognosis of DCM with SNPs and the corresponding haplotype located on BTNL2.
Materials and methods
Clinical characteristics of study population
In this cohort study, 82 DCM patients (55 males and 27 females, mean age: 50.5 ± 13.5) and 75 healthy controls (50 males and 25 female, mean age: 53.1 ± 13.9) were recruited from the Department of Cardiology at Xijing Hospital (Xi’an, China) between May 2010 and December 2013. All the eligible participants were Han Chinese with no kinship, and the age and sex of both DCM patients and healthy controls were well matched. The DCM patients were diagnosed strictly in light with American Heart Association Scientific Statement on the Classification of Cardiomyopathy [23]. DCM cases would be excluded if they simultaneously suffered from the following disorders: 1) ischemic dysfunctions (e.g. coronary artery disease, chronic hypertension, valvulopathy and tachyarrhythmia); 2) toxins (e.g. chemotherapy drug, alcohol and deficiency of trace elements); 3) endocrine and metabolic disturbances (e.g. pheochromocytoma, diabetes and thyroid disease); 4) infections (e.g. viral myocarditis); 5) autoimmune diseases (e.g. systemic lupus erythematosus, scleroderma and dermatomyositis); 6) peripartum cardiomyopathy. Subsequently, the whole DCM cases were classified into 21 grade I-II cases, 30 grade III cases and 31 grade IV cases in light of New York Heart Association (NYHA) published in 1994 [24]. Clinical characteristics of the study subjects were displayed in Table 1. The protocol of this study was approved by Ethics committee of Xijing Hospital (Xi’an, China), and each participant has signed informed consent before their participations in the study. The study strictly complied with the the Declaration of Helsinki.
Table 1.
Primers of BTNL2 genetic polymorphisms for PCR amplification
| RS ID | PCR amplification primer | |
|---|---|---|
| rs9405098 | F: 5’-AATGAGGGATGGCACGTTTGAT-3’ | R: 3’-CTTGGGTCGGTTTCAGGTGAAC-5’ |
| rs3763317 | F: 5’-GGTATGGATGTTGTGGGGCTTT-3’ | R: 3’-TCAAAGTCCACCGTTCCACGAG-5’ |
| rs3763313 | F: 5’-TCAGAAGCTACATTGAAGCCGAACT-3’ | R: 3’-TGTCGTCCCCGAAACACTTTCA-5’ |
| rs9268494 | F: 5’-GAGTGGAGTCCAGCTTGTGTGC-3’ | R: 3’-CAGTAGTCGGAGGGTCCAGTCA-5’ |
| rs9268492 | F: 5’-GCAGTACCGCTGCCTTTTTGAA-3’ | R: 3’-AGGAGGTCACACACTTTGACAGT-5’ |
| rs2076523 | F: 5’-ACACCCAACCCCATTTTCACAT-3’ | R: 3’-TGAACTGTAACGAGGACTCGACC-5’ |
| rs2076530 | F: 5’-CTGGAGAGCAGATGGCAGAGTAC-3’ | R: 3’-GGAGGTCACACACTTTGACAGTA-5’ |
| rs9268402 | F: 5’-CTAGTTTTAGGATGCTCCACCTGCCAAA-3’ | R: 3’-GGTAATGGTCTGAAGAGTTGCCTTTGGA-3’ |
| rs1555115 | F: 5’-AAACTGGAAGGTTAGGTAAAACTGTGGT-3’ | R: 3’-GTAGGTGAAGAGTATGTGGAAATAAAGA-3’ |
SNP, single-nucleotide polymorphism; PCR, polymerase chain reaction; F, forward; and R, reverse.
SNP selection
The assessment of genotyping data with respect to Han Chinese population were in accordance with International HapMap Project Databases (Hapmap Data Rel 24/Phase II Nov08, on NCBI B36, dbSNP b126). Haploview 4.0 (Broad Institute, Cambridge, MA, USA) was employed to select the tag-SNPs analyzed and SHesis software was applied to carry out haplotype analysis according to LDs among tag-SNPs. Tag-SNPs would be included in our study if minor allele frequency (MAF) was greater than 0.05 and r2 was bigger than 0.75. Several potentially DCM-associated SNPs within BTNL2 were examined based on previous studies as well [25-27].
SNP genotyping
Genomic DNAs were extracted through DNA blood kit (Watson Biotechnologies, Inc) from peripheral blood samples gathered from DCM patients and healthy controls in strict accordance with the manufacturer’s protocols. Genotyping of SNPs was performed by means of Sequenom MassARRAY®-IPLEX (Sequenom Inc., San Diego, CA, USA) on the basis of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) (SpectroREADER, Squenom). Primers designed for PCR (polymerase chain reaction) amplication and extension were prepared with usage of MassARRAY Assay Design Version 3.1 (Sequenom Inc., San Diego, CA, USA) (Table 1). PCR amplication was achieved with a reaction system containing 0.1 U Taq DNA polymerase, 5 ng genomic DNA, 2.5 pM PCR primer and 2.5 mM dNTP. The mixture was subjected to a following procedure: 95°C for 2 min; 45 cycles (at 95°C for 20 s, at 56°C for 30 s, at 72°C for 60 s); 75°C for 5 min. The remaining dNTPs would be removed by addition of 0.3 U alkaline phosphatase into the system, after which the plates were incubated at 37°C for 40 min and further placed at 85°C for 5 min. In addition, before the extension reaction, the mixed components for single base extension reaction should be managed: 5.4 pM extension primers, 50 μM dNTP/ddNTP mixture and 0.5 U thermosequenase DNA polymerase. Then the reaction condition was dealt with as follows: at 94°C for 2 min, 40 cycles (at 94°C for 5 s, at 50°C for 5 s, at 72°C for 5 s) and a final extension step at 72°C for 3 min. Afterwards the samples were desalted by resin treatment for 15 min, spotted into SpectroCHIP (Sequenom, Inc., San Diego, CA), analyzed by mass spectrometer and ultimately interpreted on SpectroTYPER 4.0 software (Sequenom, Inc., San Diego, CA). The DNA samples would be selected if the success rate for genotyping exceeded 95%.
Color Doppler echocardiography detection
The echocardiography (two-dimensional ultrasound and Doppler ultrasound) detection were implemented strictly following the requirements of the American Association of Ultrasound based on Philips Sonos7500 and iE33 echocardiography system (Philips Medical Systems, Bothell, WA) with a S5-1 broadband phased-array transducer (1-5 MHz) [28]. To sum up, the detection indicators included left ventricular end-diastolic volume (LVEDV, ml), left ventricular endsystolic volume (LVESV, ml), left ventricular stroke volume (LVSV, ml), left ventricular ejection fraction (LVEF, mm), fractional shortening (FS, %), left ventricular end diastolic diameter (LVEDD, mm) and left atrial diameter (LAD, mm).
