Abstract
E2F transcription factors regulate a wide range of biological processes, including cell cycle, apoptosis and DNA damage response. In the present study, we examined whether E2F2 is related to the poor prognosis of NSCLC and its role in progress of NSCLC. Firstly, we analyzed 86 NSCLC samples by immunohistochemistry and found that E2F2 expression was markedly increased in 62.8% (54/86) of all samples compared with the normal tissues. Further study showed that E2F2 expression was closely associated with clinical stage (P = 0.039) and tumor size (P = 0.045). Furthermore, Kaplan-Meier analysis indicated that high Bad expression was significantly correlated to overall survival (P = 0.045) but not disease-free survival (P = 0.288). In addition, our results showed that knockdown E2F2 expression could reduce cell viability and colony formation in NSCLC cells. The results in our study for the first time revealed that E2F2 act as an activator in tumor progress of NSCLC and could become a promising marker for the prognosis of patients with NSCLC.
Keywords: E2F2, proliferation, NSCLC, prognosis, survival
Introduction
Non-small cell lung cancer (NSCLC) is the main form of lung cancer and accounts for more than 85% of all the lung cancer cases [1]. NSCLC is a highly malignant and aggressive neoplasm type of lung cancer and showed a poor 5-year survival rate [2]. The NSCLC can be divided into 3 subgroups: squamous cell carcinoma, large cell carcinoma and adenocarcinoma [3]. Traditionally, surgery is regarded as the best treatment for NSCLC patients whereas only 20-25% of tumors are suitable for potentially curative resection and despite complete surgical resection, the recurrence rate remains high (30-70%) [4]. Thus, identification of biomarkers for early detection, prognostic stratification, and novel therapeutic interventions are therefore urgently needed for effective management of NSCLC.
The E2F is a cellular factor which was identified from adenovirus and could initiate E2 gene transcription [5,6]. As so far, there have been found 8 genes involved in encoding the transcription of the E2F family factors which named as E2F1-E2F8 and the genes are responsible for encoding 9 different proteins [7,8]. According to the transcriptional activities, structures and their binding proteins, the E2F family members were classified into four groups. The binding proteins of E2F family members including the pocket proteins Retinoblastoma protein (pRB), Retinoblastoma-like protein 1 (p107) and Retinoblastoma-like protein 2 (p130) [7]. The first subgroup members including E2F1, E2F2 and E2F3a which only bind pRB and they are the ‘activator’ E2Fs. The first subgroup members are necessary factors in the regulation of cell cycle into S phase. E2F4 and E2F5 represent the second subgroup which are performing the repressive function in related to pRB family member. E2F3b and E2F4-E2F8 are considered the ‘repressor’ E2Fs since they are capable of repressing the expression of mostly overlapping sets of target genes while E2F6-E2F8 repress transcription in a pocket protein-independent manner as they lack the pocket protein binding domain [7,9-12]. The E2F family of transcription factors play and important role in a plethora of biological processes related to cell cycle, differentiation, apoptosis and response to DNA damage [8,13,14]. Previous studies have demonstrated that E2F1 was involved in DNA damage response upon treatment with DNA damaging agents [15-18]. In addition, other studies have showed that when the DNA damage occurs, the protein level of E2F4 is decreased while the levels of E2F3a, E2F7 and E2F8 are upregulated which indicated that the E2F family members involved in different regulatory mechanism [19-21].
E2F2 is an important member of the E2F family. Ivey-Hoyle et al. first identified the E2F2 gene which is located on 1p36 and encodes a 47.5 kDa protein [22]. After that, many studies have focused on E2F2 function and revealed that E2F2 regulates lots of cell progresses such as cell cycle, proliferation and tumorigenesis of ovarian cancer [23,24]. Recently, Suzuki et al. have confirmed that E2F2 is overexpressed in glioblastoma and E2F2 knockdown can inhibit human embryonic stem cells proliferation and tumorigenicity without significantly harming stemness [25]. Other members of E2F family also have been studied in many cancers. Previous studies have showed that E2F1 is overexpressed in non-small cell lung carcinoma in both protein and mRNA levels. The overexpression of E2F1 has been demonstrated contributing to the development of NSCLC and indicating worse patient prognosis [26,27]. In addition, increased E2F1 has been reported in breast cancer, ovarian caner small cell lung carcinoma and thyroid cancer but absent in prostate cancer and related to a poor prognosis [23,28-32]. E2F3 has been reported overexpression in prostate cancer and patients with prostate cancer exhibiting immunohistochemically detectable nuclear E2F3 expression have poorer overall survival and E2F3 expression could be as an independent factor predicting overall survival [33]. In urothelial carcinomas of the bladder, the overexpression of E2F3 was related to invasive tumor growth and rapid tumor cell proliferation [34]. In ovarian cancer, E2F4, E2F7 and E2F8 expression is reportedly increased. Furthermore, the overexpression of E2F4 and E2F7 has been related to better overall and disease-free survival, respectively, however the overexpression of E2F8 was associated with worse overall survival [23,24,35]. In contrast to the other member of E2F family, E2F2 has only recently been investigated in colorectal adenocarcinoma [36]. The expression of E2F2 was very low in colorectal tissues and found without any relationship to clinical parameters which indicated that E2F2 expression does not contribute in colorectal carcinogenesis.
