Abstract
Aberrant expression of microRNA-133b (miR-133b) has been frequently reported in some cancers excluding ovarian cancer (OC). The role and its molecular mechanism of miR-133b in OC have not been reported. In this study, we explored the effects of miR-133b overexpression on proliferation and invasion in OC cells. The mRNA level of miR-133b in OC cell lines was determined by real-time PCR. The miR-133b mimic was transiently transfected into OC cells using Lipofectamine™ 2000 reagent. Subsequently, cell proliferation and invasion were assessed by MTT, Brdu-ELISA and Transwell assays. Moreover, the effects of miR-133b overexpression on the MAPK and PI3K/Akt signaling pathways were determined by Western blot. Protein level of EGFR was also measured by Western blotting. Meanwhile, luciferase assays were performed to validate EGFR as miR-133b target in OC cells. Our results showed that the mRNA level of miR-133b was remarkably decreased in OC cell lines compared with normal colon epithelium cells, whereas the protein expression of EGFR was significantly increased. Up-regulation of miR-133b inhibited the proliferation and invasion of OC cells. We also found that miR-133b overexpression evidently decreased the phosphorylation of Erk1/2 and Akt. Bioinformatics analysis predicted that the EGFR was a potential target gene of miR-133b. Luciferase reporter assay demonstrated that miR-133b could directly target EGFR. Altogether, our results indicated that miR-133b overexpression was shown to inhibit proliferation and invasion of OC cells through suppression of the MAPK and PI3K/Akt signaling pathways by targeting EGFR.
Keywords: MicroRNA-133b, ovarian cancer, proliferation, invasion, MAPK and PI3K/Akt signaling pathway, epidermal growth factor receptor
Introduction
Ovarian cancer (OC) is the seventh most common cancer and the eighth leading cause of cancer-related deaths in women [1,2]. Although diagnostic method, surgical technique and new chemotherapy regimens have been improved, therapy of OC is still a challenge [3]. Moreover, 5-year survival rate is still low because of the development of drug resistance, and only 45% patients with OC are diagnosed at advanced stage and can survive more than 5 years [4]. Due to the poor survival of patients with OC, it is essential and of great interest to identify the molecular mechanism underling the initiation and progression of OC. In recent years, miRNAs offer a novel molecular approach, and it has been confirmed to be associated with pathogenesis of OC.
Many studies have reported that microRNAs (miRNAs) are small (about in length), non-coding RNAs of 22 nucleotides [5], and play important roles in regulation of the biological and metabolic processes of cancers [6]. They generally function as oncogenes or tumor suppressors during the progression of cancers. Moreover, a large body of research has demonstrated that miRNAs are involved in tumorigenesis and metastasis by targeting many types of molecules [7]. In recent years, it is reported that numerous miRNAs are aberrantly expressed in plenty of cancers such as ovarian cancer. The expression of miR-25 was up-regulated in OC tissues, and down-regulation of miR-25 significantly suppressed proliferation, migration and invasion of OC cells by targeting large tumor suppressor 2 (LATS2) [8]. Lu et al. demonstrated miR-200c expression was inversely associated with lymph node metastasis of OC, and down-regulation of miR-200c inhibited metastasis in OC cells by targeting zinc finger E-box-binding homeobox 2 (ZEB2) expression [9]. These two miRNAs act as oncogene, whereas some tumor suppressor miRNAs were also studied in OC. For example, miR-212 acted as a tumor suppressor and inhibited proliferation, migration and invasion of ovarian cancer cells through directly targeting heparin-binding epidermal growth factor (HBEGF) [10]. Overexpression of miR-34a suppressed ovarian cancer cell proliferation and motility via inhibition of its target gene AXL [11]. miR-148a was markedly decreased in OC tissues, and inhibited SKOV3 cell migration and invasion by targeting sphingosine-1-phosphate receptor 1 (S1PR1) [12]. miR-498 is involved in the regulation of forkhead box O3 (FOXO3) to inhibit proliferation of OC cells [13].
MiR-133b has been involved in regulators of many cellular processes such as cell proliferation, apoptosis, migration and invasion [14]. In recent decades, miR-133b was considered as a tumor suppressor and down-regulated in multiple human cancers, including bladder, gastric and colon cancers [15-17]. Chen et al. reported that miR-133b inhibited proliferation and induced apoptosis of bladder cancer cells by targeting Bcl-w and Akt1 [15]. Furthermore, miR-133b was down-regulated in gastric cancer tissues, and acted as a tumor suppressor to inhibit gastric cancer cell growth and invasion by down-regulation of fascin actin-bundling protein 1 (FSCN1) [16]. Xiang and his colleague demonstrated that miR-133b also acted as a tumor suppressor through inhibiting proliferation in colon cancer cells by regulation of TATA box-binding protein-like protein 1 (TBPL1) [17].
