Abstract
MicroRNAs have been implicated in cancer cells proliferation, migration and invasion, including clear-cell renal cell carcinoma (ccRCC). DNA methylation, a major epigenetic change associated with cancer, may lead to transcriptional silencing of tumor suppressor genes, including miRNAs, which may be a possible mechanism in carcinogenesis. In this study, we aim to investigate the role of miR-10b in ccRCC and its possible epigenetic mechanism. By qPCR and MSP, we found that miR-10b was significantly decreased in ccRCC tissues and cells, and exhibited heavy methylation on promoter. Upregulation of miR-10b by transfecting with lentivirus highly expressed miR-10b or by exogenous demethylation agents was capable of inhibiting cell proliferation, migration and invasion measured by CCK-8 cell proliferation assay, scratch assay, transwell assay, and flow cytometric analysis of the cell cycle. Our results suggest that miR-10b plays a tumor-suppressive role in ccRCC. Demethylation of miR-10b may be therapeutically beneficial for ccRCC treatment.
Keywords: Clear-cell renal cell carcinoma, epigenetic, methylation, miR-10b, microRNA, invasion
Introduction
Clear-cell renal cell carcinoma (ccRCC), the leading lethal cause of renal cell carcinoma with steadily rising in the world, is a highly treatment-resistant tumor type [1]. Emerging evidence is elucidating the molecular mechanisms underlying ccRCC and promising therapeutic targets [2]. In ccRCC, mutations result in the up-regulation or down-regulation of many coding RNAs or non-coding RNAs, such as microRNAs (miRNAs), that necessary for tumor growth, survival and angiogenesis [3].
MiRNAs are a class of short (~22 nt) noncoding regulatory RNAs, which have been demonstrated to be promise diagnostic biomarkers and therapeutic targets for cancer. MiRNAs act as oncogenes or tumor suppressors depending on their target genes [4]. Several miRNAs, such as miR-185 [5], miR-200 [6] and miR-10b [7], have been found to be dysregulated in renal cell carcinoma. MiR-10b is significantly upregulated in non-small cell lung cancer (NSCLC) tissues as well as in A549 cell line, and positively correlated with TNM stage and regional lymph node involvement [8]. It was reported that miR-10b was significantly upregulated in bladder cancer cell lines and metastatic tissues, enhanced bladder cancer cell migration and invasion by targeting KLF4 and HOXD10 [9]. However, MiR-10b is significantly downregulated in gastric cancer cell lines and tissues. Overexpression of miR-10b in gastric cancer cell lines dramatically suppresses cell proliferation, migration, invasion, and induced apoptosis; furthermore, 5-aza-2’-deoxycytidine (5-Aza) increases miR-10b expression by decreasing the methylation level of miR-10b in the CpG islands [10]. It was found that miR-21 and miR-10b were differentially expressed in ccRCC nuclear grades. And the high ratio of miR-21/miR10b correlated significantly with poor prognosis in metastasis-free patients [11]. These findings indicate that miR-10b exhibits different functions depending on cancer types. The precise mechanisms underlying regulation of miR-10b in ccRCC remain unclear. DNA methylation, a major epigenetic change associated with cancer, may lead to transcriptional silencing of tumor suppressor genes, including miRNAs, which may be a possible mechanism in carcinogenesis. And DNA methylation has also been used to identify tumor suppressor genes. Moreover, DNA methylation markers have been used for cancer diagnosis, and prognosis.
In this study, we aim to investigate the role of miR-10b in ccRCC and its possible epigenetic mechanism. We demonstrate that heavy methylation on promoter CpGs of miR-10b in ccRCC is a regulator of multiple aspects of cancer biology. Upregulation of miR-10b is capable of inhibiting cell proliferation, migration and invasion. Our results suggest that miR-10b has a tumor-suppressive role in ccRCC. Demethylation of miR-10b may be therapeutically beneficial for treating ccRCC.
Materials and methods
Tissue samples
Total 9 cases of ccRCC tissues and the matched adjacent normal tissues used in the study were collected from 9 patients between 2010 and 2014 in our hospital. Informed consents were signed by all patients and approved by the Independent Ethical Committee of Central South University. All samples were stored at -80°C until tissue analysis.
Cells culture
HEK293, A498, ACHN, 786-O and Caki-1 cells were obtained from American type culture collection (ATCC). All the cells are cultured in RPMI1640 (Gibco) basic medium supplemented with 10% FBS at the conditions: 95% air and 5% CO2 at 37°C.