E2F2 can efficiently activate DNA synthesis in quiescent fibroblasts and its expression only accumulates as cells enter G1 phase [37]. Further, the combined ablation of E2F-2 prevents entry into S phase and its overexpression overrides the effects of growth inhibitory proteins, such as p16, p21, and p27 [38]. However, there is no study confirmed the expression of E2F2 in patient with NSCLC and its role in prognosis. Further study should be done to investigate that E2F2 could be a novel biomarker and prognostic predictor for making individual therapy strategies in patients with NSCLC.
In the present study, we detected the expression of E2F2 in 86 NSCLC patients by immunohistochemistry. The relationship between E2F2 expression and the clinicopathological features was also assessed and the prognostic value of NSCLC in NSCLC was further determined. In addition, we the first time demonstrated that E2F2 play an important role in NSCLC cell proliferation and DNA damage response.
Materials and methods
Cell lines
The NSCLC cells H1299 and A549 were purchased form ATCC (Manassas, VA) and all cells were cultured in DMEM (Gibco, Carlsbad, CA) supplemented with 10% FBS (Invitrogen Life Technologies, Carlsbad, CA) and 1% penicillin/streptomycin (Gibco). The cells were cultured for 2 weeks in drug-free medium before experiments.
Patients and tissue specimens
A total of 86 NSCLC patient specimens were obtained from The First Affiliated Hospital of Nanchang University between January 2007 and July 2010. 8 paired NSCLC and corresponding adjacent normal tissues after surgical resection were used to detect the E2F2 expression using Western blot. All tissues were immediately stored at -80°C until the experiments started. The 86 patients aged from 31 to 71 years (median age is 52). The median follow-up period was 27 months (range: 1-60 months). We defined the tumor stage according to tumor-node metastasis (TNM) classification of the American Joint Committee on International Union against Cancer. All protocols concerning human subjects were approved by the Regional Ethical Committee of Nan chang University.
Western blot
The Western blot was used to detect the protein level in NSCLC tissues and cells. Briefly, collecting tissues and cells were lysed with lysis buffer (pH 7.4, containing 1% Triton X-100 and 0.2% SDS) and equal amounts of protein (50 μg) were resolved by SDS-PAGE and transferred onto PVDF membranes. The membranes were blocked with 5% nonfat milk and incubated with primary antibody against E2F2 (at a 1:1000 dilution, Cell signaling technology, USA) and GAPDH (1:1000, Santa Cruz, USA) at 4°C for overnight. After washing three times by TBST, the membranes were incubated with horseradish peroxides-conjugated secondary antibody (1:10000 dilution for rabbit antibody and mouse antibody) for 1 h at room temperature. Eventually, the protein-antibody complex was detected by enhanced chemiluminescence detection system (Amersham, NJ). GAPDH was served as a loading control.
Plasmid construction and stable transfection
The E2F2 shRNA and scrambled shRNA were purchased from GenePharma Co, Ltd (Shanghai). After confirmed the plasmids by sequencing, we constructed the plasmids into A549 and H1299 cells by lentiviral transduction. Briefly, we used 4 μg of plasmids transfecting into A549 and H1299 cells by mixing with Lipofectamine™ 2000 (Invitrogen; Carlsbad, CA, USA) reagent according to the instructions of manufacturer. After 48 hours, the cells were added 1 mg/ml of puromycin (Sigma Aldrih, MA, USA) for selection and the concentration of puromycin was elevated step by step until 10 mg/ml. we collected the cells lysis every 2 week to test the expression level of E2F2. Finally, the puromycin resistant cell population was determined down-expression of E2F2 by western blot as previously described.
Immunohistochemistry (IHC)
We performed immunohistochemistry (IHC) analysis to detect E2F2 expression and location according to previous report [20]. Briefly, all slides were baked at 60°C for 4 h and then deparaffinized and rehydrated and boiled the slides in Ethylene Diamine Tetraacetic Acid (EDTA; 1 mmol/L; PH 8.0) for antigen retrieval. After incubated overnight at 4°C with E2F2 antibody (1:500 dilution), the slides were rinsed with PBS and incubated with a secondary antibody for half hour at room temperature. Stained cell proportions were scored depending on the methods mentioned in previous study. The receptor score was calculated as following: (positively stained cell proportion x stained intensity). We classified the E2F2 expression into two groups: high expression (at scores of ≥ 4) and low expression (at scores of < 4), respectively [21].
MTT assay
The cell viability was determined by MTT assay. Briefly, cells of 3-5 × 103 were seeded in 96-well plates and allowed to attach overnight. At a certain time (12, 24, 36, 48, 60, 72, 84, 96 h), 20 μL MTT (5 mg/mL) was added to each cell for 4 h (37°C), and then DMSO (120 μL/well) was added to dissolve the formazan. Finally, the density was measured at 540 nm by Model 550 Microplate Reader (Bio-Rad, USA). Experiments were performed at least three times.
Statistical analysis
Statistical analyses were performed using the SPSS 16.0 software (SPSS, Chicago, IL, USA). The Student t test was used for comparison between groups. The x2 test was performed to analyze the correlation between Bad expression and clinic pathological parameters. The Kaplan-Meier method (the log-rank test) was used for survival curves. Cox regression model with stepwise manner (forward, likelihood ratio) was utilized to perform a multivariate analysis. P < 0.05 (two-tailed) was considered statistically significant.