In this paper, we determined frequent down-regulation of miR-133b in ovarian cancer cell lines. Up-regulation of miR-133b inhibited cell proliferation and invasion of OC cells, which was closely related to inhibition of the MAPK and PI3K/Akt signaling pathways. Moreover, we found that EGFR was the direct target of miR-133b in OC and confirmed that miR-133b functioned as a tumor suppressor by down-regulation of EGFR. Therefore, our results showed important roles for miR-133b in the pathogenesis of OC and suggested its possible application in tumor treatment.
Materials and methods
Cell culture and miRNA transfection
Human OC cell lines SKOV-3, A2780, HO-8910, CAOV3, ES-2, OVCAR3 and an immortalized normal human fallopian tube epithelial cell line FTE187 cells were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). SKOV-3, A2780, HO-8910, CAOV3, ES-2 and OVCAR3 were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco Co., New York, USA) containing 10% fetal bovine serum (FBS) (Gibco Co., New York, USA), 100 mg/ml penicillin and 100 mg/ml streptomycin. FTE187 cell line was maintained in cell culture medium consisting of 1:1 Medium 199 and MCDB105 medium (Sigma-Aldrich) with 10% FBS and 10 ng/ml EGF (Sigma-Aldrich). All cells were cultured at 37°C in a humidified atmosphere of 5% CO2 on 0.1% gelatin-coated culture flasks. To enhance the expression of miR-133b in SKOV-3 and OVCAR3 cells, both cells were transfected with miR-133b mimic, which served as the miR-133b group. SKOV-3 and OVCAR3 cells transfected with miR-negative control (miR-NC) were used as miR-NC group. One day before transfection, cells at about 40 to 60% confluency were changed to the antibiotic-free media. After 24 h, cells were transfected with 50 nM miR-133b mimic using LipofectaminTM 2000 reagent (Invitrogen, USA) following manufacturer’s protocol.
Reverse transcription polymerase chain reaction
Total RNA of SKOV-3 and OVCAR3 cells was extracted by using Trizol reagent (Life Technologies, Carlsbad, CA). Two microgram RNA was used for gene-specific reverse transcription polymerase chain reaction (RT-PCR) using one-step RT-PCR kit (Qiagen, Venlo, the Netherlands) following the manufacturer’s protocols. The following primers were used: miR-133b (Accession number MI0000822), forward: 5’-GAACCAAGCCGCCCGAGA-3’ and reverse: 5’-CCGCCCTGCTGTGCTGGT-3’; U6, forward: 5’-CTCGCTTCGGCAGCACA-3’ and reverse: 5’-AACGCTTCACGAATTTGCGT-3’. U6 snRNA were used to normalize. Each sample was assessed in triplicate.
Cell viability assay
SKOV-3 and OVCAR3 cells (1×104 cells/100 μl) were seeded in 96-well plates for 24 h. After that, cells were transfected with miR-133B and miR-NC for 24 h. Then, the number of viable cells was determined using 3-(4, 5-dimethylthiazol-2-yl) 2, 5-diphenyltetrazolium bromide (MTT) reagent (Sigma Chemical Co., USA) according to the manufacturer’s instructions. Briefly, 10 μl MTT reagent (5 mg/ml) was added to the 100 μL medium, and incubated at 37°C for 4 h. The medium was removed and dimethyl sulfoxide (DMSO, 150 μl/well) was added to solubilize the formazan crystals. OD values of the medium at 490 nm were measured with Biotek Elx-800 plate reader.
Cell proliferation assay
To explore the effect of miR-133b mimic on proliferation of SKOV-3 and OVCAR3 cells, 1×104 cells were seeded in 96-well plate and allowed to grow for 24 h in complete medium. The medium was then removed and the cells were transfected with miR-133b mimic or miR-NC for 24 h at 37°C. Cell Proliferation ELISA-BrdU (colorimetric) Kit (Roche Diagnostics, USA) was used to detect the cells proliferation according to the manufacturer’s protocols.
Transwell invasion assay
Transwell matrigel invasion assay using Transwell chambers (8-mm pore size; Corning) precoated with Matrigel (BD Biosciences) that contained extracellular matrix proteins was used to evaluate cell invasion. In brief, after serum-starved for 24 h, 2×105 cells were suspended in 100 μl serum-free DMEM, and then were seeded in the top chamber, and 600 ml DMEM containing 10% FBS was added to the lower chamber. After 24 h incubation at 37°C in a 5% CO2 atmosphere, cells that remained in the upper surface of the membrane were removed by cotton swabs and penetrating cells were fixed in methanol, and then stained with 0.1% crystal violet. Cell invasion was quantified by counting cells on the lower surface using phase contrast microscopy.