Lentivirus infection
In order to investigate the role of miR-10b in ccRCC cells, 786-O and ACHN cells were infected with negative control lentivirus (Lv-NC) or miR-10b lentivirus (miR-10b) for 36 h by using Lipofectamine 2000 (Invitrogen), and the untreated cells were used as mock control (Con). For overexpression of Twist1 in 786-O and ACHN cells, negative control lentivirus (NC) and Twist1 lentivirus (Twist1) were transfected into these cells by using Lipofectamine 2000 (Invitrogen), and the untreated cells were used as mock control (Con). QPCR and western blot were used to detect the expression of Twist1.
Epigenetic drug treatment of cells
786-O and ACHN cells were divided as following three groups: 1. Control; 2. treated with demethylation drug 5-Aza-2’-deoxycytidine (Aza, Sigma, USA) 15.55 nM for 48 h; 3. Treated with histone deacetylase inhibitor 4-Phenylbutyric acid (PBA, Sigma) 1.5 nM for 48 h.
Quantitative PCR (qPCR) analysis
Trizol reagent (Invitrogen, USA) was used to extract total RNA from the indicated cells according to the manufacturer’s instructions. The specific primers for miRNA-10b and U6 were purchased from GeneCopoeia. miScript SYBR® Green PCR Kit (Qiagen) was used to detect the expression of miR-10b. Expression of U6 was used to normalize the expression of miR-10b as an endogenous control. FastLane Cell SYBR® Green Kit (Qiagen, Valencia, CA) was used for real time PCR to detect the expression of Twist1. The primers of Twist1 and β-actin are as follow: Twist1, sense, TTCTCGGTCTGGAGGATGGA, antisense, CCCACGCCCTGTTTCTTTGAA; β-actin, sense, AGGGGCCGGACTCGTCATACT, antisense, GGCGGCACCACCATGTACCCT. Twist1 expression was normalized by β-actin. All the qPCR data were processed using 2-ΔΔCT method.
Western blot
RIPA lysis buffer (Boster, Wuhan, China) was used to extract the total protein was extracted from the tissues or cells. BCA Protein Assay Kit (Thermo, Waltham, MA) was used to measure the protein concentration. 60 μg of total protein was then separated with 10% SDS-PAGE and transferred to a nitrocellulose membrane. The membranes were blocked and incubated with indicated primary antibody (mouse monoclonal, anti-Twist1 from Abcam, 1:2000; Rabbit polyclonal, anti-β-actin from Abcam, 1:3000) overnight at 4°C. The membrane was washed and incubated with secondary antibody for 90 min at 37°C. The signals on the membrane were detected by enhanced chemiluminescence reagent. Data was analyzed by densitometry using Image-Pro plus software 6.0 and normalized to internal control expression (β-actin).
CCK-8 cell proliferation assay
Cell growth was measured by CCK-8 assay. 2000 indicated cells were seeded in each 96-well plate for 12 h, and further incubated for 0h, 24 h, 48 h and 72 h respectively. 1 hour before the ending of incubation, 10 μl CCK-8 reagents (Boster, Wuhan, China) were added to each well. OD 570 nm value in each well was determined by an enzyme immunoassay analyzer.
Scratch assay
Cells in each group were collected and resuspended in completed medium containing 10% FBS. Then, each well of a 6-well plate was seeded with 5×105 cells. When the cells were cultured into about 100% confluence, and were scratched with the head of a 200 μl tip, and washed these cells with serum-free medium. Then, these cells were further cultured for 24 h in serum-free medium. After that, serum-free medium were replaced with H-DMEM medium containing 10% FBS and continued to culture for 72 h, then these cells in each group were photographed for analysis.
Transwell assay
The indicated cells were starved in serum-free medium for 24 h. The cells were collected and then resuspended in serum-free medium. The cells were added to the upper chamber, while the lower chamber was filled with completed medium containing 10% FBS. After 24 hours incubation, cells attached to the bottom of upper chamber were fixed and stained with crystal violet for 30 min at 37°C, and dried in air. Acetic acid was used to dissolve the crystal violet. The optical density (OD) at 570 nm was detected by an enzyme immunoassay analyzer.
Flow cytometric analysis of the cell cycle
Cells from each group were trypsinizated and washed with cold PBS, and then stained by BD CycletestTM Plus (BD Biosciences) according to the manufacturer’s instructions. The cell cycle was analyzed by flow cytometry (Beckman Coulter, USA). The experiments were independently performed in triplicate.