Results
E2F2 expression is upregulated in fresh NSCLC tissues
Firstly, we detected the expression level of E2F2 in 8 paired NSCLC tissues by western blot. According to the results, the expression of E2F2 was noticeably higher in NSCLC tissues than that in the neighboring normal tissues in 5 of 8 cases (Figure 1A). Additionally, the relevant densities of western blot bands also indicated that the E2F2 expression is upregulated in tumor tissues (Figure 1B).
Figure 1.
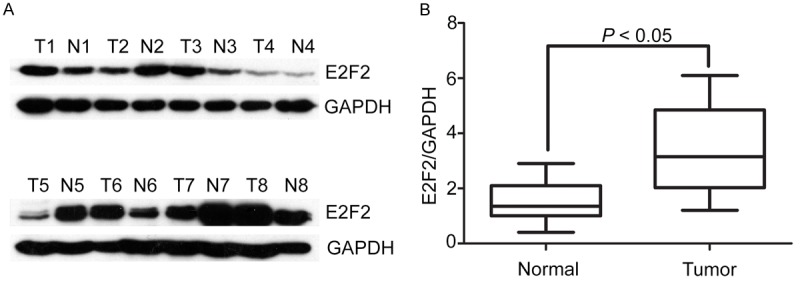
Expression of E2F2 in NSCLC tissue samples by western blot. A. Protein levels of E2F2 in NSCLC tissue samples by western blot. Representative images of E2F2 expression were presented. The ratio of E2F2/GAPDH was indicated below. B. Relative intensity of E2F2 normalized to GAPDH was calculated (n = 8).
High E2F2 expression is associated with the poor clinic pathological parameters
Then we performed IHC to assess the expression of E2F2 in 86 paraffin-embedded NSCLC tissues from the First Affiliated Hospital of Nanchang University. The IHC results showed that E2F2 was mainly located in the cytoplasm in both tumor tissues and normal tissues. In tumor tissues, there was also scattered staining of E2F2 in the nuclear (Figure 2A-E). Furthermore, we found that E2F2 exhibited higher expression in NSCLC tissues compared to neighboring tissues as shown in Figure 2F. In addition, the clinical relationship between E2F2 expression and chinicopathologic factors was examined. According to the results, a significant difference was observed in clinical stage (P = 0.039) and tumor size (P = 0.045) but there was no statistical relationship between E2F2 expression and the rest clinic pathological parameters, such as age, gender, tumor size or tumor recurrence (Table 1).
Figure 2.
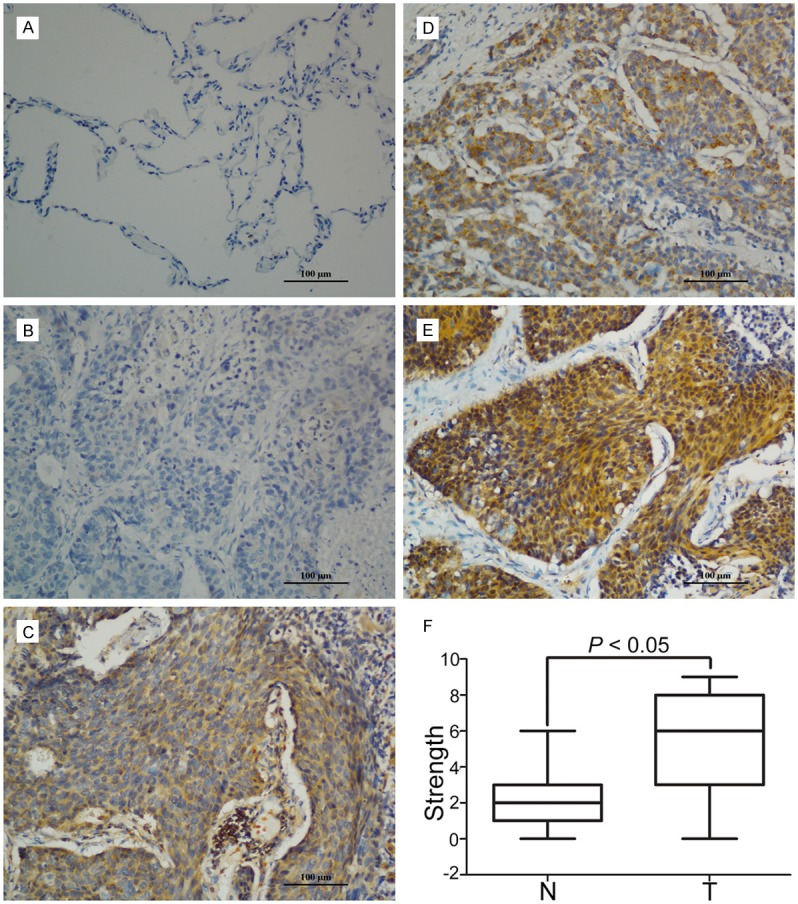
Expression of E2F2 in N SCLC tissues by IHC. A. Micrographs showed the staining of E2F2 in normal lung tissues. B-E. Micrographs showed the staining of E2F2 in tumor tissues (negative, weak, modern, strong). F. Reproducibility of the measurement in all 86 patients was calculated using the Wilcoxon matched paired test.
Table 1.