Western blot analysis
To extract the proteins, SKOV-3 and OVCAR3 cells were washed twice in cold PBS, and then lysed in RIPA lysis buffer (Beyotime Institute of Biotechnology Jiangsu, China) with protease inhibitor cocktail (Merk, Germany). The protein concentration of cell lysates was quantified by BCA Kit (Beyotime Institute of Biotechnology Jiangsu, China), and equal quantities (50 μg) of proteins were separated by SDS-PAGE on 10 % gels, and then transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, USA). The membranes were blocked in 5% shimmed milk diluted with Tri Buffered Saline Tween-20 (TBST) (in mmol/L: Tris-HCl 20, NaCl 150, PH 7.5, 0.1% Tween 20) at room temperature for 1 h and incubated overnight at 4°C with primary antibody respectively: anti-EGFR, anti-phospho-Erk1/2, anti-total-Erk, phospho-Akt and total-Akt (1:1000; Cell Signaling Technology Inc, MA, USA). The membranes were then incubated with a goat anti-rabbit IgG conjugated to horseradish peroxidase secondary antibody (1:1000; Santa Cruz Biotechnology, CA, USA) for 2 h. The proteins were visualized using ECL-plus reagents (Amersham Biosciences Corp., USA). The density of the bands was measured using the Image J software (USA), and values were normalized to the densitometric values of α-tubulin (1:1000; Sigma, USA) in each sample.
Luciferase reporter assay
SKOV-3 and OVCAR3 cells (2×105/well) were seeded in 24-well plates and incubated overnight before transfection. Cells were cotransfected with pMIR-EGFR-3’UTR wild-type or mutant reporter plasmid, miR-133b mimic or miR-NC, and pRL-SV40 renilla plasmid (Promega, USA) using Lipofectamine 2000. At 48 h after co-transfection, both firefly and renilla luciferase activities were quantified using a dual luciferase reporter system (Promega, USA) following the manufacturer’s protocols. Each treatment was performed in triplicate in three independent experiments.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 5.0 (GraphPad Software, Inc., USA). Data from each group were expressed as mean ± standard error of the mean (S.E.M.) and statistically analyzed by Student’s t test. Differences were considered statistically significant at a p value of <0.05.
Results
MiR-133b was down-regulated in ovarian cancer (OC) cell lines
It has been reported that miR-133b was down-regulated in bladder, gastric and colon cancers [15-17]. However, the expression of miR-133b in OC remains unknown. Therefore, to detect the level of miR-133b in OC cells, expression of miR-133b was detected by RT-PCR in six OC cell lines (SKOV-3, A2780, HO-8910, CAOV3, ES-2 and OVCAR3) and an immortalized normal human fallopian tube epithelial cell line FTE187. It showed that miR-133b expression was significantly down-regulated in all OC cell lines compared to that in normal human fallopian tube epithelial cell line FTE187, as shown in Figure 1A. Moreover, we found that EGFR, which contributed to the malignant cell growth, was predicted by using the online database (TargetScan 6.2) to be a direct target of miR-133b. Then, among these OC cell lines, SKOV-3 and OVCAR3 cells were used to explore further. We found that the protein level of EGFR in SKOV-3 and OVCAR3 cells was evidently up-regulated in contrast with FTE187 cell (Figure 1B).
Figure 1.
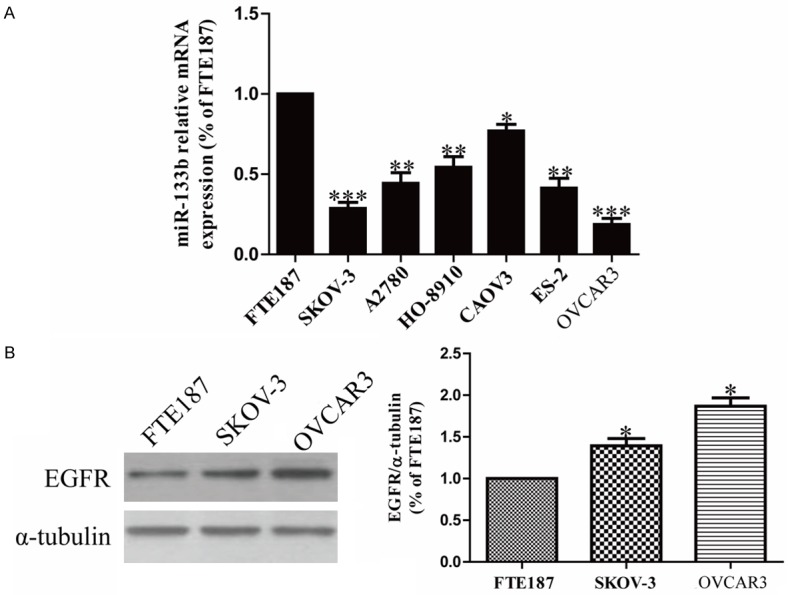
The altered expression of miR-133b and EGFR in OC cell lines. A. The relative expression of miR-133b in OC cell lines and FTE187 cell line by real-time PCR. B. EGFR protein level in SKOV-3 and OVCAR3 cells compared with FTE187 cell were determined by Western blotting. α-tubulin was detected as a loading control. All data are presented as mean ± SEM, n=6. *P<0.05, **P<0.01, ***P<0.1 vs. FTE187.