Measurement of miR-10b promoter CpG island methylation status by bisulfite genomic sequencing PCR (BSP) and methylation specific PCR (MSP).
TaKaRa Genomic DNA Extraction Kit (TaKaRa Co., China) was used to extract Genomic DNA. Total 1 μg of Genomic DNA was modified with bisulfite using the Epitect Bisulfite Kit Protocol (Qiagen) according to the manufacturer’s instructions. The primers used to amplify were as following: miR-10b forward, AATTATGTGGGAGGTAGGAAGG, and reverse, TATCTACTCCTTTTCCTACTAC. The PCR products were gel extracted (Qiagen) to confirm that a single band had been obtained and were then sequenced by Beijing Genomics Institute (Wuhan, China).
Methylation-specific PCR (MS-PCR) was performed on bisulfate-treated DNA. The primers used were Un-methylated miR-10b forward, GTATTGTATAGTGTTGAGGTGTTGG, and reverse, ATCCAAACTAACCTCTCCATTCCAC; and methylated miR-10b forward: TAAGTATTGT ATAGTGTCGAGGCGT, and reverse, ATCCGAACTAACCTCTCCGTTCCGC. The annealing temperature was 58°C for Methylated-PCR, and Unmethylated-PCR, with 35 cycles used for each.
Statistical analysis
Student t-tests or one-way ANOVA were used to analyze statistical data by Graphpad prism5 software, depending on the experimental conditions. Compared with respective controls, statistical significance was considered when P values of <0.05.
Results
Methylation of CpG islands and expression of miR-10b in ccRCC tissues and RCC cell lines
The expression of miR-10b was significantly decreased in the ccRCC samples from the 9 ccRCC patients compared with the matched adjacent tissues, as indicated by the RT-qPCR (Figure 1A). And the similar results were observed in the ccRCC cell lines (Figure 1B). We further investigated the underlying mechanisms of miR-10b silence by MSP. A significantly higher level of methylation was observed in 8 ccRCC tissues compared with the matched adjacent tissues (Figure 1C).
Figure 1.
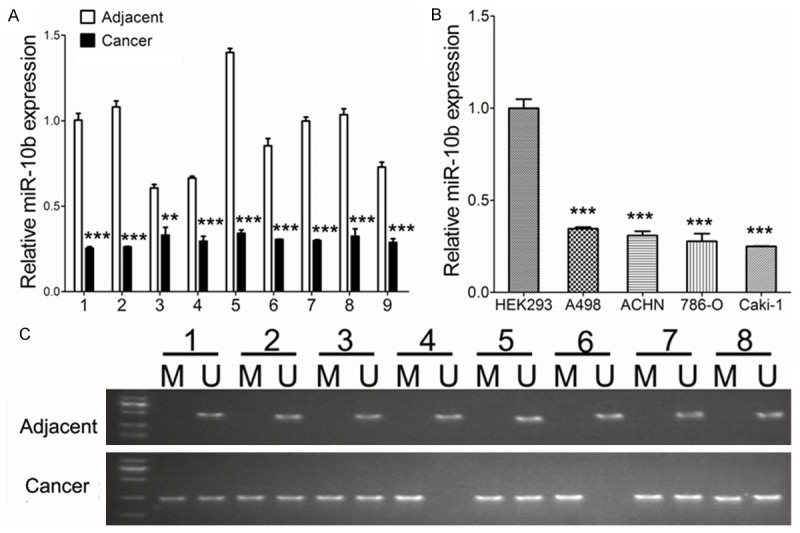
MiR-10b expression and CpG island methylation. A: MiR-10b expression levels were significantly lower in ccRCC tissues than that of in matched adjacent tissues. B: MiR-10b expression levels were significantly lower in renal cancer cell lines than those in HEK293 controls. C: MSP test results indicated that methylation of miR-10b promoter was higher in ccRCC tissues than that of in matched adjacent tissues. Data are presented by means ± s.d, *P<0.05, **P<0.01, ***P<0.001 vs. control.