Correlation of clinicopathological parameters and E2F2 expression in the NSCLC (n = 86)
| Variable | All cases | Low expression | High expression | P value |
|---|---|---|---|---|
| Age (years) | 0.719 | |||
| < 49 | 33 | 15 (57.6%) | 18 (42.3%) | |
| ≥ 49 | 53 | 22 (50.0%) | 31 (50.0%) | |
| Gender | 0.927 | |||
| Male | 46 | 20 (52.4%) | 26 (47.6%) | |
| Female | 40 | 17 (65.0%) | 23 (35.0%) | |
| Lymph node metastasis | 0.186 | |||
| Yes | 25 | 8 (54.3%) | 17 (45.7%) | |
| No | 61 | 29 (49.3%) | 32 (50.7%) | |
| Stage | 0.039 | |||
| I-II | 52 | 27 (51.8%) | 25 (48.2%) | |
| III-IV | 34 | 10 (62.5%) | 24 (37.5%) | |
| Tumor size (cm) | 0.045 | |||
| < 4 | 19 | 12 (41.5%) | 7 (58.5%) | |
| ≥ 4 | 67 | 25 (56.9%) | 42 (43.1%) | |
| Metastasis | 0.826 | |||
| Yes | 57 | 25 (50.2%) | 32 (49.8%) | |
| No | 29 | 12 (58.9%) | 17 (41.1%) | |
| Location | 0.579 | |||
| Left | 32 | 15 (47.7%) | 17 (52.3%) | |
| Right | 54 | 22 (60.5%) | 32 (39.5%) | |
| Tumor recurrence | 0.917 | |||
| Yes | 54 | 23 (63.8%) | 31 (36.2%) | |
| No | 32 | 14 (50.7%) | 18 (49.3%) |
High expression of E2F2 was related to the poor prognosis
We detected the correlation between E2F2 expression level and survival time of NSCLC patients using Kaplan-Meier survival analysis. Our results revealed that NSCLC patients with high E2F2 expression was tend to show a significant shorter overall survival (P = 0.045, Figure 3A). However, we did not observe a significant difference between the expression of E2F2 and disease-free survival (P = 0.288, Figure 3B).
Figure 3.
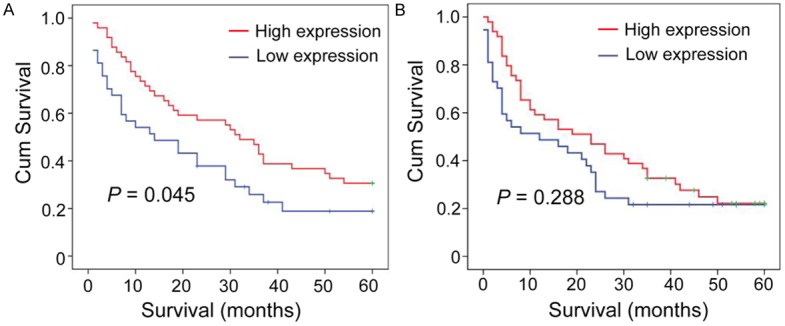
Relationship between E2F2 expression and NSCLC prognosis. E2F2 protein level showed prognostic role in overall survival (A), disease-free survival (B) a, as indicated by Kaplan-Meier analysis. Statistical significance was assessed with the log-rank test. (n = 86).
Univariate and multivariate analyses of prognostic variables in NSCLC patients
Next we performed univariate analysis to further evaluate the E2F2 expression and other clinicopathologic parameters on prognosis of NSCLC patients. Results indicated that only E2F2 expression and tumor size was responsible for efficacy of surgical treatment in HCC patient, by showing that E2F2 expression was significantly associated with overall survival (Table 2). Furthermore, the results of multivariate analysis suggested that E2F2 remained to be an independent predictor for overall survival (HR: 0.589, 95% CI: 0.483-0.717, P < 0.0001) of NSCLC patients (Table 2).
Table 2.
Univariate and multivariate analysis of clinicopathological and E2F2 for overall and disease-free survival in NSCLC (n = 86)
| Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
|
|
|
|||
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Overall survival | ||||
| Age (< 49 vs. ≥ 49 years) | 0.711 (0.392-1.290) | 0.262 | ||
| Gender (female vs. male) | 0.962 (0.546-1.697) | 0.895 | ||
| Lymph node metastasis (yes vs. no) | 1.053 (0.578-1.918) | 0.865 | ||
| Tumor size (< 4 vs. ≥ 4 cm) | 0.460 (0.236-0.895) | 0.022 | 0.452 (0.246-0.831) | 0.011 |
| Tumor recurrence (yes vs. no) | 1.319 (0.701-2.482) | 0.391 | ||
| Tumor location | 0.654 (0.368-1.161) | 0.147 | ||
| Stage (I-II vs. III-IV) | 0.942 (0.535-1.660) | 0.837 | ||
| E2F2 expression (low vs. high) | 0.407 (0.224-0.740) | 0.003 | 0.490 (0.282-0.853) | 0.012 |
| Disease-free survival | ||||
| Age (< 49 vs. ≥ 49 years) | 0.719 (0.396-1.304) | 0.278 | ||
| Gender (female vs. male) | 0.902 (0.511-1.592) | 0.721 | ||
| Lymph node metastasis (yes vs. no) | 1.133 (0.624-2.057) | 0.683 | ||
| Tumor size (< 5 vs. ≥ 5 cm) | 0.593 (0.307-1.147) | 0.120 | ||
| Tumor recurrence (yes vs. no) | 1.174 (0.632-2.183) | 0.611 | ||
| Tumor location | 0.794 (0.448-1.408) | 0.431 | ||
| Stage (I-II vs. III-IV) | 0.870 (0.489-1.545) | 0.633 | ||
| E2F2 expression (low vs. high) | 0.799 (0.472-1.351) | 0.402 | ||
Downregulation E2F2 suppress cell viability of NSCLC cells
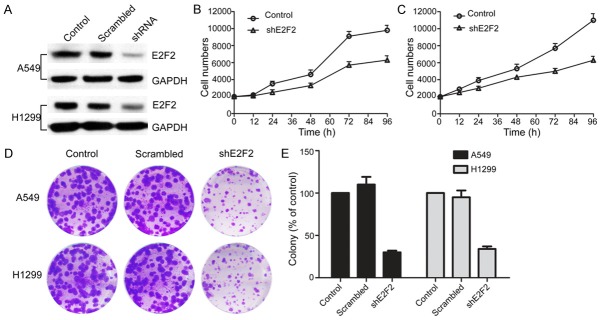