Overexpression of miR-133b suppressed proliferation and invasion of SKOV-3 and OVCAR3 cells
However, the reduced expression of miR-133b in OC cell lines implied that miR-133b might act as a tumor suppressor in OC. Our results showed that miR-133b displayed evident up-regulation of mRNA levels in miR-133b mimic group compared to miR-NC group (Figure 2A). These outcomes indicated that we availably increased miR-133b expression in SKOV-3 and OVCAR3 cells. To test the function of miR-133b in proliferation of OC cells, SKOV-3 and OVCAR3 cells were transfected with miR-133b mimic or miR-NC. Results from MTT assay showed that up-regulation of miR-133b significantly inhibited the viabilities of SKOV-3 and OVCAR3 cells (Figure 2B). Besides, we also observed anti-proliferative effect in cells transfected with miR-133b mimic, as assessed by the Brdu-ELISA assay (Figure 2C). These findings suggested that overexpression of miR-133b had available anti-proliferative effect in both SKOV-3 and OVCAR3 cells.
Figure 2.
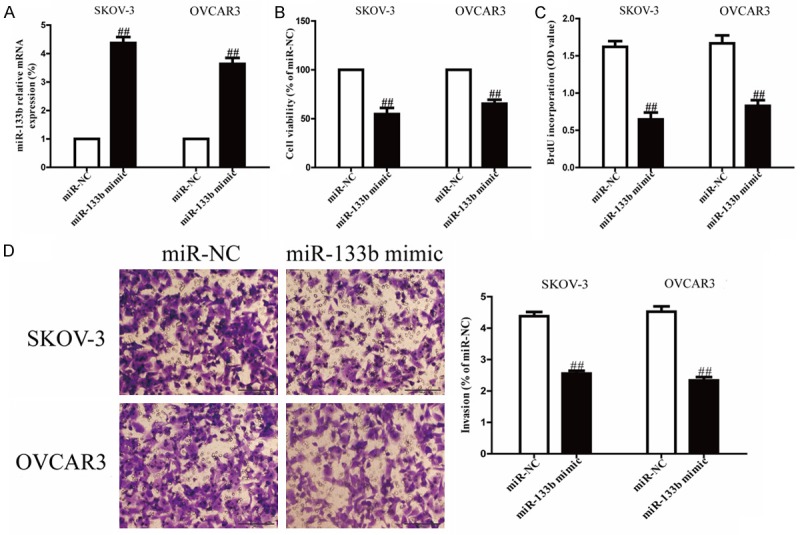
Effects of miR-133b overexpression on proliferation and invasion of SKOV-3 and OVCAR3 cells. SKOV-3 and OVCAR3 cells were transfected with miR-133b mimic or miR-NC. A. The mRNA levels of miR-133b in SKOV-3 and OVCAR3 cells were detected by real-time PCR. B. Cell viability was estimated by MTT assay. C. Cell proliferation was assessed by BrdU-ELISA assay. D. Cell invasion was assessed by Transwell assay. All data are presented as mean ± SEM, n=6. ##P<0.01 vs. miR-NC.
Next, to know whether overexpression of miR-133b possesses a negative effect on invasion of OC cells, we further transfected a miR-133b mimic into SKOV-3 and OVCAR3 cells, and the invasion of SKOV-3 and OVCAR3 cells were evaluated by Transwell invasion assay. The results from Transwell assay showed that the number of SKOV-3 and OVCAR3 cells invading through the Transwell membrane was significantly lower in miR-133b group compared to miR-NC group (Figure 2D). Our results suggested that overexpression of miR-133b inhibited the invasion of OC cells.
Up-regulation of miR-133b inhibited MAPK and PI3K/Akt signaling pathways in OC cells
To explore the molecular mechanisms by which miR-133b regulated the proliferation and invasion of OC cells, we studied the effects of miR-133b overepxression on the MAPK and PI3K/Akt signaling pathways. Hence, in order to investigate the effects of miR-133b mimic on both signaling pathways, Western blot analysis was used to detect the expressions of phospho-Erk1/2, total-Erk1/2, phospho-Akt and total-Akt. After transfection with miR-133b mimic for 24 h, the phosphorylation of Erk1/2 and Akt in both SKOV-3 and OVCAR3 cells were detected by Western blotting. Our results showed that up-regulation of miR-133b significantly reduced phosphorylation of Erk1/2 and Akt to inhibit the both MAPK and PI3K/Akt signaling pathways in SKOV-3 and OVCAR3 cells (Figure 3). But there were no effects for miR-133b mimic on total expressions of Erk1/2 and Akt. Altogether, our data suggested that overexpression of miR-133b inhibited the proliferation and invasion of OC cells by inactivation of both MAPK and PI3K/Akt signaling pathways.