Follow-up
Totally, 82 DCM patients were included in the follow-up process via telephone communications until January, 2015. The clinical follow-up regarding the genetic status of DCM patients was carried out in single-blind fashion with the end point of cardiac death.
Statistical analysis
The experimental data were all analyzed with assistance of software SPSS 19.0 (SPSS Inc., Chicago, IL, USA). The chi-square test was utilized to compare the difference in gender between DCM patients and healthy controls while the student’s t test was performed to figure out if there existed distinctions in age, SBP (systolic pressure), DBP (diastolic blood pressure), LVEDD, LVEDV, LVESV, LAD, LVSV, LVSF and FS. In SNP genotypes, three genetic models were established to assess the genotype distributions, including the dominant model (WM+MM vs. WW), recessive model (MM vs. WW+WM) and allelic model (W vs. M). Odds ratio (OR) and respective 95% confidence intervals (CIs) were demonstrated to assess the influence of any difference between genotypes, alleles and haplotypes. Probability values of 0.05 or less were regarded as statistically significant in patients with DCM in comparison with control subjects. Cox proportional hazard models (multivariable models) and Kaplan-Meier curves (univariable analysis) were utilized to test associations of SNPs/haplotypes with survival time. When the multivariable models were conducted, covariates, such as SBP, DBP, LVEDD, LVEDV, LVESV, LAD, LVSV, LVSF and FS, would be controlled.
Results
Clinical characteristics of study population
The clinical characteristics of 82 DCM patients and 75 healthy controls were shown in Table 2. No striking variation in age or gender was observed between DCM patients and healthy controls. However, remarkably higher values could be found DCM patients in terms of SBP (P = 0.008), DBP (P < 0.001), beats (P < 0.001), LVEDV (P < 0.001), LVESV (P < 0.001), LVEDD (P < 0.001) and LAD (P < 0.001) than the control group, while notably lower LVSV (P = 0.042), LVEF (P < 0.001) and FS (P < 0.001) might be observed in the DCM patients when compared with the control group.
Table 2.
Comparison of dilated cardiomyopathy (DCM) patients and controls by selective clinical characteristics
| Clinical characteristics | DCM patients (n = 82) | Healthy controls (n = 75) | P value |
|---|---|---|---|
| Age, years | 50.5 ± 13.5 | 53.1 ± 13.9 | 0.236 |
| Gender (men/women) | 55/27 | 50/25 | 0.960 |
| SBP, mmHg | 132.32 ± 18.85 | 126.11 ± 7.49 | 0.008 |
| DBP, mmHg | 80.81 ± 15.60 | 75.65 ± 6.17 | 0.008 |
| Beats, bpm | 84.06 ± 16.34 | 73.38 ± 8.48 | < 0.001 |
| LVEDV, ml | 246.96 ± 77.12 | 92.56 ± 20.43 | < 0.001 |
| LVESV, ml | 188.82 ± 83.39 | 33.97 ± 11.36 | < 0.001 |
| LVSV, ml | 53.87 ± 18.20 | 59.32 ± 14.73 | 0.042 |
| LVEDD, mm | 61.67 ± 8.91 | 47.40 ± 3.60 | < 0.001 |
| LAD, mm | 45.09 ± 4.34 | 32.8 ± 1.54 | < 0.001 |
| LVEF, mm | 25.55 ± 10.64 | 65.84 ± 2.80 | < 0.001 |
| FS, % | 25.31 ± 5.18 | 41.56 ± 6.23 | < 0.001 |
SBP: systolic pressure; DBP: diastolic blood pressure; LVEDV: left ventricular end-diastolic volume; LVESV: Left Ventricular End-systolic Volume; LVEDD: left ventricular end diastolic diameter; LAD: left atrial diameter; LVSV: left ventricular stroke volume; LVEF: left ventricular ejection fraction; FS: fractional shortening.
Association analysis of genotype and haplotypes of BTNL2 SNP with the DCM susceptibility
As displayed in the linkage disequilibrium map (Figure 1), 9 tag-SNPs within or near BTNL2 were finally included in the study, namely, rs9405098 (G > A), rs3763317 (T > C), rs3763313 (C > A), rs9268494 (C > A), rs9268492 (G > C), rs2076523 (G > A), rs2076530 (G > A), rs9268402 (G > A), rs1555115 (G > C). The frequency distributions of genotypes and alleles of the 9 SNPs, all complying with Hardy-Weinberg equilibrium (HWE) model, were evaluated between healthy controls and DCM patients (Table 3). In the polymorphisms of rs3763313 (A > C), subjects with heterozygote CA and homozygote CC appeared to have a lower risk of DCM than those with homozygote AA (OR = 0.30, 95% CI = 0.09-0.96, P = 0.041) in the dominant model (CC+CA vs. AA). The allele distributions of rs9268492 (C > G) also varied dramatically between cases and controls, manifested as the tendency that subjects carrying GG and GC genotypes were more likely to develop DCM than CC carriers (OR = 0.31, 95% CI = 0.10-0.99, P = 0.047). Furthermore, C alleles of rs9268494 (C > A) seemed to decrease the genetic susceptibility to DCM compared with A alleles (OR = 0.62, 95% CI = 0.40-0.97, P = 0.042). The genotype distribution of rs9268402 polymorphisms (A > G) indicated that DCM patients had a remarkably higher frequency of G alleles than A alleles (OR = 0.62, 95% CI = 0.40-0.97, P = 0.041). Nevertheless, no associated relationship could be found between rs9405098, rs3763317, rs2076523, rs2076530 and rs1555115 and the susceptibility to DCM in both dominant and allelic models (all P > 0.05).
Figure 1.
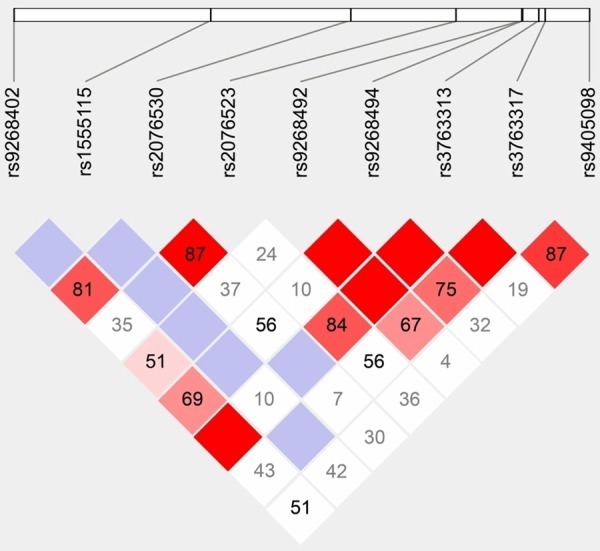
Linkage disequilibrium map of 9 SNPs within or near BTNL2.