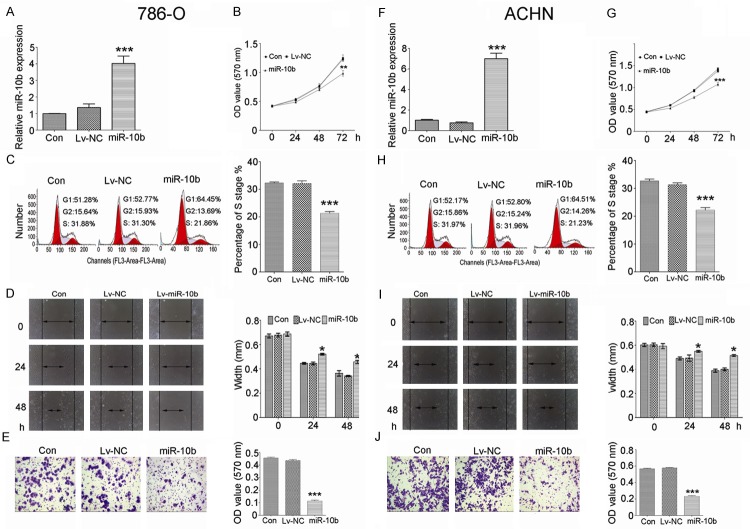
Effects of miR-10b on cell proliferation, cell cycle, migration and invasion
In order to investigate the role of miR-10b in ccRCC cells, we infected the 786-O and ACHN cells with lentivirus to overexpress miR-10b (Figure 2A and 2F). CCK-8 was used to detect the 786-O and ACHN cells proliferation. We found that inhibition of proliferation was induced by upregulation of miR-10b (Figure 2B and 2G). Flow cytometric analysis was used to analyze cell cycle alterations. The results revealed that upregulation of miR-10b induced cell cycle arrest with a significant decrease in S phase in 786-O and ACHN cells (Figure 2C and 2H). Scratch assay was used to measure the migration ability of 786-O and ACHN cells after miR-10b treatment. The results indicated that the migration ability was significantly decreased by miR-10b compared to negative control (Figure 2D and 2I). Transwell assay was used to measure the invasive ability of 786-O and ACHN cells after miR-10b treatment. The results indicated that the invasive ability was significantly decreased by miR-10b compared to negative control (Figure 2E and 2J).
Figure 2.
Overexpression of miR-10b inhibits cell proliferation, cell cycle, cell migration and invasion in 786-O and ACHN cells. The expression of miR-10b was significantly increased in 786-O (A) and ACHN (F) cells after transfecting with miR-10b lentivirus. Overexpression of miR-10b dramatically reduced cell growth in 786-O (B) and ACHN (G) cells. Overexpression of miR-10b arrested cell cycle at S phase in 786-O (C) and ACHN (H) cells, significantly inhibited cell migration in 786-O (D) and ACHN (I) cells, and repressed cell invasion in 786-O (E) and ACHN (J) cells. Data are presented by means ± s.d, *P<0.05, **P<0.01, ***P<0.001 vs. negative control (NC).
Upregulated expression of miR-10b was induced by demethylation agents and Twist1
In order to investigate the underlying mechanisms of miR-10b silence in ccRCC, ccRCC cell lines were treated with demethylation agents. The CpG island methylation of miR-10b was examined using BSP after demethylation agents treatment. The results indicated that methylation of miR-10b was decreased after 15.55 nM 5-Aza or 1.5 nM PBA treatment (Figure 3A). A significant increase expression of miR-10b was observed in 786-O and ACHN cells treated with 15.55 nM 5-Aza and 1.5 nM PBA (Figure 3B). Furthermore, in line with previous report that Twist1 was a well known regulator of miR-10b [12], we also confirmed that overexpression of Twist1 (Figure 3C and 3D) dramatically upregulated miR-10b expression in ccRCC cells (Figure 3E).
Figure 3.