Previous studies have demonstrated that E2F2 is associated with cell proliferation in cancers. To investigate the role of E2F2 in NSCLC cell proliferation, we chose two NSCLC cells for our research: the A549 cell and H1299 cell which are overexpressing E2F2 protein. First of all, we down-regulated E2F2 expression in A549 and H1299 cells stably with shE2F2. The down-regulation of E2F2 expression was confirmed by Western blot analysis (Figure 4A). The cell viability assay results demonstrated that the downregulation of E2F2 expression in both A549 and H1299 cells significantly inhibited cell viability compared to control cells (Figure 4B and 4C). Our results were consisted to the previous study that knockdown E2F2 expression can slow down the cell proliferation. Furthermore, we assessed the effect of E2F2 on long-term cellular colony formation potential, Normal control, scrambled and knockdown shE2F2 clones were plated in soft agar for 2 weeks. The colony formation data showed that the cell growth in shE2F2 cells was less than 20% of control in both A549 and H1299 cells (Figure 4D and 4E). These data indicated that ablation of E2F2 significantly block cell survival and colony formation in NSCLC cells.
Figure 4.
E2F2 shRNA significantly decreased cell proliferation in NSCLC cells. E2F2 shRNA noticeably down regulated expression of E2F2 protein in A549 and H1299 cells (A). Scramble and E2F2 shRNA were transected into NSCLC cells for 24 h. Relative E2F2 expressions were detected by Western blot. The cell viabilities were determined using MTT assays (B, C). A549 and H1299 knockdown cells were plated in soft agar and colonies were counted 2 wk later. shControl represents the parental line after transduction of shE2F2 represents independent constructs against E2F2 (D). **, P < 0.01, in comparison to the control (two-tailed t test). Error bars represent three independent experiments that were done in triplicate.
Discussion
In the present study, we discussed the relationship between E2F2 expression and clinical significance of NSCLC patient. Our data revealed for the first time that E2F2 was upregulated in NSCLC patients and indicates few worse outcomes. Furthermore, we found that E2F2 deficient cells showed a slow growth rate. However, we didn’t observe a statically significance between E2F2 expression and the survival time. A further study should be done to determine the role of E2F2 in NSCLC progression.
In the past decades, many reports revealed that the E2Fs family members’ activity play a crucial role in tumor growth and worse outcomes [26,33,39]. Previous studies indicated that the ‘activator’ E2Fs (E2F1, E2F2 and E2F3) determine the progress of some types of cancer such as non-small cell lung carcinoma and esophageal squamous cell carcinoma [26,39]. In the present study, we found that high E2F2 expression is associated with increasing tumor size and advanced clinical stage which indicated that E2F2 expression might be served as a promising hallmark of lung cancer outcomes. However, we did not observe a significant relationship between E2F2 expression and other clinical characters such as tumor recurrence, metastasis or lymph node metastasis. We can put forward that monitoring the expression dynamics of E2F2 may contribute to define patients as follow-up of patients in need of treatment. In addition, many studies have determined the relationship between E2F2 expression with clinical pathological parameters in other tumors such as hepatocellular carcinoma and Ovarian Cancer [23,40]. Their results also depicted that E2F2 was over-expressed in tumor tissues and the over-expression of E2F2 closely correlated with larger tumor size and worse outcomes which is consistent with our findings. Elsewhere, the low E2F2 expression was demonstrated to be significantly associated with better outcomes in our study.
For further study, we analyzed the relationship of E2F2 expression with overall survival (OS) and disease-free survival time (DFS) by Kaplan-Meier survival analysis. The results showed that patients with high E2F2 expression tend to be a shorter overall survival compared with that of low E2F2 expression. The similar result has obtained from other cancers. It is found that high expression of E2F2 protein indicate a worse prognosis in ovarian cancer [23]. Furthermore, E2F2 remains to be an independent prognostic factor for overall survival by Cox regression analysis, which suggesting its pivotal role in tumor generation and progression. In addition, recent study provided a comprehensive understanding of the role of E2F2 in tumorigenesis in human glioblastoma cells [41]. In summary, high expression of E2F2 in tumor tissues may promote tumor growth and progression, indicating an adverse prognosis in non-small cell lung carcinoma.