Table 3.
Allele and genotype distributions of BTNL2 genetic polymorphisms in dilated cardiomyopathy patients (n = 82) and controls (n = 75)
| SNP | Group | Genotype | Dominant model | Allele frequency | Allelic model | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| 11 | 12 | 22 | OR (95% CI) | P | 1 | 2 | MAF | OR (95% CI) | P | ||
| rs9405098G > A | Case | 57 | 22 | 3 | 0.72 (0.12, 4.44) | 0.724 | 136 | 28 | 0.17A | 0.83 (0.45, 1.53) | 0.644 |
| Control | 55 | 18 | 2 | Ref. | - | 128 | 22 | 0.15A | Ref. | - | |
| rs3763317 T > C | Case | 27 | 40 | 15 | 0.60 (0.25, 1.49) | 0.375 | 94 | 70 | 0.43C | 0.90 (0.57, 1.40) | 0.648 |
| Control | 24 | 42 | 9 | Ref. | .- | 90 | 60 | 0.40C | Ref. | - | |
| rs3763313 A > C | Case | 48 | 21 | 13 | 0.30 (0.09, 0.96) | 0.041 | 117 | 47 | 0.29C | 0.55 (0.32, 0.93) | 0.033 |
| Control | 52 | 19 | 4 | Ref. | - | 123 | 27 | 0.18C | Ref. | - | |
| rs9268494C > A | Case | 16 | 35 | 31 | 0.68 (0.35, 1.33) | 0.312 | 67 | 97 | 0.41C | 0.62 (0.40, 0.97) | 0.042 |
| Control | 26 | 27 | 22 | Ref. | - | 79 | 71 | 0.52C | Ref. | - | |
| rs9268492C > G | Case | 34 | 44 | 14 | 0.31 (0.10, 0.99) | 0.047 | 100 | 64 | 0.39G | 0.57 (0.35, 0.90) | 0.023 |
| Control | 39 | 32 | 4 | Ref. | 110 | 40 | 0.26G | Ref. | |||
| rs2076523 G > A | Case | 15 | 44 | 23 | 0.81 (0.40, 1.66) | 0.590 | 74 | 90 | 0.45G | 0.84 (0.60, 1.47) | 0.821 |
| Control | 13 | 44 | 18 | Ref. | 70 | 80 | 0.47G | Ref. | |||
| rs2076530 G > A | Case | 24 | 24 | 34 | 0.94 (0.50, 1.78) | 0.872 | 72 | 92 | 0.44G | 0.97 (0.62, 1.51) | 0.910 |
| Control | 22 | 23 | 30 | Ref. | 67 | 83 | 0.46G | Ref. | |||
| rs9268402A > G | Case | 16 | 30 | 36 | 0.53 (0.27, 1.03) | 0.070 | 62 | 102 | 0.41A | 0.62 (0.40, 0.98) | 0.041 |
| Control | 21 | 32 | 22 | Ref. | 74 | 76 | 0.49A | Ref. | |||
| rs1555115 G > C | Case | 77 | 4 | 2 | 0.54 (0.05, 6.08) | 1.000 | 158 | 7 | 0.04C | 0.93 (0.30, 2.82) | 1.000 |
| Control | 71 | 4 | 1 | Ref. | 146 | 6 | 0.04C | Ref. | |||
11, wild genotype; 12, heterozygote mutant genotype; 22, homozygote mutant genotype; HWE, Hardy-Weinberg equilibrium; OR, odds ratio; CI, confidence interval; MAF, minor allele frequency.
Moreover, DCM patients were categorized into four level groups in line with NYHA grading systems (Table 4) and the correlations between DCM development and four SNPs (rs3763313, rs9268494, rs9268492 and rs9268402) were estimated in three groups (NYHA I-II, NYHA III and NYHA IV), respectively. The stratified analyses showed that in the polymorphism of rs3763313 (A > C), AA and AC carriers were less inclined to suffer from the fourth grade of DCM when compared with CC carriers (OR = 0.19, 95% CI: 0.05-0.72, P = 0.014). Similarly, lower prevalence of DCM could be noticed in CC and CA carriers of rs9268494, CC and CG carriers of rs9268492, as well as AA and AG carriers of rs9268402 than carriers of homozygote AA, GG and GG, separately [OR = 0.30 (95% CI: 0.13-0.72; P = 0.008), OR = 0.16 (95% CI: 0.05-0.59; P = 0.005) and OR = 0.34 (95% CI: 0.14-.81; P= 0.016)]. Additionally, the allelic distributions of rs3763313, rs9268494, rs9268492 and rs9268402 also suggested that A, C, C and A alleles could be negatively associated with elevated susceptibility to DCM (NYHA IV) in comparison with C, A, G and G alleles, respectively [OR = 0.43 (95% CI: 0.22, 0.84; P = 0.018), OR = 0.49 (95% CI: 0.27-0.91; P = 0.024), OR = 0.50 (95% CI: 0.27-0.94; P = 0.035) and OR = 0.53 (95% CI: 0.28-0.97; P = 0.048)].
Table 4.