Figure 3.
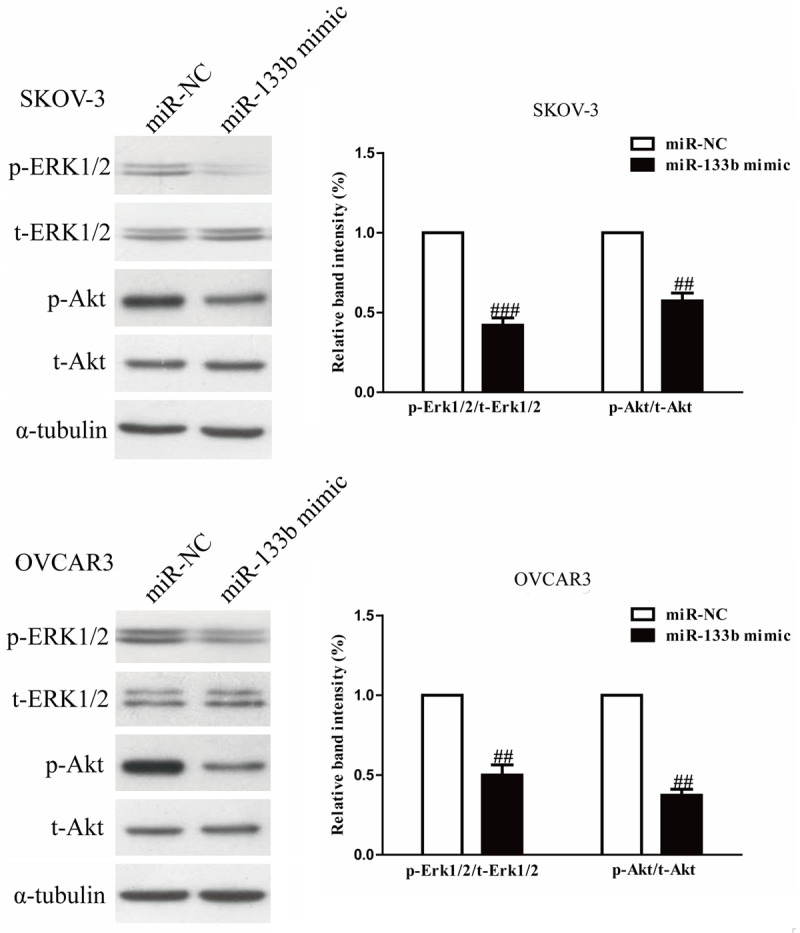
Overexpression of miR-133b inhibited the MAPK and PI3K/Akt signaling pathways. SKOV-3 and OVCAR3 cells were transfected with miR-133b mimic or miR-NC, the p-Erk1/2, t-Erk1/2, p-Akt and t-Akt protein expression levels were analyzed by Western blotting, respectively. α-tubulin was detected as a loading control. All data are presented as mean ± SEM, n=6. ##P<0.01, ###P<0.001 vs. miR-NC.
EGFR is a direct target of miR-133b in OC cells
Because EGFR was a binding target of miR-133b predicted by the online database, TargetScan 6.2, Western blotting were used to determine the expression of EGFR on protein level in SKOV-3 and OVCAR3 cells transfected with miR-133b mimic. We found that expression of EGFR wa evidently decreased after up-regulation of miR-133b (Figure 4A). To further confirm whether EGFR was a direct target of miR-133b, EGFR 3’-UTR was cloned into a luciferase reporter vector and the putative miR-133b binding site in the EGFR 3’-UTR was mutated (Figure 4B). The effect of miR-133b was determined using luciferase reporter assay. The results showed that overexpression of miR-24 significantly inhibited the luciferase activity of pMir-EGFR 3’-UTR WT (Figure 4C). Mutation of the miR-133b-binding site in the EGFR 3’-UTR abolished the effect of miR-133b, which indicated that EGFR was directly and negatively regulated by miR-133b.
Figure 4.