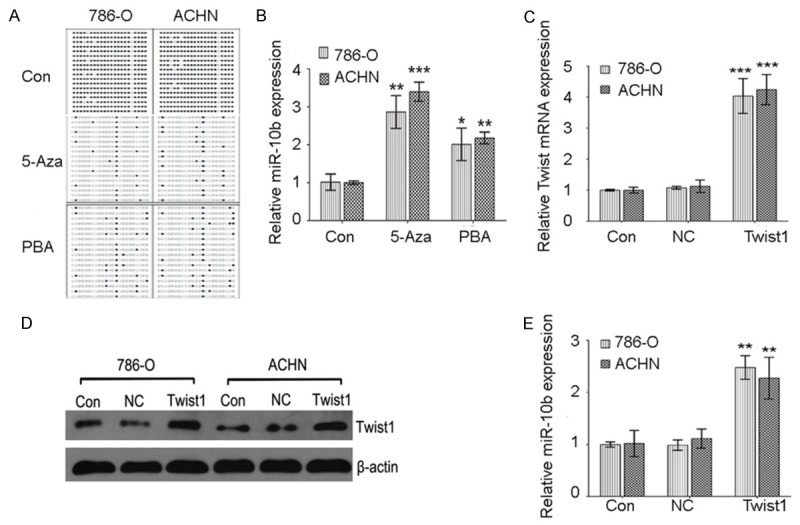
MiR-10b expression and CpG island methylation after epigenetic drugs treatment and Twist1 upregulation. A: The BSP detection results demonstrated that methylation of miR-10b was decreased after 5-Aza or PBA treatment. B: MiR-10b expression levels were significantly increased by 5-Aza or PBA treatment in 786-O and ACHN cells. C: QPCR was used to detect Twist1 mRNA expression after lentivirus transfection. D: Western blot was used to detect Twist1 protein expression after lentivirus transfection. E: MiR-10b expression levels were significantly induced by Twist1 upregulation in 786-O and ACHN cells. Data are presented by means ± s.d, *P<0.05, **P<0.01, ***P<0.001 vs. control.
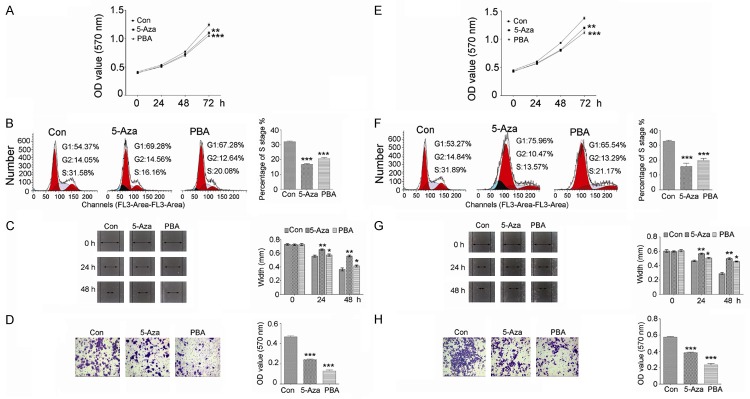
Effects of demethylation agents on cell proliferation, cell cycle, migration and invasion
CCK-8 was used to detect the 786-O and ACHN cells proliferation. We found that inhibition of proliferation was induced by 5-Aza or PBA treatment (Figure 4A and 4E). Flow cytometric analysis was used to analyze cell cycle alterations. The results revealed that 5-Aza or PBA treatment in 786-O and ACHN cells induced cell cycle arrest with a decrease in S phase (Figure 4B and 4F). Scratch assay was used to measure the migration ability of 786-O and ACHN cells after 5-Aza or PBA treatment. The results indicated that the migration ability was significantly decreased by 5-Aza or PBA compared to control group (Figure 4C and 4G). Transwell assay was used to measure the invasive ability of 786-O and ACHN cells after 5-Aza or PBA treatment. The results indicated that the invasive ability was significantly decreased by 5-Aza or PBA compared to control group (Figure 4D and 4H).
Figure 4.
The effects of epigenetic drugs on cell proliferation, cell cycle, cell migration and invasion. Cells proliferation was inhibited by 5-Aza or PBA treatment in 786-O (A) and ACHN (E) cells. Cell cycle was arrested at S phase in 786-O (B) and ACHN (F) cells after 5-Aza or PBA treatment. Cell migration was repressed by 5-Aza or PBA treatment in 786-O (C) and ACHN (G) cells. Cells invasive ability was reduced by 5-Aza or PBA treatment in 786-O (D) and ACHN (H) cells. Data are presented by means ± s.d, *P<0.05, **P<0.01, ***P<0.001 vs. control.
Discussion
Aberrant expression of miR-10b has been described in several types of cancer, including nasopharyngeal carcinoma [13], breast cancer [14], esophageal squamous cell carcinoma [15], and ccRCC [16]. MiR-10b is a bi-functional gene either as oncogene or tumor suppressor. High level miR-10b expression has been found in colorectal cancer, and associated with lymphatic invasion and poor prognosis [17]. And increased miR-10b expression in pancreatic ductal adenocarcinoma (PDAC) has been identified as a marker of disease aggressiveness [18]. Severino P et al also demonstrated that miR-10b played an oncogenic role in head and neck squamous cell carcinomas involved in cell cycle progression [19]. These results indicate that miR-10b acts as an oncogene in these cancers. However, down-regulation of miR-10b expression is also observed in ccRCC, bladder cancer, endometrioid endometrial carcinoma and breast cancer [20-23], supporting the tumor suppressor role of miR-10b in these cancers. In our recent study, we find that miR-10b is significantly downregulated in ccRCC tissues and renal cancer cell lines compared to those in the matched adjacent tissues and HEK293 cell lines, respectively. Our results are in line with the report that expression level of miR-10b was lower in the tumors of RCC patients who developed tumor relapse and metastatic tumors than those of relapse-free RCC patients [16].