Stratified analyses of rs3763313, rs9268494, rs9268492 and rs9268402 in DCM patients classified by NYHA and controls
| SNP | Subgroup | N | Allelic model | Dominant model | Recessive model | |||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |||
| rs3763313 (A > C) | Control | 75 | Ref. | Ref. | Ref. | |||
| Case | ||||||||
| NYHA I-II | 21 | 0.80 (0.35, 1.88) | 0.656 | 0.54 (0.09, 3.15) | 0.609 | 0.88 (0.32, 2.48) | 0.796 | |
| NYHA III | 30 | 0.56 (0.28, 1.12) | 0.132 | 0.37 (0.09, 1.57) | 0.221 | 0.58 (0.24, 1.39) | 0.258 | |
| NYHA IV | 31 | 0.43 (0.22, 0.84) | 0.018 | 0.19 (0.05, 0.72) | 0.014 | 0.54 (0.23, 1.27) | 0.182 | |
| rs9268494 (C > A) | Control | 75 | Ref. | Ref. | Ref. | |||
| Case | ||||||||
| NYHA I-II | 21 | 0.74 (0.37, 1.48) | 0.485 | 1.04 (0.36, 3.02) | 1.000 | 0.44 (0.14, 1.46) | 0.196 | |
| NYHA III | 30 | 0.69 (0.38, 1.26) | 0.285 | 1.36 (0.51, 3.64) | 0.633 | 0.21 (0.06, 0.76) | 0.015 | |
| NYHA IV | 31 | 0.49 (0.27, 0.91) | 0.024 | 0.30 (0.13, 0.72) | 0.008 | 0.77 (0.31, 1.92) | 0.654 | |
| rs9268492 (C > G) | Control | 75 | Ref. | Ref. | Ref. | |||
| Case | ||||||||
| NYHA I-II | 21 | 0.65 (0.31, 1.36) | 0.254 | 0.53 (0.09, 3.15) | 0.609 | 0.57 (0.21, 1.53) | 0.326 | |
| NYHA III | 30 | 0.59 (0.31, 1.10) | 0.099 | 0.37 (0.09, 1.57) | 0.221 | 0.63 (0.26, 1.55) | 0.373 | |
| NYHA IV | 31 | 0.50 (0.27, 0.94) | 0.035 | 0.16 (0.05, 0.59) | 0.005 | 0.67 (0.29, 1.55) | 0.397 | |
| rs9268402 (A > G) | Control | 75 | Ref. | Ref. | Ref. | |||
| Case | ||||||||
| NYHA I-II | 21 | 0.70 (0.35, 1.40) | 0.383 | 0.83 (0.30, 2.34) | 0.790 | 0.43 (0.11, 1.61) | 0.261 | |
| NYHA III | 30 | 0.68 (0.37, 1.26) | 0.284 | 0.62 (0.26, 1.51) | 0.357 | 0.64 (0.23, 1.80) | 0.466 | |
| NYHA IV | 31 | 0.53 (0.28, 0.97) | 0.048 | 0.34 (0.14, 0.81) | 0.016 | 0.75 (0.28, 2.00) | 0.635 | |
OR, odds ratio; CI, confidence interval; NYHA, New York Heart Association.
Association analysis between haplotypes of BTNL2 and susceptibility to DCM
The haplotypes made up of 4 SNPs (rs3763317, rs3763313, rs9268494 and rs9268492) were investigated in the current study since there existed outstanding (approximately 100%) LDs among the 4 SNPs in the Chinese population. Subsequently, 16 haplotypes were obtained after randomly combining the 4 SNPs based on SHesis software, but 6 haplotypes were finally excluded for their low frequency in the studied crowd (each lower than 3%). It was finally indicated in the haplotype analyses that subjects carrying haplotype C-C-A-T were inversely linked with DCM risk compared with carriers of all other haplotypes (OR = 0.50, 95% CI = 0.26-0.97, P = 0.038) (Table 5).
Table 5.
Haplotype structure and frequency analysis of BTNL2 genetic polymorphisms between dilated cardiomyopathy (DCM) patients and controls
| Haplotype | Frequency | x2-test | OR (95% CI) | P-value | |
|---|---|---|---|---|---|
|
| |||||
| DCM (2n = 164) | Control (2n = 150) | ||||
| C-A-A-C | 18 (0.1098) | 17 (0.1165) | 0.042 | 1.07 (0.53, 2.20) | 0.837 |
| C-A-C-T | 10 (0.0595) | 6 (0.0383) | 1.10 | 1.73 (0.61, 4.92) | 0.295 |
| G-C-A-T | 11 (0.0647) | 10 (0.0665) | 0.069 | 1.13 (0.46, 2.75) | 0.792 |
| G-A-A-C | 12 (0.0702) | 6 (0.0409) | 2.24 | 2.13 (0.78, 5.87) | 0.132 |
| G-A-A-T | 15 (0.0931) | 9 (0.0614) | 2.22 | 1.78 (0.75, 4.23) | 0.186 |
| G-C-A-C | 8 (0.0488) | 6 (0.0443) | 0.33 | 1.38 (0.46, 4.08) | 0.563 |
| C-A-A-T | 23 (0.1456) | 26 (0.1747) | 0.17 | 0.88 (0.47, 1.63) | 0.677 |
| C-C-A-T | 16 (0.1012) | 29 (0.1893) | 4.28 | 0.50 (0.26, 0.97) | 0.038 |
| C-C-A-C | 12 (0.0763) | 19 (0.1262) | 1.67 | 0.61 (0.28, 1.30) | 0.196 |
| C-C-C-T | 7 (0.0413) | 6 (0.0415) | 0.097 | 1.20 (0.39, 3.66) | 0.760 |
OR, odds ratio; CI, confidence interval.
Association analysis between 4 SNPs within or near BTNL2 and prognosis of DCM
Altogether 82 DCM patients were followed in the survival analyses and they possessed a mean survival period of 27 ± 12 months. Among the participants, 2 NYHA I-II cases, 12 NYHA III cases and 24 NYHA IV cases eventually died of cardiac death during follow-up, in which period the whole patients received constant medical treatment, but without heart transplantation.
In the present study, the potential correlations between 4 SNPs as well as haplotype C-C-A-T and prognosis of DCM patients classified by NYHA grading were investigated. As illustrated in the Kaplan-Meier curve (Figure 2), a longer life span could be expected in AA and AG carriers (NYHA IV) than in GG carriers in terms of rs9268402 (Long-Rank P = 0.029) and the trend remained remarkable when analyzed with a multivariate Cox proportional hazard model after the adjustment of SBP, DBP, LVEDD, LVEDV, LVESV, LAD, LVSV, LVSF and FS (AA/GA vs. GG, HR: 0.241, 95% CI: 0.089-0.650, P = 0.005). Analogously, C-C-A-T carriers (NYHA IV) tended to obtain more favorable prognosis than subjects carrying other haplotypes without or with consideration of the parameters mentioned above, such as SBP, DBP, LVEDD and so on (Long-Rank P = 0.036; HR = 0.126, 95% CI: 0.035-0.457, P = 0.002) (Figure 3; Table 6). Nonetheless, for the polymorphism of rs3763313, notable correlations could only be observed between CC carriers and poor prognosis of DCM patients (NYHA IV) (P = 0.009), as expressed in the Kaplan-Meier curve (Figure 4).
Figure 2.