E2F2 is involved in many biological progresses of cell cycle, DNA synthesis and apoptosis by binding to Rb protein to form the Rb/E2F complexes [42,43]. E2F2 is belonging to the activator group of E2Fs family. It has been demonstrated that E2F2 can promote cell proliferation by repressing cell cycle regulators and Myc-induced lymphomagenesis [44,45]. Interestingly, DeGregori et al. found that E2F2 may act as either a tumor suppressor or activator in different cancer types [46]. For instance, loss of E2F2 can accelerate cell proliferation in T cells but reduce the cell viability in viable tumor cells [25,45]. Timmers et al. has demonstrated that E2F2 regulated cell cycle by influencing cell cycle regulators such as cyclin, cyclin dependent kinases (CDKs) and checkpoint signals [47]. However, it is little known about the roles of E2F2 in non-small cell lung carcinoma. In our study, when we explored the roles of E2F2 on the proliferation of non-small cell lung carcinoma cells, we found that when knockdown E2F2 expression, the cell proliferation and colony formation were ablation compared to the control group. The results showed that E2F2 is involved in the cell growth of NSCLC. Elsewhere, Infante et al. found that E2F2 acts as a suppressor by repressing cell cycle regulators to maintain quiescence [44]. Nevertheless, little is known about roles of E2F2 in lung cancer cells, and here we validated for the first time that E2F2 acts as a tumor activator in non-small cell lung carcinoma.
Taken together, our study suggests that E2F2 outperformed an independent indicator for overall survival in NSCLC patients. Increased E2F2 expression is noteworthy related to larger tumor size and advanced clinical stage. Our present study may provide a promising biomarker for predicting the postsurgical prognosis of patients who suffer from this deadly disease.
Acknowledgements
This work was supported by Educational Commission of Nanchang University.
Disclosure of conflict of interest
None.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Xu MM, Mao GX, Liu J, Li JC, Huang H, Liu YF, Liu JH. Low expression of the FoxO4 gene may contribute to the phenomenon of EMT in non-small cell lung cancer. Asian Pac J Cancer Prev. 2014;15:4013–4018. doi: 10.7314/apjcp.2014.15.9.4013. [DOI] [PubMed] [Google Scholar]
- 3.Bonomi P. Novel approaches for the treatment of non-small cell lung cancer. Semin Oncol. 2001;28:45–49. doi: 10.1016/s0093-7754(01)90059-4. [DOI] [PubMed] [Google Scholar]
- 4.Arriagada R, Auperin A, Burdett S, Higgins JP, Johnson DH, Le Chevalier T, Le Pechoux C, Parmar MK, Pignon JP, Souhami RL, Stephens RJ, Stewart LA, Tierney JF, Tribodet H, van Meerbeeck J. Adjuvant chemotherapy, with or without postoperative radiotherapy, in operable non-small-cell lung cancer: two meta-analyses of individual patient data. Lancet. 2010;375:1267–1277. doi: 10.1016/S0140-6736(10)60059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kovesdi I, Reichel R, Nevins JR. Identification of a cellular transcription factor involved in E1A trans-activation. Cell. 1986;45:219–228. doi: 10.1016/0092-8674(86)90386-7. [DOI] [PubMed] [Google Scholar]
- 6.Kovesdi I, Reichel R, Nevins JR. Role of an adenovirus E2 promoter binding factor in E1A-mediated coordinate gene control. Proc Natl Acad Sci U S A. 1987;84:2180–2184. doi: 10.1073/pnas.84.8.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevens C, La Thangue NB. E2F and cell cycle control: a double-edged sword. Arch Biochem Biophys. 2003;412:157–169. doi: 10.1016/s0003-9861(03)00054-7. [DOI] [PubMed] [Google Scholar]
- 8.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9:785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cartwright P, Muller H, Wagener C, Holm K, Helin K. E2F-6: a novel member of the E2F family is an inhibitor of E2F-dependent transcription. Oncogene. 1998;17:611–623. doi: 10.1038/sj.onc.1201975. [DOI] [PubMed] [Google Scholar]
- 10.Gaubatz S, Wood JG, Livingston DM. Unusual proliferation arrest and transcriptional control properties of a newly discovered E2F family member, E2F-6. Proc Natl Acad Sci U S A. 1998;95:9190–9195. doi: 10.1073/pnas.95.16.9190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Stefano L, Jensen MR, Helin K. E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J. 2003;22:6289–6298. doi: 10.1093/emboj/cdg613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen J, Cloos P, Toftegaard U, Klinkenberg D, Bracken AP, Trinh E, Heeran M, Di Stefano L, Helin K. Characterization of E2F8, a novel E2F-like cell-cycle regulated repressor of E2F-activated transcription. Nucleic Acids Res. 2005;33:5458–5470. doi: 10.1093/nar/gki855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bagchi S, Raychaudhuri P, Nevins JR. Adenovirus E1A proteins can dissociate heteromeric complexes involving the E2F transcription factor: a novel mechanism for E1A trans-activation. Cell. 1990;62:659–669. doi: 10.1016/0092-8674(90)90112-r. [DOI] [PubMed] [Google Scholar]