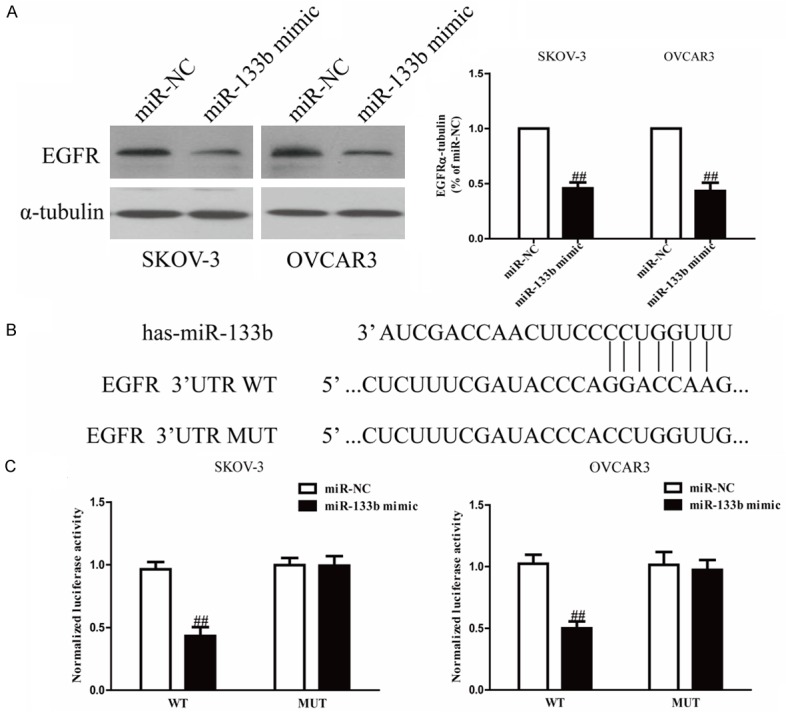
EGFR was a direct target of miR-133b. SKOV-3 and OVCAR3 cells were transfected with miR-133b mimic or miR-NC. A. The protein expression of EGFR was determined by Western blot. α-tubulin was detected as a loading control. B. Schematic representation of EGFR 3’UTRs showing putative miRNA target site. C. The analysis of the relative luciferase activities of EGFR-WT, EGFR-MUT in OC cells. All data are presented as mean ± SEM, n=6. ##P<0.01 vs. miR-NC.
Discussion
miRNAs have been recognized as crucial regulators of carcinogenesis and progression of malignant cancers through regulation of tumor suppressor genes and oncogenes [18,19]. It has been reported that miR-133b inhibits tumor progression and metastasis in bladder, gastric, colon and other cancers [14-17]. In the present study, we explored the expression and biological function of miR-133b in OC. Our results confirmed that miR-133b was frequently down-regulated in six OC cell lines including SKOV-3, A2780, HO-8910, CAOV3, ES-2 and OVCAR3 compared to normal human fallopian tube epithelial cell line. From these findings, we speculated that miR-133b might be a potential tumor suppressor in OC. As expected, up-regulation of miR-133b significantly inhibited proliferation and invasion of SKOV-3 and OVCAR3 cells. Our results suggested that miR-133b played critical roles in regulation of proliferation and invasion in OC and may be potential diagnostic and predictive biomarkers.
It has been reported that EGFR is involved in many types of cancers, including pancreas, breast, lung, colon, ovarian and skin cancer [20-25], which is comprehensively overexpressed in these cancer cells and regulates cell proliferation, invasion and apoptosis through downstream signal transduction pathways [26]. Recently, targeting the EGFR and its downstream pathways, such as JAK/STAT, PI3K/AKT and ERK1/2, is considered as the novel treatment method. Molecularly, although miR-133b has been shown to decrease EGFR expression in cancers of the prostate and non-small-cell lung [27,28], there has been no previous report that demonstrates the function of miR-133b in the regulation of EGFR expression in OC. In this study, we found that the level of EGFR was inversely correlated with the expression of miR-133b. High expression of EGFR in OC cells was accompanied by a low level of expression of miR-133b. Furthermore, Western blot analysis showed that up-regulation of miR-133b decreased the protein expression of EGFR in human OC cell lines. Then, we performed a bioinformatic analysis and predicted that EGFR cDNA contained hsa-miR-133b targeting sequences. Next, the luciferase report assay was used to confirm the interaction between miR-133b and the EGFR 3’UTR in human OC cell lines.
Besides, the possible molecular mechanisms of OC cell proliferation and invasion inhibited by miR-133b were investigated. Our findings revealed that miR-133b might be associated with EGFR downstream signaling pathways. It has been reported that abnormal activation of ERK1/2 and Akt signaling is closely related to progression and metastasis of OC [29-31]. Our results indicated that miR-133b inactivated Erk1/2 and Akt signaling pathways by reducing the phosphorylation of Erk1/2 and Akt in both SKOV-3 and OVCAR3 cells, which are classical signal transduction pathways and play important roles in tumor progression and metastasis. Altogether, we confirmed that miR-133b exerted its biological functions through suppression of both Erk1/2 and Akt signaling pathways, which might also clarify the molecular mechanisms of miR-133b on suppression of OC cell proliferation and invasion.