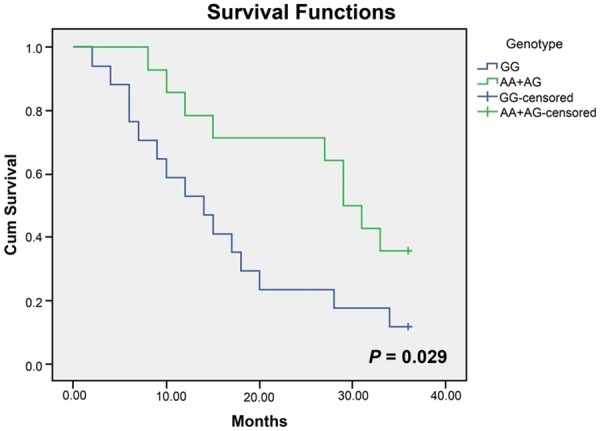
Kaplan-Meier survival curves free of cardiac death for GG homozygotes and (AA+AG) carriers (grade IV by NYHA) of rs9268402.
Figure 3.
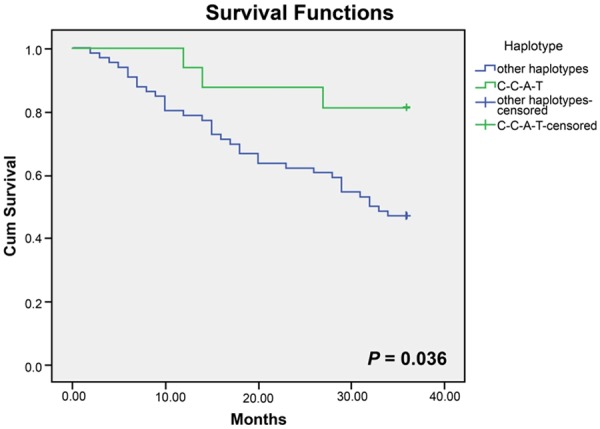
Kaplan-Meier survival curves free of cardiac death for haplotype C-C-A-T and other haplotypes.
Table 6.
Association between the BTNL2 SNPs in the dominant model and patient’s survival
| Overall survival | |||||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| Characteristics | NYHA | Univariate survival analysis | Multivariate survival analysis* | ||||
|
|
|||||||
| HR | 95% CI | P | HR | 95% CI | P | ||
| SNP | |||||||
| rs3763313 | III | 0.555 | (0.121, 2.546) | 0.449 | 0.227 | (0.039, 1.334) | 0.101 |
| (AA/AC vs. CC) | IV | 0.280 | (0.108, 0.727) | 0.009 | 0.238 | (0.054, 1.054) | 0.059 |
| rs9268494 | III | 0.465 | (0.140, 1.549) | 0.212 | 0.152 | (0.021, 1.079) | 0.060 |
| (CC/CA vs. AA) | IV | 0.520 | (0.225, 1.201) | 0.126 | 0.727 | (0.285, 1.858) | 0.505 |
| rs9268492 | III | 0.538 | (0.118, 2.464) | 0.425 | 1.242 | (0.175, 8.793) | 0.828 |
| (CC/GC vs. GG) | IV | 1.140 | (0.423, 3.073) | 0.795 | 1.048 | (0.326, 3.369) | 0.937 |
| rs9268402 | III | 0.471 | (0.151, 1.467) | 0.194 | 0.700 | (0.203, 2.415) | 0.573 |
| (AA/GA vs. GG) | IV | 0.407 | (0.176, 0.944) | 0.029 | 0.241 | (0.089, 0.650) | 0.005 |
| Haplotype | |||||||
| C-C-A-T | I-IV | 0.282 | (0.087, 0.919) | 0.036 | 0.126 | (0.035, 0.457) | 0.002 |
HR, hazard ratio; CI, confidence interval.
Figure 4.
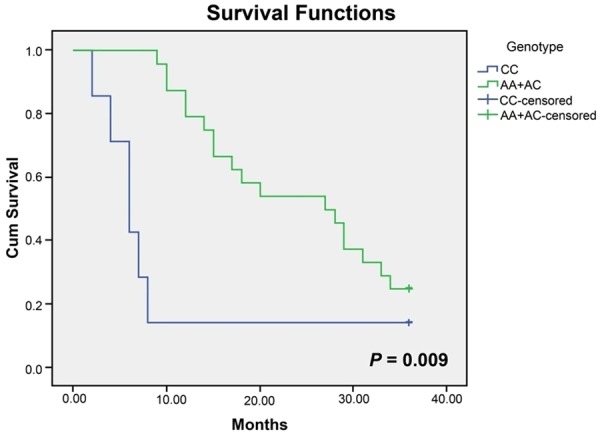
Kaplan-Meier survival curves free of cardiac death for CC homozygotes and (AA+AC) carriers (grade IV by NYHA) of rs3763313.
Discussion
Associations of nine SNPs within or near BTNL2 with DCM development were examined in a Chinese Han population in the current study. Results were obtained that genetic mutants located within C6orf10~BTNL2 (rs9268402) and BTNL2 (rs3763313, rs9268494 and rs9268492) might be associated with DCM (grade IV by NYHA) susceptibility and identification of two SNPs therein could aid in estimation of unfavorable prognosis of DCM patients (grade IV by NYHA).
The risk SNP rs9268402 has been identified by GWAS as a regulatory locus regarding CAD, one disease of cardiac failure, in a Chinese Han population [29]. The untranslated region (UTR), where rs9268402 was situated, shared the common 6q21.3 region with major histocompatibility complex (MHC). Genetic polymorphisms on leukocyte-binding MHC, also called human leukocyte antigen (HLA), have been shown to be tightly relevant to susceptibility to multiple immunity-associated disorders, such as osteoarthritis, rheumatoid arthritis, kawasaki disease and ankylosing spondylitis [30]. Furthermore, variants of candidate C6orf10 within 6p21.32 could elevate susceptibility to psoriasis [31] and the gene could be activated by tumor necrosis factor (TNF)-α, one vital proinflammatory cytokine, found in keratinocyte [32]. The above findings suggested that variant of rs9268402 could interact with its surrounding SNPs to regulate immune responses, contributing to the occurrence of myocardial diseases, such as DCM.
It should also be noteworthy that BTNL2, situated ~170 kb from HLA and not far from rs9268402, was illustrated as the first butyrophilin family member in modulation of inflammatory immune diseases, such as myositis and sarcoidosis [15,18]. More specifically, BTNL2 abundantly expressed in intestine and lymphoid organs could be involved in powerful inhibition of T cell proliferation and modest suppression of IL-2 production [19,33], demonstrating its possible linkage with constraint of immune responses as a negative costimulatory molecule [18,34]. Thus, tag-SNPs on BTNL2 (rs1555115, rs2076530, rs2076523, rs9268492, rs9268494, rs3763313, rs3763317 and rs9405098) were potently candidate polymorphisms related with the presence of immunity-causing DCM, especially ones that displayed high/moderate LD with rs9268402, a CAD-associated and immunity-related SNP confirmed by GWAS. In fact, the associations of rs3763313, rs9268494 and rs9268492 with tuberculosis, hepatitis B vaccination, ulcerative colitis and Crohn disease have been confirmed [27,35-37].