- 14.Lavia P, Jansen-Durr P. E2F target genes and cell-cycle checkpoint control. Bioessays. 1999;21:221–230. doi: 10.1002/(SICI)1521-1878(199903)21:3<221::AID-BIES6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 15.Huang Y, Ishiko T, Nakada S, Utsugisawa T, Kato T, Yuan ZM. Role for E2F in DNA damage-induced entry of cells into S phase. Cancer Res. 1997;57:3640–3643. [PubMed] [Google Scholar]
- 16.Blattner C, Sparks A, Lane D. Transcription factor E2F-1 is upregulated in response to DNA damage in a manner analogous to that of p53. Mol Cell Biol. 1999;19:3704–3713. doi: 10.1128/mcb.19.5.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Connor DJ, Lu X. Stress signals induce transcriptionally inactive E2F-1 independently of p53 and Rb. Oncogene. 2000;19:2369–2376. doi: 10.1038/sj.onc.1203540. [DOI] [PubMed] [Google Scholar]
- 18.Lin WC, Lin FT, Nevins JR. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 2001;15:1833–1844. [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Y, Freeman SN, Cress WD. E2F4 deficiency promotes drug-induced apoptosis. Cancer Biol Ther. 2004;3:1262–1269. doi: 10.4161/cbt.3.12.1239. [DOI] [PubMed] [Google Scholar]
- 20.Martinez LA, Goluszko E, Chen HZ, Leone G, Post S, Lozano G, Chen Z, Chauchereau A. E2F3 is a mediator of DNA damage-induced apoptosis. Mol Cell Biol. 2010;30:524–536. doi: 10.1128/MCB.00938-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zalmas LP, Zhao X, Graham AL, Fisher R, Reilly C, Coutts AS, La Thangue NB. DNA-damage response control of E2F7 and E2F8. EMBO Rep. 2008;9:252–259. doi: 10.1038/sj.embor.7401158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivey-Hoyle M, Conroy R, Huber HE, Goodhart PJ, Oliff A, Heimbrook DC. Cloning and characterization of E2F-2, a novel protein with the biochemical properties of transcription factor E2F. Mol Cell Biol. 1993;13:7802–7812. doi: 10.1128/mcb.13.12.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reimer D, Sadr S, Wiedemair A, Goebel G, Concin N, Hofstetter G, Marth C, Zeimet AG. Expression of the E2F family of transcription factors and its clinical relevance in ovarian cancer. Ann N Y Acad Sci. 2006;1091:270–281. doi: 10.1196/annals.1378.073. [DOI] [PubMed] [Google Scholar]
- 24.Reimer D, Sadr S, Wiedemair A, Stadlmann S, Concin N, Hofstetter G, Muller-Holzner E, Marth C, Zeimet AG. Clinical relevance of E2F family members in ovarian cancer--an evaluation in a training set of 77 patients. Clin Cancer Res. 2007;13:144–151. doi: 10.1158/1078-0432.CCR-06-0780. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki DE, Nakahata AM, Okamoto OK. Knockdown of E2F2 inhibits tumorigenicity, but preserves stemness of human embryonic stem cells. Stem Cells Dev. 2014;23:1266–1274. doi: 10.1089/scd.2013.0592. [DOI] [PubMed] [Google Scholar]
- 26.Gorgoulis VG, Zacharatos P, Mariatos G, Kotsinas A, Bouda M, Kletsas D, Asimacopoulos PJ, Agnantis N, Kittas C, Papavassiliou AG. Transcription factor E2F-1 acts as a growth-promoting factor and is associated with adverse prognosis in non-small cell lung carcinomas. J Pathol. 2002;198:142–156. doi: 10.1002/path.1121. [DOI] [PubMed] [Google Scholar]
- 27.Imai MA, Oda Y, Oda M, Nakanishi I, Kawahara E. Overexpression of E2F1 associated with LOH at RB locus and hyperphosphorylation of RB in non-small cell lung carcinoma. J Cancer Res Clin Oncol. 2004;130:320–326. doi: 10.1007/s00432-003-0538-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han S, Park K, Bae BN, Kim KH, Kim HJ, Kim YD, Kim HY. E2F1 expression is related with the poor survival of lymph node-positive breast cancer patients treated with fluorouracil, doxorubicin and cyclophosphamide. Breast Cancer Res Treat. 2003;82:11–16. doi: 10.1023/B:BREA.0000003843.53726.63. [DOI] [PubMed] [Google Scholar]
- 29.Zacharatos P, Kotsinas A, Evangelou K, Karakaidos P, Vassiliou LV, Rezaei N, Kyroudi A, Kittas C, Patsouris E, Papavassiliou AG, Gorgoulis VG. Distinct expression patterns of the transcription factor E2F-1 in relation to tumour growth parameters in common human carcinomas. J Pathol. 2004;203:744–753. doi: 10.1002/path.1582. [DOI] [PubMed] [Google Scholar]
- 30.Alonso MM, Fueyo J, Shay JW, Aldape KD, Jiang H, Lee OH, Johnson DG, Xu J, Kondo Y, Kanzawa T, Kyo S, Bekele BN, Zhou X, Nigro J, McDonald JM, Yung WK, Gomez-Manzano C. Expression of transcription factor E2F1 and telomerase in glioblastomas: mechanistic linkage and prognostic significance. J Natl Cancer Inst. 2005;97:1589–1600. doi: 10.1093/jnci/dji340. [DOI] [PubMed] [Google Scholar]