In conclusion, we demonstrated that miR-133b was significantly decreased in OC cell lines. Up-regulation of miR-133b evidently suppressed proliferation and invasion of SKOV-3 and OVCAR3 cells through inhibition of both MAPK and PI3K/Akt signaling pathways by targeting EGFR. Consequently, miR-133b may act as a tumor suppressor and be the potential molecular target for OC treatment.
Disclosure of conflict of interest
None.
References
- 1.Vargas-Hernández VM, Moreno-Eutimio MA, Acosta-Altamirano G, Vargas-Aguilar VM. Management of recurrent epithelial ovarian cancer. Gland Surg. 2014;3:198–202. doi: 10.3978/j.issn.2227-684X.2013.10.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clarke T, Galaal K, Bryant A, Naik R. Evaluation of follow-up strategies for patients with epithelial ovarian cancer following completion of primary treatment. Cochrane Database Syst Rev. 2014;9:CD006119. doi: 10.1002/14651858.CD006119.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marcus CS, Maxwell GL, Darcy KM, Hamilton CA, McGuire WP. Current approaches and challenges inmanaging andmonitoring treatment response in ovarian cancer. J Cancer. 2014;5:25–30. doi: 10.7150/jca.7810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li L, He L, Zhao JL, Xiao J, Liu M, Li X, Tang H. MiR-17-5p up-regulates YES1 to modulate the cell cycle progression and apoptosis in ovarian cancer cell lines. J Cell Biochem. 2015;116:1050–1059. doi: 10.1002/jcb.25060. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. Micrornas: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina M, Spruce T, Parenti AR, Daidone MG, Bicciato S, Piccolo S. A microRNA targeting diacer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 7.Uematsu K, He B, You L, Xu Z, McCormick F, Jablons DM. Activation of the Wnt pathway in non small cell lung cancer: evidence of disheveled overexpression. Oncogene. 2003;22:7218–7221. doi: 10.1038/sj.onc.1206817. [DOI] [PubMed] [Google Scholar]
- 8.Feng S, Pan W, Jin Y, Zheng J. MiR-25 promotes ovarian cancer proliferation and motility by targeting LATS2. Tumour Biol. 2014;35:12339–12344. doi: 10.1007/s13277-014-2546-0. [DOI] [PubMed] [Google Scholar]
- 9.Lu YM, Shang C, Ou YL, Yin D, Li YN, Li X, Wang N, Zhang SL. miR-200c modulates ovarian cancer cell metastasis potential by targeting zinc finger E-box-binding homeobox 2 (ZEB2) expression. Med Oncol. 2014;38:134. doi: 10.1007/s12032-014-0134-1. [DOI] [PubMed] [Google Scholar]
- 10.Wei LQ, Liang HT, Qin DC, Jin HF, Zhao Y, She MC. MiR-212 exerts suppressive effect on SKOV3 ovarian cancer cells through targeting HBEGF. Tumour Biol. 2014;35:12427–12434. doi: 10.1007/s13277-014-2560-2. [DOI] [PubMed] [Google Scholar]
- 11.Li R, Shi X, Ling F, Wang C, Liu J, Wang W, Li M. MiR-34a suppresses ovarian cancer proliferation and motility by targeting AXL. Tumour Biol. 2015 doi: 10.1007/s13277-015-3445-8. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 12.Wen Z, Zhao S, Liu S, Liu Y, Li X, Li S. MicroRNA-148a inhibits migration and invasion of ovarian cancer cells via targeting sphingosine-1-phosphate receptor 1. Mol Med Rep. 2015;12:3775–80. doi: 10.3892/mmr.2015.3827. [DOI] [PubMed] [Google Scholar]
- 13.Liu R, Liu F, Li L, Sun M, Chen K. MiR-498 regulated FOXO3 expression and inhibited the proliferation of human ovarian cancer cells. Biomed Pharmacother. 2015;72:52–57. doi: 10.1016/j.biopha.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Zhao H, Li M, Li L, Yang X, Lan G, Zhang Y. MiR-133b is down-regulated in human osteosarcoma and inhibits osteosarcoma cells proliferation, migration and invasion, and promotes apoptosis. PLoS One. 2013;8:e83571. doi: 10.1371/journal.pone.0083571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen XN, Wang KF, Xu ZQ, Li SJ, Liu Q, Fu DH, Wang X, Wu B. MiR-133b regulates bladder cancer cell proliferation and apoptosis by targeting Bcl-w and Akt1. Cancer Cell Int. 2014;14:70. doi: 10.1186/s12935-014-0070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo L, Bai H, Zou D, Hong T, Liu J, Huang J, He P, Zhou Q, He J. The role of microRNA-133b and its target gene FSCN1 in gastric cancer. J Exp Clin Cancer Res. 2014;33:99. doi: 10.1186/s13046-014-0099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiang KM, Li XR. MiR-133b acts as a tumor suppressor and negatively regulates TBPL1 in colorectal cancer cells. Asian Pac J Cancer Prev. 2014;15:3767–3772. doi: 10.7314/apjcp.2014.15.8.3767. [DOI] [PubMed] [Google Scholar]