By using MSP, we find that a significantly higher level of methylation of miR-10b promoter is observed in ccRCC tissues compared to the matched adjacent tissues. Previous studies have demonstrated that miR-10b is regulated by Twist1 in breast cancer [12,24], and we also confirm this case in ccRCC. DNA methylation of miR-10b has been implicated in cancer cell growth, invasion and metastasis, and chemoresistance [25]. Hypermethylated CpG sites in miR-10a are found in hepatocellular carcinoma [26]. And miR-10b promoter is heavily methylated in gastric cancers [27]. Additionally, the expression of miR-10b is down-regulated in ccRCC metastatic tissue samples compared with the normal tissue [11], and a higher ratio of miR-21/miR-10b indicates a poorer prognosis [28]. Thus, hypermethylation of miR-10b promoter may contribute to ccRCC progression.
By gain of functional experiments, we find that overexpression of miR-10b in ccRCC cell lines significantly inhibits cell growth, arrests cell cycle at S phase, represses migration and invasion. These results are confirmed by demethylation drugs (5-Aza and PBA) treatment in ccRCC cell lines. We analyze methylation and expression of miR-10b in ccRCC cells in the presence of 5-Aza and PBA, and find that methylation of miR-10b shows a dramatic reduction, whereas miR-10b expression exhibits a significant increase after 5-Aza and PBA treatment in two of the cell lines. Furthermore, 5-Aza and PBA treatment significantly induce inhibition of cell proliferation, arrest of cell cycle, and suppression of cell migration and invasion. Previous studies have shown that 5-aza-2’-deoxycytidine and trichostain A increase miR-10b expression in gastric cancer cells [10], and renal cell carcinoma cell lines [22]. Taken together, our data strongly suggest that miR-10b expression is relevant to epigenetic regulation such as promoter methylation, at least in renal cancer cells tested in this study. Although we have revealed that epigenetic modulation as an upstream regulator regulated the expression of miR-10b, a tumor suppressor, in ccRCC, more efforts are needed to explore the downstream target genes of miR-10b. And some targets of miR-10b have been identified, such as microtubule-associated protein, RP/EB family, member 1 (MAPRE1) in gastric cancer [27], homemobox D10 in ovarian cancer [29], and CCNA2 in breast cancer [23]. The target of miR-10b in ccRCC remains to be demonstrated.
In conclusion, we find that miR-10b expression is frequently silenced in ccRCC by its promoter methylation and that miR-10b may act as a tumor suppressor. Thus, we suggest that modulation of miR-10b may represent a therapeutic benefit for ccRCC management.
Disclosure of conflict of interest
None.