- 31.Eymin B, Gazzeri S, Brambilla C, Brambilla E. Distinct pattern of E2F1 expression in human lung tumours: E2F1 is upregulated in small cell lung carcinoma. Oncogene. 2001;20:1678–1687. doi: 10.1038/sj.onc.1204242. [DOI] [PubMed] [Google Scholar]
- 32.Onda M, Nagai H, Yoshida A, Miyamoto S, Asaka S, Akaishi J, Takatsu K, Nagahama M, Ito K, Shimizu K, Emi M. Up-regulation of transcriptional factor E2F1 in papillary and anaplastic thyroid cancers. J Hum Genet. 2004;49:312–318. doi: 10.1007/s10038-004-0146-3. [DOI] [PubMed] [Google Scholar]
- 33.Foster CS, Falconer A, Dodson AR, Norman AR, Dennis N, Fletcher A, Southgate C, Dowe A, Dearnaley D, Jhavar S, Eeles R, Feber A, Cooper CS. Transcription factor E2F3 overexpressed in prostate cancer independently predicts clinical outcome. Oncogene. 2004;23:5871–5879. doi: 10.1038/sj.onc.1207800. [DOI] [PubMed] [Google Scholar]
- 34.Oeggerli M, Tomovska S, Schraml P, Calvano-Forte D, Schafroth S, Simon R, Gasser T, Mihatsch MJ, Sauter G. E2F3 amplification and overexpression is associated with invasive tumor growth and rapid tumor cell proliferation in urinary bladder cancer. Oncogene. 2004;23:5616–5623. doi: 10.1038/sj.onc.1207749. [DOI] [PubMed] [Google Scholar]
- 35.De Meyer T, Bijsmans IT, Van de Vijver KK, Bekaert S, Oosting J, Van Criekinge W, van Engeland M, Sieben NL. E2Fs mediate a fundamental cell-cycle deregulation in high-grade serous ovarian carcinomas. J Pathol. 2009;217:14–20. doi: 10.1002/path.2452. [DOI] [PubMed] [Google Scholar]
- 36.Xanthoulis A, Kotsinas A, Tiniakos D, Fiska A, Tentes AA, Kyroudi A, Kittas C, Gorgoulis V. The relationship between E2F family members and tumor growth in colorectal adenocarcinomas: A comparative immunohistochemical study of 100 cases. Appl Immunohistochem Mol Morphol. 2014;22:471–477. doi: 10.1097/PAI.0b013e3182598198. [DOI] [PubMed] [Google Scholar]
- 37.DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci U S A. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu L, Timmers C, Maiti B, Saavedra HI, Sang L, Chong GT, Nuckolls F, Giangrande P, Wright FA, Field SJ, Greenberg ME, Orkin S, Nevins JR, Robinson ML, Leone G. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001;414:457–462. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- 39.Ebihara Y, Miyamoto M, Shichinohe T, Kawarada Y, Cho Y, Fukunaga A, Murakami S, Uehara H, Kaneko H, Hashimoto H, Murakami Y, Itoh T, Okushiba S, Kondo S, Katoh H. Over-expression of E2F-1 in esophageal squamous cell carcinoma correlates with tumor progression. Dis Esophagus. 2004;17:150–154. doi: 10.1111/j.1442-2050.2004.00393.x. [DOI] [PubMed] [Google Scholar]
- 40.Zhan L, Huang C, Meng XM, Song Y, Wu XQ, Miu CG, Zhan XS, Li J. Promising roles of mammalian E2Fs in hepatocellular carcinoma. Cell Signal. 2014;26:1075–1081. doi: 10.1016/j.cellsig.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 41.Nakahata AM, Suzuki DE, Rodini CO, Fiuza ML, Okamoto OK. RNAi-mediated knockdown of inhibits tumorigenicity of human glioblastoma cells. Oncol Lett. 2014;8:1487–1491. doi: 10.3892/ol.2014.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dimova DK, Dyson NJ. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24:2810–2826. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- 43.McClellan KA, Ruzhynsky VA, Douda DN, Vanderluit JL, Ferguson KL, Chen D, Bremner R, Park DS, Leone G, Slack RS. Unique requirement for Rb/E2F3 in neuronal migration: evidence for cell cycle-independent functions. Mol Cell Biol. 2007;27:4825–4843. doi: 10.1128/MCB.02100-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Infante A, Laresgoiti U, Fernandez-Rueda J, Fullaondo A, Galan J, Diaz-Uriarte R, Malumbres M, Field SJ, Zubiaga AM. E2F2 represses cell cycle regulators to maintain quiescence. Cell Cycle. 2008;7:3915–3927. doi: 10.4161/cc.7.24.7379. [DOI] [PubMed] [Google Scholar]
- 45.Opavsky R, Tsai SY, Guimond M, Arora A, Opavska J, Becknell B, Kaufmann M, Walton NA, Stephens JA, Fernandez SA, Muthusamy N, Felsher DW, Porcu P, Caligiuri MA, Leone G. Specific tumor suppressor function for E2F2 in Myc-induced T cell lymphomagenesis. Proc Natl Acad Sci U S A. 2007;104:15400–15405. doi: 10.1073/pnas.0706307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeGregori J. The genetics of the E2F family of transcription factors: shared functions and unique roles. Biochim Biophys Acta. 2002;1602:131–150. doi: 10.1016/s0304-419x(02)00051-3. [DOI] [PubMed] [Google Scholar]
- 47.Timmers C, Sharma N, Opavsky R, Maiti B, Wu L, Wu J, Orringer D, Trikha P, Saavedra HI, Leone G. E2f1, E2f2, and E2f3 control E2F target expression and cellular proliferation via a p53-dependent negative feedback loop. Mol Cell Biol. 2007;27:65–78. doi: 10.1128/MCB.02147-05. [DOI] [PMC free article] [PubMed] [Google Scholar]