- 18.Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibé B, Bouix J, Caiment F, Elsen JM, Eychenne F, Larzul C, Laville E, Meish F, Milenkovic D, Tobin J, Charlier C, Georges M. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 19.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, Qin LX, Man K, Lo CM, Lee J, Ng IO, Fan J, Tang ZY, Sun HC, Wang XW. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baldwin AS. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-kappaB. J Clin Invest. 2001;107:241–246. doi: 10.1172/JCI11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bera A, Zhao S, Cao L, Chiao PJ, Freeman JW. Oncogenic K-Ras and loss of Smad4 mediate invasion by activating an EGFR/NF-κB Axis that induces expression of MMP9 and uPA in human pancreas progenitor cells. PLoS One. 2013;8:e82282. doi: 10.1371/journal.pone.0082282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Normanno N, Campiglio M, Maiello MR, De Luca A, Mancino M, Gallo M, D’Alessio A, Menard S. Breast cancer cells with acquired resistance to the EGFR tyrosine kinase inhibitor gefitinib show persistent activation of MAPK signaling. Breast Cancer Res Treat. 2008;112:25–33. doi: 10.1007/s10549-007-9830-2. [DOI] [PubMed] [Google Scholar]
- 23.Gadgeel SM, Ali S, Philip PA, Ahmed F, Wozniak A, Sarkar FH. Response to dual blockade of epidermal growth factor receptor (EGFR) and cycloxygenase-2 in nonsmall cell lung cancer may be dependent on the EGFR mutational status of the tumor. Cancer. 2007;110:2775–2784. doi: 10.1002/cncr.23100. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu M, Deguchi A, Lim JT, Moriwaki H, Kopelovich L, Weinstein IB. (-)-Epigallocatechin gallate and polyphenon E inhibit growth and activation of the epidermal growth factor receptor and human epidermal growth factor receptor-2 signaling pathways in human colon cancer cells. Clin Cancer Res. 2005;11:2735–2746. doi: 10.1158/1078-0432.CCR-04-2014. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Ling MT, Feng H, Wong YC, Tsao SW, Wang X. Id-I stimulates cell proliferation through activation of EGFR in ovarian cancer cells. Br J Cancer. 2004;91:2042–2047. doi: 10.1038/sj.bjc.6602254. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Singh F, Gao D, Lebwohl MG, Wei H. Shikonin modulates cell proliferation by inhibiting epidermal growth factor receptor signaling in human epidermoid carcinoma cells. Cancer Lett. 2003;200:115–121. doi: 10.1016/s0304-3835(03)00239-8. [DOI] [PubMed] [Google Scholar]
- 27.Tao J, Wu D, Xu B, Qian W, Li P, Lu Q, Yin C, Zhang W. microRNA-133 inhibits cell proliferation, migration and invasion in prostate cancer cells by targeting the epidermal growth factor receptor. Oncol Rep. 2012;27:1967–1975. doi: 10.3892/or.2012.1711. [DOI] [PubMed] [Google Scholar]
- 28.Liu L, Shao X, Gao W, Zhang Z, Liu P, Wang R, Huang P, Yin Y, Shu Y. MicroRNA-133b inhibits the growth of non-small-cell lung cancer by targeting the epidermal growth factor receptor. FEBS J. 2012;279:3800–2812. doi: 10.1111/j.1742-4658.2012.08741.x. [DOI] [PubMed] [Google Scholar]
- 29.Zhou X, Hu Y, Dai L, Wang Y, Zhou J, Wang W, Di W, Qiu L. MicroRNA-7 inhibits tumor metastasis and reverses epithelial-mesenchymal transition through AKT/ERK1/2inactivation by targeting EGFR in epithelial ovarian cancer. PLoS One. 2014;9:e96718. doi: 10.1371/journal.pone.0096718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W, Ren F, Wu Q, Jiang D, Li H, Shi H. MicroRNA-497 suppresses angiogenesis by targeting vascular endothelial growth factor A through the PI3K/AKTand MAPK/ERK pathways in ovarian cancer. Oncol Rep. 2014;32:2127–2133. doi: 10.3892/or.2014.3439. [DOI] [PubMed] [Google Scholar]
- 31.Yu H, Zhang L, Liu P. CXCR7 signaling induced epithelial-mesenchymal transition by AKT and ERK pathways in epithelial ovariancarcinomas. Tumour Biol. 2015;36:1679–1683. doi: 10.1007/s13277-014-2768-1. [DOI] [PubMed] [Google Scholar]