References
- 1.Jonasch E, Gao J, Rathmell WK. Renal cell carcinoma. BMJ. 2014;349:g4797. doi: 10.1136/bmj.g4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dabestani S, Marconi L, Hofmann F, Stewart F, Lam TB, Canfield SE, Staehler M, Powles T, Ljungberg B, Bex A. Local treatments for metastases of renal cell carcinoma: a systematic review. Lancet Oncol. 2014;15:e549–e561. doi: 10.1016/S1470-2045(14)70235-9. [DOI] [PubMed] [Google Scholar]
- 3.Al-Ali BM, Ress AL, Gerger A, Pichler M. MicroRNAs in renal cell carcinoma: implications for pathogenesis, diagnosis, prognosis and therapy. Anticancer Res. 2012;32:3727–3732. [PubMed] [Google Scholar]
- 4.Maher ER. Genomics and epigenomics of renal cell carcinoma. Semin Cancer Biol. 2013;23:10–17. doi: 10.1016/j.semcancer.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Ma X, Shen D, Li H, Zhang Y, Lv X, Huang Q, Gao Y, Li X, Gu L, Xiu S, Bao X, Duan J, Zhang X. MicroRNA-185 inhibits cell proliferation and induces cell apoptosis by targeting VEGFA directly in von Hippel-Lindau-inactivated clear cell renal cell carcinoma. Urol Oncol. 2015;33:169, e1–11. doi: 10.1016/j.urolonc.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Gao C, Peng FH, Peng LK. MiR-200c sensitizes clear-cell renal cell carcinoma cells to sorafenib and imatinib by targeting heme oxygenase-1. Neoplasma. 2014;61:680–689. doi: 10.4149/neo_2014_083. [DOI] [PubMed] [Google Scholar]
- 7.Wu X, Weng L, Li X, Guo C, Pal SK, Jin JM, Li Y, Nelson RA, Mu B, Onami SH, Wu JJ, Ruel NH, Wilczynski SP, Gao H, Covarrubias M, Figlin RA, Weiss LM, Wu H. Identification of a 4-microRNA signature for clear cell renal cell carcinoma metastasis and prognosis. PLoS One. 2012;7:e35661. doi: 10.1371/journal.pone.0035661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Xu L, Yang Z, Lu H, Hu D, Li W, Zhang Z, Liu B, Ma S. MicroRNA-10b indicates a poor prognosis of non-small cell lung cancer and targets E-cadherin. Clin Transl Oncol. 2015;17:209–214. doi: 10.1007/s12094-014-1213-7. [DOI] [PubMed] [Google Scholar]
- 9.Xiao H, Li H, Yu G, Xiao W, Hu J, Tang K, Zeng J, He W, Zeng G, Ye Z, Xu H. MicroRNA-10b promotes migration and invasion through KLF4 and HOXD10 in human bladder cancer. Oncol Rep. 2014;31:1832–1838. doi: 10.3892/or.2014.3048. [DOI] [PubMed] [Google Scholar]
- 10.Li Z, Lei H, Luo M, Wang Y, Dong L, Ma Y, Liu C, Song W, Wang F, Zhang J, Shen J, Yu J. DNA methylation downregulated mir-10b acts as a tumor suppressor in gastric cancer. Gastric Cancer. 2015;18:43–54. doi: 10.1007/s10120-014-0340-8. [DOI] [PubMed] [Google Scholar]
- 11.Fritz HK, Lindgren D, Ljungberg B, Axelson H, Dahlback B. The miR (21/10b) ratio as a prognostic marker in clear cell renal cell carcinoma. Eur J Cancer. 2014;50:1758–1765. doi: 10.1016/j.ejca.2014.03.281. [DOI] [PubMed] [Google Scholar]
- 12.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 13.Allaya N, Khabir A, Sallemi-Boudawara T, Sellami N, Daoud J, Ghorbel A, Frikha M, Gargouri A, Mokdad-Gargouri R, Ayadi W. Over-expression of miR-10b in NPC patients: correlation with LMP1 and Twist1. Tumour Biol. 2015;36:3807–14. doi: 10.1007/s13277-014-3022-6. [DOI] [PubMed] [Google Scholar]
- 14.Eissa S, Matboli M, Shehata HH, Essawy NO. MicroRNA-10b and minichromosome maintenance complex component 5 gene as prognostic biomarkers in breast cancer. Tumour Biol. 2015;36:4487–94. doi: 10.1007/s13277-015-3090-2. [DOI] [PubMed] [Google Scholar]
- 15.Dong W, Shen R, Cheng S. Reduction of TIP30 in esophageal squamous cell carcinoma cells involves promoter methylation and microRNA-10b. Biochem Biophys Res Commun. 2014;453:772–777. doi: 10.1016/j.bbrc.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 16.Slaby O, Redova M, Poprach A, Nekvindova J, Iliev R, Radova L, Lakomy R, Svoboda M, Vyzula R. Identification of MicroRNAs associated with early relapse after nephrectomy in renal cell carcinoma patients. Genes Chromosomes Cancer. 2012;51:707–716. doi: 10.1002/gcc.21957. [DOI] [PubMed] [Google Scholar]