Obviously, rs3763313 revealed higher LD (close to 100%) with rs9268402 when compared with rs2076530 (81%), suggesting that rs3763313 could be a more promising SNP than rs2076530 with respect to participation in the immune dysfunctions that contributed to myocardial damage. Interestingly, among the 7 SNPs investigated (rs9268402 and rs3763313 excluded), simply rs9268492 and rs9268494 achieved moderate/high LD with both rs9268402 and rs3763313 (all LD > 50%), and the couple of SNPs were found associated with DCM in the current study accordingly. The above results might indicate that rs9268402 and rs3763313 were pivotal polymorphisms linked with the appearance of DCM, and SNPs that showed relatively apparent LD with the two target polymorphisms might also potentially predispose to DCM.
However, although rs2076530 A allele has largely been confirmed to be correlated with incremental susceptibility to inflammatory disorders (e.g. tuberculosis) [38,39], the genetic predisposition to DCM was absent in the current study, which would be interpreted as ethnic distinctions. To be specific, correlations between rs2076530 and specific immune diseases, taking tuberculosis for instance, could be merely observed among European and South African populations, rather than in Chinese Han populations [27]. Furthermore, the A allele of rs2076530 could elevate risk of sarcoidosis primarily in white population and Japanese population as well [25,40,41], but the finding might not be appropriate for the Chinese population with consideration of genetic discrepancy. With respect to polymorphisms of rs9405098, rs3763317, rs2076523 and rs1555115, even though their associations with certain immunity-related disorders have been estimated [25-27], the sophisticated reactions inside each specific disease might differ, which could account for the irrelevance of the SNPs with risk of DCM.
The haplotype analyses (Figure 3) integrating 4 SNPs of fairly high LD (rs9268492, rs9268494, rs3763313 and rs3763317) also implied that C-C-A-T carriers were less inclined to suffer from DCM and were 1.67-fold less susceptible to perish within 36 months than carriers of other haplotypes. Similarly, AA and AC carriers (grade IV by NYHA) of rs3763313 as well as AA and AG carriers (grade IV by NYHA) of rs9268402 were approximately 1.38-fold and 3.5-fold more likely to survive than CC carriers and GG carriers, respectively (Figures 2, 4). The prognosis condition, therefore, further validated the positive association of heterozygotes CC/GG with DCM susceptibility.
Conclusively, our data showed that the genetic polymorphisms of BTNL2 (rs3763313, rs9268494 and rs9268492) and C6orf10~BTNL2 (rs9268402) were associated with DCM (grade IV by NYHA) susceptibility in a Chinese population. Among the four SNPs, mutants of rs3763313 and rs9268402 were deemed to be tightly correlated with undesirable prognosis of DCM (grade IV by NYHA), indicating the possibility of SNPs within or near BTNL2 as genetic markers to identify the DCM risk group and to predict their prognosis. Nonetheless, as the sample number and ethnicity studied in this investigation appeared to be finite and the SNPs studied were based on previously published studies, more meticulous investigations covering additional SNPs need to be furthered in terms of DCM-causing genes.
Disclosure of conflict of interest
None.
References
- 1.Hershberger RE, Morales A, Siegfried JD. Clinical and genetic issues in dilated cardiomyopathy: a review for genetics professionals. Genet Med. 2010;12:655–667. doi: 10.1097/GIM.0b013e3181f2481f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manolio TA, Baughman KL, Rodeheffer R, Pearson TA, Bristow JD, Michels VV, Abelmann WH, Harlan WR. Prevalence and etiology of idiopathic dilated cardiomyopathy (summary of a National Heart, Lung, and Blood Institute workshop. Am J Cardiol. 1992;69:1458–1466. doi: 10.1016/0002-9149(92)90901-a. [DOI] [PubMed] [Google Scholar]
- 3.Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, Arnett D, Moss AJ, Seidman CE, Young JB, American Heart Association; Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; Council on Epidemiology and Prevention Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation. 2006;113:1807–1816. doi: 10.1161/CIRCULATIONAHA.106.174287. [DOI] [PubMed] [Google Scholar]
- 4.Jefferies JL, Towbin JA. Dilated cardiomyopathy. Lancet. 2010;375:752–762. doi: 10.1016/S0140-6736(09)62023-7. [DOI] [PubMed] [Google Scholar]
- 5.Mestroni L, Rocco C, Gregori D, Sinagra G, Di Lenarda A, Miocic S, Vatta M, Pinamonti B, Muntoni F, Caforio AL, McKenna WJ, Falaschi A, Giacca M, Camerini Familial dilated cardiomyopathy: evidence for genetic and phenotypic heterogeneity. Heart Muscle Disease Study Group. J Am Coll Cardiol. 1999;34:181–190. doi: 10.1016/s0735-1097(99)00172-2. [DOI] [PubMed] [Google Scholar]
- 6.Burkett EL, Hershberger RE. Clinical and genetic issues in familial dilated cardiomyopathy. J Am Coll Cardiol. 2005;45:969–981. doi: 10.1016/j.jacc.2004.11.066. [DOI] [PubMed] [Google Scholar]
- 7.McNally EM, Golbus JR, Puckelwartz MJ. Genetic mutations and mechanisms in dilated cardiomyopathy. J Clin Invest. 2013;123:19–26. doi: 10.1172/JCI62862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herman DS, Lam L, Taylor MR, Wang L, Teekakirikul P, Christodoulou D, Conner L, DePalma SR, McDonough B, Sparks E, Teodorescu DL, Cirino AL, Banner NR, Pennell DJ, Graw S, Merlo M, Di Lenarda A, Sinagra G, Bos JM, Ackerman MJ, Mitchell RN, Murry CE, Lakdawala NK, Ho CY, Barton PJ, Cook SA, Mestroni L, Seidman JG, Seidman CE. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ingles J, Doolan A, Chiu C, Seidman J, Seidman C, Semsarian C. Compound and double mutations in patients with hypertrophic cardiomyopathy: implications for genetic testing and counselling. J Med Genet. 2005;42:e59. doi: 10.1136/jmg.2005.033886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Girolami F, Ho CY, Semsarian C, Baldi M, Will ML, Baldini K, Torricelli F, Yeates L, Cecchi F, Ackerman MJ, Olivotto I. Clinical features and outcome of hypertrophic cardiomyopathy associated with triple sarcomere protein gene mutations. J Am Coll Cardiol. 2010;55:1444–1453. doi: 10.1016/j.jacc.2009.11.062. [DOI] [PubMed] [Google Scholar]
- 11.Staab J, Ruppert V, Pankuweit S, Meyer T. Polymorphisms in genes encoding nonsarcomeric proteins and their role in the pathogenesis of dilated cardiomyopathy. Herz. 2012;37:836–841. doi: 10.1007/s00059-012-3698-6. [DOI] [PubMed] [Google Scholar]
- 12.Ruppert V, Meyer T, Struwe C, Petersen J, Perrot A, Posch MG, Ozcelik C, Richter A, Maisch B, Pankuweit S German Heart Failure Network. Evidence for CTLA4 as a susceptibility gene for dilated cardiomyopathy. Eur J Hum Genet. 2010;18:694–699. doi: 10.1038/ejhg.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franz WM, Muller OJ, Katus HA. Cardiomyopathies: from genetics to the prospect of treatment. Lancet. 2001;358:1627–1637. doi: 10.1016/S0140-6736(01)06657-0. [DOI] [PubMed] [Google Scholar]
- 14.Schonberger J, Seidman CE. Many roads lead to a broken heart: the genetics of dilated cardiomyopathy. Am J Hum Genet. 2001;69:249–260. doi: 10.1086/321978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stammers M, Rowen L, Rhodes D, Trowsdale J, Beck S. BTL-II: a polymorphic locus with homology to the butyrophilin gene family, located at the border of the major histocompatibility complex class II and class III regions in human and mouse. Immunogenetics. 2000;51:373–382. doi: 10.1007/s002510050633. [DOI] [PubMed] [Google Scholar]
- 16.Sharpe AH, Freeman GJ. The B7-CD28 superfamily. Nat Rev Immunol. 2002;2:116–126. doi: 10.1038/nri727. [DOI] [PubMed] [Google Scholar]
- 17.Agostini C, Trentin L, Perin A, Facco M, Siviero M, Piazza F, Basso U, Adami F, Zambello R, Semenzato G. Regulation of alveolar macrophage-T cell interactions during Th1-type sarcoid inflammatory process. Am J Physiol. 1999;277:L240–250. doi: 10.1152/ajplung.1999.277.2.L240. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen T, Liu XK, Zhang Y, Dong C. BTNL2, a butyrophilin-like molecule that functions to inhibit T cell activation. J Immunol. 2006;176:7354–7360. doi: 10.4049/jimmunol.176.12.7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valentonyte R, Hampe J, Huse K, Rosenstiel P, Albrecht M, Stenzel A, Nagy M, Gaede KI, Franke A, Haesler R, Koch A, Lengauer T, Seegert D, Reiling N, Ehlers S, Schwinger E, Platzer M, Krawczak M, Muller-Quernheim J, Schurmann M, Schreiber S. Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nat Genet. 2005;37:357–364. doi: 10.1038/ng1519. [DOI] [PubMed] [Google Scholar]
- 20.Mitsunaga S, Hosomichi K, Okudaira Y, Nakaoka H, Kunii N, Suzuki Y, Kuwana M, Sato S, Kaneko Y, Homma Y, Kashiwase K, Azuma F, Kulski JK, Inoue I, Inoko H. Exome sequencing identifies novel rheumatoid arthritis-susceptible variants in the BTNL2. J Hum Genet. 2013;58:210–215. doi: 10.1038/jhg.2013.2. [DOI] [PubMed] [Google Scholar]
- 21.Lu X, Wang L, Chen S, He L, Yang X, Shi Y, Cheng J, Zhang L, Gu CC, Huang J, Wu T, Ma Y, Li J, Cao J, Chen J, Ge D, Fan Z, Li Y, Zhao L, Li H, Zhou X, Chen L, Liu D, Chen J, Duan X, Hao Y, Wang L, Lu F, Liu Z, Yao C, Shen C, Pu X, Yu L, Fang X, Xu L, Mu J, Wu X, Zheng R, Wu N, Zhao Q, Li Y, Liu X, Wang M, Yu D, Hu D, Ji X, Guo D, Sun D, Wang Q, Yang Y, Liu F, Mao Q, Liang X, Ji J, Chen P, Mo X, Li D, Chai G, Tang Y, Li X, Du Z, Liu X, Dou C, Yang Z, Meng Q, Wang D, Wang R, Yang J, Schunkert H, Samani NJ, Kathiresan S, Reilly MP, Erdmann J Coronary ARtery DIsease Genome-Wide Replication And Meta-Analysis (CARDIoGRAM) Consortium. Peng X, Wu X, Liu D, Yang Y, Chen R, Qiang B, Gu D. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nat Genet. 2012;44:890–894. doi: 10.1038/ng.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato H, Sugimura T, Akagi T, Sato N, Hashino K, Maeno Y, Kazue T, Eto G, Yamakawa R. Long-term consequences of Kawasaki disease. A 10- to 21-year follow-up study of 594 patients. Circulation. 1996;94:1379–1385. doi: 10.1161/01.cir.94.6.1379. [DOI] [PubMed] [Google Scholar]
- 23.Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, Arnett D, Moss AJ, Seidman CE, Young JB American Heart Association; Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; Council on Epidemiology and Prevention. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation. 2006;113:1807–1816. doi: 10.1161/CIRCULATIONAHA.106.174287. [DOI] [PubMed] [Google Scholar]
- 24.Bennett JA, Riegel B, Bittner V, Nichols J. Validity and reliability of the NYHA classes for measuring research outcomes in patients with cardiac disease. Heart Lung. 2002;31:262–270. doi: 10.1067/mhl.2002.124554. [DOI] [PubMed] [Google Scholar]
- 25.Spagnolo P, Sato H, Grutters JC, Renzoni EA, Marshall SE, Ruven HJ, Wells AU, Tzouvelekis A, van Moorsel CH, van den Bosch JM, du Bois RM, Welsh KI. Analysis of BTNL2 genetic polymorphisms in British and Dutch patients with sarcoidosis. Tissue Antigens. 2007;70:219–227. doi: 10.1111/j.1399-0039.2007.00879.x. [DOI] [PubMed] [Google Scholar]