- 17.Nishida N, Yamashita S, Mimori K, Sudo T, Tanaka F, Shibata K, Yamamoto H, Ishii H, Doki Y, Mori M. MicroRNA-10b is a prognostic indicator in colorectal cancer and confers resistance to the chemotherapeutic agent 5-fluorouracil in colorectal cancer cells. Ann Surg Oncol. 2012;19:3065–3071. doi: 10.1245/s10434-012-2246-1. [DOI] [PubMed] [Google Scholar]
- 18.Ouyang H, Gore J, Deitz S, Korc M. microRNA-10b enhances pancreatic cancer cell invasion by suppressing TIP30 expression and promoting EGF and TGF-beta actions. Oncogene. 2014;33:4664–4674. doi: 10.1038/onc.2013.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Severino P, Bruggemann H, Andreghetto FM, Camps C, Klingbeil MF, de Pereira WO, Soares RM, Moyses R, Wunsch-Filho V, Mathor MB, Nunes FD, Ragoussis J, Tajara EH. MicroRNA expression profile in head and neck cancer: HOX-cluster embedded microRNA-196a and microRNA-10b dysregulation implicated in cell proliferation. BMC Cancer. 2013;13:533. doi: 10.1186/1471-2407-13-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaravinos A, Radojicic J, Lambrou GI, Volanis D, Delakas D, Stathopoulos EN, Spandidos DA. Expression of miRNAs involved in angiogenesis, tumor cell proliferation, tumor suppressor inhibition, epithelial-mesenchymal transition and activation of metastasis in bladder cancer. J Urol. 2012;188:615–623. doi: 10.1016/j.juro.2012.03.122. [DOI] [PubMed] [Google Scholar]
- 21.Tsukamoto O, Miura K, Mishima H, Abe S, Kaneuchi M, Higashijima A, Miura S, Kinoshita A, Yoshiura K, Masuzaki H. Identification of endometrioid endometrial carcinoma-associated microRNAs in tissue and plasma. Gynecol Oncol. 2014;132:715–721. doi: 10.1016/j.ygyno.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 22.Wotschofsky Z, Liep J, Meyer HA, Jung M, Wagner I, Disch AC, Schaser KD, Melcher I, Kilic E, Busch J, Weikert S, Miller K, Erbersdobler A, Mollenkopf HJ, Jung K. Identification of metastamirs as metastasis-associated microRNAs in clear cell renal cell carcinomas. Int J Biol Sci. 2012;8:1363–1374. doi: 10.7150/ijbs.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biagioni F, Bossel BN, Fontemaggi G, Canu V, Mori F, Antoniani B, Di Benedetto A, Santoro R, Germoni S, De Angelis F, Cambria A, Avraham R, Grasso G, Strano S, Muti P, Mottolese M, Yarden Y, Domany E, Blandino G. miR-10b*, a master inhibitor of the cell cycle, is down-regulated in human breast tumours. EMBO Mol Med. 2012;4:1214–1229. doi: 10.1002/emmm.201201483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsukerman P, Yamin R, Seidel E, Khawaled S, Schmiedel D, Bar-Mag T, Mandelboim O. MiR-520d-5p directly targets TWIST1 and downregulates the metastamiR miR-10b. Oncotarget. 2014;5:12141–12150. doi: 10.18632/oncotarget.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Y, Yao J, Yu J, Wei Q, Cao X. The association between abnormal microRNA-10b expression and cancer risk: a meta-analysis. Sci Rep. 2014;4:7498. doi: 10.1038/srep07498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen J, Wang S, Zhang YJ, Kappil MA, Chen WH, Kibriya MG, Wang Q, Jasmine F, Ahsan H, Lee PH, Yu MW, Chen CJ, Santella RM. Genome-wide aberrant DNA methylation of microRNA host genes in hepatocellular carcinoma. Epigenetics. 2012;7:1230–1237. doi: 10.4161/epi.22140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim K, Lee HC, Park JL, Kim M, Kim SY, Noh SM, Song KS, Kim JC, Kim YS. Epigenetic regulation of microRNA-10b and targeting of oncogenic MAPRE1 in gastric cancer. Epigenetics. 2011;6:740–751. doi: 10.4161/epi.6.6.15874. [DOI] [PubMed] [Google Scholar]
- 28.Fritz HK, Lindgren D, Ljungberg B, Axelson H, Dahlback B. The miR (21/10b) ratio as a prognostic marker in clear cell renal cell carcinoma. Eur J Cancer. 2014;50:1758–1765. doi: 10.1016/j.ejca.2014.03.281. [DOI] [PubMed] [Google Scholar]
- 29.Nakayama I, Shibazaki M, Yashima-Abo A, Miura F, Sugiyama T, Masuda T, Maesawa C. Loss of HOXD10 expression induced by upregulation of miR-10b accelerates the migration and invasion activities of ovarian cancer cells. Int J Oncol. 2013;43:63–71. doi: 10.3892/ijo.2013.1935. [DOI] [PubMed] [Google Scholar]