- 26.Hsueh KC, Lin YJ, Chang JS, Wan L, Tsai FJ. BTNL2 gene polymorphisms may be associated with susceptibility to Kawasaki disease and formation of coronary artery lesions in Taiwanese children. Eur J Pediatr. 2010;169:713–719. doi: 10.1007/s00431-009-1099-5. [DOI] [PubMed] [Google Scholar]
- 27.Lian Y, Yue J, Han M, Liu J, Liu L. Analysis of the association between BTNL2 polymorphism and tuberculosis in Chinese Han population. Infect Genet Evol. 2010;10:517–521. doi: 10.1016/j.meegid.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 28.Schiller NB, Shah PM, Crawford M, DeMaria A, Devereux R, Feigenbaum H, Gutgesell H, Reichek N, Sahn D, Schnittger I, et al. Recommendations for quantitation of the left ventricle by two-dimensional echocardiography. American Society of Echocardiography Committee on Standards, Subcommittee on Quantitation of Two-Dimensional Echocardiograms. J Am Soc Echocardiogr. 1989;2:358–367. doi: 10.1016/s0894-7317(89)80014-8. [DOI] [PubMed] [Google Scholar]
- 29.Lu X, Wang L, Chen S, He L, Yang X, Shi Y, Cheng J, Zhang L, Gu CC, Huang J, Wu T, Ma Y, Li J, Cao J, Chen J, Ge D, Fan Z, Li Y, Zhao L, Li H, Zhou X, Chen L, Liu D, Duan X, Hao Y, Lu F, Liu Z, Yao C, Shen C, Pu X, Yu L, Fang X, Xu L, Mu J, Wu X, Zheng R, Wu N, Zhao Q, Liu X, Wang M, Yu D, Hu D, Ji X, Guo D, Sun D, Wang Q, Yang Y, Liu F, Mao Q, Liang X, Ji J, Chen P, Mo X, Li D, Chai G, Tang Y, Li X, Du Z, Dou C, Yang Z, Meng Q, Wang D, Wang R, Yang J, Schunkert H, Samani NJ, Kathiresan S, Reilly MP, Erdmann J, Peng X, Chen R, Qiang B, Gu D. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nat Genet. 2012;44:890–894. doi: 10.1038/ng.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trowsdale J, Knight JC. Major histocompatibility complex genomics and human disease. Annu Rev Genomics Hum Genet. 2013;14:301–323. doi: 10.1146/annurev-genom-091212-153455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng BJ, Sun LD, Soltani-Arabshahi R, Bowcock AM, Nair RP, Stuart P, Elder JT, Schrodi SJ, Begovich AB, Abecasis GR, Zhang XJ, Callis-Duffin KP, Krueger GG, Goldgar DE. Multiple Loci within the major histocompatibility complex confer risk of psoriasis. PLoS Genet. 2009;5:e1000606. doi: 10.1371/journal.pgen.1000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banno T, Gazel A, Blumenberg M. Pathway-specific profiling identifies the NF-kappa B-dependent tumor necrosis factor alpha-regulated genes in epidermal keratinocytes. J Biol Chem. 2005;280:18973–18980. doi: 10.1074/jbc.M411758200. [DOI] [PubMed] [Google Scholar]
- 33.Price P, Santoso L, Mastaglia F, Garlepp M, Kok CC, Allcock R, Laing N. Two major histocompatibility complex haplotypes influence susceptibility to sporadic inclusion body myositis: critical evaluation of an association with HLA-DR3. Tissue Antigens. 2004;64:575–580. doi: 10.1111/j.1399-0039.2004.00310.x. [DOI] [PubMed] [Google Scholar]
- 34.Arnett HA, Escobar SS, Gonzalez-Suarez E, Budelsky AL, Steffen LA, Boiani N, Zhang M, Siu G, Brewer AW, Viney JL. BTNL2, a butyrophilin/B7-like molecule, is a negative costimulatory molecule modulated in intestinal inflammation. J Immunol. 2007;178:1523–1533. doi: 10.4049/jimmunol.178.3.1523. [DOI] [PubMed] [Google Scholar]
- 35.Yang C, Pan L, Zhang L, Wu X, Zhu X, Yan B, Xu A, Li H, Liu Y. BTNL2 associated with the immune response to hepatitis B vaccination in a Chinese Han population. J Med Virol. 2014;86:1105–1112. doi: 10.1002/jmv.23934. [DOI] [PubMed] [Google Scholar]
- 36.Juyal G, Prasad P, Senapati S, Midha V, Sood A, Amre D, Juyal RC, Bk T. An investigation of genome-wide studies reported susceptibility loci for ulcerative colitis shows limited replication in north Indians. PLoS One. 2011;6:e16565. doi: 10.1371/journal.pone.0016565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murdoch TB, Bernstein CN, El-Gabalawy H, Stempak JM, Sargent M, Elias B, Xu W, Pathan S, Silverberg MS. Prevalence of genetic variants associated with inflammatory bowel disease in a healthy First Nations cohort. CMAJ. 2012;184:E435–441. doi: 10.1503/cmaj.110613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson CM, Traherne JA, Jamieson SE, Tremelling M, Bingham S, Parkes M, Blackwell JM, Trowsdale J. Analysis of the BTNL2 truncating splice site mutation in tuberculosis, leprosy and Crohn’s disease. Tissue Antigens. 2007;69:236–241. doi: 10.1111/j.1399-0039.2006.00795.x. [DOI] [PubMed] [Google Scholar]
- 39.Xie Y, Chen Y, Lin M, Wen J, Fan G, Wu Y. High-performance liquid chromatographic method for the determination and pharmacokinetic study of oxypeucedanin hydrate and byak-angelicin after oral administration of Angelica dahurica extracts in mongrel dog plasma. J Pharm Biomed Anal. 2007;44:166–172. doi: 10.1016/j.jpba.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Levin AM, Iannuzzi MC, Montgomery CG, Trudeau S, Datta I, McKeigue P, Fischer A, Nebel A, Rybicki BA. Association of ANXA11 genetic variation with sarcoidosis in African Americans and European Americans. Genes Immun. 2013;14:13–18. doi: 10.1038/gene.2012.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki H, Ota M, Meguro A, Katsuyama Y, Kawagoe T, Ishihara M, Asukata Y, Takeuchi M, Ito N, Shibuya E, Nomura E, Uemoto R, Nishide T, Namba K, Kitaichi N, Morimoto S, Kaburaki T, Ando Y, Takenaka S, Nakamura J, Saeki K, Ohno S, Inoko H, Mizuki N. Genetic characterization and susceptibility for sarcoidosis in Japanese patients: risk factors of BTNL2 gene polymorphisms and HLA class II alleles. Invest Ophthalmol Vis Sci. 2012;53:7109–7115. doi: 10.1167/iovs.12-10491. [DOI] [PubMed] [Google Scholar]


